Abstract
Rpb9, a non-essential subunit of RNA polymerase II, mediates a transcription-coupled repair (TCR) subpathway in Saccharomyces cerevisiae. This subpathway initiates at the same upstream site as the previously identified Rad26 subpathway. However, the Rpb9 subpathway operates more effectively in the coding region than in the region upstream of the transcription start site, whereas the Rad26 subpathway operates equally in the two regions. Rpb4, another non-essential subunit of RNA polymerase II, plays a dual role in regulating the two subpathways, suppressing the Rpb9 subpathway and facilitating the Rad26 subpathway. Simultaneous deletion of RPB9 and RAD26 genes completely abolishes TCR in both the coding and upstream regions, indicating that no other TCR subpathway exists in RNA polymerase II-transcribed genes.
Keywords: nucleotide excision repair/Rad26/Rpb4/Rpb9/transcription coupled
Introduction
Nucleotide excision repair (NER) is capable of removing a variety of helix-distorting lesions, including UV-induced cis–syn cyclobutane pyrimidine dimers (CPDs) and 6–4 photoproducts, as well as bulky chemical adducts (for a review see Hanawalt, 2001). NER in DNA is heterogeneous in the genome. Damage in a transcriptionally silent gene and in the non-transcribed strand (NTS) of an actively transcribed gene is repaired by the so-called global genomic repair (GGR) pathway, which in Saccharomyces cerevisiae is dependent on both Rad7 and Rad16 (Verhage et al., 1994). On the other hand, damage in the transcribed strand (TS) of an actively transcribed gene is repaired more rapidly than in the NTS (Hanawalt, 2001). This pathway of NER, termed transcription-coupled repair (TCR), has been shown to function in Escherichia coli (Mellon and Hanawalt, 1989), S.cerevisiae (Smerdon and Thoma, 1990), and mammalian cells (Mellon et al., 1987). Putative transcription-repair coupling factors, i.e. Mfd (Selby and Sancar, 1993), Rad26 (van Gool et al., 1994), and CSA and CSB (van Hoffen et al., 1993), have been identified in E.coli, yeast and human, respectively.
In eukaryotic mRNA genes, TCR is believed to be triggered by stalled RNA polymerase II (Pol II) (Friedberg et al., 1995; Hanawalt, 2001). Pol II of the yeast S.cerevisiae is composed of 12 subunits designated Rpb1–12. Rpb4 exhibits some unique features distinguishing it from the other subunits. The stoichiometry of Rpb4 is dependent on growth conditions. In optimally growing cells, the fraction of Pol II molecules containing Rpb4 is ∼20%, and it gradually increases following the shift to post-logarithmic phases (Kolodziej et al., 1990; Choder and Young, 1993). Thus, in stationary phase, virtually all Pol II molecules contain Rpb4 (Choder and Young, 1993), and these molecules, unlike Pol II molecules obtained from log-phase cells, can form high-quality two-dimensional crystals (Asturias et al., 1997; Jensen et al., 1998).
Rpb4 is not essential for cell viability (Woychik and Young, 1989). Under optimal growth conditions at moderate temperatures (18–22°C), cells lacking Rpb4 have growth indistinguishable from their wild-type counterparts (Choder and Young, 1993). Consistently, under these conditions, the global transcriptional activity in rpb4 cells is comparable with that in the wild-type strains. However, rpb4 cells rapidly lose the capacity for efficient growth and global transcription as they experience temperature extremes (<12°C and >32°C) (Choder and Young, 1993; Miyao et al., 2001).
As Rpb4 is present at substoichiometric levels in logarithmically growing cells, the most homogeneous preparations of Pol II are obtained from rpb4 cells (Darst et al., 1991). Comparison of a lower resolution structure of the entire 12 subunit Pol II with the high resolution structure of the Pol II obtained from rpb4 cells revealed a difference in conformation, i.e. the wild-type Pol II favors a closed conformation and the mutant enzyme favors an open conformation (Asturias et al., 1997; Jensen et al., 1998).
Rpb9 is another Pol II subunit that is not essential for cell viability (Woychik et al., 1991). In yeast, deletion of Rpb9 results in mild temperature sensitivity and relatively normal levels of transcription in vivo. Rpb9 is located at the tip of the so-called ‘jaws’ of Pol II, which are thought to function by clamping the DNA downstream of the active site (Cramer et al., 2001; Gnatt et al., 2001). It has been shown that Rpb9 regulates transcription initiation and elongation (Hull et al., 1995; Awrey et al., 1997; Hemming et al., 2000; van Mullem et al., 2002).
There seems to be only one TCR pathway in E.coli, and the mechanism has been well demonstrated (Selby and Sancar, 1993; Park et al., 2002). In contrast, the TCR mechanism is much more elusive in eukaryotes. Several studies suggest that there may be alternative TCR subpathway(s) in addition to the Rad26-mediated one in yeast. First, rad16 rad26 or rad7 rad26 double mutants are not as sensitive to UV as are cells that are completely defective for NER (such as rad14 cells) (Verhage et al., 1996). Secondly, in RAD26-deleted cells, a residual or even substantial extent of TCR still exists in certain genes, such as RPB2 (Bhatia et al., 1996; Verhage et al., 1996, 1997; Gregory and Sweder, 2001) and MFA2 (Teng and Waters et al., 2000). The residual TCR in the RPB2 gene in RAD26-deleted cells apparently is not dependent on a functional RAD28 gene, the homolog of the human CSA gene, since essentially the same kinetics of NER of the TS were observed in a rad7 rad26 double mutant and in a rad7 rad26 rad28 triple mutant (Bhatia et al., 1996).
We attempted to identify possible alternative TCR pathway(s) in S.cerevisiae. The data clearly indicate that an alternative TCR subpathway does exist, which is mediated by Pol II itself via its Rpb9 subunit. Furthermore, the results indicate that association of Rpb4 with the polymerase complex significantly affects the Rpb9 and Rad26 TCR subpathways.
Results
Requirement of Rad26 for TCR is cell growth dependent
We first analyzed NER in the yeast GAL1 gene, which is highly inducible by galactose (Bash and Lohr, 2001). RAD+, rad16 and rad16 rad26 cells were grown to log phase, or stationary phase, in minimal medium containing galactose and irradiated with 50 J/m2 (254 nm) UV light. Genomic DNA was isolated from the cells immediately after UV irradiation or after incubating the irradiated cells at 28°C for different periods of time in repair medium. The DNA was digested with restriction enzymes and incised at CPD sites with excess T4 endonuclease V (Lloyd, 1999). The incised fragments were end-labeled strand specifically, resolved on DNA sequencing gels and exposed to phosphorimager screens, as previously described (Li and Waters, 1996; Li et al., 2000).
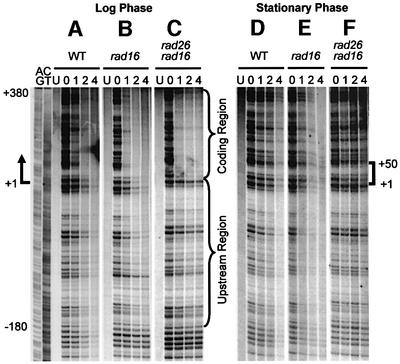
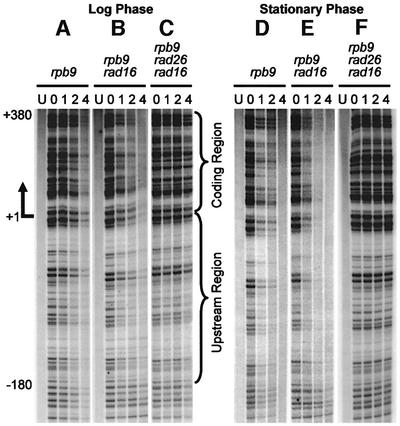
In log-phase cells, fast repair can be seen in the TS of the GAL1 gene (Figure 1A–C), starting at ∼180 nucleotides upstream of the transcription start site (+1). Deletion of the RAD16 gene, which is essential for GGR but dispensable for TCR (Verhage et al., 1994), eliminates NER in the NTS and in the region >180 nucleotides upstream of the transcription start site of the TS, thereby making the TCR more apparent. Furthermore, in rad16 cells, TCR rates are faster in the coding region (+1 to +380) than in the upstream region (–1 to –180) (Figures 1B and 2C). Deletion of the RAD26 gene did not significantly compromise repair in the coding region, while it significantly slowed down repair in the upstream region (compare Figures 1B and C, and 2C and E).
Fig. 1. Gels showing TCR in the GAL1 gene in wild-type, rad16 and rad16 rad26 cells. The left two lanes are for Maxam–Gilbert sequencing markers (AG and CT). The other lanes are DNA samples from unirradiated (U) and irradiated cells following 0, 1, 2 and 4 h of repair incubation. The arrow indicates the transcription start site of the GAL1 gene. The bracket on the right of (F) indicates the short region where residual TCR can still be seen in the stationary phase rad16 rad26 cells.
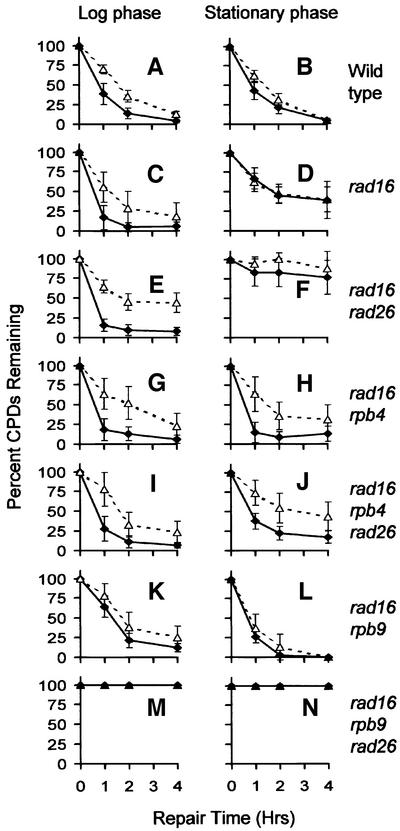
Fig. 2. Percentage of CPDs remaining in the coding (+1 to +380 nucleotides) (filled diamonds) and upstream (–1 to –180 nucleotides) (open triangles) regions in the TS of the GAL1 gene following different times of repair incubation. The plots show the average ± SD calculated from all the CPD sites quantified in the respective regions.
In stationary wild-type and rad16 cells, TCR can also be seen starting at ∼180 nucleotides upstream of the transcription start site (Figure 1D and E). However, the repair rates in the coding region of stationary cells slowed down significantly, making these rates comparable with those in the upstream region (compare Figures 1B and E, and 2C and D). Surprisingly, repair was virtually absent in the stationary phase rad16 rad26 cells (Figures 1F and 2F), except for a short (∼50 nucleotide) region immediately downstream of the transcription start site (Figure 1F, region marked with a bracket), where a small degree of repair can still be seen.
Deletion of the RPB4 gene, encoding a non-essential subunit of Pol II, reinstates TCR in stationary rad16 rad26 cells
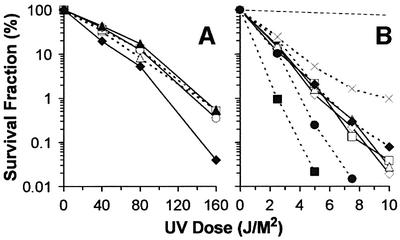
The robust TCR in log-phase rad16 rad26 cells implies that an alternative Rad26-independent TCR subpathway may exist in yeast. This alternative TCR subpathway may be suppressed when the cells are in stationary phase. Substoichiometric amounts of Rpb4 exist in Pol II in log-phase cells, but are associated with virtually all Pol II molecules in stationary cells (Kolodziej et al., 1990; Choder and Young, 1993). This unique feature of Rpb4 makes it an appealing candidate for suppressing the possible alternative TCR subpathway. To test this notion, RPB4 deletions were made in wild-type, rad16 and rad26 backgrounds. In agreement with a previous report (Woychik and Young, 1989), rpb4 mutants grow almost normally at temperatures between 20 and 30°C, except for having a longer lag phase (data not shown). Similar to the deletion of RAD26, deletion of RPB4 results in increased UV sensitivity only when RAD16 is also deleted (Figure 3). Furthermore, rad16 rad26, rad16 rpb4 and rad16 rad26 rpb4 cells have the same UV sensitivity (Figure 3B), indicating that Rpb4 and Rad26 may be involved in the same TCR subpathway.
Fig. 3. UV sensitivity of different deletion mutants. (A) Wild-type (open squares), rpb4 (open triangles), rad26 (open circles), rpb9 (filled triangles) and rad26 rpb9 (filled diamonds). (B) Wild-type (dotted line), rad16 (crosses), rad16 rpb4 (open squares), rad16 rpb9 (filled diamonds), rad16 rad26 (open diamonds), rad16 rad26 rpb4 (open triangles), rad16 rad26 rpb9 (filled squares) and rad1 (filled circles).
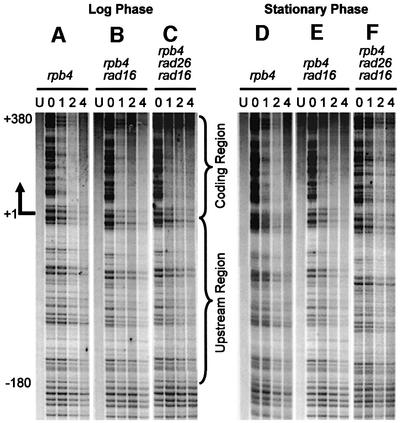
Deletion of RPB4 did not significantly change TCR kinetics in log-phase rad16 and rad16 rad26 cells (Figures 1–4; compare Figure 2C and G, and E and I), reflecting that only a small portion of Pol II engaged in GAL1 transcription may actually contain Rpb4 in log-phase RPB4+ cells. In contrast, RPB4 deletion resulted in significant enhancement in TCR in the coding region of the stationary phase rad16 cells (compare Figures 1E and 4E, and 2D and H). This indicates that Rpb4 present in the stationary cultures may indeed suppress at least a portion of TCR. More strikingly, deletion of RPB4 reinstated TCR in the stationary phase rad16 rad26 cells, both in the upstream region and in the coding region (compare Figures 1F and 4F, and 2F and J). These results suggest that RPB4 deletion releases the otherwise suppressed Rad26 independent TCR subpathway.
Fig. 4. Gels showing TCR in the GAL1 gene in rpb4, rpb4 rad16 and rpb4 rad26 rad16 cells. See Figure 1 legend for details.
The Rad26-independent TCR subpathway is mediated by Rpb9, another non-essential subunit of Pol II
Stably arrested Pol II at a pause site can be reactivated for chain elongation by transcription elongation factor S-II (TFIIS). It has been proposed that TFIIS may be involved in TCR by a similar mechanism (Donahue et al., 1994). However, it was found later that TFIIS is not required for TCR in wild-type or rad26 rad7 mutant yeast cells (Verhage et al., 1997). It is possible that some redundancy for chain elongation factors exists in yeast. The C-terminal zinc-binding domain of Rpb9, another non-essential subunit of Pol II, shares 30% sequence identity with that of TFIIS (Kaine et al., 1994). Rpb9 is required, along with TFIIS, to effect transcription through blocks to elongation encoded by the DNA template (Awrey et al., 1997; Hemming et al., 2000).
We wondered if Rpb9, instead of TFIIS, is required for TCR, and constructed RPB9 deletions in RAD+, rad16 and rad26 backgrounds. RPB9 deletion mutants show a wider range of temperature tolerance than the RPB4 deletion mutants (data not shown), in agreement with previous reports (Woychik and Young, 1989; Woychik et al., 1991). Although rpb9 and rad26 single mutant cells are not sensitive to UV, rpb9 rad26 double mutants are slightly more sensitive to UV than wild-type cells (Figure 3A). Furthermore, rpb9 rad16 rad26 triple mutants are much more UV sensitive than rad16 rad26 or rad16 rpb9 double mutants. In fact, the UV sensitivity of the triple mutants is even more severe than that of the isogenic RAD1 deletion mutants (Figure 3B), where NER is completely abolished (Friedberg et al., 1995). These results suggest that: (i) simultaneous deletion of RAD16, RAD26 and RPB9 may completely abolish NER; (ii) if Rpb9 is indeed involved in TCR, this TCR subpathway is distinct from that mediated by Rad26; and (iii) in addition to NER, one or more of the three proteins (i.e. Rad16, Rad26 and Rpb9) may play additional roles that affect viability.
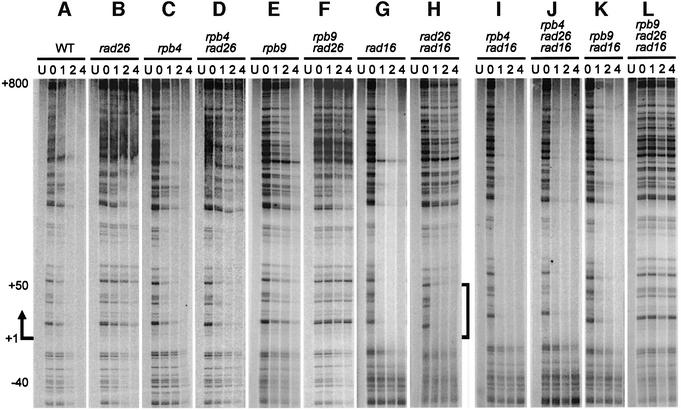
To test these hypotheses, NER was also examined in the GAL1 gene of these mutants. As can be seen, absolutely no repair occurs in any region of the GAL1 gene in the rad16 rad26 rpb9 triple mutant cells (Figures 2M and N, and 5C and F). Thus, Rpb9 indeed mediates the Rad26-independent TCR subpathway.
Fig. 5. Gels showing TCR in the GAL1 gene in rpb9, rpb9 rad16 and rpb9 rad26 rad16 cells. See Figure 1 legend for details.
Rad26- and Rpb9-dependent TCR subpathways initiate at similar upstream sites but have different efficiencies in different regions
Both Rad26- and Rpb9-dependent TCR subpathways start to occur at ∼180 nucleotides upstream of the transcription start site of the GAL1 gene (Figures 1, 4 and 5). This suggests that both TCR subpathways can operate not only when Pol II is in the elongation mode, but also when it is in the initiation, or even pre-initiation, state.
In the rad16 rpb9 cells, where only Rad26-mediated TCR takes place, the repair rates are not significantly different between the coding region and the upstream region (Figures 2K and L, and 5B and E). These results suggest that the Rad26-dependent TCR subpathway is equally effective in the two regions. This notion is supported by the observations that in stationary phase wild-type and rad16 cells, where Rpb9-dependent TCR is virtually inactive due to saturation of Pol II with Rpb4, TCR rates are also similar between the coding and upstream regions (Figures 1E, and 2B and D).
In the rad16 rad26 rpb4 cells, where only the unsuppressed (due to RPB4 deletion) Rpb9-mediated TCR occurs, repair rates are higher in the coding region than in the upstream region (Figures 2I and J, and 4C and F). This indicates that the Rpb9-dependent TCR subpathway, if not suppressed, is more effective in the coding region than in the upstream region. This notion is supported by the observations that rpb4 rad16 cells (Figure 2G and H), log-phase rad16 cells (where only a portion of Pol II contains Rpb4) (Figure 2C) and log-phase rad16 rad26 cells (Figure 2E) show faster repair in the coding region than in the upstream region.
Similar TCR trends exist in the constitutively expressed RPB2 gene
The GAL1 gene has several distinct features, such as: (i) being completely inactive in glucose media and highly inducible with galactose; and (ii) sharing an upstream activating sequence with the divergent GAL10 gene (Bash and Lohr, 2001). Therefore, we asked how general are the repair trends we observed in the GAL1 gene. To address this question, TCR was analyzed in the constitutively expressed RPB2 gene, using the same DNA preparations as used for analyzing repair in the GAL1 gene. The overall TCR trends in the RPB2 gene (Figure 6; data for stationary cultures not shown) are similar to those in the GAL1 gene (Figures 1, 4 and 5), indicating that the TCR features we observe in the GAL1 gene are general. Importantly, deletion of RPB4 enhances TCR in rad26 cells (compare Figure 6B and D, and H and J), indicating that Rpb4 also suppresses Rpb9-mediated TCR in this gene. Furthermore, simultaneous deletion of Rad26 and Rpb9 completely abolishes TCR in this gene (compare Figure 6B, E and F, and H, K and L), indicating that Rad26 and Rpb9 mediate two (and only two) alternative TCR pathways.
Fig. 6. Gels showing TCR in the RPB2 gene of log-phase cultures. The lanes are DNA samples from unirradiated (U) and irradiated cells following 0, 1, 2 and 4 h repair incubation. The arrow indicates the transcription start site of the RPB2 gene. The bracket on the right of (H) indicates the region where robust Rad26-independent TCR occurs.
There are also some differences in TCR between the GAL1 and RPB2 genes. First, TCR initiates at ∼40 nucleotides upstream of the transcription start site of the RPB2 gene, which is less than that in the GAL1 gene (∼180 nucleotides upstream of the transcription start site). This indicates that the sequence where Pol II is recruited during transcription initiation (or pre-initiation) may be closer to the transcription start site in the RPB2 gene than that in the GAL1 gene. Secondly, TCR in log-phase rad16 rad26 cells is much slower in the RPB2 gene (compare Figures 1C and 6H), except for the short (∼50 nucleotide) region immediately downstream of the transcription start site.
Effects of Rpb4 and Rpb9 on TCR are not due to aberrant transcription
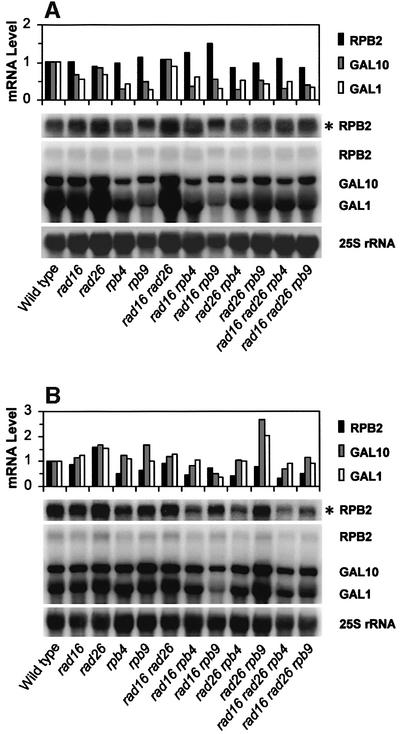
We wished to determine if the effects of Rpb4 and Rpb9 on TCR are due to the direct action of the two Pol II subunits, or resulted from a transcriptional change when the subunits are deleted. To address this question, mRNA levels were measured in both log-phase and stationary-phase cultures (i.e. the same as for NER analyses). In log-phase cells, deletion of RPB4 or RPB9 does not dramatically affect RPB2 mRNA levels (Figure 7A). Furthermore, although the mRNA levels of GAL1 and GAL10 are ∼2- to 3-fold lower in the rpb4 and rpb9 cells than in wild-type cells, relatively high levels of mRNA of these two genes are still observed. Indeed, the levels of GAL1 and GAL10 mRNA in these mutant cells are still much higher than those of RPB2 mRNA (Figure 7A), indicating that the GAL1-10 genes are still strongly induced by galactose in these mutants, albeit to a lesser extent than in the wild-type.
Fig. 7. mRNA levels of GAL1, GAL10 and RPB2 genes in (A) log-phase and (B) stationary-phase cultures. Gels show northern blots of RNA hybridized with probes generated using DNA fragments of RPB2, GAL1-10 and rDNA. As the mRNA level of the RPB2 gene is much lower than those of the GAL1 and GAL10 genes, the RPB2 mRNA band is also shown with a 10-fold higher exposure (see asterisks). Plots above each gel show the relative levels of GAL1, GAL10 and RPB2 mRNA (relative to wild-type cells, which is designated as 1). Loading variations of different samples are normalized using the signal intensities of the 25S rRNA as an internal control.
In stationary cultures, the levels of RPB2 mRNA in rpb4 or rpb9 cells are about one- to two-thirds of those in wild-type cells (Figure 7B). Furthermore, the GAL1 and GAL10 mRNA levels appear to be less affected by deletion of RPB4 or RPB9 in stationary- (Figure 7B) than in log-phase (Figure 7A) cultures.
Importantly, deletion of the RAD26 gene in rpb9 cells does not seem to decrease the mRNA levels of the GAL1, GAL10 or RPB2 genes further (Figure 7, compare mRNA levels in rpb9 and rad26 rpb9 cells, and those in rad16 rpb9 and rad16 rad26 rpb9 cells). This indicates that the lack of TCR in rad26 rpb9 double mutant cells (Figures 5C and F, and 6F and L) is due to elimination of coupling between transcription and NER, rather than complete shut down of transcription. This is not unexpected, since the RPB2 gene is essential for survival (Woychik et al., 1991), and GAL1 and GAL10 genes are indispensable for cell growth in galactose media (Bash and Lohr, 2001). Furthermore, deletion of RPB4 in rad26 cells results in similar or decreased levels of GAL1, GAL10 or RPB2 mRNA (Figure 7, compare mRNA levels in rad26 and rad26 rpb4 cells, and in rad16 rad26 and rad16 rad26 rpb4 cells), indicating enhanced TCR caused by deletion of RPB4 in rad26 cells (compare Figures 1F and 4F, 6B and D, and H and J) is due to release of Rpb9-mediated TCR, rather than increased transcription.
Discussion
In the present study, we identified a Rpb9-mediated TCR subpathway in yeast, which appears to be the only alternative subpathway besides that previously identified as being mediated by Rad26. We also show that Rpb4 may serve as a ‘switch’ in regulating the two TCR subpathways.
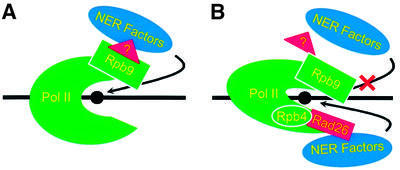
The biochemical mechanism of Rpb9-mediated TCR remains to be elucidated. One scenario is that Rpb9 directly recruits NER factors to the damaged site. Alternatively, Rpb9 may exert its role on TCR via an as yet unidentified match-maker (Figure 8). It has been shown that Rpb9 interacts with a plethora of factors involved in transcription elongation and histone modifications, including TFIIS (Awrey et al., 1997; Hemming et al., 2000), TFIIE, SAGA and elongator histone acetyltransferases (van Mullem et al., 2002).
Fig. 8. A model for two alternative TCR subpathways in yeast. (A) In the absence of Rpb4, Pol II has a more open conformation, which allows Rpb9-mediated TCR to take place. (B) In the presence of Rpb4, Pol II exists in a closed conformation, which suppresses Rpb9-mediated TCR, while facilitating Rad26-mediated TCR. The damage on DNA is shown as a solid circle. A possible ‘match-maker’ protein between Rpb9 and NER factors is shown as a triangle with a question mark.
Rpb4 interacts with Rpb7, a small but essential subunit of Pol II (Woychik, 1998). This subunit pair can dissociate from Pol II upon biochemical purification, and deletion of the RPB4 gene from yeast cells results in no association of the Rpb7 subunit with Pol II. Thus, it is possible that the role of Rpb4 in TCR may be indirect (i.e. via its interaction with Rpb7). The Rpb4–Rpb7 complex is located downstream of the catalytic site in the center of the Pol II cleft (Jensen et al., 1998; Cramer et al., 2000). Compared with the wild-type 12 subunit Pol II, the Rpb4–Rpb7 Pol II clamps DNA in a more open, less stable conformation (Asturias et al., 1997; Jensen et al., 1998). Rpb9 is located on the tip of the Pol II ‘upper jaw’ (Cramer et al., 2001). It is possible that Rpb4 suppresses Rpb9-dependent TCR by simply closing up the ‘jaws’ of Pol II, leaving the damage inaccessible to NER factors recruited by Rpb9. On the other hand, it is also possible that the conformational change of Pol II caused by the association of Rpb4 somehow renders Rpb9 unable to recruit NER factors (Figure 8). A further means of elucidating the role of Rpb4 in suppressing Rpb9-mediated TCR would be by analyzing a rad16 rpb4 rpb9 triple mutant. However, despite extensive efforts, we were unable to obtain rpb4 rpb9 or rad16 rpb4 rpb9 mutants, presumably because cells deleted for both RPB4 and RPB9 are not viable. To our knowledge, no RPB4 RPB9 double deletion mutants have been created successfully.
Rpb4, by itself or via the interaction with Rpb7, may play a dual role during TCR. While suppressing Rpb9-mediated TCR, the interaction of Rpb4 with other subunits of Pol II may change the conformation of the polymerase complex. This, in turn, may improve the interactions with Rad26 protein (Figure 8). Several lines of evidence support this notion. First, as with the RAD26 deletion, the RPB4 deletion does not cause any detectable UV sensitivity in the RAD+ background (Figure 3A), indicating that Rpb4 may not have a significant role in GGR. Secondly, deletion of the RPB4 gene does not cause additional UV sensitivity to the rad26 mutant, and vice versa (Figure 3B), indicating the two proteins may be involved in the same TCR subpathway. Thirdly, except for the rad16 rpb9 cells, stationary cultures of all the strains show TCR rates that are equivalent or slower than those in log-phase cultures (Figure 2). In contrast, rad16 rpb9 cells, where only Rad26-dependent TCR takes place, show faster repair in stationary cultures than in log-phase cultures (compare Figures 2K and L, and 5B and E), indicating that the increased amount of Rpb4-associated Pol II complexes in stationary cells may facilitate Rad26-dependent TCR. A more direct clarification of the role of Rpb4 in facilitating Rad26-mediated TCR would be by comparing TCR between rpb9 and rpb4 rpb9, or between rad16 rpb9 and rad16 rpb4 rpb9 mutants. However, as mentioned above, we were unable to combine RPB4 and RPB9 deletions in the same cells.
Some factors are shared by the essential general transcription factor TFIIH and the DNA repair machinery (Qiu et al., 1993; Feaver et al., 1993; Guzder et al., 1994a,b; Wang et al., 1994, 1995; Svejstrup et al., 1995). The obligatory loading of TFIIH onto promoter sites during transcription initiation suggests an obvious mechanism for directly coupling Pol II transcription to NER at sites of damage in the TS. TCR in the yeast URA3 gene becomes Rad26 dependent 30–40 nucleotides downstream from the transcription start site (Tijsterman et al., 1997). In the human JUN gene, TCR becomes CSA and CSB dependent at about +20 nucleotides into the coding region (Tu et al., 1997, 1998). It has been proposed that this coupling factor-independent TCR close to the transcription start site can be due to the association of TFIIH with Pol II, as TFIIH is believed to be released from Pol II 30–60 nucleotides downstream from the start site (for a recent review see Svejstrup, 2002). We also observed very efficient TCR close to the transcription start site in the GAL1 (Figure 1C) and RPB2 (Figure 6H, marked with a bracket) genes in log-phase rad16 rad26 cells. Further more, in stationary phase rad16 rad26 cells, although TCR is suppressed in most regions of the GAL1 gene, residual TCR can still be seen in a short region immediately downstream of the transcription start site (Figure 1F, marked with a bracket). However, simultaneous deletion of RPB9 and RAD26 genes also completely abolishes TCR in these regions (Figures 5C and F, and 6F and L), indicating that the Rad26-independent TCR in these regions is Rpb9 dependent. Thus, it is likely that Rpb9-mediated TCR is less suppressed by Rpb4 in the short region immediately downstream of the transcription start site. Furthermore, TFIIH may not directly recruit NER machinery to the damaged site in this region of the TS.
It was proposed that TCR occurs when RNA polymerase is stalled at a lesion during transcriptional elongation (Mellon et al., 1987). Indeed, in the human PGK1 gene, preferential repair of the TS began just downstream of the transcription start site but was most pronounced beginning at nucleotide +140 (Gao et al., 1994). In the yeast URA3 gene, TCR is seen only in the coding region (Tijsterman et al., 1997). Furthermore, as discussed above, TCR becomes Rad26 or CSA/CSB dependent at a certain site downstream of the transcription start site (20–40 nucleotides) in the yeast URA3 and human JUN genes, respectively (Tijsterman et al., 1997; Tu et al., 1997, 1998). These studies suggest that TCR occurs only when Pol II is in elongation mode. We show that both Rpb9- and Rad26-mediated TCR pathways initiate upstream of the transcription start site (–180 and –40 nucleotides for GAL1 and RPB2 genes, respectively), although Rpb9-mediated TCR is less effective in the upstream region than in the coding region. Thus, it is possible that the sites where Pol II is loaded during transcription pre-initiation are more upstream in the GAL1 or RPB2 gene than in the yeast URA3 or human JUN genes. Once loaded onto a promoter, Pol II may be able to mediate TCR.
DNA in eukaryotic cells is packaged in chromatin, which may affect NER (for example, see Smerdon and Conconi, 1999). The GAL structural genes are inactive without galactose but are highly transcribed in its presence (Bash and Lohr, 2001). In the inactive state, GAL genes demonstrate a characteristic promoter chromatin organization, which helps inhibit gene expression in this state. Induction of GAL expression triggers Gal4p-dependent upstream nucleosome disruption. Regulator-mediated competition between nucleosomes and the TATA-binding protein complex for the TATA region is probably a central aspect of GAL regulation and a focal point for the numerous factors that contribute to it (Bash and Lohr, 2001). Our mRNA measurements show that the GAL1 gene is still strongly induced in rpb4 and rpb9 mutants (Figure 7), albeit to a lesser extent than in wild-type cells. This indicates that nucleosome structure in the GAL1 gene is also disrupted upon galactose induction in these mutant cells. Therefore, it is possible that Rpb4 and Rpb9 may indirectly influence TCR by affecting chromatin remodeling during transcription.
In log-phase wild-type cells, the relative contribution of Rad26 and Rpb9 subpathways to TCR may be different from gene to gene. For the URA3 gene, Rad26 seems to be absolutely required, except for a short region close to the transcription start site (Tijsterman et al., 1997), indicating that TCR is accomplished primarily by the Rad26 subpathway. In agreement with previous reports (Bhatia et al., 1996; Verhage et al., 1996, 1997; Gregory and Sweder, 2001), Rad26 is partially required for TCR in the RPB2 gene (Figure 6H), indicating that both subpathways contribute to TCR in this gene. For the GAL1 gene, Rad26 is almost dispensable, especially in the coding region (Figure 1C), indicating that TCR in this gene of log-phase cultures is fulfilled primarily by the Rpb9 subpathway. One plausible explanation for this difference is that Pol II engaged in robustly transcribed genes (such as the induced GAL1 gene) has a lower content of Rpb4 than those engaged in slowly transcribed genes (such as the constitutively expressed URA3 gene). Therefore, Rpb9-mediated TCR may be less suppressed by Rpb4 in highly transcribed genes than in slowly transcribed genes.
The subunits of Pol II are well conserved between yeast and human cells (Woychik and Hampsey, 2002). Seven Pol II small subunits from S.cerevisiae, namely Rpb4 (Khazak et al., 1998), Rpb6, Rpb7, Rpb8, Rpb9, Rpb10 and Rpb12 (McKune et al., 1995), are functionally interchangeable with the human homologs. Importantly, NER pathways are also believed to be conserved (Friedberg et al., 1995), and our findings raise several questions. First, is there a Rpb9-dependent TCR subpathway in human cells, in addition to the one mediated by CSA and CSB? Secondly, does Rpb4 also regulate different TCR pathways in higher eukaryotes? The yeast Rpb4 is absolutely required for Pol II transcription at elevated temperature (37°C) (Miyao et al., 2001). It was proposed that the open conformation of Pol II containing no Rpb4–Rpb7 may not be able to form a stable association with the single-stranded DNA template or nascent RNA transcript at the non-permissive temperature (Miyao et al., 2001). If this feature of the yeast Pol II is true for human cells, the human Pol II must contain a Rpb4–Rpb7 subcomplex to enable normal transcription at 37°C. Thus, it is possible that, in normal human cells, the Rpb9-mediated TCR is more suppressed by Rpb4 in regions other than those close to the transcription start site.
Trichothiodystrophy (TTD) is a rare human genetic disorder characterized by hair dysplasia and associated with numerous symptoms affecting mainly organs derived from the neuroectoderm (Bergmann and Egly, 2001). These deficiencies may result from mutations in XPB or XPD, two subunits of TFIIH. At present, the gene responsible for a third group of photosensitive TTD, TTD-A, has not been identified (Bergmann and Egly, 2001). Beyond a deficiency in NER pathways, it is hypothesized that the TTD phenotypes may also result from a decreased basal level of transcription (Bergmann and Egly, 2001). It seems feasible to propose that mutations of RPB4 or RPB9 may result in human disorders that are related to deficiencies in transcription and/or TCR. Thus, an appealing candidate for such a disorder is TTD-A.
Materials and methods
Yeast strains
The wild-type yeast strain Y452 (MATα, ura3-52, his3–1, leu2-3, leu2-112, cir°) was supplied by Dr Louise Prakash (University of Texas Medical Branch, Galveston, TX). All deletion mutants were made in Y452 background. The cells were transformed with linearized plasmids bearing the respective genes to be deleted, with a portion of their gene replaced by the yeast URA3 or LEU2 gene. Nucleotides (with respect to the starting codon ATG) +345 to +1755, +58 to +2297, –8 to +588, +11 to +366, and –212 to +3853 were deleted for the RAD16, RAD26, RPB4, RPB9 and RAD1 genes, respectively. The transformed cells were selected on SD plates containing no uracil or leucine at 28°C. In order to introduce a second deletion using a plasmid bearing the gene of interest replaced by the URA3 gene, the previously introduced URA3 gene that had replaced the first gene deletion was knocked-out. The URA3 knock-out was done by transforming the cells with a linearized plasmid bearing a truncated (with the sequence between the sites of StuI and EcoRV removed) URA3 gene, and selecting the cells on SD plates containing 5-fluoro-orotic acid (Boeke et al., 1984). All the deletions were confirmed by Southern blot analysis, and verified further by assessing the phenotypic restoration following transformation of a strain with a single copy centromeric plasmid bearing the respective intact gene.
UV irradiation, repair incubation and DNA isolation
Yeast cells were grown at 28°C in minimal medium containing 2% galactose to log phase (A600 ∼1.0) or stationary phase, harvested, and washed with ice-cold 2% galactose. The washed cells were resuspended in 2% galactose and irradiated with 50 J/m2 UV light. One-tenth volume of a stock solution containing 10% yeast extract and 20% peptone was added immediately to the irradiated cell suspension, and the cells were incubated for various times in the dark at 28°C before being pelleted. To a sample of ∼2 × 109 pelleted cells were added 2 ml of ice-cold NIB (50 mM Tris–HCl, 2 mM MgCl2, 150 mM NaCl, 17% glycerol, 0.5 mM spermine, 0.15 mM spermidine pH 8.0) and 2 ml of acid-washed glass beads (Sigma, 425–600 µm). The mixtures were vortexed for 30 s and kept on ice for 2–5 min. This procedure was repeated three times to break up the cells completely. The samples were mixed with 10 ml of 50 mM Tris–HCl, 400 mM NaCl, 2% SDS, 2 mM EDTA pH 8.0 and incubated at 65°C for >30 min. After cooling to room temperature, the samples were mixed with 8 ml of 5 M NaCl and left on ice overnight. The samples were then centrifuged at 4°C and the supernatant collected. The DNA was precipitated with ethanol, treated with RNase A and extracted with phenol/chloroform. After re-precipitation, the DNA was dissolved in H2O and stored at –20°C before using.
Mapping of NER
The GAL1 and RPB2 fragments were end labeled with [α-32P]dATP using the procedure described previously (Li and Waters, 1996; Li et al., 2000), with slight modifications. Briefly, ∼1 µg of genomic DNA was digested with restriction endonuclease(s) to release the fragments of interest and incised at CPD sites with an excess amount of purified T4 endonuclease V (kindly provided by Dr R.Stephen Lloyd, University of Texas Medical Branch, Galveston, TX). Excess copies of biotinylated oligonucleotides, which are complementary to the 3′ end of the fragment to be labeled, were mixed with the sample. The T-tract in the biotinylated oligonucleotides, described previously (Li and Waters, 1996; Li et al., 2000), was changed to short runs (4–5) of Ts separated by Gs. This change ensured full-length incorporation of radioactive dAMPs opposite the Ts contained in the oligonucleotides, and eliminated hybridization between the oligonucleotides and contaminating poly(A) tails of mRNA. The mixture was heated to 95°C for 5 min to denature the DNA and then cooled to an annealing temperature. The annealed fragments were attached to streptavidin magnetic beads (Dynal, Inc.) and the other fragments were removed by washing the beads at the annealing temperature. The attached fragments were labeled using [α-32P]dATP (NEN Life Sciences) and non-radioactive dCTP, rather than using [α-32P]dATP alone (Li and Waters, 1996; Li et al., 2000). The labeled fragments were eluted from the beads and resolved on sequencing gels. The dried gels were then exposed to PhosphorImager screens (Molecular Dynamics) and the band intensity quantified as described previously (Li and Waters, 1996; Li et al., 2000).
Northern blot analysis
Cells were cultured to log or stationary phase under the same conditions as those used for NER analysis. Total RNA was isolated using the glass beads method, as described (Collart and Oliviero, 1996). The RNA was fractionated by electrophoresis on formaldehyde–agarose gels (Sambrook and Russell, 2001), followed by transfer to Hybond N+ membranes (Amersham-Pharmacia). A 2 kb GAL1-10 DNA fragment encompassing the upstream activating sequence and 5′ portions (0.7 kb) of each of the genes, a 1.15 kb DraI fragment of RPB2 corresponding to the 5′ portion of the gene, and a 2.9 kb EcoRI rDNA fragment were used for generating probes for GAL1, GAL10, RPB2 mRNA and 25S rRNA, respectively. Each probe was 32P labeled using the Prime-It® II Random Primer Labeling Kit (Stratagene). Membranes were first hybridized with probes to GAL1-10 and RPB2 mRNA. After exposure to PhosphorImager screens, the probes were stripped off by boiling the membranes in 1% SDS. The stripped membranes were re-hybridized with probes to the 25S rRNA and exposed to PhosphorImager screens. The signal intensities of the 25S rRNA were used as loading controls, assuming that deletions of RPB4 and RPB9 do not affect the rRNA level. The rationale for this assumption is that: (i) rRNA is not degraded rapidly; and (ii) Rpb4 and Rpb9 are not shared by RNA polymerase I (Pol I) and Pol II (Cramer, 2002). Quantification of RNA levels was performed using ImageQuant software (Molecular Dynamics).
Acknowledgments
Acknowledgements
We thank Dr Fritz Thoma for helpful comments on the manuscript, Dr R.Stephen Lloyd for supplying purified T4 endonuclease V, and Deirdre Fahy for technical assistance. We also thank members of the Smerdon laboratory for critical discussions and technical help. This study was supported by NIH grant ES04106 from the National Institute of Environmental Health Sciences.
References
- Asturias F.J., Meredith,G.D., Poglitsch,C.L. and Kornberg,R.D. (1997) Two conformations of RNA polymerase II revealed by electron crystallography. J. Mol. Biol., 272, 536–540. [DOI] [PubMed] [Google Scholar]
- Awrey D.E., Weilbaecher,R.G., Hemming,S.A., Orlicky,S.M., Kane,C.M. and Edwards,A.M. (1997) Transcription elongation through DNA arrest sites. A multistep process involving both RNA polymerase II subunit RPB9 and TFIIS. J. Biol. Chem., 272, 14747–14754. [DOI] [PubMed] [Google Scholar]
- Bash R. and Lohr,D. (2001) Yeast chromatin structure and regulation of GAL gene expression. Prog. Nucleic Acid Res. Mol. Biol., 65, 197–259. [DOI] [PubMed] [Google Scholar]
- Bergmann E. and Egly,J.M. (2001) Trichothiodystrophy, a transcription syndrome. Trends Genet., 17, 279–286. [DOI] [PubMed] [Google Scholar]
- Bhatia P.K., Verhage,R.A., Brouwer,J. and Friedberg,E.C. (1996) Molecular cloning and characterization of Saccharomyces cerevisiae RAD28, the yeast homolog of the human Cockayne syndrome A (CSA) gene. J. Bacteriol., 178, 5977–5988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeke J.D., LaCroute,F. and Fink,G.R. (1984) A positive selection for mutants lacking orotidine-5′-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol. Gen. Genet., 197, 345–346. [DOI] [PubMed] [Google Scholar]
- Choder M. and Young,R.A. (1993) A portion of RNA polymerase II molecules has a component essential for stress responses and stress survival. Mol. Cell. Biol., 13, 6984–6991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collart M.A. and Oliviero,S. (1996) Preparation of yeast RNA. In Ausubel,F.M., Brent,R., Kingston,R.E., Moore,D.D., Seidman,J.G., Smith,J.A. and Struhl,K. (eds), Current Protocols in Molecular Biology. Vol. 2. Wiley Interscience, New York, NY, pp. 13.12.1–13.12.5.
- Cramer P. (2002) Multisubunit RNA polymerases. Curr. Opin. Struct. Biol., 12, 89–97. [DOI] [PubMed] [Google Scholar]
- Cramer P. et al. (2000) Architecture of RNA polymerase II and implications for the transcription mechanism. Science, 288, 640–649. [DOI] [PubMed] [Google Scholar]
- Cramer P., Bushnell,D.A. and Kornberg,R.D. (2001) Structural basis of transcription: RNA polymerase II at 2.8 Å resolution. Science, 292, 1863–1876. [DOI] [PubMed] [Google Scholar]
- Darst S.A., Kubalek,E.W., Edwards,A.M. and Kornberg,R.D. (1991) Two-dimensional and epitaxial crystallization of a mutant form of yeast RNA polymerase II. J. Mol. Biol., 221, 347–357. [DOI] [PubMed] [Google Scholar]
- Donahue B.A., Yin,S., Taylor,J.S., Reines,D. and Hanawalt,P.C. (1994) Transcript cleavage by RNA polymerase II arrested by a cyclobutane pyrimidine dimer in the DNA template. Proc. Natl Acad. Sci. USA, 91, 8502–8506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feaver W.J., Svejstrup,J.Q., Bardwell,L., Bardwell,A.J., Buratowski,S., Gulyas,K.D., Donahue,T.F., Friedberg,E.C. and Kornberg,R.D. (1993) Dual roles of a multiprotein complex from S.cerevisiae in transcription and DNA repair. Cell, 75, 1379–1387. [DOI] [PubMed] [Google Scholar]
- Friedberg E.C., Walker,G.C. and Siede,W. (1995) DNA Repair and Mutagenesis. ASM Press, Washington, DC.
- Gao S., Drouin,R. and Holmquist,G.P. (1994) DNA repair rates mapped along the human PGK1 gene at nucleotide resolution. Science, 263, 1438–1440. [DOI] [PubMed] [Google Scholar]
- Gnatt A.L., Cramer,P., Fu,J., Bushnell,D.A. and Kornberg,R.D. (2001) Structural basis of transcription: an RNA polymerase II elongation complex at 3.3 Å resolution. Science, 292, 1876–1882. [DOI] [PubMed] [Google Scholar]
- Gregory S.M. and Sweder,K.S. (2001) Deletion of the CSB homolog, RAD26, yields Spt(–) strains with proficient transcription-coupled repair. Nucleic Acids Res., 29, 3080–3086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzder S.N., Qiu,H., Sommers,C.H., Sung,P., Prakash,L. and Prakash,S. (1994a) DNA repair gene RAD3 of S.cerevisiae is essential for transcription by RNA polymerase II. Nature, 367, 91–94. [DOI] [PubMed] [Google Scholar]
- Guzder S.N., Sung,P., Bailly,V., Prakash,L. and Prakash,S. (1994b) RAD25 is a DNA helicase required for DNA repair and RNA polymerase II transcription. Nature, 369, 578–581. [DOI] [PubMed] [Google Scholar]
- Hanawalt P.C. (2001) Controlling the efficiency of excision repair. Mutat. Res., 485, 3–13. [DOI] [PubMed] [Google Scholar]
- Hemming S.A., Jansma,D.B., Macgregor,P.F., Goryachev,A., Friesen,J.D. and Edwards,A.M. (2000) RNA polymerase II subunit Rpb9 regulates transcription elongation in vivo. J. Biol. Chem., 275, 35506–35511. [DOI] [PubMed] [Google Scholar]
- Hull M.W., McKune,K. and Woychik,N.A. (1995) RNA polymerase II subunit RPB9 is required for accurate start site selection. Genes Dev., 9, 481–490. [DOI] [PubMed] [Google Scholar]
- Jensen G.J., Meredith,G., Bushnell,D.A. and Kornberg,R.D. (1998) Structure of wild-type yeast RNA polymerase II and location of Rpb4 and Rpb7. EMBO J., 17, 2353–2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaine B.P., Mehr,I.J. and Woese,C.R. (1994) The sequence and its evolutionary implications, of a Thermococcus celer protein associated with transcription. Proc. Natl Acad. Sci. USA, 91, 3854–3856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khazak V., Estojak,J., Cho,H., Majors,J., Sonoda,G., Testa,J.R. and Golemis,E.A. (1998) Analysis of the interaction of the novel RNA polymerase II (pol II) subunit hsRPB4 with its partner hsRPB7 and with pol II. Mol. Cell. Biol., 18, 1935–1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolodziej P.A., Woychik,N., Liao,S.M. and Young,R.A. (1990) RNA polymerase II subunit composition, stoichiometry and phosphorylation. Mol. Cell. Biol., 10, 915–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S. and Waters,R. (1996) Nucleotide level detection of cyclobutane pyrimidine dimers using oligonucleotides and magnetic beads to facilitate labelling of DNA fragments incised at the dimers and chemical sequencing reference ladders. Carcinogenesis, 17, 1549–1552. [DOI] [PubMed] [Google Scholar]
- Li S., Waters,R. and Smerdon,M.J. (2000) Low and high resolution mapping of DNA damage at specific sites. Methods, 22, 170–179. [DOI] [PubMed] [Google Scholar]
- Lloyd R.S. (1999) The initiation of DNA base excision repair of dipyrimidine photoproducts. Prog. Nucleic Acid Res. Mol. Biol., 62, 155–175. [DOI] [PubMed] [Google Scholar]
- McKune K., Moore,P.A., Hull,M.W. and Woychik,N.A. (1995) Six human RNA polymerase subunits functionally substitute for their yeast counterparts. Mol. Cell. Biol., 15, 6895–6900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellon I. and Hanawalt,P.C. (1989) Induction of the Escherichia coli lactose operon selectively increases repair of its transcribed DNA strand. Nature, 342, 95–98. [DOI] [PubMed] [Google Scholar]
- Mellon I., Spivak,G. and Hanawalt,P.C. (1987) Selective removal of transcription-blocking DNA damage from the transcribed strand of the mammalian DHFR gene. Cell, 51, 241–249. [DOI] [PubMed] [Google Scholar]
- Miyao T., Barnett,J.D. and Woychik,N.A. (2001) Deletion of the RNA polymerase subunit RPB4 acts as a global, not stress-specific, shut-off switch for RNA polymerase II transcription at high temperatures. J. Biol. Chem., 276, 46408–46413. [DOI] [PubMed] [Google Scholar]
- Park J.S., Marr,M.T. and Roberts,J.W. (2002) E.coli transcription repair coupling factor (Mfd protein) rescues arrested complexes by promoting forward translocation. Cell, 109, 757–767. [DOI] [PubMed] [Google Scholar]
- Qiu H., Park,E., Prakash,L. and Prakash,S. (1993) The Saccharomyces cerevisiae DNA repair gene RAD25 is required for transcription by RNA polymerase II. Genes Dev., 7, 2161–2171. [DOI] [PubMed] [Google Scholar]
- Sambrook J. and Russell,D.W. (2001) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Selby C.P. and Sancar,A. (1993) Molecular mechanism of transcription– repair coupling. Science, 260, 53–58. [DOI] [PubMed] [Google Scholar]
- Smerdon M.J. and Conconi,A. (1999) Modulation of DNA damage and DNA repair in chromatin. Prog. Nucleic Acid Res. Mol. Biol., 62, 227–255. [DOI] [PubMed] [Google Scholar]
- Smerdon M.J. and Thoma,F. (1990) Site-specific DNA repair at the nucleosome level in a yeast minichromosome. Cell, 61, 675–684. [DOI] [PubMed] [Google Scholar]
- Svejstrup J.Q., Wang,Z., Feaver,W.J., Wu,X., Bushnell,D.A., Donahue,T.F., Friedberg,E.C. and Kornberg,R.D. (1995) Different forms of TFIIH for transcription and DNA repair: holo-TFIIH and a nucleotide excision repairosome. Cell, 80, 21–28. [DOI] [PubMed] [Google Scholar]
- Svejstrup J.Q. (2002) Mechanisms of transcription-coupled DNA repair. Nat. Rev. Mol. Cell. Biol., 3, 21–29. [DOI] [PubMed] [Google Scholar]
- Teng Y. and Waters,R. (2000) Excision repair at the level of the nucleotide in the upstream control region, the coding sequence and in the region where transcription terminates of the Saccharomyces cerevisiae MFA2 gene and the role of RAD26. Nucleic Acids Res., 28, 1114–1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tijsterman M., Verhage,R.A., van de Putte,P., Tasseron-de Jong,J.G. and Brouwer,J. (1997) Transitions in the coupling of transcription and nucleotide excision repair within RNA polymerase II-transcribed genes of Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA, 94, 8027–8032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu Y., Bates,S. and Pfeifer,G.P. (1997) Sequence-specific and domain-specific DNA repair in xeroderma pigmentosum and Cockayne syndrome cells. J. Biol. Chem., 272, 20747–20755. [DOI] [PubMed] [Google Scholar]
- Tu Y., Bates,S. and Pfeifer,G.P. (1998) The transcription–repair coupling factor CSA is required for efficient repair only during the elongation stages of RNA polymerase II transcription. Mutat. Res., 400, 143–151. [DOI] [PubMed] [Google Scholar]
- van Gool A.J., Verhage,R., Swagemakers,S.M., van de Putte,P., Brouwer,J., Troelstra,C., Bootsma,D. and Hoeijmakers,J.H. (1994) RAD26, the functional S.cerevisiae homolog of the Cockayne syndrome B gene ERCC6. EMBO J., 13, 5361–5369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hoffen A., Natarajan,A.T., Mayne,L.V, van Zeeland,A.A., Mullenders,L.H. and Venema,J. (1993) Deficient repair of the transcribed strand of active genes in Cockayne’s syndrome cells. Nucleic Acids Res., 21, 5890–5895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Mullem V., Wery,M., Werner,M., Vandenhaute,J. and Thuriaux,P. (2002) The Rpb9 subunit of RNA polymerase II binds transcription factor TFIIE and interferes with the SAGA and elongator histone acetyltransferases. J. Biol. Chem., 277, 10220–10225. [DOI] [PubMed] [Google Scholar]
- Verhage R., Zeeman,A.M., de Groot,N., Gleig,F., Bang,D.D., van de Putte,P. and Brouwer,J. (1994) The RAD7 and RAD16 genes, which are essential for pyrimidine dimer removal from the silent mating type loci, are also required for repair of the nontranscribed strand of an active gene in Saccharomyces cerevisiae.Mol. Cell. Biol., 14, 6135–6142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhage R.A., van Gool,A.J., de Groot,N., Hoeijmakers,J.H., van de Putte,P. and Brouwer,J. (1996) Double mutants of Saccharomyces cerevisiae with alterations in global genome and transcription-coupled repair. Mol. Cell. Biol., 16, 496–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhage R.A., Heyn,J., van de Putte,P. and Brouwer,J. (1997) Transcription elongation factor S-II is not required for transcription-coupled repair in yeast. Mol. Gen. Genet., 254, 284–290. [DOI] [PubMed] [Google Scholar]
- Wang Z., Svejstrup,J.Q., Feaver,W.J., Wu,X., Kornberg,R.D. and Friedberg,E.C. (1994) Transcription factor b (TFIIH) is required during nucleotide-excision repair in yeast. Nature, 368, 74–76. [DOI] [PubMed] [Google Scholar]
- Wang Z., Buratowski,S., Svejstrup,J.Q., Feaver,W.J., Wu,X., Kornberg,R.D., Donahue,T.F. and Friedberg,E.C. (1995) The yeast TFB1 and SSL1 genes, which encode subunits of transcription factor IIH, are required for nucleotide excision repair and RNA polymerase II transcription. Mol. Cell. Biol., 15, 2288–2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woychik N.A. (1998) Fractions to functions: RNA polymerase II thirty years later. Cold Spring Harbor Symp. Quant. Biol., 63, 311–317. [DOI] [PubMed] [Google Scholar]
- Woychik N.A. and Hampsey,M. (2002) The RNA polymerase II machinery: structure illuminates function. Cell, 108, 453–463. [DOI] [PubMed] [Google Scholar]
- Woychik N.A. and Young,R.A. (1989) RNA polymerase II subunit RPB4 is essential for high- and low-temperature yeast cell growth. Mol. Cell. Biol., 9, 2854–2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woychik N.A., Lane,W.S. and Young,R.A. (1991) Yeast RNA polymerase II subunit RPB9 is essential for growth at temperature extremes. J. Biol. Chem., 266, 19053–19055. [PubMed] [Google Scholar]