Abstract
DNA base flipping is an important mechanism in molecular enzymology, but its study is limited by the lack of an accessible and reliable diagnostic technique. A series of crystalline complexes of a DNA methyltransferase, M.HhaI, and its cognate DNA, in which a fluorescent nucleobase analogue, 2-aminopurine (AP), occupies defined positions with respect the target flipped base, have been prepared and their structures determined at higher than 2 Å resolution. From time-resolved fluorescence measurements of these single crystals, we have established that the fluorescence decay function of AP shows a pronounced, characteristic response to base flipping: the loss of the very short (∼100 ps) decay component and the large increase in the amplitude of the long (∼10 ns) component. When AP is positioned at sites other than the target site, this response is not seen. Most significantly, we have shown that the same clear response is apparent when M.HhaI complexes with DNA in solution, giving an unambiguous signal of base flipping. Analysis of the AP fluorescence decay function reveals conformational heterogeneity in the DNA–enzyme complexes that cannot be discerned from the present X-ray structures.
INTRODUCTION
The DNA double helix is a dynamic structure that undergoes conformational change in response to interaction with agents such as enzymes and drugs. A particularly remarkable example of localized conformational distortion is the phenomenon of base flipping, induced by DNA methyltransferase enzymes (Figure 1a). This involves 180° rotation of the target nucleotide around the phosphate backbone, out of the DNA helix and into the reactive site of the enzyme. Base flipping was first observed by X-ray crystallography for the bacterial C5-cytosine methyltransferase, M.HhaI (1) and subsequently for other methyltransferases [M.HaeIII (2) and M.TaqI (3)] and various DNA repair enzymes (4), including recently the human proteins AGT (5) and oxoG interacting with undamaged DNA (6).
Figure 1.
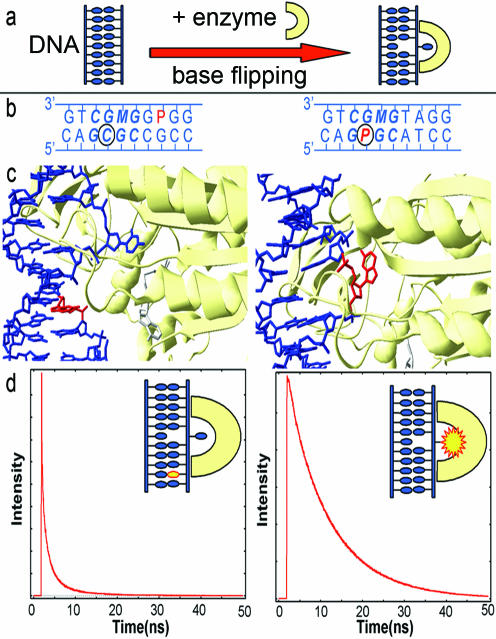
Establishing the response of the AP fluorescence decay function to base flipping. (a) A schematic representation of base flipping. (b) The sequences of the 10 bp at the centre of the APout duplex (left) and APtarget duplex (right). Bases in the M.HhaI recognition sequence are shown in bold/italic; the target base is circled; AP is denoted P and is in red; M is 5-methyl cytosine (used to direct enzyme binding to the opposite strand of the duplex). (c) Crystal structures of the complexes of the M.HhaI enzyme with the APout duplex (left) and the APtarget duplex (right), showing the molecular structure in the vicinity of the recognition sequence. (d) Fluorescence decay curves of the crystalline complexes of M.HhaI with APout (left) and APtarget (right); cartoons show the AP base highlighted. (Fluorescence intensity is shown normalized to the number of counts in the maximum channel.)
Recognition and modification of specific residues in DNA is a key event in many cellular processes. The methylation by DNA methyltransferases of cytosine and adenine nucleotides within specific DNA sequences is found in many organisms from bacteria to man and fulfils many functions including gene regulation, genomic imprinting, chromatin remodelling and the marking of host chromosomal DNA at specific target sequences. This last process is widespread in bacteria and protects the bacterial DNA from degradation by host restriction endonucleases that destroy invading viral DNA containing unmethylated target sequences (7). Aberrations in cytosine methylation correlate with human genetic disease and, therefore, methyltransferases are potent candidate targets for developing new therapies (8). All methyltransferases have a common catalytic core, and it is believed that they will all use base flipping to gain access to the nucleotide targeted for methylation (9).
Base flipping seems likely to be a fundamental mechanism of DNA–enzyme interaction in situations where bases need to be covalently modified or removed, or where the DNA helix is to be opened up for replication or transcription. To date, X-ray crystallography of protein–DNA complexes has been relied upon for absolute proof of base flipping. However, co-crystallization of proteins with their DNA substrates is difficult and time consuming, and in some cases might be impossible; the number of crystal structures determined remains small. An alternative method to unambiguously detect base flipping in aqueous solution is essential for extending studies of this phenomenon.
The fluorescent analogue of adenine, 2-aminopurine (AP), would appear to be an ideal probe of DNA–enzyme interactions. The fluorescence of AP is strongly quenched within the structure of double-stranded DNA, but is enhanced if the base stacking or base pairing is perturbed. Such perturbations of duplex structure are caused by protein binding and AP emission intensity has been used to monitor DNA polymerase reactions (10), helicase activity (11) and the action of DNA repair enzymes (12–15) and methyltransferases (16–20). However, to date, AP has proved less than ideal as a probe of base flipping. Some methyltransferases cause a dramatic increase in fluorescence intensity when they bind to a DNA duplex containing AP at the target site for methylation (16–20), whilst others do not (21,22). In some cases, AP located at sites other than the methylation site show changes in fluorescence intensity even though these sites should not undergo base flipping (21,22). The ambiguity of these results reflects the fact that a change in AP emission intensity merely indicates some local distortion of the DNA helix and is not specific to a particular type of structural change, such as base flipping. In contrast, the form of the fluorescence decay function is a precise and responsive probe of the local molecular environment and, as we demonstrate here, can be used as an unequivocal indicator of base flipping of AP.
We have solved X-ray structures and made the first time-resolved fluorescence measurements of single crystals of AP-labelled-DNA duplexes complexed with M.HhaI. This has allowed us to identify a pronounced, characteristic response of the fluorescence decay function of AP to base flipping. Furthermore, we have shown that this response constitutes an unambiguous signature of base flipping in solution.
MATERIALS AND METHODS
Materials
M.HhaI methyltransferase was overexpressed and purified as described previously (23). 2AP-labelled oligonucleotides were obtained from Fermentas (Lithuania) and MWG Biotech (Germany).
The sequences of the oligonucleotides used in the solution phase studies were 5′-GACTGGTACAGTATCAGGPGCTGACCCACAACATCCG/5′-TCGGATGTTGTGGGTCAGMGCCTGATACTGTACCAGT (APtarget); 5′-GACAGTATCAGGCGCCGCCCCACAA/5′-GTTGTGGGGPGGMGCCTGATACTGT (APout); 5′-ACTGGTACAGTATCAGGCGCTGACCCACAACATCTG/5′-CAGATGTTGTGGGTCAGMPCCTGATACTGTACCAGT (APopp) and 5′-GACTGGTACAGTATCAGPCGCTGACCCACAACATCCG/5′-TCGGATGTTGTGGGTCAGMGCCTGATACTGTACCAGT (APadj); where P is AP and M is 5-methyl cytosine. The target site of M.HhaI is underlined. Oligonucleotide strands were annealed using a 10% excess of the non-fluorescent strand to ensure that none of the AP-labelled strand was left unbound in the final solution. In order to ensure complete binding of the DNA by M.HhaI, samples were prepared containing 1 µM DNA duplex, 3 µM M.HhaI (T250G mutant) and 100 µM cofactor [Kd for the ternary DNA–HhaI–AdoHcy complex is 4.2 pM (24)]. The buffer used was 10 mM Tris–HCl, 0.5 mM EDTA and 50 mM NaCl, pH of 7.4. The cofactor S-adenosyl methionine (AdoMet) was used in experiments on the APtarget duplex, whereas the cofactor analogue S-adenosyl homocysteine (AdoHcy) was used with the APout duplex in order to prevent methylation of the target cytosine base and subsequent dissociation of the enzyme from the duplex.
Crystals were grown using sitting-drop vapour diffusion at 19°C. The enzyme (wild-type for APout, APadj and APopp crystals and T250G mutant for APtarget crystal) was mixed with an oligonucleotide duplex and AdoHcy at a 1:1.2:1.5 molar ratio and a final protein concentration of 7 mg/ml. The sequences of the duplexes were 5′-TGTCAGPGCATCC/5′-TGGATGMGCTGAC (APtarget), 5′-TGTCAGCGCCGCC/5′-TGGPGGMGCTGAC (APout), 5′-TGTCAGCGCATCC/5′-TGGATGMPCTGAC (APopp) and 5′-TGTCAPCGCATCC/5′-TGGATGMGCTGAC (APadj). Samples were mixed with an equal volume of well solution (50 mM sodium citrate, pH 5.6, 1.2–2.0 M ammonium sulfate and 0–15% glucose). Single rhombohedral crystals were harvested by washing thoroughly with the well solution. For X-ray diffraction studies crystals were soaked for several seconds in a cryobuffer containing 25% glycerol and flash-frozen at 90 K in a gaseous nitrogen stream.
Diffraction data collection and structure determination
Diffraction datasets were collected from single crystals at 90 K at the ESRF, Grenoble, France or at the DORIS storage ring, EMBL-DESY, Hamburg, Germany. Data were processed with MOSFLM (25) and further processed using SCALA (26) and TRUNCATE (27) from CCP4 (28) package. The structure was solved by molecular replacement using the protein backbone of the previously solved ternary complex (PDB entry 3mht) as the initial model. For initial phasing, 7.0 Å spheres around the key structural elements (residue 250 and the AP base) were omitted from phase calculation to avoid model bias. Only elements present in the omitted densities were built back into the model. Models were adjusted manually using O (29) in SIGMAA-weighted (30) maps with phases calculated from the models and refined with the CNS package (31). Data collection and refinement statistics are summarized in Table 1.
Table 1.
Data collection and refinement statistics
| D2A6, M.HhaI(T250G)–APtarget–AdoHcy | D75A6, M.HhaI–APadj–AdoHcy | 2D2, M.HhaI–APopp–AdoHcy | D80A2, M.HhaI–APout–AdoHcy | |
|---|---|---|---|---|
| Data collection | ||||
| Space group | H32 | H32 | H32 | H32 |
| Cell dimensions | ||||
| a, b, c (Å) | 95.6, 95.6, 315.8 | 95.1, 95.1, 312.2 | ||
| α, β, γ (°) | 90, 90, 120 | 90, 90, 120 | 90, 90, 120 | 90, 90, 120 |
| Resolution (Å) | 29.5–1.9 (2.00–1.90)a | 23.2–1.9 (1.95–1.90) | 56.8–1.7 (1.79–1.70) | 31.0–1.85 (1.96–1.85) |
| Rmerge (%) | 6.7 (23.9) | 5.0 (22.0) | 7.1 (33.9) | 6.7 (20.4) |
| I/σI | 8.4 (2.8) | 30.0 (18.4) | 7.5 (2.2) | 8.5 (3.7) |
| Completeness (%) | 99.9 (99.6) | 99.4 (97.4) | 99.6 (97.6) | 98.7 (94.5) |
| Redundancy | 6.0 (6.0) | 17.4 (16.6) | 11.8 (7.5) | 8.7 (3.6) |
| Refinement | ||||
| Resolution (Å) | 1.90 | 1.90 | 1.70 | 1.85 |
| No. reflections | 44234 | 43361 | 59771 | 45796 |
| Rwork/Rfree | 20.0 (23.7)/21.9 (24.5) | 19.4 (21.7)/21.5 (22.7) | 19.6 (24.8)/22.0 (25.1) | 18.9 (21.3)/21.2 (23.7) |
| Coordinate error (Å) | 0.17 | 0.14 | 0.14 | 0.14 |
| No. of atoms | 3389 | 3549 | 3380 | 3585 |
| Ligand/ion | 566 | 572 | 573 | 588 |
| Water | 220 | 342 | 186 | 376 |
| B-factors | ||||
| Protein | 16.3 | 15.9 | 16.5 | 13.1 |
| Ligand/ion | 27.2 | 25.9 | 25.3 | 23.3 |
| Water | 25.2 | 29.9 | 22.1 | 25.4 |
| R.m.s deviations | ||||
| Bond lengths (Å) | 0.006 | 0.006 | 0.006 | 0.006 |
| Bond angles (degrees) | 1.2 | 1.3 | 1.2 | 1.3 |
Wavelengths used for data collection: D2A6 (λ = 0.9755 Å, T = 100 K, detector MarCCD, beamline ID13, ESRF, Grenoble); D75A6 (λ = 0.8120 Å, T = 100 K, detector Mar345, beamline DORIS/X11, EMBL-DESY, Hamburg); 2D2 (λ = 0.8120 Å, T = 100 K, detector MarCCD, beamline DORIS/X11, EMBL-DESY, Hamburg); D80A2 (λ = 1.050 Å, T = 100 K, detector Mar345, beamline DORIS/X31, EMBL-DESY, Hamburg).
aHighest resolution shell is shown in parentheses.
Time-resolved fluorescence measurements
Time-resolved fluorescence spectroscopy was performed using the technique of time-correlated single photon counting. The solution and crystal phase samples were measured in an Edinburgh Instruments spectrometer equipped with TCC900 photon counting electronics. Single crystals were mounted in quartz capillary tubes of 1 mm diameter (Hampton Research). Solution samples were contained in fused silica microcuvettes of volume 0.16 ml (Optiglass Ltd). The excitation source was a Ti-Sapphire femtosecond laser system (Coherent, 10 W Verdi and Mira Ti-Sapphire) producing pulses of ∼200 fs at 76 MHz repetition rate. The output of the Ti-Sapphire laser was passed through a pulse picker to reduce the repetition rate to 4.75 MHz and then frequency tripled to give an output at 320 nm. The emission from the sample was collected orthogonal to the excitation direction through a polarizer set at the magic angle with respect to the vertically polarized excitation. The fluorescence was passed through a monochromator (bandpass 10 nm), then detected by a Hamamatsu microchannel plate photomultiplier (R3809U-50). The instrument response of the system, measured using a Ludox scatterer for solution phase measurements and a frosted quartz plate for the crystalline samples, was 50 ps FWHM.
Fluorescence decay curves were recorded on a timescales of 20 and 50 ns, resolved into 4096 channels, to a total of 10 000 counts in the peak channel. Decay curves were analysed using a standard iterative reconvolution method, assuming a multiexponential decay function in the following equation:
| 1 |
where Ai is the fractional amplitude and τi is the fluorescence lifetime of the i-th decay component.
Fluorescence was excited at 320 nm and decay curves recorded at three emission wavelengths, 370, 390 and 410 nm. The three decays were analysed globally using Edinburgh Instruments Level 2 software, i.e. they were fitted simultaneously, with lifetimes, τi, as common parameters. The quality of fit was judged on the basis of the reduced χ2-statistic, χ2, and the randomness of residuals.
RESULTS
Crystal structures
Crystal structures were determined for ternary complexes of M.HhaI, a cofactor analogue (AdoHcy) and four different AP-labelled DNA duplexes, APout, APtarget, APadj and APopp. In APout, the AP was inserted at a position outside the M.HhaI recognition sequence; in APtarget, AP was placed at the target site for base flipping; in APadj, AP was adjacent to the target base; in APopp, AP was the pairing partner of the target base on the opposite strand. The sequences of the 10 bp at the centre of the duplexes are shown in Figures 1b and 3a. The recognition sequence of M.HhaI, 5′-GCGC-3′, is palindromic so that the enzyme will methylate the 5′ cytosine residue in the recognition sequence on either of the DNA strands. Therefore, to direct enzyme binding to the target base, a methylated cytosine base is used in the other strand.
Figure 3.
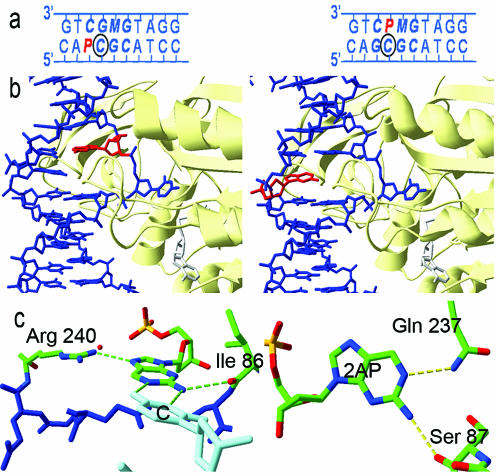
Interaction of M.HhaI with duplexes where AP is opposite or adjacent to the target base. (a) The sequences of the 10 bp at the centre of the APadj duplex (left) and APopp duplex (right). Bases in the M.HhaI recognition sequence are shown in boldface/italic; the target base is circled; AP is denoted P and is in red; M is 5-methyl cytosine (used to direct enzyme binding to the opposite strand of the duplex). (b) Crystal structures of the complexes of the M.HhaI enzyme with the APadj duplex (left) and the APopp duplex (right), showing the molecular structure in the vicinity of the recognition sequence. (c) Detailed view of the H-bond interactions between the M.HhaI enzyme and the APadj duplex (left) and the APopp duplex (right).
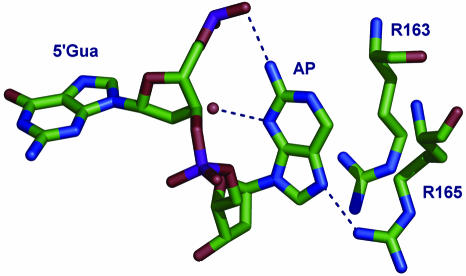
The crystal structures of the APout–M.HhaI(wild-type)–AdoHcy and APtarget–M.HhaI(T250G mutant)–AdoHcy complexes are shown in Figure 1c. When AP is outside the recognition sequence it remains stacked and paired within the DNA duplex, while the cytosine target base is flipped out. When AP is at the target site, it is flipped out of the DNA helix and into the catalytic cleft of the bilobal enzyme. The flipped out AP base is bound in a slightly different position than that observed with cytosine, uracil or adenine at the target (32). The enzyme embraces and stabilizes the duplex structure around the flipped out base, using its mobile catalytic loop such that the flipped out AP is tightly locked in a polar cavity with no access to bulk solvent. The base makes contacts to the side chains of R165, R163, the phosphate of the 5′Gua on the same strand and a bound water molecule, as shown in Figure 2.
Figure 2.
Detailed view of the H-bond interactions between the M.HhaI enzyme and the APtarget duplex. DNA and protein residues are shown as sticks, a bound solvent molecule (presumed water) is shown as a red ball.
The structures of the APadj–M.HhaI(wild-type)–AdoHcy and APopp–M.HhaI(wild type)–AdoHcy complexes are shown in Figure 3b. In both cases, the AP base remains intrahelical when the neighbouring cytosine is flipped out. In the APopp complex, the enzyme infiltrates the duplex via the glutamine 237 and serine 87 residues which hydrogen bond with the N1 and N2 atoms of the orphan (unpaired) AP, respectively, as shown in Figure 3c. In the normal recognition sequence, where the orphan base is guanine, the O6 and N1 atoms of the base are mainly involved in interactions with glutamine 237 (1). In the APadj complex, the AP forms a ‘wobble’ base pair with cytosine. The enzyme maintains the fidelity of this base pair during flipping, through interaction of arginine 240 and isoleucine 86 with the AP, as shown in Figure 3c.
Establishing the response of AP to base flipping
To establish the photophysical response of AP to base flipping, we compare the fluorescence decay of the APout complex, characteristic of the unflipped AP base, with that of the APtarget complex, characteristic of AP flipped out of the DNA helix into the enzyme active site. There is a dramatic qualitative difference between the two decay functions (Figure 1d) illustrating the sensitivity of the AP probe to its molecular environment.
The decay of the APout complex (unflipped AP) is multiexponential, requiring four lifetime components to give a satisfactory fit (Table 2 and Supplementary Table S1). Although the crystal is ostensibly homogeneous, with each identical duplex containing a single AP base in the same sequence position, the 4-component decay shows that AP experiences a heterogeneous environment. This signifies the existence of at least four conformational states of DNA in the crystal. There may be additional shorter decay components that are beyond the time resolution of our experiments, but these make negligible contribution to the fluorescence intensity. The A-factors (Ai in Equation 1) indicate the fraction of the emitting AP population with a given lifetime and hence the fractional population of each conformational state. Modelling of the decay function by a continuous distribution of fluorescence lifetimes confirms the existence of four well-defined emitting populations whose mean lifetimes are well represented by the individual exponential decay components. It is, therefore, physically realistic to describe the AP decays by discrete lifetimes, recognizing that each lifetime represents a distribution.
Table 2.
Fluorescence lifetimes (τi) and their fractional amplitudes (Ai) for the crystalline DNA–M.HhaI–AdoHcy complexes
| DNA–M.HhaI complex | τ1/ns | τ2/ns | τ3/ns | τ4/ns | A1 | A2 | A3 | A4 |
|---|---|---|---|---|---|---|---|---|
| APout–M.HhaI(wild-type)–AdoHcy | 0.07 | 0.53 | 2.1 | 7.4 | 0.64 | 0.19 | 0.14 | 0.03 |
| APtarget–M.HhaI(T250G)–AdoHcy | — | 1.1 | 6.3 | 10.9 | — | 0.07 | 0.17 | 0.76 |
| APadj–M.HhaI(wild-type)–AdoHcy | 0.19 | 0.91 | 3.5 | 10.1 | 0.54 | 0.32 | 0.10 | 0.04 |
| APopp–M.HhaI(wild-type)–AdoHcy | 0.15 | 0.94 | 3.4 | 9.4 | 0.50 | 0.30 | 0.15 | 0.05 |
Decays collected at three emission wavelengths were analysed globally to give the reported lifetimes (Supplementary Data). The fractional amplitudes (A factors) show little variation with emission wavelength and those for 390 nm emission are reported throughout.
One conformation of APout is dominant, occupied by ∼65% of the duplexes, and has a very short fluorescence lifetime of 70 ps. This can be identified with a strongly stacked structure in which AP is efficiently quenched by electron transfer from adjacent guanine bases (33). Intrastrand electron transfer from guanine to excited AP occurs in tens of picoseconds (34), consistent with the measured decay time. The longest lifetime component of 7.4 ns is similar to that of free AP in moderately polar solution (e.g. in ethanol) (35,36) and is characteristic of AP free from interbase interactions, in an extrahelical environment. Only 3% of duplexes are in this conformation and, therefore, make negligible contribution to the X-ray structure. The conformational states with intermediate lifetimes remain undefined, although they must be imperfectly stacked as they are not subject to rapid electron transfer quenching. The crystal structure clearly shows that the AP base is paired with guanine and stacked within the DNA helix, consistent with the observation that most of the AP bases undergo a short 70 ps decay. Although the structure is of the highest resolution (1.9 Å) reported for any methyltransferase–DNA complex to date, it is unable to reveal the minor, imperfectly stacked conformational states (37).
The fluorescence decay parameters of the APtarget complex (Table 2 and Supplementary Table S2) very clearly report the change in environment of the AP base when it is flipped into the catalytic cleft of the enzyme (Figure 2). The very short decay component characteristic of strongly stacked intrahelical AP is absent, and the majority (77%) of flipped out AP bases show a long lifetime of 10.9 ns, very similar to that of free AP in aqueous solution. However, the flipped out AP is not entirely free from quenching. The presence of two other decay components with shorter lifetimes indicates some conformational heterogeneity in the flipped-out complex. In these minor conformations, the AP appears to be subject to quenching interactions with amino acid residues in the enzyme pocket or the backbone of the 5′-adjacent Gua nucleotide (Figure 2).
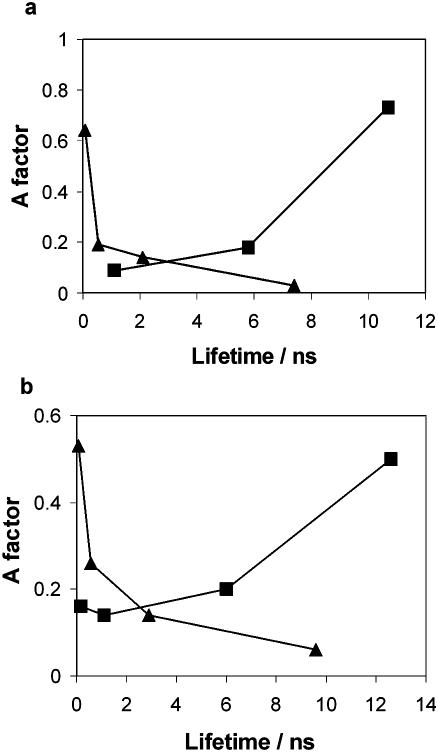
In the crystalline state, the fluorescence decay parameters of AP unquestionably show a definitive response to base flipping, as shown graphically in Figure 4a. The key features of this response are the disappearance of the very short decay component (base-stacked AP) and the predominance of a long lifetime component (unquenched, flipped-out AP). If this is to be a useful indicator of base flipping, the same distinctive response must be observable in solution.
Figure 4.
Graphical representation of the characteristic response of the AP fluorescence decay parameters to base flipping. (a) Plot of fractional amplitude (A factor) versus lifetime for the crystalline complexes APout–M.HhaI–AdoHcy (triangles) and APtarget–M.HhaI–AdoHcy (squares). (b) Plot of A factor versus lifetime for the free APtarget duplex (triangles), the APtarget–M.HhaI (T250G)–AdoMet complex (squares) in aqueous solution.
Base flipping in solution
The interaction of the M.HhaI enzyme with APout and APtarget duplexes was investigated in aqueous solution. Fluorescence decays were recorded for the duplex alone and when bound to the enzyme and cofactor. The fluorescence decay parameters of unbound APout, given in Table 3 and Supplementary Table S3, are consistent with those reported previously for AP-labelled duplexes (13,38–40). They are remarkably similar to those of the crystalline APout complex. This indicates that in solution the DNA duplex exists in essentially the same conformational states, with similar populations, as those in the crystal. The only significant difference between the solution and crystalline systems is in the value of the longest lifetime, reflecting the different extrahelical environments. The longer value of 11 ns measured for APout in solution is characteristic of AP free in aqueous solution (35,36,41,42). Complexation with the enzyme and cofactor leaves the APout decay parameters essentially unchanged (Table 3 and Supplementary Table S4), indicating that flipping of the target cytosine base has negligible effect on the duplex conformation in the vicinity of the AP probe, away from the recognition sequence.
Table 3.
Fluorescence lifetimes (τi) and their fractional amplitudes (Ai) for the free DNA duplexes, binary complexes with M.HhaI, and ternary complexes with M.HhaI and cofactor, in aqueous solution
| Solution composition | τ1/ns | τ2/ns | τ3/ns | τ4/ns | A1 | A2 | A3 | A4 |
|---|---|---|---|---|---|---|---|---|
| APout alone | 0.04 | 0.50 | 2.9 | 11.0 | 0.70 | 0.12 | 0.10 | 0.08 |
| Apout+M.HhaI(wild-type)+AdoHcy | 0.03 | 0.47 | 2.8 | 10.3 | 0.70 | 0.12 | 0.08 | 0.10 |
| APtarget alone | 0.08 | 0.58 | 2.9 | 9.6 | 0.53 | 0.27 | 0.14 | 0.06 |
| Aptarget+M.HhaI(T250G) | 0.14 | 1.0 | 5.3 | 12.6 | 0.19 | 0.16 | 0.19 | 0.46 |
| Aptarget+M.HhaI(T250G)+AdoMet | 0.17 | 1.1 | 6.0 | 12.6 | 0.16 | 0.14 | 0.20 | 0.50 |
| APadja alone | 0.04 | 0.45 | 2.6 | 10.3 | 0.84 | 0.09 | 0.05 | 0.02 |
| APadja+M.HhaI(wild-type)+AdoHcy | 0.08 | 0.32 | 2.4 | 9.9 | 0.79 | 0.15 | 0.04 | 0.02 |
| Apopp alone | 0.05 | 0.50 | 3.0 | 9.8 | 0.64 | 0.16 | 0.10 | 0.10 |
| Apopp+M.HhaI(wild-type)+AdoHcy | 0.06 | 0.44 | 2.7 | 9.4 | 0.65 | 0.18 | 0.09 | 0.08 |
aThe sequence of this APadj duplex differs from that in the crystalline complex; in this duplex the base 5′ to AP is guanine.
For the APtarget duplex in the absence of enzyme interaction, the decay parameters (Table 3 and Supplementary Table S5) are very similar to those of APout; small differences suggest subtle variations in the local environment of AP in the two duplexes. Binding to the enzyme (T250G mutant) and cofactor causes a pronounced change in the APtarget decay parameters (Table 3 and Supplementary Table S6), that is strikingly similar to the response to base flipping observed in the crystal studies, as shown in Figure 4. Therefore, we have a definitive signal of base flipping in solution. In the absence of the cofactor, interaction with the enzyme produces essentially the same fluorescence response (Table 3 and Supplementary Table S7), confirming that base flipping also occurs in the binary M.HhaI–DNA complex, as inferred from previous fluorescence intensity measurements (19).
Confirming the specificity of the AP response
To determine the specificity of the response of AP to base flipping, we investigated the fluorescence decay of the duplexes with AP opposite or adjacent to the target base. In these duplexes, flipping of the proximate cytosine might be expected to significantly perturb the immediate environment of the AP probe, and thus induce a fluorescence response that could be mistaken for base flipping of the AP itself.
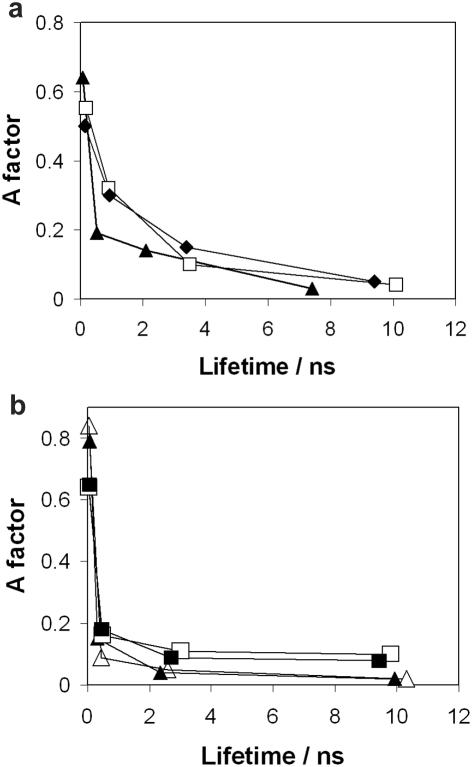
The crystal structures (Figure 3b) show that in both the APadj and APopp complexes, the AP remains within the helix when the neighbouring cytosine is flipped out. The fluorescence decay parameters of these crystalline complexes (Table 2 and Supplementary Tables S8 and S9) are clearly characteristic of intrahelical AP, resembling those of the APout complex, as illustrated in Figure 5a. The predominant decay component has a short lifetime, <200 ps, owing to base-stacked AP, and in only ∼5% of duplexes is AP in an extrahelical environment (a similar proportion to the 3% found in APout). The somewhat longer lifetime of the short decay component in APadj and APopp, compared with APout, reflects the absence of AP-guanine base stacking and hence reduced charge-transfer quenching in these duplexes.
Figure 5.
Graphical representation of decay parameters showing that there is no base-flipping signal when AP is opposite or adjacent to the target base. (a) Plot of A factor versus lifetime for the crystalline complexes APadj–M.HhaI–AdoHcy (squares) and APopp–M.HhaI–AdoHcy (diamonds), in comparison with APout–M.HhaI–AdoHcy (triangles). (b) Plot of A factors versus lifetimes for the solution phase systems APadj free duplex (open triangles); APadj ternary complex (closed triangles); APopp free duplex (open squares); and APopp ternary complex (closed squares).
In solution, the decay parameters of the APadj and APopp duplexes remain essentially unchanged on binding of the enzyme and cofactor (Table 3 and Supplementary Tables S10–S13), and they are unambiguously indicative of unflipped AP, as illustrated in Figure 5b. There is certainly no false-positive response to base flipping here; neither unstacking on one face in APadj nor removing the pairing partner in APopp gives the flipping signal.
DISCUSSION
In both crystalline and solution-phase complexes of M.HhaI with AP-labelled duplexes, we observe an unambiguous response of the AP fluorescence decay to base flipping. The similarity of the fluorescence decay functions of the duplexes in the corresponding crystalline and solution-phase systems is reassuring confirmation that the DNA–enzyme interactions occurring in solution are faithfully captured in the crystalline complexes. The time-resolved fluorescence measurements reveal conformational heterogeneity in the crystals at room temperature that is not apparent in the X-ray structure. The fluorescence decay can detect the existence of species that constitute only a few percent of the excited state population and is sensitive to transient conformational states that exist on the timescale of the excited state lifetime. Small changes in conformational geometry may significantly affect interbase interactions and hence the fluorescence lifetime. The crystal structure shows the average or dominant conformational geometry at 90 K, within the limits of the available structural resolution.
In solution, the excited state decay of the AP-labelled duplexes is probably influenced by base dynamics. Barton and coworkers (43,44) have demonstrated that electron transfer from guanine to AP in DNA is mediated by base motion. They propose a model in which base motion, during the AP excited state lifetime, enables the initially excited conformational population to sample geometries that have greater propensity for charge-transfer quenching by guanine. Recently, O'Neill and Barton (45) have reported fluorescence intensity measurements on AP-labelled DNA duplexes rendered rigid in a frozen matrix at 77 K. They show a dramatic increase in fluorescence intensity in the rigid helix, reflecting the loss of quenching of excited AP when base motion is prevented. For a duplex with AP immediately adjacent to guanine, the quantum yield measured relative to free AP in the same matrix, Φrel, increased ∼20-fold between 298 and 77 K, to a value of 0.45. It is informative to compare this with the Φrel value of 0.06 for the APout duplex in the crystalline state [where Φrel is the ratio of the number-average lifetime of APout, ΣAiτi, to the lifetime of AP riboside in aqueous solution (36)]. This suggests that in the crystal, at room temperature, the duplex is far from rigid and the base motions that facilitate quenching of AP remain largely uninhibited.
While the decay parameters of the crystalline and solution phase systems are generally very similar, there are small differences that reflect the greater conformational mobility of the complexes in solution. For the unflipped systems, the lifetimes of the highly quenched, intrahelical conformations (τ1 and τ2) are significantly shorter in solution than in the crystal. This is consistent with greater dynamic freedom and consequent enhanced charge-transfer quenching, in line with Barton's model. The difference between crystal and solution systems is most apparent for the base-flipped APtarget complex. In the crystalline complex, the very short decay component (τ1), characteristic of intrahelical base-stacked AP is absent, whereas in solution this component is detectable (with much reduced amplitude) after base flipping. This indicates that the target AP base continues to sample a base-stacked conformational state in the solution-phase complex, although with much reduced probability than in the free duplex. This is consistent with previous studies of base flipping in M.HhaI–DNA that have shown evidence of a dynamic equilibrium between flipped-out and stacked states of the target base (19,23). The concentrations of protein and DNA used in the solution studies ensure that all the DNA is bound to M.HhaI.
In the APopp and APadj duplexes, the constancy of the AP fluorescence decay function, when the neighbouring cytosine is flipped out, is striking, and demonstrates the specificity with which M.HhaI flips out the target base and the effectiveness of the enzyme in supporting the surrounding duplex structure during base flipping. The hydrogen bonding interactions between the enzyme and the AP successfully mimic the normal intrahelical environment of the base. The base-flipping event appears to have no significant impact on the conformational structure or dynamics of the duplex, as seen by the AP probe. The AP remains demonstrably intrahelical and does not show any increased tendency to occupy an extrahelical conformation.
The clear response of the AP decay function to base flipping is not restricted to the M.HhaI enzyme system. In preliminary studies of other methyltransferases, M.TaqI and M.EcoKI, we have observed a similar response (A.C. Jones, D.T.F. Dryden and E. Weinhold, unpublished data), clearly indicative of base flipping, and we expect that this will apply generally to DNA methyltransferase and repair enzymes. Thus, the use of AP time-resolved fluorescence as a probe of base flipping will enable the scope of investigation of this mechanism to be greatly expanded.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Supplementary Material
Acknowledgments
This research was supported by the Scottish Higher Education Funding Council, the Engineering and Physical Sciences Research Council, the Royal Society ESEP, the Howard Hughes Medical Institute, the Max Planck Society and the Lithuanian Science and Study Foundation. EMBL beamline access was supported by EC FP6 Structuring the European Research Area Programme. Funding to pay the Open Access publication charges for this article was provided by JISC.
Conflict of interest statement. None declared.
REFERENCES
- 1.Klimasauskas S., Kumar S., Roberts R.J., Cheng X.D. Hhal methyltransferase flips its target base out of the DNA helix. Cell. 1994;76:357–369. doi: 10.1016/0092-8674(94)90342-5. [DOI] [PubMed] [Google Scholar]
- 2.Reinisch K.M., Chen L., Verdine G.L., Lipscomb W.N. The crystal structure of HaeIII methyltransferase covalently complexed to DNA: an extrahelical cytosine and rearranged base pairing. Cell. 1995;82:143–153. doi: 10.1016/0092-8674(95)90060-8. [DOI] [PubMed] [Google Scholar]
- 3.Goedecke K., Pignot M., Goody R.S., Scheidig A.J., Weinhold E. Structure of the N6-adenine DNA methyltransferase M.TaqI in complex with DNA and a cofactor analog. Nature Struct. Biol. 2001;8:121–125. doi: 10.1038/84104. [DOI] [PubMed] [Google Scholar]
- 4.Stivers J.T., Jiang Y.L. A mechanistic perspective on the chemistry of DNA repair glycosylases. Chemical Rev. 2003;103:2729–2759. doi: 10.1021/cr010219b. [DOI] [PubMed] [Google Scholar]
- 5.Daniels D.S., Woo T.T., Luu K.X., Noll D.M., Clarke N.D., Pegg A.E., Tainer J.A. DNA binding and nucleotide flipping by the human DNA repair protein AGT. Nature Struct. Mol. Biol. 2004;11:714–720. doi: 10.1038/nsmb791. [DOI] [PubMed] [Google Scholar]
- 6.Banerjee A., Yang W., Karplus M., Verdine G.L. Structure of a repair enzyme interrogating undamaged DNA elucidates recognition of damaged DNA. Nature. 2005;434:612–618. doi: 10.1038/nature03458. [DOI] [PubMed] [Google Scholar]
- 7.Bickle T.A., Kruger D.H. Biology of DNA restriction. Microbiol. Rev. 1993;57:434–450. doi: 10.1128/mr.57.2.434-450.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Laird P.W. Cancer epigentics. Hum. Mol. Genet. 2005;14:R65–R76. doi: 10.1093/hmg/ddi113. [DOI] [PubMed] [Google Scholar]
- 9.Cheng X.D., Roberts R.J. AdoMet-dependent methylation, DNA methyltransferases and base flipping. Nucleic Acids Res. 2001;29:3784–3795. doi: 10.1093/nar/29.18.3784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beechem J.M., Otto M.R., Bloom L.B., Eritja R., Reha-Krantz L.J., Goodman M.F. Exonuclease-polymerase active site partitioning of primer-template DNA strands and equilibrium Mg2+ binding properties of bacteriophage T4 DNA polymerase. Biochemistry. 1998;37:10144–10155. doi: 10.1021/bi980074b. [DOI] [PubMed] [Google Scholar]
- 11.Raney K.D., Sowers L.C., Millar D.P., Benkovic S.J. A fluorescence-based assay for monitoring helicase activity. Proc. Natl Acad. Sci. USA. 1994;91:6644–6648. doi: 10.1073/pnas.91.14.6644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McCullough A.K., Dodson M.L., Scharer O.D., Lloyd R.S. The role of base flipping in damage recognition and catalysis by T4 endonuclease V. J. Biol. Chem. 1997;272:27210–27217. doi: 10.1074/jbc.272.43.27210. [DOI] [PubMed] [Google Scholar]
- 13.Rachofsky E.L., Seibert E., Stivers J.T., Osman R., Ross J.B.A. Conformation and dynamics of abasic sites in DNA investigated by time-resolved fluorescence of 2-aminopurine. Biochemistry. 2001;40:957–967. doi: 10.1021/bi001665g. [DOI] [PubMed] [Google Scholar]
- 14.Stivers J.T. 2-Aminopurine fluorescence studies of base stacking interactions at abasic sites in DNA: metal-ion and base sequence effects. Nucleic Acids Res. 1998;26:3837–3844. doi: 10.1093/nar/26.16.3837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stivers J.T., Pankiewicz K.W., Watanabe K.A. Kinetic mechanism of damage site recognition and uracil flipping by Escherichia coli uracil DNA glycosylase. Biochemistry. 1999;38:952–963. doi: 10.1021/bi9818669. [DOI] [PubMed] [Google Scholar]
- 16.Allan B.W., Reich N.O., Beechem J.M. Measurement of the absolute temporal coupling between DNA binding and base flipping. Biochemistry. 1999;38:5308–5314. doi: 10.1021/bi9900020. [DOI] [PubMed] [Google Scholar]
- 17.Allan B.W., Reich N.O. Targeted base stacking disruption by the EcoRI DNA methyltransferase. Biochemistry. 1996;35:14757–14762. doi: 10.1021/bi9615708. [DOI] [PubMed] [Google Scholar]
- 18.Allan B.W., Beechem J.M., Lindstrom W.M., Reich N.O. Direct real time observation of base flipping by the EcoRI DNA methyltransferase. J. Biol. Chem. 1998;273:2368–2373. doi: 10.1074/jbc.273.4.2368. [DOI] [PubMed] [Google Scholar]
- 19.Vilkaitis G., Dong A., Weinhold E., Cheng X., Klimasauskas S. Functional roles of the conserved threonine 250 in the target recognition domain of HhaI DNA methyltransferase. J. Biol. Chem. 2000;275:38722–38730. doi: 10.1074/jbc.m005278200. [DOI] [PubMed] [Google Scholar]
- 20.Holz B., Klimasauskas S., Serva S., Weinhold E. 2-Aminopurine as a fluorescent probe for DNA base flipping by methyltransferases. Nucleic Acids Res. 1998;26:1076–1083. doi: 10.1093/nar/26.4.1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gowher H., Jeltsch A. Molecular enzymology of the EcoRV DNA-(Adenine-N6)-methyltransferase: kinetics of DNA binding and bending, kinetic mechanism and linear diffusion of the enzyme on DNA. J. Mol. Biol. 2000;303:93–110. doi: 10.1006/jmbi.2000.4127. [DOI] [PubMed] [Google Scholar]
- 22.Reddy Y.V.R., Rao D.N. Binding of EcoP15I DNA methyltransferase to DNA reveals a large structural distortion within the recognition sequence. J. Mol. Biol. 2000;298:597–610. doi: 10.1006/jmbi.2000.3673. [DOI] [PubMed] [Google Scholar]
- 23.Klimasauskas S., Szyperski T., Serva S., Wuthrich K. Dynamic modes of the flipped-out cytosine during HhaI methyltransferase–DNA interactions in solution. EMBO J. 1998;17:317–324. doi: 10.1093/emboj/17.1.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vilkaitis G., Merkiene E., Serva S., Weinhold E., Klimasauskas S. The mechanism of DNA cytosine-5 methylation. Kinetic and mutational dissection of HhaI methyltransferase. J. Biol. Chem. 2001;276:20924–20934. doi: 10.1074/jbc.M101429200. [DOI] [PubMed] [Google Scholar]
- 25.Leslie A.G.W. Crystallographic Computing. Oxford, UK: Oxford University Press; 1990. [Google Scholar]
- 26.Evans P.R. 1997. Scala. Joint CCP4 and ESF-EACBM Newsletter 33; pp. 22–24. [Google Scholar]
- 27.French G.S., Wilson K.S. On the treatment of negative intensity observations. Acta Crystallogr. 1978;A34:517–525. [Google Scholar]
- 28.Collaborative Computational Project Number 4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. 1994;D50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 29.Jones T.A., Zou J.Y., Cowan S.W., Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. 1991;A47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 30.Read R. Improved Fourier coefficients for maps using phases from partial structures with errors. Acta Crystallogr. 1986;A42:140–149. [Google Scholar]
- 31.Brunger A.T., Adams P.D., Clore G.M., Delano W.L., Gros P., Grosse-Kunstleve R.W., Jiang J.S., Kuszewski J., Nilges M., Pannu N.S., et al. Crystallography and NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. 1998;D54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 32.O'Gara M., Horton J.R., Roberts R.J., Cheng X.D. Structures of HhaI methyltransferase complexed with substrates containing mismatches at the target base. Nature Struct. Biol. 1998;5:872–877. doi: 10.1038/2312. [DOI] [PubMed] [Google Scholar]
- 33.Kelley S.O., Barton J.K. Electron transfer between bases in double helical DNA. Science. 1999;283:375–381. doi: 10.1126/science.283.5400.375. [DOI] [PubMed] [Google Scholar]
- 34.Wan C., Fiebig T., Schiemann O., Barton J.K., Zewail A.H. Femtosecond direct observation of charge transfer between bases in DNA. Proc. Natl Acad. Sci. USA. 2000;97:14052–14055. doi: 10.1073/pnas.250483297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rachofsky E.L., Osman R., Ross J.B.A. Probing structure and dynamics of DNA with 2-aminopurine: effects of local environment on fluorescence. Biochemistry. 2001;40:946–956. doi: 10.1021/bi001664o. [DOI] [PubMed] [Google Scholar]
- 36.Neely R.K., Magennis S.W., Dryden D.T.F., Jones A.C. Evidence of tautomerism in 2-aminopurine from fluorescence lifetime measurements. J. Phys. Chem. B. 2004;108:17606–17610. [Google Scholar]
- 37.DePristo M.A., de Bakker P.I.W., Blundell T.L. Heterogeneity and inaccuracy in protein structures solved by X-ray crystallography. Structure. 2004;12:831–838. doi: 10.1016/j.str.2004.02.031. [DOI] [PubMed] [Google Scholar]
- 38.Nordlund T.M., Andersson S., Nilsson L., Rigler R. Structure and dynamics of a fluorescent DNA oligomer containing the EcoRI recognition sequence: fluorescence, molecular dynamics, and NMR studies. Biochemistry. 1989;28:9095–9103. doi: 10.1021/bi00449a021. [DOI] [PubMed] [Google Scholar]
- 39.Guest C.R., Hochstrasser R.A., Sowers L.C., Millar D.P. Dynamics of mismatched base pairs in DNA. Biochemistry. 1991;30:3271–3279. doi: 10.1021/bi00227a015. [DOI] [PubMed] [Google Scholar]
- 40.Hochstrasser R.A., Carver T.E., Sowers L.C., Millar D.P. Melting of a DNA helix terminus within the active site of a DNA polymerase. Biochemistry. 1994;33:11971–11979. doi: 10.1021/bi00205a036. [DOI] [PubMed] [Google Scholar]
- 41.Rachofsky E.L., Sowers L., Hawkins M.L., Balis F.M., Laws W.R., Ross J.B.A. Emission kinetics of fluorescent nucleoside analogs. Proc. SPIE. 1998;3256:68–75. [Google Scholar]
- 42.Holmen A., Norden B., Albinsson B. Electronic transition moments of 2-aminopurine. J. Am. Chem. Soc. 1997;119:3114–3121. [Google Scholar]
- 43.O'Neill M.A., Barton J.K. DNA charge transport: conformationally gated hopping through stacked domains. J. Am. Chem. Soc. 2004;126:11471–11483. doi: 10.1021/ja048956n. [DOI] [PubMed] [Google Scholar]
- 44.O'Neill M.A., Becker H.C., Wan C.Z., Barton J.K., Zewail A.H. Ultrafast dynamics in DNA-mediated electron transfer: base gating and the role of temperature. Angew. Chem. Int. Ed. 2003;42:5896–5900. doi: 10.1002/anie.200352831. [DOI] [PubMed] [Google Scholar]
- 45.O'Neill M.A., Barton J.K. DNA-mediated charge transport requires conformational motion of the DNA bases: elimination of charge transport in rigid glasses at 77 K. J. Am. Chem. Soc. 2004;126:13234–13235. doi: 10.1021/ja0455897. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.