Abstract
Background
Regulatory functions of nitric oxide (NO•) that bypass the second messenger cGMP are incompletely understood. Here, cGMP-independent effects of NO• on gene expression were globally examined in U937 cells, a human monoblastoid line that constitutively lacks soluble guanylate cyclase. Differentiated U937 cells (>80% in G0/G1) were exposed to S-nitrosoglutathione, a NO• donor, or glutathione alone (control) for 6 h without or with dibutyryl-cAMP (Bt2cAMP), and then harvested to extract total RNA for microarray analysis. Bt2cAMP was used to block signaling attributable to NO•-induced decreases in cAMP.
Results
NO• regulated 110 transcripts that annotated disproportionately to the cell cycle and cell proliferation (47/110, 43%) and more frequently than expected contained AU-rich, post-transcriptional regulatory elements (ARE). Bt2cAMP regulated 106 genes; cell cycle gene enrichment did not reach significance. Like NO•, Bt2cAMP was associated with ARE-containing transcripts. A comparison of NO• and Bt2cAMP effects showed that NO• regulation of cell cycle genes was independent of its ability to interfere with cAMP signaling. Cell cycle genes induced by NO• annotated to G1/S (7/8) and included E2F1 and p21/Waf1/Cip1; 6 of these 7 were E2F target genes involved in G1/S transition. Repressed genes were G2/M associated (24/27); 8 of 27 were known targets of p21. E2F1 mRNA and protein were increased by NO•, as was E2F1 binding to E2F promoter elements. NO• activated p38 MAPK, stabilizing p21 mRNA (an ARE-containing transcript) and increasing p21 protein; this increased protein binding to CDE/CHR promoter sites of p21 target genes, repressing key G2/M phase genes, and increasing the proportion of cells in G2/M.
Conclusion
NO• coordinates a highly integrated program of cell cycle arrest that regulates a large number of genes, but does not require signaling through cGMP. In humans, antiproliferative effects of NO• may rely substantially on cGMP-independent mechanisms. Stress kinase signaling and alterations in mRNA stability appear to be major pathways by which NO• regulates the transcriptome.
Background
Nitric oxide (NO•) plays a pivotal role in vascular biology through both cGMP-dependent and -independent mechanisms. In health, NO• regulates vascular tone by activating soluble guanylate cyclase [1-3]. However, other important effects of NO• in the vasculature such as cytoprotection and anti-adhesion appear to occur independent of cGMP signaling [4-6]. Likewise, NO• regulation of inflammation has frequently been associated with signal transduction events that do not involve cGMP [7,8]. NO• induces TNFα in human cells by decreasing intracellular levels of cAMP, thereby removing cAMP-mediated repression of the TNFα promoter through a proximal Sp element [9,10]. Analogs of cAMP and Sp site mutation both block, while antagonists of cAMP-dependent protein kinase simulate the effect of NO• on TNFα. [9,11]. In contrast to TNFα, NO• induces interleukin-8 (IL-8) [12] through a distinct post-transcriptional mechanism that is both cGMP- and cAMP-independent. IL-8 mRNA is stabilized by NO• activation of p38 MAPK, increasing its half-life and translation [13]. These and other reports [14-16]. suggest that cGMP-independent gene regulation by NO• occurs through multiple pathways.
Similar to the regulation of blood pressure and inflammatory responses, NO• regulation of cell proliferation is of central importance to circulatory health. Failure of this regulatory pathway has been linked to atherosclerosis and other forms of vascular dysfunction [17-19]. Despite extensive investigation, the relative contribution of cGMP-independent NO• signaling in the regulation of cell cycle genes remains controversial. In rats, NO• has been shown to activate transcription through cGMP-dependent effects on AP-1 promoter sites [20]. Also in rodents, a NO•-cGMP-PKA-ERK1/2 signal transduction pathway has been described that inhibits cell proliferation [21,22] and increases expression of p21/Waf1/Cip1 [23,24]. A master regulatory gene, p21 directly inhibits Cdk complexes [25,26] and represses the transcription of many cell cycle genes through CDE/CHR (cell cycle dependent element/cell cycle gene homology region) promoter elements [27,28]. In contrast to rodents, NO• regulation of cell cycle genes in humans, including regulation of p21, appears to occur, at least in part, independent of cGMP [19,29]. However, a global examination of cGMP-independent NO• effects on the transcriptome in general or on cell cycle genes specifically has not been undertaken in either rodents or humans.
Here, oligonucleotide microarrays and human U937 cells that lack soluble guanylate cyclase [9,30] were used to globally characterize the cGMP-independent effects of NO• on gene expression. Differentiation with PMA was employed to render cells capable of cytokine production [9]. This treatment also forced >80% of cells into the G0/G1 phase of the cell cycle, which facilitated the analysis of cell cycle gene regulation. Since NO• lowers cAMP levels in U937 cells [9] and cAMP is known to affect cell proliferation, NO• effects were also tested in the absence and presence of a cell permeable cAMP analog. For genes affected by NO•-induced decreases in cAMP, cAMP analog would be expected to produce an apposite effect. Hypotheses generated from microarray results were further investigated by examining downstream changes in protein expression and signal transduction pathways.
Results
Functional distribution of NO•-regulated genes and hypothesis generation
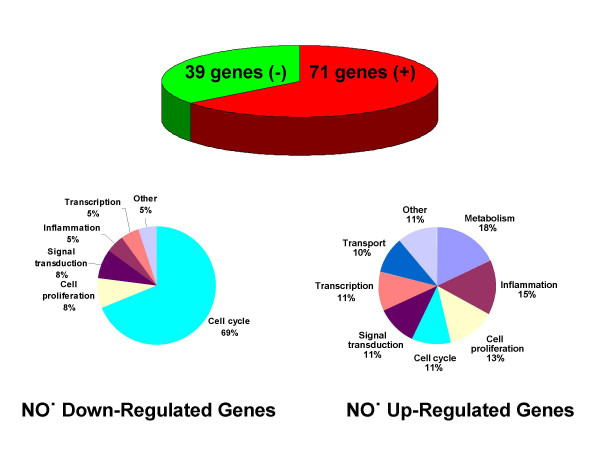
Of 110 NO•-responsive genes, 71 were induced, and 39 were repressed; the majority were not previously known to be NO•-responsive. Both naïve and differentiated U937 cells lack NO•-sensitive soluble guanylate cyclase [9,30], and therefore gene regulation by NO• in these cells can be attributed to cGMP-independent mechanisms. Genes were annotated into functional categories (Fig. 1) [see Additional files 1 and 2 for complete gene lists]. NO• had broad biological effects independent of cGMP. Heme oxygenase 1 (HMOX1), a known NO•-responsive gene, had the second largest fold change among up-regulated genes. TNFα and IL-8, cytokines previously associated with specific cGMP-independent mechanisms of NO• regulation [8,9,12], were also detected as differentially regulated by the microarray analysis. NO•-regulated genes annotated disproportionately to the cell cycle [[31] of 106 (29%) compared to 407 of 4870 genes (8%) on the microarray, as annotated in the Gene Ontology [GO] Biological Process database; P = 0.0001] (Table 1). In particular, a large majority of NO• down-regulated genes annotated specifically to the cell cycle (21/38, as annotated in the GO Biological Process database; P = 0.0001). Additional annotation using PubMed identified 47 of 110 genes (43%) as cell cycle or cell proliferation related. Of 39 down-regulated transcripts, 27 (69%) were ultimately annotated specifically to the cell cycle.
Figure 1.
Distribution of NO•-regulated genes. Of 110 differentially regulated genes, 71 were up-regulated (red) and 39 (green) were down-regulated. Genes were classified into functional categories using NIH-DAVID [83] and PubMed [52]. Data are from seven independent microarray experiments.
Table 1.
NO•-regulated cell cycle related genes
| GenBank | Symbol | Functional category | Fold changea |
| G1/S | |||
| J04111 | JUN | Regulation of cell cycle | 3.68 ± 0.76 |
| U09579 | CDKN1A (p21) | CDK inhibitor | 2.06 ± 0.58 |
| U77949 | CDC6 | DNA replication | 1.95 ± 0.48 |
| M74093 | CCNE1 | Cell cycle control | 1.77 ± 0.31 |
| X89398 | UNG | DNA repair | 1.68 ± 0.30 |
| S49592 | E2F1 | G1 phase of mitotic cell cycle | 1.60 ± 0.19 |
| X61123 | BTG1 | Negative regulation of cell proliferation | 1.60 ± 0.28 |
| L13689 | BMI1 | Modifies chromatin | 0.67 ± 0.09 |
| U67369 | GFI1 | G1/S-specific transcription in mitotic cell cycle | 0.54 ± 0.06 |
| U22376 | c-Myb | Cell cycle control | 0.30 ± 0.10 |
| G2 | |||
| U66838 | CCNA1 | Regulation of CDK activity | 0.60 ± 0.12 |
| Z36714 | CCNF | Regulation of cell cycle | 0.60 ± 0.11 |
| J04088 | 0.57 ± 0.14 | ||
| L47276 | TOP2A b | Spindle assembly, chromsome condensation | 0.42 ± 0.07 |
| X05360 | CDC2 | Start control point of mitotic cell cycle | 0.57 ± 0.11 |
| U28386 | KPNA2 | Cytoskeleton organization and biogenesis | 0.51 ± 0.05 |
| D14678 | KIFC1 | Spindle assembly, chromsome condensation | 0.44 ± 0.10 |
| U14518 | CENPA | Chromosome organization and biogenesis | 0.38 ± 0.06 |
| G2/M | |||
| U48807 | DUSP4 | Regulation of cell cycle | 1.40 ± 0.35 |
| U63743 | KIF2C | Microtubule motor activity | 0.72 ± 0.10 |
| U83115 | AIM1 | Tumor supressor, cytoskeleton | 0.71 ± 0.07 |
| M34458 | LMNB1 | Cytoskeletal anchoring | 0.66 ± 0.16 |
| D63880 | CNAP1 | Mitotic surveilance | 0.63 ± 0.05 |
| X65550 | MKI67 | Chromatin/chromosome structure | 0.61 ± 0.09 |
| S78187 | CDC25B | Cell cycle control | 0.60 ± 0.05 |
| D38553 | BRRN1 | Chromatid separation | 0.59 ± 0.08 |
| U73379 | UBE2C | Protein degradation | 0.54 ± 0.08 |
| U30872 | CENPF | Mitosis | 0.53 ± 0.07 |
| Z15005 | CENPE | Mitotic chromosome movement | 0.53 ± 0.10 |
| U29343 | HMMR | Mitotic surveilance, cell motility | 0.51 ± 0.11 |
| U05340 | CDC20 | Ubiquitin-dependent protein degradation | 0.49 ± 0.07 |
| M86699 | TTK | Spindle assembly/mitotic checkpoint | 0.47 ± 0.05 |
| U01038 | PLK | Mitosis | 0.44 ± 0.04 |
| M25753 | CCNB1 | Mitotic checkpoint | 0.43 ± 0.07 |
| D38751 | KIF22 | Mitosis | 0.40 ± 0.18 |
| U04810 | TROAP | Cell adhesion | 0.24 ± 0.03 |
a Fold change comparing glutathione (GSH) to S-nitrosoglutathione (GSNO)-treated cells, expressed as the mean ± SE (N = 7)
b Represented by more than one probe set on the microarray that reached statistical significance; each result is shown
Previously, we demonstrated that NO• activates p38 MAPK and thereby stabilizes IL-8 mRNA through effects on AU-rich elements [ARE] in 3' untranslated regions [UTR] [13]. Therefore, the ARE database http://rc.kfshrc.edu.sa/ared was used to identify ARE-containing genes among those regulated by NO•. Twenty-two of 110 genes contained ARE (20%) compared to 540 ARE genes of 5086 on the microarray (11%; P = 0.008). An additional 11 ARE-containing genes were identified in PubMed for a total of 33 (Table 2). Nearly half of these genes (14/33; 42%) have been reported to be p38 MAPK regulated (Table 2). Importantly, for these 14 genes, p38 MAPK activation produces responses that are in the same direction as those observed here for NO•. The broad influence of NO• on cell cycle-related genes and ARE-containing transcripts independent of cGMP was unexpected, as was the strong association of these effects with p38 MAPK. Therefore, further experiments were performed to confirm these results and to define underlying regulatory mechanisms that might link NO• effects on the cell cycle with post-transcriptional gene regulation through ARE sites.
Table 2.
NO•-regulated genes containing AU-rich elements
| GenBank | Symbol | Regulation by NO• | Reported regulation by p38 MAPKa |
| X54150 | FCAR | UP | |
| L06633 | PSCDBP | UP | |
| K03195 | SLC2A1 | UP | |
| X59834 | GLUL | UP | |
| U03398 | TNFSF9 | UP | |
| M59465 | TNFAIP3 | UP | UP [77] |
| U15174 | BNIP3 | UP | |
| D86962 | GRB10 | UP | |
| M60278 | DTR | UP | UP [77] |
| M69043 | NFKBIA | UP | |
| M92357 | TNFAIP2 | UP | |
| U48807 | DUSP4 | UP | |
| M16750 | PIM1 | UP | UP [77] |
| X89398 | UNG | UP | UP [77] |
| S49592 | E2F1 | UP | UP [66] |
| U70426 | RGS16 | UP | |
| X02910 | TNF | UP | UP [87, 88] |
| X04500 | IL1B | UP | UP [77, 87] |
| M20681 | SLC2A3 | UP | |
| M28130 | IL8 | UP | UP [77, 89] |
| M57731 | GRO2 | UP | UP [77] |
| U71203 | RIT | UP | |
| S81914 | IER3 | UP | |
| J04076 | EGR2 | UP | |
| D90070 | PMAIP1 | UP | UP [77] |
| J04111 | JUN | UP | UP [90] |
| U09579 | CDKN1A | UP | UP [42] |
| D16532 | VLDLR | UP | |
| U14518 | CENPA | DOWN | |
| U67369 | GFI1 | DOWN | |
| U22376 | c-Myb | DOWN | |
| U66838 | CCNA1 | DOWN | DOWN [91] |
| M25753 | CCNB1 | DOWN | DOWN [74] |
a As reported in the literature; citations shown in parentheses
Validation of NO•-regulated genes
To determine whether NO• and cAMP effects on mRNA, as measured by microarray, produced downstream effects on secreted protein, TNFα, IL-8 and IL-1β were measured in supernatants collected from parallel cell cultures incubated for 24 h. S-nitrosoglutathione (GSNO) significantly increased TNFα, IL-8, and IL-1β protein [see Additional file 3, part A (P < 0.0001 for all)]. In contrast, cAMP decreased TNFα (P < 0.0001), increased IL-1β (P = 0.003), and had no significant effect on IL-8. NO•-induced changes in transcript abundance as determined by microarray were consistent with these results (see Additional file 3, part B].
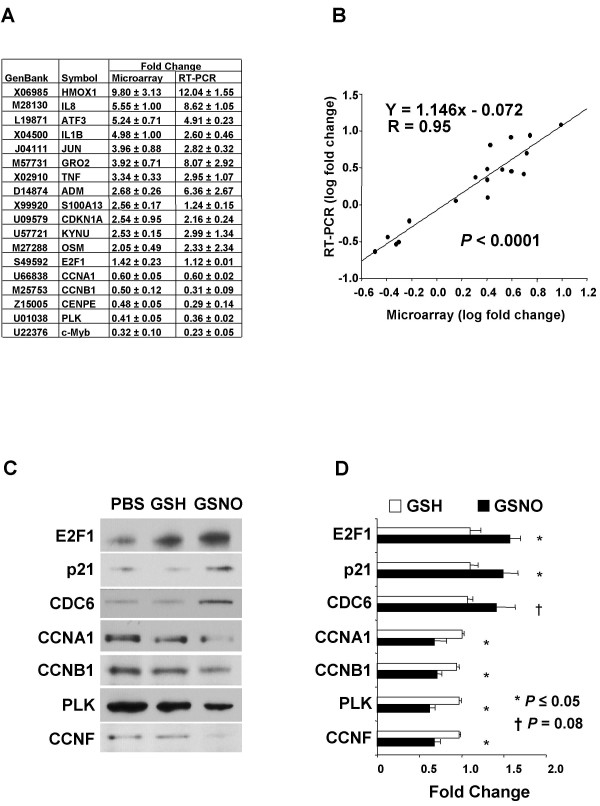
Real-time RT- PCR (TaqMan®) was used to validate NO•-mediated changes in mRNA levels (Fig. 2A and 2B). Of 18 selected genes, 13 were NO• up-regulated, and 5 were down-regulated. Fold changes from microarray experiments strongly correlated with results from RT-PCR (R = 0.95, P < 0.0001).
Figure 2.
Validation of microarray results by real-time, reverse transcription (RT) PCR and Western blotting. (A) Fold changes for microarray and RT-PCR at 6 h comparing S-nitrosoglutathione (GSNO; 400 μM) to glutathione (GSH; 400 μM) incubated cells. (B) Correlation of fold change comparing the two methods after logarithmic transformation. Results are means ± SE of the last three microarray experiments for which material was available for RT-PCR (performed in triplicate). (C) Representative Western blots of cell cycle genes detected by specific antibody and enhanced chemiluminescence. Differentiated U937 cells (1 × 107) were incubated with PBS, GSH (400 μM) or GSNO (400 μM) for 12 h and then lysed for Western blotting. (D) Western blot results quantified with laser densitometry and expressed as ratios relative to PBS control values. Data are means ± SE of three or four independent experiments.
Western blotting of key cell cycle genes regulated by NO• was performed to test whether microarray results accurately predicted changes in protein expression (Fig. 2C and 2D). Three induced genes, E2F transcription factor 1 (E2F1), p21/Waf1/Cip1 (Cdk inhibitor; CDKN1A), and cell division cycle 6 (CDC6) were examined. E2F1 and p21 are well-characterized master regulatory proteins that control the cell cycle. Four repressed genes, cyclin A1 (CCNA1), cyclin B1 (CCNB1), polo-like kinase (PLK) and cyclin F (CCNF) were also measured by Western blotting. In all cases, directional changes in protein expression were consistent with the differential effect of NO• on corresponding transcripts as determined by microarray analysis.
NO•-regulation of cell cycle genes independent of cAMP
NO• induces TNFα by decreasing intracellular cAMP; dibutyryl-cAMP (Bt2cAMP), a cell permeable cAMP analog, blocks this effect [9,10]. Moreover, cAMP is an omnipresent second messenger that affects cell proliferation and the cell cycle in a variety of contexts [31-33]. Therefore, Bt2cAMP was added to some conditions to test for the cAMP-dependence of NO•-mediated effects. Of 106 Bt2cAMP-responsive genes, 16 of 103 (16%) compared to 407 of 4870 on the microarray (8%), as annotated in the GO Biological Process database, were cell cycle related, but this effect was not statistically significant (P = 0.5). Only one additional cAMP-responsive gene was subsequently annotated to the cell cycle by searching PubMed [see Additional file 4]. However, like NO•, cAMP-regulated genes did contain ARE more frequently than expected [[24] of 106 (23%) compared to 540 of 5086 (11%) on the microarray; P = 0.0003]. This finding was consistent with the known ability of cAMP to stabilize transcripts that contain ARE [34].
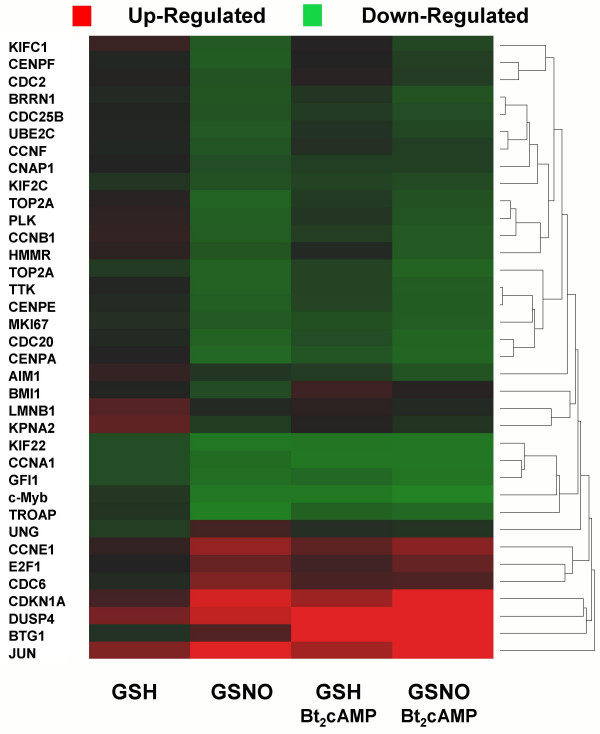
To further compare the effects of NO• and cAMP, a hierarchical cluster analysis was performed using the 35 cell cycle genes regulated by NO• (Fig. 3). For each of the 5 cell cycle genes significantly affected by both NO• and Bt2cAMP [c-Myb, B-cell translocation gene 1 (BTG1), dual specificity phosphatase 4 (DUSP4), growth factor independent 1 (GFI1), and cyclin A1 (CCNA1)], the direction of regulation was the same. Further, for cell cycle genes regulated by NO•, cAMP analog either had no effect on or produced expression changes that were similar to and additive with those observed for NO• (Fig. 3). These results suggest that NO• effects on cell cycle genes are independent of its interference with cAMP signaling, since cAMP analog (the opposite signal) was not antagonistic to the actions of NO•.
Figure 3.
Heat map of NO• and cAMP effects on the expression of NO•-regulated cell cycle genes. Differentiated U937 cells were incubated with glutathione (GSH; 400 μM) or S-nitrosoglutathione (GSNO; 400 μM) in the absence or presence of dibutyryl-cAMP (Bt2cAMP; 100 μM). Color intensity reflects fold change from differentiated cells at 0 h; up-regulation is shown in red and down-regulation in green. Fold changes were computed from the mean results of seven independent microarray experiments.
Analysis of NO• effects on the cell cycle
NO• causes arrest in either the G1 or G2/M phase of the cell cycle [19,35-37]. However, the mechanisms underlying this effect are not well characterized. Annotation of NO•-regulated genes to their respective phase of the cell cycle revealed that expression changes were not random (Table 1). Most NO• up-regulated genes (7/8) were G1/S associated, while down-regulated genes were strikingly G2 and G2/M phase associated (24/27). The latter included topoisomerase II alpha (TOP2A), cyclin B1, PLK, and CDC25B, genes that are necessary factors for mitosis. These results show that NO• suppresses the cell cycle in early G2/M by triggering a highly integrated program of gene regulation that does not require soluble guanylate cyclase or cGMP.
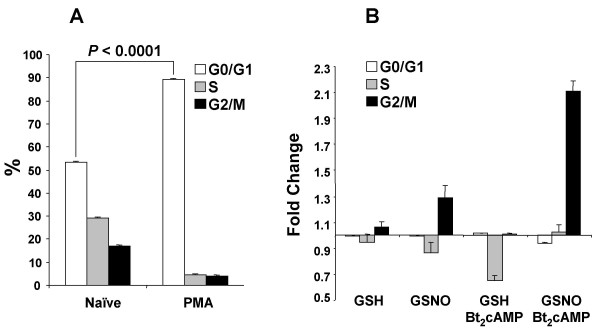
To further test this hypothesis, cell cycle analysis was performed on U937 cells using flow cytometry. PMA-differentiation significantly increased the portion of cells in G0/G1 (P < 0.0001), while decreasing cells in S (P = 0.0007) and G2/M (P = 0.003) compared to a naïve, undifferentiated cell population (Fig. 4A). Differentiated cells were then treated with glutathione (GSH) or GSNO in the absence or presence of Bt2cAMP. Consistent with NO•-induced changes in mRNA expression at 6 h, cell cycle analysis at 24 h demonstrated that NO• increased the portion of cells in G2/M (P = 0.0004), and in combination with Bt2cAMP, NO• synergistically increased G2/M phase cells (Fig. 4B; P = 0.008).
Figure 4.
Cell cycle analysis of U937 cells. (A) U937 cells (1 × 106) were differentiated with PMA (100 nM) for 48 h and then compared with naïve cells for cell cycle distribution using propidium iodide staining. The cell cycle distribution of stained cells was examined by flow cytometry. The percentage of cells in G0/G1, S, and G2/M was determined using ModFit software. (B) U937 cells (1 × 106) differentiated as in A were treated for 24 h with medium alone, medium with glutathione (GSH; 400 μM) or medium with S-nitrosoglutathione (GSNO; 400 μM) in the absence or presence of dibutyryl-cAMP (Bt2cAMP; 100 μM). Cell cycle distribution data, presented as fold change (percentage of cells in each phase of the cell cycle relative to medium alone), are means ± SE of four independent experiments.
NO• induction of p21, a master cell cycle regulator; dependence on p38 MAPK and role of mRNA stabilization
The Cdk inhibitor, p21 is known to induce cell cycle arrest in late G1 or early G2/M, [38-40] effects similar to those of NO•. NO• increased both p21 mRNA and protein expression in the current experiments. We have previously shown that NO• activates p38 MAPK in U937 and THP-1 cells [13,41]. Activation of p38 MAPK induces p21 in other cell types, [42] and like NO• here, can trigger G2/M cell arrest [40]. We therefore reasoned that NO• might up-regulate p21 by activating p38 MAPK in the present system.
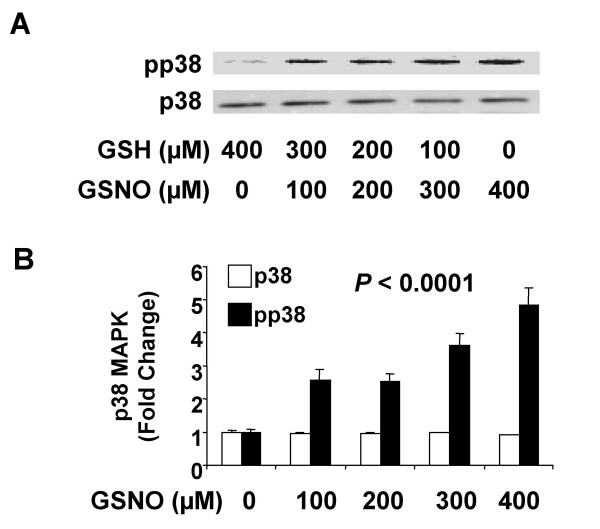
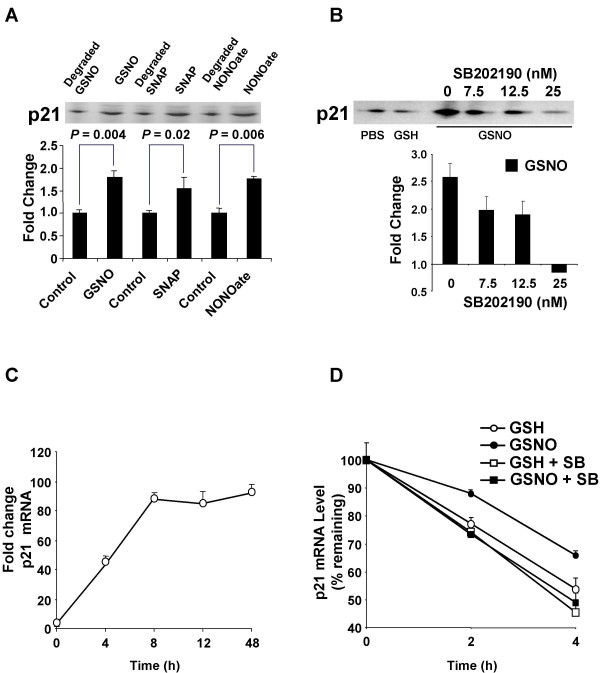
GSNO was first confirmed in PMA-differentiated U937 cells to dose-dependently increase p38 MAPK activation. This effect reached significance at the lowest (100 μM) GSNO concentration tested (P ≤ 0.0001; Fig. 5). Next, three chemically distinct NO• donors were tested for their ability to up-regulate p21 protein in PMA-differentiated U937 cells. GSNO, S-nitroso-N-acetylpenicillamine (SNAP), and DETA-NONOate similarly increased p21 expression in these cells compared to degraded controls (Fig. 6A; P < 0.05 for all). Thus, independent of cGMP and type of donor molecule, NO• consistently increased the expression of p21 protein in U937 cells. A specific p38 MAPK inhibitor (SB202190) was used to determine whether blocking this pathway could prevent NO• induction of p21. As shown in Fig. 6B, SB202190 dose-dependently reduced NO•-induced p21 protein expression (P = 0.0005). Collectively, these results suggest that NO• induces p21 through p38 MAPK activation. Finally, we investigated the effects of NO• and p38 MAPK inhibition on the stability of p21 mRNA, which harbors ARE in its 3' UTR [43]. After 8 h of PMA exposure, p21 expression increased almost 100 fold compared to naïve U937 cells (Fig. 6C). NO• stabilized p21 mRNA in the absence of SB202190 (Fig. 6D;P = 0.004), but had no effect in the presence of SB202190 (P = 0.5).
Figure 5.
Effect of NO• on p38 MAPK phosphorylation. Differentiated U937 cells (5 × 10 6) were incubated with S-nitrosoglutathione (GSNO; 0–400 μM) for 30 min; cells were then lysed for Western blotting to detect total (p38) and phosphorylated forms (pp38) p38 MAPK. (A) Representative gel for Western blotting. (B) Western blotting results were quantified with laser densitometry and expressed as ratios relative to control values (GSNO = 0 μM). Data are means ± SE of three independent experiments.
Figure 6.
Effect of NO• donors and p38 MAPK inhibition on p21 expression and mRNA stabilization. (A) Differentiated U937 cells (1 × 107) were incubated with NO• donors, S-nitrosoglutathione (GSNO; 400 μM), S-nitroso-N-acetylpenicillamine (SNAP; 400 μM), or DETA-NONOate (1 mM) or their degraded controls. Western blotting was then performed to detect p21 expression after 12 h of incubation. Results were quantified with laser densitometry and expressed as ratios relative to their appropriate degraded control. Data are means ± SE of four independent experiments. (B) Differentiated U937 cells (1 × 107) were incubated with increasing concentrations of the p38 inhibitor SB202190 (0 nM to 25 nM) for 30 min, then exposed to PBS, glutathione (GSH; 400 μM) or GSNO (400 μM) for 12 h. Western blotting was performed to detect p21 expression. Results were quantified with laser densitometry. Data, presented as fold change relative to PBS control values, are means ± SE of three independent experiments. Next, TaqMan® RT-PCR was used to quantify p21 mRNA levels normalized to GAPDH mRNA. (C) Changes in p21 mRNA levels during differentiation of U937 cells (1 × 107) with PMA. Data, presented as fold change relative to mean mRNA level in naïve cells, are means ± SE of three independent experiments. (D) NO• stabilization of p21 mRNA is dependent on p38 MAPK. U937 cells (1 × 107) were differentiated with PMA for 8 h. After 30 min pretreatment with actinomycin D (2.5 μg/ml) without and with SB202190 (0.1 μM), cells were further incubated with GSH (400 μM) or GSNO (400 μM) for 2 to 4 h. At the specific time points, cells were harvested for total RNA extraction. Data, presented relative to mRNA level at 0 h (arbitrarily set to 100%), are means ± SE of three independent experiments.
NO• regulation of the cell cycle through E2F1 and p21
The E2F family of transcription factors and p21 act as master regulatory switches that control the cell cycle. E2F1 regulates target genes through E2F-binding sites and thereby plays an essential role in DNA synthesis and the G1/S transition [44-47]. Some p21 effects are mediated by inhibition of E2F factor binding, while other downstream targets contain cell cycle dependent element/cell cycle gene homology region [CDE/CHR] repressor sites within their promoters [27,28,39,48-50]. Protein binding to CDE/CHR sites, triggered by p21 expression, causes repression of a diverse group of cell cycle genes and subsequent late G1 or early G2/M phase arrest, responses that are highly similar to NO• effects shown here. The ability of NO• to increase the expression of E2F1 and p21 may explain much of its broad control over the cell cycle that ultimately involves dozens of gene products. We therefore identified NO•-regulated genes that contain E2F or CDE/CHR promoter sites by searching TRANSFAC [51] and PubMed[52]. Sixteen NO•-regulated genes contain apparent E2F sites (Table 3A). Seven of these are annotated to the G1/S phase of the cell cycle, six of which have reported E2F1 responses that are concordant with NO• effects in the current experiment. Notably, of 10 NO• down-regulated transcripts with possible E2F-binding sites, only c-Myb is a G1/S phase gene. Further, 5 of these genes are known to also contain a CDE/CHR binding site that appears to be functionally dominant (Table 3B). Moreover, of the 27 cell cycle genes down-regulated by NO•, 8 are known targets of p21 repression (all G2/M associated), including 6 genes with putative CDE/CHR sites (Table 3B) and two others, lamin B1 (LMNB1) and centromere protein F (CENPF) [39].
Table 3A.
Specific promoter elements associated with NO•-regulated cell cycle genes
| 3A. Genes with E2F sites | ||||
| GenBank | Symbol | Cell cycle phase | Regulation by NO• | Regulation by E2F1a |
| J04111 | JUN | G1/S | Up | Up [63] |
| U09579 | CDKN1A | G1/S | Up | Up [59] |
| U77949 | CDC6 | G1/S | Up | Up [61] |
| M74093 | CCNE1 | G1/S | Up | Up [63] |
| X89398 | UNG | G1/S | Up | Up [64] |
| S49592 | E2F1 | G1/S | Up | Up [61] |
| U22376 | c-Myb | G1/S | Down | Up [63, 92] |
| U66838 | CCNA1 b | G2 | Down | Up [93] |
| J04088 | TOP2Ab | G2 | Down | Up [62] |
| L47276 | TOP2A b | G2 | Down | Up [62] |
| X05360 | CDC2 b | G2 | Down | Up [62, 64] |
| X65550 | MKI67 | G2M | Down | Up [63] |
| U14518 | CENPA b | G2/M | Down | |
| S78187 | CDC25B | G2/M | Down | |
| M86699 | TTK | G2/M | Down | |
| U01038 | PLK b | G2/M | Down | |
| M25753 | CENPE | G2/M | Down | |
a As reported in the literature; citations shown in parentheses
b These genes also contain CDE/CHR sites as shown in Table 3B
Table 3B.
Specific promoter elements associated with NO•-regulated cell cycle genes
| 3B. Genes with CDE/CHR sites | ||||
| GenBank | Symbol | Cell cycle phase | Regulation by NO• | Reported regulation by p21a |
| U66838 | CCNA1 b | G2 | Down | Down |
| J04088 | TOP2A b | G2 | Down | Down |
| L47276 | TOP2A b | G2 | Down | Down |
| X05360 | CDC2 b | G2 | Down | Down |
| U14518 | CENPA b | G2/M | Down | Down |
| U01038 | PLK b | G2/M | Down | Down |
| M25753 | CCNB1 | G2/M | Down | Down |
a As reported in the literature [39]
b These genes also contain E2F sites as shown in Table 3A
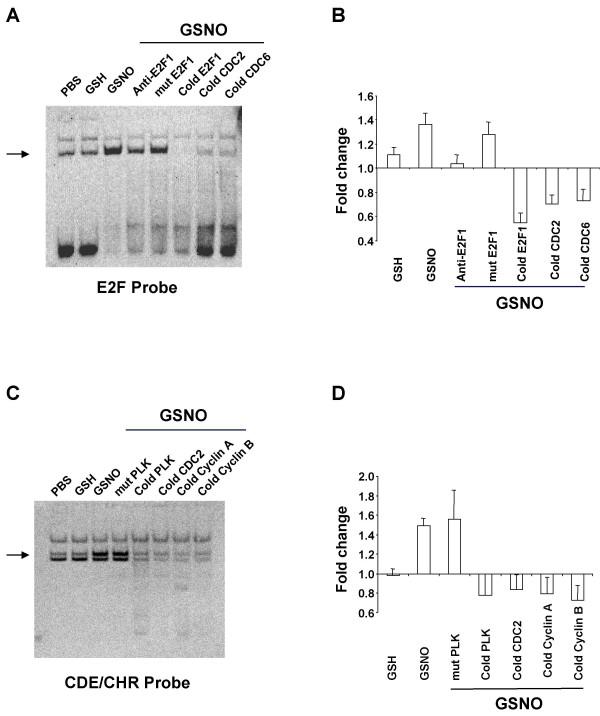
Next, electrophoretic mobility shift assays (EMSA) were performed to test whether NO• altered protein binding to E2F and CDE/CHR consensus sequences. PMA-differentiated U937 cells were treated with PBS, GSH, or GSNO followed by preparation of nuclear extract. NO• increased binding to both E2F (Fig. 7A and 7B) and CDE/CHR probes (Fig. 7C and 7D). Site-directed mutagenesis of each consensus sequence abolished competition (Fig. 7) and E2F1-directed antibody blocked complex formation with labeled E2F probe (Fig. 7A and 7B).
Figure 7.
NO• increases protein binding to E2F and CDE/CHR promoter sites. (A) Representative gel for protein binding to the E2F probe detected by ECL. (B) Results for the E2F probe quantified with laser densitometry. Data, presented as ratios relative to PBS values (set to 1), are means ± SE of six independent experiments. (C) Representative gel for protein binding to CDE/CHR probe detected by ECL. (D) Results for the CDE/CHR probe quantified with laser densitometry. Data, presented as ratios relative to PBS values (set to 1), are means ± SE of six independent experiments. Differentiated U937 cells were incubated with glutathione (GSH; 400 μM) or S-nitrosoglutathione (GSNO; 400 μM) for 3 h. Nuclear extract (15 μg) was prepared and incubated with double-stranded, biotin-N4-CTP labeled DNA probe representing the E2F or CDE/CHR concensus sequence from the E2F1 or PLK promoter, respectively. Protein binding was then determined by electrophoretic mobility shift assay. Complexes were competed with 100-fold molar excess of cold probes, site-directed mutagenesis of the consensus sequence, and for the E2F probe, E2F1-directed antibody as indicated to test for binding specificity.
Summary
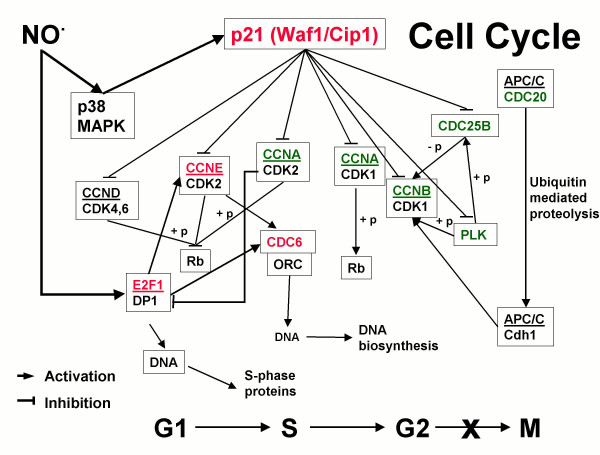
NO•, independent of cGMP, regulated a diverse subset of genes involved in inflammation, metabolism, apoptosis, the cell cycle, proliferation, signal transduction, and transport. Notably, genes associated with the cell cycle and proliferation, including the master cell cycle regulatory genes E2F1 and p21, were over-represented. Further, NO•-regulated transcripts had ARE (post-transcriptional regulatory sites) in their 3' UTR and were p38 MAPK responsive more frequently than expected. E2F1 induction by NO• was associated with up-regulation of several genes involved in G1/S transition that contain E2F-binding sites. NO• also induced p21, an ARE-containing gene, through p38 MAPK activation and mRNA stabilization. This was associated with the down-regulation of G2/M phase genes, at least in part, through changes in protein binding to CDE/CHR promoter sites. Collectively, these results demonstrate that NO•, independent of cGMP and cAMP, triggers a specific and highly coordinated genetic program that alters the G1/S transition and induces arrest in early G2/M (Fig. 8). MAPK pathways and mRNA stability are major mechanisms by which NO• regulates the transcriptome.
Figure 8.
Schematic representation of NO• regulation of the cell cycle. Genes differentially regulated in this investigation are shown in color; red signifies up- and green down-regulation. At the bottom, NO• is depicted as allowing progression to G2 where it induces cell cycle arrest. KEGG pathway [86] reproduced and Modified with permission.
Discussion
NO• has potent anti-tumor and anti-atherosclerotic effects that are closely associated with its ability to block cell proliferation [18,53]. This activity of NO• has been ascribed to both cGMP-dependent and -independent mechanisms. Experiments in rodents have found, with a few notable exceptions [54,55], that NO• controls the cell cycle through cGMP. These studies have focused on the importance of a NO•-cGMP-PKA-ERK 1/2 signal transduction pathway [22-24]. Accordingly, cAMP itself has also been reported to inhibit cell proliferation through activation of PKA and/or ERK 1/2 with the up-regulation of p27 or p21 in a cell-specific manner [31-33,56]. In contrast, the anti-proliferation effects of NO• in human cells have been frequently associated with cGMP-independent signaling [19,29]. Here, a transcriptome-wide approach revealed that NO• exerts broad control over the cell cycle through p38 MAPK activation and mRNA stabilization.
In a previous study, we found that NO• up-regulates TNFα by decreasing cAMP, an effect antagonized by cAMP analogs. Therefore Bt2cAMP was used in this investigation to explore whether some effects of NO• on the transcriptome could be attributed to its interaction with cAMP signaling. However, our results indicate that NO•-cAMP signaling appears to be a minor pathway, regulating less than 6 of the affected transcripts in U937 cells (data not shown). These included TNFα, as well as pim-1 oncogene (PIM1), TNFα-induced protein 2 (TNFAIP2), and glutathione reductase (GSR). Importantly, for cell cycle genes, NO• and Bt2cAMP consistently had the same directional effect on transcripts, although NO• regulated more genes overall. Thus, decreases in intracellular cAMP appear unrelated to NO• effects on the cell cycle. Furthermore, treatment with both NO• and Bt2cAMP synergistically provoked cell cycle arrest in G2/M, suggesting that NO•-induced decreases in cAMP may attenuate some of its effects on the cell cycle. Although this experiment also provides useful intormation on gene regulation by cAMP in U937 cells, the reader should keep in mind that Bt2cAMP was the only analog studied and some effects may have been caused by its butyryl component.
U937 cells were PMA-differentiated in the current experiments to render them capable of producing TNFα and IL-8, two cytokines previously identified as NO•-responsive [7-13]. Further, this treatment also reduced cell proliferation and forced >80% of the cells into the G0/G1 phase of the cell cycle, allowing for a more coherent analysis of cell cycle regulation (Fig. 4). However, PMA itself had large effects on NO•-regulated genes such as p21 (Fig. 6D) and the findings here cannot be extrapolated directly to naïve U937 cells. Fortunately, Turpev and colleagues have recently reported selected microarray results from NO• exposure of undifferentiated U937 and Mono Mac 6 cells [57]. Of interest, a number of key genes identified by this group were also found to be similarly regulated by NO• in PMA-differentiated cells including HMOX1, IL-8, activating transcription factor 4 (ATF4), BCL2/Adenovirus E1B 19 kD-interacting protein 3 (BNIP3), and importantly p21/Waf1/Cip1.
The NO• donor GSNO was found to activate p38 MAPK in U937 cells, which was consistent with our previous result using SNAP, another NO• donor [41]. Furthermore, three different NO• donors were shown here to consistently increase p21 protein expression, indicating that this effect is NO•-specific and donor independent. Importantly, very low concentrations of the p38 MAPK inhibitor SB202190 were found to block the induction of p21 protein by NO•, further establishing the role of p38 MAPK as an intermediary signal transduction event. Finally, p21 mRNA was measured serially by RT-PCR after transcriptional blockade in the absence or presence of SB202190 showing that this transcript is stabilized by NO• through a p38 MAPK-dependent mechanism.
Others have found that NO• increases p21 mRNA and protein expression in human vascular smooth muscle independent of cGMP [58]. In addition, p38 MAPK activation has been shown to increase p21 expression by both transcriptional activation and protein stabilization [42]. E2F1 is also known to induce p21 transcription [59] providing another mechanism by which NO• may have increased p21 expression in the current experiment. Conversely, as already discussed, NO• decreases cAMP, reducing the ability of Sp1 to bind to GC box elements and thereby repressing the transcription of Sp1-dependent genes such as eNOS [11]. Interestingly, p21 is highly dependent on Sp1 for transcription [60] and is induced by cAMP [56], findings consistent with the possibility that p21 may be transcriptionally repressed by NO•-cAMP-Sp1 signal transduction. Nonetheless, NO• induction of p21 demonstrates that other mechanisms dominate over any negative effects of NO• on Sp1 binding to the p21 promoter. Here, we focused on mRNA stabilization, because of the strong indication in our microarray data that this is a major mechanism of gene regulation by NO•. However, other mechanisms such as changes in transcription, translation, or protein stability may have contributed substantially to the net effects of NO• on p21 expression.
The E2F family of transcription factors play important roles in G1/S phase transition. E2F1 up-regulates many G1/S phase genes including itself, cyclin E1 (CCNE1), CDC6, uracil-DNA glycosylase (UNG), JUN, p21 and c-Myb [46,47,61-65]. Except for c-Myb, all were up-regulated by NO• in the present study, suggesting that NO• may drive differentiated U937 cells through the G1/S transition by inducing E2F1 expression. This conclusion is further supported by EMSA showing that NO• increases E2F1 binding to E2F consensus sequence. However, increased E2F1 expression may not be the only mechanism contributing to these observed changes in DNA binding. NO• activates p38 MAPK in U937 cells and p38 MAPK has been shown to increase E2F1 binding to E2F sites [66]. Further, cyclin A1 was down-regulated by NO• and has been shown to turn off E2F1 target genes by decreasing E2F1 DNA binding [47,67].
Notably, c-Myb and a number of G2 or G2/M phase genes that contain E2F sites were down-regulated by NO•. E2F sites can function as repressors in some genes and their disruption by mutation leads to promoter activation [46,61,68]. Further, NO•-responsive genes with both E2F elements and CDE/CHR repressor sites were uniformly down-regulated. Promoters with CDE/CHR motifs are repressed by p21 [28,39], which was also induced by NO•. Therefore, even for promoters activated by E2F1, repression through CDE/CHR sites appears to be the dominant action of NO• in this cellular context. Moreover, E2F and CHR sites may cooperate as co-repressors [69]. Although CDC25B lacks an identifiable CDE/CHR site, it does have a proximal repressor and its regulation is similar to CDE/CHR-containing genes [70].
Cell cycle arrest induced by p21 occurs in late G1/S [40,71] or early G2/M [38] and is mediated, at least in part, by the repression of target genes with CDE/CHR sites [28]. CDE/CHR sites are present in the promoters of cyclin A1 [50], CDC2 [48], cyclin B1 [72], and TOP2A [49], centromere protein A (CENPA) [73] and PLK [27]. All of these genes are G2 or G2/M related and are down-regulated by p21, results consistent with the effects of NO• observed here. EMSA confirmed the hypothesis that NO• regulates protein binding to CDE/CHR sites. Collectively, these findings suggest that NO• regulates many G2/M phase cell cycle genes through p21. However, NO• may also regulate some of these downstream p21 targets through additional mechanisms. For example, c-Myb, cyclin A1, cyclin B1, and CENPA have ARE in their 3' UTR, indicating that NO• might alter the stability of these transcripts. Notably, cyclin A1 and cyclin B1 are down-regulated by p38 MAPK, a signal transduction pathway that was activated by NO• in the current experiments. Importantly, p38 MAPK has been shown to induce cell cycle arrest at the G2 checkpoint through mechanisms that were only partially dependent on p21 [74].
ARE in 3' UTR have been implicated in the control of transcript stability and have an important post-transcriptional impact on transcriptome content [11,75-77]. We previously demonstrated that independent of cGMP, NO• up-regulates IL-8, but not TNFα post-transcriptionally through p38 MAPK activation [13]. In the current investigation, ARE- containing genes including IL-8 and p21 were over-represented among NO•-regulated genes. Nearly half of these ARE genes have been reported to be regulated by p38 MAPK (Table 2). Notably, NO• responses were all in the same direction as those reported for p38 MAPK activation. Previous microarray experiments that globally tested mRNA stability found that 10% of transcripts were associated with p38 MAPK-dependent regulation [77]. The over-representation of p38 MAPK-regulated genes in our experiments indicates that this stress kinase is an important target of NO•.
Conclusion
The present investigation was focused on understanding cGMP-independent gene regulation by NO•. Major themes within the identified gene list were the predominance of cell cycle-related genes and ARE-containing transcripts. NO• was found to trigger a specific and coordinated cell cycle arrest independent of both cGMP and cAMP. E2F1 induction up-regulated target genes involved in G1/S transition through E2F sites. NO• stabilization of p21 mRNA was p38 MAPK dependent and led to increased protein binding to CDE/CHR promoter sites and the down-regulation of G2/M phase genes. The cell cycle is a major target of NO•-mediated gene regulation. Importantly, p38 MAPK and mRNA stability are major intermediary mechanisms through which NO• affects the human transcriptome.
Methods
Reagents and cell culture
PMA, GSNO, S-nitroso-N-SNAP, Bt2cAMP and SB202190 were purchased from Calbiochem (San Diego, CA). DETA-NONOate was obtained from Cayman (Ann Arbor, Michigan); actinomycin D and GSH were from Sigma-Aldrich (St. Louis, MO). U937 cells (ATCC, Rockville, MD), a human monoblastoid line devoid of NO• sensitive guanylate cyclase, [9,30] were cultivated and then differentiated with PMA (100 nM) for 48 h as described previously [9].
Microarray experiments
NO• donor, GSNO (400 μM), or its precursor GSH (400 μM) was added into differentiated U937 cells in the absence or presence of Bt2cAMP (100 μM) followed by incubation at 37°C for 6 h (N = 7). Cells were then washed three times with ice cold PBS. Total RNA was extracted using RNeasy Mini kits (Qiagen, Valencia, CA) and reverse transcribed (10 μg) using the SuperScript II® custom kit (Invitrogen, Carlsbad, CA). Resulting cDNA (1 μg) was in vitro transcribed into biotin-labeled cRNA using the BioArray high yield RNA transcript labeling kit (Enzo Life Sciences, Farmingdale, NY). After fragmentation, biotin-labeled cRNA (20 μg) was hybridized to Affymetrix HuGeneFL 6800® microarrays [> 5,000 unique transcripts after masking uninformative probe sets [Affymetrix Website, #106] following the Affymetrix protocol [78]. After staining with streptavidin phycoerythrin (Molecular Probes) and enhancing with anti-streptavidin (0.5 mg/ml, Vector Laboratories, Burlingame, CA), microarrays were scanned using Agilent GeneArray Scanner.
Cytokine assay and TaqMan® Real time RT-PCR
Supernatants were collected from duplicate cell cultures after 24 h of incubation. TNFα, IL-8 and IL-1β production were measured using ELISA kits (R & D Systems, Minneapolis, MN) according to the manufacturer's instructions.
The TaqMan® Real time RT-PCR system (Applied Biosystems, ABI, Rockville, MD) was employed to quantify mRNA levels. Gene-specific TaqMan®probes and PCR primers were designed using Primer Express 1.0 (ABI) and their sequences are provided in supplemental data [see Additional file 5]. The High-capacity cDNA Archive kit (ABI, Foster City, CA) was employed to prepare cDNA from 2 μg of total RNA. The resulting cDNA was used for RT-PCR in triplicate according to the standard ABI protocol. The average quantities of target gene mRNA relative to GAPDH mRNA was determined for each sample. The target gene/GAPDH ratio in GSH treated cells was arbitrarily set at 1 and results from all other samples were expressed relative to that standard.
Western blotting
Polyclonal antibodies detecting p21, cyclin A1, cyclin B1, CDC6, E2F1 and cyclin F were obtained from Santa Cruz Biotechnology (Santa Cruz, CA). PLK antibody was purchased from BD Transduction Laboratories (San Diego, CA). Aliquots of 1 × 107 cells were incubated with PBS, GSH (400 μM), or GSNO (400 μM) for 12 h to prepare whole cell lysates. Separate experiments were conducted to detect total and phosphorylated p38 MAPK using anti-p38 and anti-pp38 (Promega, WI). All Western blotting was performed using 20 μg of whole cell lysate as previously described [13].
Cell cycle analysis
Cells were harvested and stained with propidium iodide and the cell cycle distribution of stained cells was determined by flow cytometry (FACS Calibur, Becton Dickinson). The percentage of cells in G0/G1, S, and G2/M was determined using ModFit (Verify Software House Inc., Topsham, ME) and expressed as relative change compared to PMA-differentiation alone. Naïve U937 cells were compared to cells (1 × 106) incubated with PMA for 48 h to examine the effects of differentiation on the cell cycle. As expected, PMA differentiation pushed cells into G0/G1 arrest (>80% of cells). These cells were then treated with GSH (400 μM) or GSNO (400 μM) without or with Bt2cAMP for 24 h and processed for cell cycle analysis as described above.
EMSA
Differentiated U937 cells were cultured for 3 h with GSH (400 μM) or GSNO (400 μM). EMSA were performed with 15 μg of nuclear extract and double-stranded DNA probes labeled with biotin-N4-CTP according to manufacturer's instructions (Pierce, Rockford, Illinois). Probes purchased from Sigma-Genosys (The Woodlands, TX) were as follows: E2F probe (5'-CGTGGCTCTTTCGCGGCAAAAAGGA-3') representing the -39 to -15 section of the E2F1 promoter and CDE/CHR probe (5'-GTTCCCAGCGCCGCGTTTGAATTC-3') representing the -10 to +14 section of human PLK promoter. Binding complexes were competed using 100-fold molar excess of cold probes. Sequences for these cold probes are available in [see Additional file 6].
Microarray data analysis, gene annotation, and statistics
Images were analyzed using Microarray Suite 4.0 (Affymetrix). Global scaling was set at 100. Data were transformed and analyzed using the MSCL Analyst's Toolbox http://affylims.cit.nih.gov written in the JMP scripting language (SAS Institute, Cary, NC). Average difference (AD) values were standardized and transformed using the Symmetric Adaptive Transform [79-81] yielding quantile-normalized, homogenous variance scaled results. Differentially regulated genes were identified from 7 independent experiments using a combination of consistency tests set at a 4% false discovery rate (FDR) and an average AD above 20 for at least one condition. One of 7 experiments was an outlier for some genes, but was not allowed to eliminate genes found significant in the other six. Fold change in gene expression was calculated directly from AD results after raising negative values to 10, and likewise adding 10 to all positive values.
Genes were annotated by searching NIH-DAVID [82,83] and PubMed [52]. Over-representation of gene categories among differentially expressed transcripts was tested using Expression Analysis Systematic Explorer [84,85]. EASE scores (penalized Fisher exact test), corrected for multiple comparisons using bootstrap resampling with 10,000 iterations, are reported as P-values. These analyses and tests of significance relied on databases within EASE and therefore did not include additional genes that were annotated to particular functional categories using PubMed.
All data not derived from microarrays are presented as mean ± standard error (SE) of at least three independent experiments. All P-values are two-sided unless noted otherwise, and considered significant if less than 0.05. To compare treatment effects on cytokine secretion, a two-way ANOVA with blocking for experiment was carried out on the logarithm of the measured concentrations for TNFα, IL-8 and IL-1β (supplemental Fig. 1A). A linear regression of RT-PCR log fold change versus microarray log fold change was generated to evaluate the validity of the microarray data (Fig. 2B). To determine whether NO• affected the protein expression of various cell cycle genes, paired t-tests, unadjusted for multiple comparisons, were performed for GSH versus GSNO, after log normalization to PBS (Fig. 2D). Log percentages of naïve and PMA-differentiated U937 cells in each phase of the cell cycle were compared by paired t-tests, unadjusted for multiple comparisons (Fig. 4A). Two-way ANOVAs with blocking were performed on log percentage of cells in each phase of the cell cycle to assess the significance of the NO• effect, cAMP effect, and their interaction (Fig. 4B). Effects of NO• on p38 MAPK phosphorylation (Fig. 5) were investigated with a one-way ANOVA comparing the log fold change of laser densitometry intensity (pp38/p38) over different concentrations of GSNO. A post-hoc Dunnett's test was carried out to determine the lowest concentration at which the effect became significant compared to control. The expression of p21 in the presence of GSNO, SNAP, or DETA-NONOate was compared to that in the presence of their respective degraded controls with paired t-tests, unadjusted for multiple comparisons (Fig. 6A). The dose effect of SB202190 on NO•-induced p21 protein expression normalized to PBS was analyzed using a one-way ANOVA (Fig. 6B). NO• stabilization of p21 mRNA over time (with and without SB202190) was assessed using constrained one-way analysis of covariance, after natural log transformation of relative mRNA amounts (Fig. 6D).
List of abbreviations
ARE: AU-rich elements; Bt2cAMP: dibutyryl-cAMP; CDE/CHR: cell cycle dependent element/cell cycle gene homology region; EMSA: electrophoretic mobility shift assays; GO: Gene Ontology; GSH: glutathione; GSNO: S-nitrosoglutathione; NO•: nitric oxide; SNAP: S-nitroso-N-acetylpenicillamine; UTR: untranslated regions.
Authors' contributions
Xiaolin Cui, Shuibang Wang, and Robert L. Danner: framing the question and designing experiments; developing downstream hypotheses; writing the manuscript. Jianhua Zhang: developing, designing, and performing the real time RT-PCR and EMSA experiments; raising questions that contributed to the scientific process. Xiaolin Cui and Penglin Ma: Designing and performing experiments to test downstream hypotheses. Performing Western blotting. Final annotation of gene lists. Daniela E. Myers, Ilana G. Goldberg and Ana del Pilar Cintron: developing methodology and performing bench work for microarray experiments including preparation of total RNA, reverse transcription, labeling, hybridization, and scanning. Preliminary annotation of gene lists. Kelly J. Sittler, Peter J. Munson and Jennifer J. Barb: developing computation tools and data normalization methods; detecting differentially regulated genes and performing statistical analysis. J. Philip McCoy: developing, designing, and performing the cell cycle analysis experiments. Editing the manuscript.
Supplementary Material
Classification of NO•-Upregulated Genes. Complete list of genes upregulated by NO• Genes are classified by function and fold change from control is shown.
Classification of NO•-Downregulated Genes. Complete list of genes downregulated by NO• Genes are classified by function and fold change from control is shown.
Confirmation of NO• effects on TNFα, IL-8 and IL-1β (A) NO• up-regulated secreted TNFα, IL-8 and IL-1β protein at 24 h as measured by ELISA. Dibutyryl cAMP (Bt2cAMP) decreased TNFα, increased IL-1β, and had no effect on IL-8. Data are means ± SE of six independent experiments. (B) NO• effect on TNFα, IL-8 and IL-1β mRNA at 6 h as measured by microarray (N = 7) were similar to changes in secreted protein.
Classification of cAMP-Regulated Genes. Complete list of genes regulated by cAMP. Genes are classified by function and fold change from control is shown.
RT-PCR Primers and Probes. List of genes tested by RT-PCR including the sequence of primers and probes used in the assays.
Electrophoretic Mobility Shift Assay (EMSA) Probes. List of genes from which E2F and CDE/CHR promoter sequences were derived for testing by electrophoretic mobility shift assay (EMSA). For each gene, the EMSA probe sequence is shown.
Acknowledgments
Acknowledgements
The authors would like to thank Joel Moss, Anthony Suffredini, and James Shelhamer for their helpful suggestions. This work was supported by intramural NIH funds from the Critical Care Medicine Department, Clinical Center and the Mathematical and Statistical Computing Laboratory, Division of Computational Bioscience, Center for Information Technology.
Contributor Information
Xiaolin Cui, Email: xcuia2003@yahoo.com.
Jianhua Zhang, Email: jzhang@mail.cc.nih.gov.
Penglin Ma, Email: plma1019@yahoo.com.
Daniela E Myers, Email: rdanner@mail.nih.gov.
Ilana G Goldberg, Email: rdanner@cc.nih.gov.
Kelly J Sittler, Email: ksitt001@umaryland.edu.
Jennifer J Barb, Email: barbj@mail.nih.gov.
Peter J Munson, Email: munson@mail.nih.gov.
Ana del Pilar Cintron, Email: acintron@ora.fda.gov.
J Philip McCoy, Email: mccoyjp@mail.nih.gov.
Shuibang Wang, Email: swang@cc.nih.gov.
Robert L Danner, Email: rdanner@mail.nih.gov.
References
- Palmer RM, Ashton DS, Moncada S. Vascular endothelial cells synthesize nitric oxide from L-arginine. Nature. 1988;333:664–666. doi: 10.1038/333664a0. [DOI] [PubMed] [Google Scholar]
- Pollock JS, Forstermann U, Mitchell JA, Warner TD, Schmidt HH, Nakane M, Murad F. Purification and characterization of particulate endothelium-derived relaxing factor synthase from cultured and native bovine aortic endothelial cells. Proc Natl Acad Sci U S A. 1991;88:10480–10484. doi: 10.1073/pnas.88.23.10480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang PL, Huang Z, Mashimo H, Bloch KD, Moskowitz MA, Bevan JA, Fishman MC. Hypertension in mice lacking the gene for endothelial nitric oxide synthase. Nature. 1995;377:239–242. doi: 10.1038/377239a0. [DOI] [PubMed] [Google Scholar]
- Gaboury J, Woodman RC, Granger DN, Reinhardt P, Kubes P. Nitric oxide prevents leukocyte adherence: role of superoxide. Am J Physiol. 1993;265:H862–7. doi: 10.1152/ajpheart.1993.265.3.H862. [DOI] [PubMed] [Google Scholar]
- Kubes P, Kanwar S, Niu XF, Gaboury JP. Nitric oxide synthesis inhibition induces leukocyte adhesion via superoxide and mast cells. Faseb J. 1993;7:1293–1299. doi: 10.1096/fasebj.7.13.8405815. [DOI] [PubMed] [Google Scholar]
- Niu XF, Smith CW, Kubes P. Intracellular oxidative stress induced by nitric oxide synthesis inhibition increases endothelial cell adhesion to neutrophils. Circ Res. 1994;74:1133–1140. doi: 10.1161/01.res.74.6.1133. [DOI] [PubMed] [Google Scholar]
- Lander HM, Sehajpal P, Levine DM, Novogrodsky A. Activation of human peripheral blood mononuclear cells by nitric oxide-generating compounds. J Immunol. 1993;150:1509–1516. [PubMed] [Google Scholar]
- Van Dervort AL, Yan L, Madara PJ, Cobb JP, Wesley RA, Corriveau CC, Tropea MM, Danner RL. Nitric oxide regulates endotoxin-induced TNF-alpha production by human neutrophils. J Immunol. 1994;152:4102–4109. [PubMed] [Google Scholar]
- Wang S, Yan L, Wesley RA, Danner RL. Nitric oxide increases tumor necrosis factor production in differentiated U937 cells by decreasing cyclic AMP. J Biol Chem. 1997;272:5959–5965. doi: 10.1074/jbc.272.9.5959. [DOI] [PubMed] [Google Scholar]
- Wang S, Wang W, Wesley RA, Danner RL. A Sp1 binding site of the tumor necrosis factor alpha promoter functions as a nitric oxide response element. J Biol Chem. 1999;274:33190–33193. doi: 10.1074/jbc.274.47.33190. [DOI] [PubMed] [Google Scholar]
- Zhang J, Wang S, Wesley RA, Danner RL. Adjacent sequence controls the response polarity of nitric oxide-sensitive Sp factor binding sites. J Biol Chem. 2003;278:29192–29200. doi: 10.1074/jbc.M213043200. [DOI] [PubMed] [Google Scholar]
- Corriveau CC, Madara PJ, Van Dervort AL, Tropea MM, Wesley RA, Danner RL. Effects of nitric oxide on chemotaxis and endotoxin-induced interleukin-8 production in human neutrophils. J Infect Dis. 1998;177:116–126. doi: 10.1086/513829. [DOI] [PubMed] [Google Scholar]
- Ma P, Cui X, Wang S, Zhang J, Nishanian EV, Wang W, Wesley RA, Danner RL. Nitric oxide post-transcriptionally up-regulates LPS-induced IL-8 expression through p38 MAPK activation. J Leukoc Biol. 2004;76:278–287. doi: 10.1189/jlb.1203653. [DOI] [PubMed] [Google Scholar]
- Peng HB, Rajavashisth TB, Libby P, Liao JK. Nitric oxide inhibits macrophage-colony stimulating factor gene transcription in vascular endothelial cells. J Biol Chem. 1995;270:17050–17055. doi: 10.1074/jbc.270.28.17050. [DOI] [PubMed] [Google Scholar]
- Marshall HE, Stamler JS. Inhibition of NF-kappa B by S-nitrosylation. Biochemistry. 2001;40:1688–1693. doi: 10.1021/bi002239y. [DOI] [PubMed] [Google Scholar]
- Von Knethen A, Brune B. Activation of peroxisome proliferator-activated receptor gamma by nitric oxide in monocytes/macrophages down-regulates p47phox and attenuates the respiratory burst. J Immunol. 2002;169:2619–2626. doi: 10.4049/jimmunol.169.5.2619. [DOI] [PubMed] [Google Scholar]
- von der Leyen HE, Gibbons GH, Morishita R, Lewis NP, Zhang L, Nakajima M, Kaneda Y, Cooke JP, Dzau VJ. Gene therapy inhibiting neointimal vascular lesion: in vivo transfer of endothelial cell nitric oxide synthase gene. Proc Natl Acad Sci U S A. 1995;92:1137–1141. doi: 10.1073/pnas.92.4.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssens S, Flaherty D, Nong Z, Varenne O, van Pelt N, Haustermans C, Zoldhelyi P, Gerard R, Collen D. Human endothelial nitric oxide synthase gene transfer inhibits vascular smooth muscle cell proliferation and neointima formation after balloon injury in rats. Circulation. 1998;97:1274–1281. doi: 10.1161/01.cir.97.13.1274. [DOI] [PubMed] [Google Scholar]
- Tanner FC, Meier P, Greutert H, Champion C, Nabel EG, Luscher TF. Nitric oxide modulates expression of cell cycle regulatory proteins: a cytostatic strategy for inhibition of human vascular smooth muscle cell proliferation. Circulation. 2000;101:1982–1989. doi: 10.1161/01.cir.101.16.1982. [DOI] [PubMed] [Google Scholar]
- Pilz RB, Suhasini M, Idriss S, Meinkoth JL, Boss GR. Nitric oxide and cGMP analogs activate transcription from AP-1-responsive promoters in mammalian cells. Faseb J. 1995;9:552–558. doi: 10.1096/fasebj.9.7.7737465. [DOI] [PubMed] [Google Scholar]
- Garg UC, Hassid A. Nitric oxide-generating vasodilators and 8-bromo-cyclic guanosine monophosphate inhibit mitogenesis and proliferation of cultured rat vascular smooth muscle cells. J Clin Invest. 1989;83:1774–1777. doi: 10.1172/JCI114081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornwell TL, Arnold E, Boerth NJ, Lincoln TM. Inhibition of smooth muscle cell growth by nitric oxide and activation of cAMP-dependent protein kinase by cGMP. Am J Physiol. 1994;267:C1405–13. doi: 10.1152/ajpcell.1994.267.5.C1405. [DOI] [PubMed] [Google Scholar]
- Gu M, Brecher P. Nitric oxide-induced increase in p21(Sdi1/Cip1/Waf1) expression during the cell cycle in aortic adventitial fibroblasts. Arterioscler Thromb Vasc Biol. 2000;20:27–34. doi: 10.1161/01.atv.20.1.27. [DOI] [PubMed] [Google Scholar]
- Gu M, Lynch J, Brecher P. Nitric oxide increases p21(Waf1/Cip1) expression by a cGMP-dependent pathway that includes activation of extracellular signal-regulated kinase and p70(S6k) J Biol Chem. 2000;275:11389–11396. doi: 10.1074/jbc.275.15.11389. [DOI] [PubMed] [Google Scholar]
- Harper JW, Adami GR, Wei N, Keyomarsi K, Elledge SJ. The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell. 1993;75:805–816. doi: 10.1016/0092-8674(93)90499-G. [DOI] [PubMed] [Google Scholar]
- Xiong Y, Hannon GJ, Zhang H, Casso D, Kobayashi R, Beach D. p21 is a universal inhibitor of cyclin kinases. Nature. 1993;366:701–704. doi: 10.1038/366701a0. [DOI] [PubMed] [Google Scholar]
- Uchiumi T, Longo DL, Ferris DK. Cell cycle regulation of the human polo-like kinase (PLK) promoter. J Biol Chem. 1997;272:9166–9174. doi: 10.1074/jbc.272.14.9166. [DOI] [PubMed] [Google Scholar]
- Zhu H, Chang BD, Uchiumi T, Roninson IB. Identification of promoter elements responsible for transcriptional inhibition of polo-like kinase 1 and topoisomerase IIalpha genes by p21(WAF1/CIP1/SDI1) Cell Cycle. 2002;1:59–66. [PubMed] [Google Scholar]
- Ishida A, Sasaguri T, Miwa Y, Kosaka C, Taba Y, Abumiya T. Tumor suppressor p53 but not cGMP mediates NO-induced expression of p21(Waf1/Cip1/Sdi1) in vascular smooth muscle cells. Mol Pharmacol. 1999;56:938–946. doi: 10.1124/mol.56.5.938. [DOI] [PubMed] [Google Scholar]
- Yan L, Wang S, Rafferty SP, Wesley RA, Danner RL. Endogenously produced nitric oxide increases tumor necrosis factor-alpha production in transfected human U937 cells. Blood. 1997;90:1160–1167. [PubMed] [Google Scholar]
- Ii M, Hoshiga M, Fukui R, Negoro N, Nakakoji T, Nishiguchi F, Kohbayashi E, Ishihara T, Hanafusa T. Beraprost sodium regulates cell cycle in vascular smooth muscle cells through cAMP signaling by preventing down-regulation of p27(Kip1) Cardiovasc Res. 2001;52:500–508. doi: 10.1016/S0008-6363(01)00411-4. [DOI] [PubMed] [Google Scholar]
- van Oirschot BA, Stahl M, Lens SM, Medema RH. Protein kinase A regulates expression of p27(kip1) and cyclin D3 to suppress proliferation of leukemic T cell lines. J Biol Chem. 2001;276:33854–33860. doi: 10.1074/jbc.M104395200. [DOI] [PubMed] [Google Scholar]
- Stork PJ, Schmitt JM. Crosstalk between cAMP and MAP kinase signaling in the regulation of cell proliferation. Trends Cell Biol. 2002;12:258–266. doi: 10.1016/S0962-8924(02)02294-8. [DOI] [PubMed] [Google Scholar]
- Short S, Tian D, Short ML, Jungmann RA. Structural determinants for post-transcriptional stabilization of lactate dehydrogenase A mRNA by the protein kinase C signal pathway. J Biol Chem. 2000;275:12963–12969. doi: 10.1074/jbc.275.17.12963. [DOI] [PubMed] [Google Scholar]
- Takagi K, Isobe Y, Yasukawa K, Okouchi E, Suketa Y. Nitric oxide blocks the cell cycle of mouse macrophage-like cells in the early G2+M phase. FEBS Lett. 1994;340:159–162. doi: 10.1016/0014-5793(94)80128-2. [DOI] [PubMed] [Google Scholar]
- Kelly MR, Geigerman CM, Loo G. Epigallocatechin gallate protects U937 cells against nitric oxide-induced cell cycle arrest and apoptosis. J Cell Biochem. 2001;81:647–658. doi: 10.1002/jcb.1097. [DOI] [PubMed] [Google Scholar]
- Pervin S, Singh R, Chaudhuri G. Nitric oxide-induced cytostasis and cell cycle arrest of a human breast cancer cell line (MDA-MB-231): potential role of cyclin D1. Proc Natl Acad Sci U S A. 2001;98:3583–3588. doi: 10.1073/pnas.041603998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulic V, Stein GH, Far DF, Reed SI. Nuclear accumulation of p21Cip1 at the onset of mitosis: a role at the G2/M-phase transition. Mol Cell Biol. 1998;18:546–557. doi: 10.1128/mcb.18.1.546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang BD, Watanabe K, Broude EV, Fang J, Poole JC, Kalinichenko TV, Roninson IB. Effects of p21Waf1/Cip1/Sdi1 on cellular gene expression: implications for carcinogenesis, senescence, and age-related diseases. Proc Natl Acad Sci U S A. 2000;97:4291–4296. doi: 10.1073/pnas.97.8.4291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavelle D, DeSimone J, Hankewych M, Kousnetzova T, Chen YH. Decitabine induces cell cycle arrest at the G1 phase via p21(WAF1) and the G2/M phase via the p38 MAP kinase pathway. Leuk Res. 2003;27:999–1007. doi: 10.1016/S0145-2126(03)00068-7. [DOI] [PubMed] [Google Scholar]
- Wang W, Wang S, Nishanian EV, Del Pilar Cintron A, Wesley RA, Danner RL. Signaling by eNOS through a superoxide-dependent p42/44 mitogen-activated protein kinase pathway. Am J Physiol Cell Physiol. 2001;281:C544–54. doi: 10.1152/ajpcell.2001.281.2.C544. [DOI] [PubMed] [Google Scholar]
- Kim GY, Mercer SE, Ewton DZ, Yan Z, Jin K, Friedman E. The stress-activated protein kinases p38 alpha and JNK1 stabilize p21(Cip1) by phosphorylation. J Biol Chem. 2002;277:29792–29802. doi: 10.1074/jbc.M201299200. [DOI] [PubMed] [Google Scholar]
- Wang W, Wang S, Yan L, Madara P, Del Pilar Cintron A, Wesley RA, Danner RL. Superoxide production and reactive oxygen species signaling by endothelial nitric-oxide synthase. J Biol Chem. 2000;275:16899–16903. doi: 10.1074/jbc.M000301200. [DOI] [PubMed] [Google Scholar]
- Johnson DG, Ohtani K, Nevins JR. Autoregulatory control of E2F1 expression in response to positive and negative regulators of cell cycle progression. Genes Dev. 1994;8:1514–1525. doi: 10.1101/gad.8.13.1514. [DOI] [PubMed] [Google Scholar]
- DeGregori J, Kowalik T, Nevins JR. Cellular targets for activation by the E2F1 transcription factor include DNA synthesis- and G1/S-regulatory genes. Mol Cell Biol. 1995;15:4215–4224. doi: 10.1128/mcb.15.8.4215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyson N. The regulation of E2F by pRB-family proteins. Genes Dev. 1998;12:2245–2262. doi: 10.1101/gad.12.15.2245. [DOI] [PubMed] [Google Scholar]
- Stevens C, La Thangue NB. E2F and cell cycle control: a double-edged sword. Arch Biochem Biophys. 2003;412:157–169. doi: 10.1016/S0003-9861(03)00054-7. [DOI] [PubMed] [Google Scholar]
- Zwicker J, Lucibello FC, Wolfraim LA, Gross C, Truss M, Engeland K, Muller R. Cell cycle regulation of the cyclin A, cdc25C and cdc2 genes is based on a common mechanism of transcriptional repression. Embo J. 1995;14:4514–4522. doi: 10.1002/j.1460-2075.1995.tb00130.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isaacs RJ, Davies SL, Sandri MI, Redwood C, Wells NJ, Hickson ID. Physiological regulation of eukaryotic topoisomerase II. Biochim Biophys Acta. 1998;1400:121–137. doi: 10.1016/s0167-4781(98)00131-6. [DOI] [PubMed] [Google Scholar]
- Muller C, Yang R, Beck-von-Peccoz L, Idos G, Verbeek W, Koeffler HP. Cloning of the cyclin A1 genomic structure and characterization of the promoter region. GC boxes are essential for cell cycle-regulated transcription of the cyclin A1 gene. J Biol Chem. 1999;274:11220–11228. doi: 10.1074/jbc.274.16.11220. [DOI] [PubMed] [Google Scholar]
- TRANSFAC [http://www.gene-regulation.com/pub/databases.html#transfac]
- PubMed [http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=PubMed]
- Hussain SP, Trivers GE, Hofseth LJ, He P, Shaikh I, Mechanic LE, Doja S, Jiang W, Subleski J, Shorts L, Haines D, Laubach VE, Wiltrout RH, Djurickovic D, Harris CC. Nitric oxide, a mediator of inflammation, suppresses tumorigenesis. Cancer Res. 2004;64:6849–6853. doi: 10.1158/0008-5472.CAN-04-2201. [DOI] [PubMed] [Google Scholar]
- Bauer PM, Buga GM, Ignarro LJ. Role of p42/p44 mitogen-activated-protein kinase and p21waf1/cip1 in the regulation of vascular smooth muscle cell proliferation by nitric oxide. Proc Natl Acad Sci U S A. 2001;98:12802–12807. doi: 10.1073/pnas.211443198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ignarro LJ, Buga GM, Wei LH, Bauer PM, Wu G, del Soldato P. Role of the arginine-nitric oxide pathway in the regulation of vascular smooth muscle cell proliferation. Proc Natl Acad Sci U S A. 2001;98:4202–4208. doi: 10.1073/pnas.071054698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi S, Morishita R, Matsushita H, Nakagami H, Taniyama Y, Nakamura T, Aoki M, Yamamoto K, Higaki J, Ogihara T. Cyclic AMP inhibited proliferation of human aortic vascular smooth muscle cells, accompanied by induction of p53 and p21. Hypertension. 2000;35:237–243. doi: 10.1161/01.hyp.35.1.237. [DOI] [PubMed] [Google Scholar]
- Turpaev K, Bouton C, Diet A, Glatigny A, Drapier JC. Analysis of differentially expressed genes in nitric oxide-exposed human monocytic cells. Free Rad Biol Med. 2005;38:1392–1400. doi: 10.1016/j.freeradbiomed.2005.02.002. [DOI] [PubMed] [Google Scholar]
- Ishida A, Sasaguri T, Kosaka C, Nojima H, Ogata J. Induction of the cyclin-dependent kinase inhibitor p21(Sdi1/Cip1/Waf1) by nitric oxide-generating vasodilator in vascular smooth muscle cells. J Biol Chem. 1997;272:10050–10057. doi: 10.1074/jbc.272.6.3324. [DOI] [PubMed] [Google Scholar]
- Hiyama H, Iavarone A, Reeves SA. Regulation of the cdk inhibitor p21 gene during cell cycle progression is under the control of the transcription factor E2F. Oncogene. 1998;16:1513–1523. doi: 10.1038/sj.onc.1201667. [DOI] [PubMed] [Google Scholar]
- Gartel AL, Ye X, Goufman E, Shianov P, Hay N, Najmabadi F, Tyner AL. Myc represses the p21(WAF1/CIP1) promoter and interacts with Sp1/Sp3. Proc Natl Acad Sci U S A. 2001;98:4510–4515. doi: 10.1073/pnas.081074898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helin K. Regulation of cell proliferation by the E2F transcription factors. Curr Opin Genet Dev. 1998;8:28–35. doi: 10.1016/S0959-437X(98)80058-0. [DOI] [PubMed] [Google Scholar]
- Ishida S, Huang E, Zuzan H, Spang R, Leone G, West M, Nevins JR. Role for E2F in control of both DNA replication and mitotic functions as revealed from DNA microarray analysis. Mol Cell Biol. 2001;21:4684–4699. doi: 10.1128/MCB.21.14.4684-4699.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller H, Bracken AP, Vernell R, Moroni MC, Christians F, Grassilli E, Prosperini E, Vigo E, Oliner JD, Helin K. E2Fs regulate the expression of genes involved in differentiation, development, proliferation, and apoptosis. Genes Dev. 2001;15:267–285. doi: 10.1101/gad.864201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polager S, Kalma Y, Berkovich E, Ginsberg D. E2Fs up-regulate expression of genes involved in DNA replication, DNA repair and mitosis. Oncogene. 2002;21:437–446. doi: 10.1038/sj.onc.1205102. [DOI] [PubMed] [Google Scholar]
- Ren B, Cam H, Takahashi Y, Volkert T, Terragni J, Young RA, Dynlacht BD. E2F integrates cell cycle progression with DNA repair, replication, and G(2)/M checkpoints. Genes Dev. 2002;16:245–256. doi: 10.1101/gad.949802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Nath N, Minden A, Chellappan S. Regulation of Rb and E2F by signal transduction cascades: divergent effects of JNK1 and p38 kinases. Embo J. 1999;18:1559–1570. doi: 10.1093/emboj/18.6.1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krek W, Xu G, Livingston DM. Cyclin A-kinase regulation of E2F-1 DNA binding function underlies suppression of an S phase checkpoint. Cell. 1995;83:1149–1158. doi: 10.1016/0092-8674(95)90141-8. [DOI] [PubMed] [Google Scholar]
- Bennett JD, Farlie PG, Watson RJ. E2F binding is required but not sufficient for repression of B-myb transcription in quiescent fibroblasts. Oncogene. 1996;13:1073–1082. [PubMed] [Google Scholar]
- Liu N, Lucibello FC, Zwicker J, Engeland K, Muller R. Cell cycle-regulated repression of B-myb transcription: cooperation of an E2F site with a contiguous corepressor element. Nucleic Acids Res. 1996;24:2905–2910. doi: 10.1093/nar/24.15.2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korner K, Jerome V, Schmidt T, Muller R. Cell cycle regulation of the murine cdc25B promoter: essential role for nuclear factor-Y and a proximal repressor element. J Biol Chem. 2001;276:9662–9669. doi: 10.1074/jbc.M008696200. [DOI] [PubMed] [Google Scholar]
- Chen WJ, Lin JK. Induction of G1 arrest and apoptosis in human jurkat T cells by pentagalloylglucose through inhibiting proteasome activity and elevating p27Kip1, p21Cip1/WAF1, and Bax proteins. J Biol Chem. 2004;279:13496–13505. doi: 10.1074/jbc.M212390200. [DOI] [PubMed] [Google Scholar]
- Wasner M, Tschop K, Spiesbach K, Haugwitz U, Johne C, Mossner J, Mantovani R, Engeland K. Cyclin B1 transcription is enhanced by the p300 coactivator and regulated during the cell cycle by a CHR-dependent repression mechanism. FEBS Lett. 2003;536:66–70. doi: 10.1016/S0014-5793(03)00028-0. [DOI] [PubMed] [Google Scholar]
- Shelby RD, Vafa O, Sullivan KF. Assembly of CENP-A into centromeric chromatin requires a cooperative array of nucleosomal DNA contact sites. J Cell Biol. 1997;136:501–513. doi: 10.1083/jcb.136.3.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garner AP, Weston CR, Todd DE, Balmanno K, Cook SJ. Delta MEKK3:ER* activation induces a p38 alpha/beta 2-dependent cell cycle arrest at the G2 checkpoint. Oncogene. 2002;21:8089–8104. doi: 10.1038/sj.onc.1206000. [DOI] [PubMed] [Google Scholar]
- Bakheet T, Frevel M, Williams BR, Greer W, Khabar KS. ARED: human AU-rich element-containing mRNA database reveals an unexpectedly diverse functional repertoire of encoded proteins. Nucleic Acids Res. 2001;29:246–254. doi: 10.1093/nar/29.1.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilusz CJ, Wormington M, Peltz SW. The cap-to-tail guide to mRNA turnover. Nat Rev Mol Cell Biol. 2001;2:237–246. doi: 10.1038/35067025. [DOI] [PubMed] [Google Scholar]
- Frevel MA, Bakheet T, Silva AM, Hissong JG, Khabar KS, Williams BR. p38 Mitogen-activated protein kinase-dependent and -independent signaling of mRNA stability of AU-rich element-containing transcripts. Mol Cell Biol. 2003;23:425–436. doi: 10.1128/MCB.23.2.425-436.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Affymetrix Website [http://www.affymetrix.com/support/technical/mask_files.affx]; [www.affymetrix.com/support/technical/manual/expression_manual.affx]
- Munson PJ. A consistency test for determining the significance of gene expression changes on replicate samples and two convenient variance-stabilizing transformations.: ; Bethesda, MD.. http://stat-www.berkeley.edu/users/terry/zarray/Affy/GL_Workshop/genelogic2001.html; 2001. [Google Scholar]
- Durbin BP, Hardin JS, Hawkins DM, Rocke DM. A variance-stabilizing transformation for gene-expression microarray data. Bioinformatics. 2002;18 Suppl 1:S105–10. doi: 10.1093/bioinformatics/18.suppl_1.s105. [DOI] [PubMed] [Google Scholar]
- Jison ML, Munson PJ, Barb JJ, Suffredini AF, Talwar S, Logun C, Raghavachari N, Beigel JH, Shelhamer JH, Danner RL, Gladwin MT. Blood mononuclear cell gene expression profiles characterize the oxidant, hemolytic, and inflammatory stress of sickle cell disease. Blood. 2004;104:270–280. doi: 10.1182/blood-2003-08-2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis GJ, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:P3. doi: 10.1186/gb-2003-4-5-p3. [DOI] [PubMed] [Google Scholar]
- NIH-DAVID [http://www.david.niaid.nih.gov]
- Hosack DA, Dennis GJ, Sherman BT, Lane HC, Lempicki RA. Identifying biological themes within lists of genes with EASE. Genome Biol. 2003;4:R70. doi: 10.1186/gb-2003-4-10-r70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- EASE [http://apps1.niaid.nih.gov/david/upload.asp>http://apps1.niaid.nih.gov/david/upload.asp]
- KEGG pathway [http://www.genome.jp/kegg/pathway/hsa/hsa04110.html] reproduced and Modified with permission
- Garcia J, Lemercier B, Roman-Roman S, Rawadi G. A Mycoplasma fermentans-derived synthetic lipopeptide induces AP-1 and NF-kappaB activity and cytokine secretion in macrophages via the activation of mitogen-activated protein kinase pathways. J Biol Chem. 1998;273:34391–34398. doi: 10.1074/jbc.273.51.34391. [DOI] [PubMed] [Google Scholar]
- Schafer PH, Wang L, Wadsworth SA, Davis JE, Siekierka JJ. T cell activation signals up-regulate p38 mitogen-activated protein kinase activity and induce TNF-alpha production in a manner distinct from LPS activation of monocytes. J Immunol. 1999;162:659–668. [PubMed] [Google Scholar]
- Marie C, Roman-Roman S, Rawadi G. Involvement of mitogen-activated protein kinase pathways in interleukin-8 production by human monocytes and polymorphonuclear cells stimulated with lipopolysaccharide or Mycoplasma fermentans membrane lipoproteins. Infect Immun. 1999;67:688–693. doi: 10.1128/iai.67.2.688-693.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beltman J, Erickson JR, Martin GA, Lyons JF, Cook SJ. C3 toxin activates the stress signaling pathways, JNK and p38, but antagonizes the activation of AP-1 in rat-1 cells. J Biol Chem. 1999;274:3772–3780. doi: 10.1074/jbc.274.6.3772. [DOI] [PubMed] [Google Scholar]
- Philips A, Roux P, Coulon V, Bellanger JM, Vie A, Vignais ML, Blanchard JM. Differential effect of Rac and Cdc42 on p38 kinase activity and cell cycle progression of nonadherent primary mouse fibroblasts. J Biol Chem. 2000;275:5911–5917. doi: 10.1074/jbc.275.8.5911. [DOI] [PubMed] [Google Scholar]
- Sala A, Nicolaides NC, Engelhard A, Bellon T, Lawe DC, Arnold A, Grana X, Giordano A, Calabretta B. Correlation between E2F-1 requirement in the S phase and E2F-1 transactivation of cell cycle-related genes in human cells. Cancer Res. 1994;54:1402–1406. [PubMed] [Google Scholar]
- Liu N, Lucibello FC, Engeland K, Muller R. A new model of cell cycle-regulated transcription: repression of the cyclin A promoter by CDF-1 and anti-repression by E2F. Oncogene. 1998;16:2957–2963. doi: 10.1038/sj.onc.1201838. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Classification of NO•-Upregulated Genes. Complete list of genes upregulated by NO• Genes are classified by function and fold change from control is shown.
Classification of NO•-Downregulated Genes. Complete list of genes downregulated by NO• Genes are classified by function and fold change from control is shown.
Confirmation of NO• effects on TNFα, IL-8 and IL-1β (A) NO• up-regulated secreted TNFα, IL-8 and IL-1β protein at 24 h as measured by ELISA. Dibutyryl cAMP (Bt2cAMP) decreased TNFα, increased IL-1β, and had no effect on IL-8. Data are means ± SE of six independent experiments. (B) NO• effect on TNFα, IL-8 and IL-1β mRNA at 6 h as measured by microarray (N = 7) were similar to changes in secreted protein.
Classification of cAMP-Regulated Genes. Complete list of genes regulated by cAMP. Genes are classified by function and fold change from control is shown.
RT-PCR Primers and Probes. List of genes tested by RT-PCR including the sequence of primers and probes used in the assays.
Electrophoretic Mobility Shift Assay (EMSA) Probes. List of genes from which E2F and CDE/CHR promoter sequences were derived for testing by electrophoretic mobility shift assay (EMSA). For each gene, the EMSA probe sequence is shown.