Abstract
Centromeres are sites of spindle attachment for chromosome segregation. During meiosis, recombination is absent at centromeres and surrounding regions. To understand the molecular basis for recombination suppression, we have comprehensively annotated the 3.5-Mb region that spans a fully sequenced rice centromere. Although transcriptional analysis showed that the 750-kb CENH3-containing core is relatively deficient in genes, the recombination-free region differs little in gene density from flanking regions that recombine. Likewise, the density of transposable elements is similar between the recombination-free region and flanking regions. We also measured levels of histone H4 acetylation and histone H3 methylation at 176 genes within the 3.5-Mb span. Active genes showed enrichment of H4 acetylation and H3K4 dimethylation as expected, including genes within the core. Our inability to detect sequence or histone modification features that distinguish recombination-free regions from flanking regions that recombine suggest that recombination suppression is an epigenetic feature of centromeres maintained by the assembly of CENH3-containing nucleosomes within the core. CENH3-containing centrochromatin does not appear to be distinguished by a unique combination of H3 and H4 modifications. Rather, the varied distribution of histone modifications might reflect the composition and abundance of sequence elements that inhabit centromeric DNA.
INTRODUCTION
The centromere is the most characteristic cytological landmark on every eukaryotic chromosome. It serves as the site for assembly of the proteinaceous kinetochore to which spindle microtubules attach at mitosis and meiosis. Centromeres are also distinctive because they are packaged into chromatin by centromere-specific nucleosomes that contain a special histone H3 variant (CENH3), together with other centromere-specific proteins, such as CENP-C and CENP-H (Amor et al., 2004).
In most eukaryotes, CENH3-containing chromatin is embedded within heterochromatin, a cytologically distinct form of chromatin that is enriched in particular histone modifications, such as dimethylated H3 lysine-9 (H3K9me2), and particular heterochromatin-associated proteins, such as HP1. Centromeric heterochromatin is late-replicating and transcriptionally inert and lacks meiotic recombination. In plants and animals, these features coincide with the highly repetitive nature of the multimegabase satellite sequence arrays that span both centromeres and pericentric heterochromatin (Schueler et al., 2001; Jin et al., 2005). The coincidence of satellite sequences with special centromeric and pericentric chromatin makes it difficult to distinguish chromatin-based features from those that depend on DNA sequence. However, some centromeres lack extensive satellite repeats, yet are entirely conventional in other ways. For example, rice (Oryza sativa) Centromere 8 (Cen8), which spans 750 kb of CENH3-rich chromatin, contains only a single ∼60-kb stretch of a 155-bp centromere-specific tandem repeat (Nagaki et al., 2004). Nevertheless, Cen8 is cytologically not different from satellite-rich centromeres found in other rice chromosomes and in other plants and animals. Furthermore, Cen8 is characterized by suppression of meiotic recombination (Harushima et al., 1998), despite the fact that it lacks satellite sequences that are typically found in pericentric heterochromatin. This makes rice Cen8 an ideal model system to study features of centromeres and pericentric heterochromatin without the complication of satellite-rich DNA.
Suppression of recombination around centromeres was first recognized in the 1930s in Drosophila melanogaster (Beadle, 1932; Mather, 1939). The same phenomenon has been reported in a wide range of eukaryotes, including Saccharomyces cerevisiae (Lambie and Roeder, 1986), Schizosaccharonyces pombe (Nakaseko et al., 1986), Neurospora crassa (Davis et al., 1994), humans (Jackson et al., 1996; Mahtani and Willard, 1998), and numerous plant species (Tanksley et al., 1992; Werner et al., 1992; Sherman and Stack, 1995; Künzel et al., 2000; Haupt et al., 2001; Anderson et al., 2003). The precise physical sizes of the recombination-free domains associated with centromeres are not known in most, if any, multicellular eukaryotes because the highly repetitive centromeric DNA hampers both physical and fine-scale genetic mapping. Integration of genetic and physical maps in several plant species indicated that the nearly recombination-free domains may span from several megabases up to almost half of the chromosomes (Werner et al., 1992; Sherman and Stack, 1995; Künzel et al., 2000; Haupt et al., 2001).
Satellite-rich centromeric and pericentric regions do not contain active genes. However, recent findings show that centromeres that lack satellites do indeed contain genes. Numerous human neocentromeres have been reported, and many of these lack highly repetitive DNA sequences (Choo, 2001; Warburton, 2004). Genes within one of these neocentromeres are transcriptionally competent, despite being embedded in regions of CENP-A (human CENH3)-containing nucleosomes (Saffery et al., 2003). Rice Cen8 has been fully sequenced (Nagaki et al., 2004; Wu et al., 2004) and found to contain several active genes within the CENH3 binding domain (Nagaki et al., 2004). These results demonstrate that centromere formation per se does not inhibit transcriptional activity.
Centromeres are therefore different from other regions of the genome in that meiotic recombination is suppressed, yet genes are active. This implies that there should be different chromatin features that are involved in suppressing meiotic recombination and in allowing gene expression to occur. To investigate the basis for this difference and to better understand the relationship between chromatin features and centromere function, we conducted an in-depth analysis of transcription and histone modifications of genes located in Cen8. An ∼3.5-Mb virtual contig spanning the recombination-free region of Cen8 and the surrounding pericentromere was constructed and annotated. We tested 198 genes within this virtual contig for transcription using an RT-PCR assay. We also measured histone modifications in Cen8 using chromatin immunoprecipitation (ChIP) analysis. Our data revealed the euchromatic nature of transcribed chromatin throughout the 3.5-Mb region around Cen8 and the close resemblance between the recombination-free region and its flanking regions in sequence and histone modifications.
RESULTS
Annotation of Cen8 Sequence That Spans the Recombination-Free Region
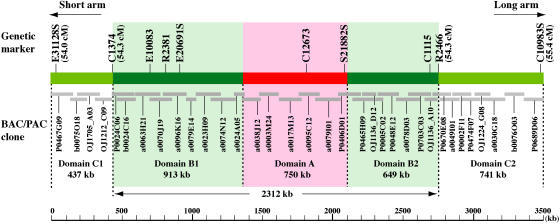
We constructed a 3,489,829-bp virtual contig from the finished sequences of 34 rice BAC and P1 artificial chromosome (PAC) clones. This virtual contig spans the genetic region from 54.0 to 55.4 centimorgan (cM) on the linkage map of rice chromosome 8 (Figure 1) based on a population that included 186 F2 plants derived from a cross between O. sativa subsp japonica var Nipponbare and O. sativa subsp indica var Kasalath (Harushima et al., 1998). This map located Cen8 at 54.3 cM by assigning individual genetically mapped DNA markers to the short or long arm using secondary and telotrisomic stocks (Singh et al., 1996; Harushima et al., 1998). The sequence map corresponding to the genetic region at 54.3 cM spans 2312 kb and includes eight cosegregated restriction fragment length polymorphism (RFLP) markers (Figure 1). We refer to this region with <1/186 recombinants as recombination-free. We previously reported that a 750-kb region in Cen8 is associated with rice CENH3 (Nagaki et al., 2004). This 750-kb region is embedded within the 2312-kb recombination-free region (Figure 1). The 3.5-Mb virtual contig is divided into five domains: domain A (750 kb) binds CENH3, domains B1 (913 kb) and B2 (649 kb) flank domain A and are likewise located within the recombination-free region, and domains C1 (437 kb) and C2 (741 kb) flank the recombination-free region (Figure 1). All 10 rice RFLP markers anchored to this 3.5-Mb region were originally derived from cDNA clones. As expected, all 10 markers were annotated as transcripts of active genes in this study.
Figure 1.
Map of the ∼3.5-Mb Region Spanning the Recombination-Free Region, Including the Centromere, of Rice Chromosome 8.
The recombination-free region is located at 54.3 cM on the genetic linkage map and spans ∼2312 kb. This region is divided into three domains. Domain A includes the ∼750-kb CENH3 binding region (in pink) (Nagaki et al., 2004). Domains B1 and B2 (in light green) flank domain A. The two distal domains, C1 and C2, flank the recombination-free region. Sequences of 34 BAC/PAC clones (gray bars) were downloaded from GenBank and assembled into the Cen8 virtual contig of 3,489,829 bp. Relative position and overlapping of these clones was determined using National Center for Biotechnology Information (NCBI) Blast2seq. The 10 genetically mapped RFLP markers in this region are shown at the top of the map.
We annotated the virtual contig for gene composition by integrating both DNA and protein homolog searching, computational predictions, and intensive manual inspection (see Methods). We identified 225 genes in the virtual contig (see Supplemental Table 1 online). For 106 genes, we were able to identify either complete (for 79 genes) or incomplete (for 27 genes) transcript sequences in the EST and full-length cDNA (fl-cDNA) collections. The other 119 genes did not match any existing ESTs or fl-cDNAs, and the prediction of these genes was based on their similarities to known proteins (for 40 genes) or solely based on computational predictions (for 79 genes).
Transcription of Cen8 Genes
Because 146 Cen8 genes match with either incomplete transcripts or no transcript at all, we first sought to verify how many of these genes are transcribed using RT-PCR. We were able to design PCR primers for 128 genes (see Supplemental Table 2 online), including all 27 genes with incomplete transcripts (gene categories A1 to A4 in Supplemental Table 3 online), all 79 genes predicted by gene finders (gene category C in Supplemental Table 3 online), and 22 of the 40 genes predicted based on protein matches (gene category B in Supplemental Table 3 online). The remaining 18 genes belong to members of gene families for which we were not able to design specific primers. We first surveyed the amplification specificity of these primers using rice genomic DNA as a template with an annealing temperature between 62 and 67°C. We observed single-band products from 111 genes and one strong band plus one or two weak bands from 16 genes. Only the two primer sets for hypothetical gene Cen8.t01683 failed to amplify the genomic DNA and were not listed in Supplemental Table 2 online. We tested the expression of all 127 genes on four cDNA samples derived from 14-d-old leaves, 14-d-old roots, 14-d-old etiolated seedlings, and calli.
For the 27 genes supported by incomplete transcripts, seven of them lack start/stop codons and another seven contain either known gaps between partial sequences of the same cDNA clones or unknown gaps between ESTs or fl-cDNAs showing similarity to the same protein in Arabidopsis thaliana or to another rice transcript mapped elsewhere in the genome. We successfully constructed tentative complete transcript sequences for all 14 of these genes by extending the original transcripts to the nearest start or stop codon and by sequencing RT-PCR products to fill the gaps (see Supplemental Table 3 online; Figure 2). Each of the other 13 genes contained a single exon that was supported by ESTs or fl-cDNAs without poly(A) tails. We verified for 12 genes that their transcript matches do represent genuine transcripts and were not derived from contamination of genomic DNA. This analysis left a single gene (Cen8.t00749) whose EST was not confirmed, and this gene model was discarded from the data set. RT-PCR analysis confirmed 14 of the 22 protein homology-based genes and 43 of the 78 hypothetical genes as being transcribed in at least one of the four tissues (Table 1, Figure 2). Two of the hypothetical genes (Cen8.t00037 and Cen8.t00948) yielded smaller-than-predicted products of <100 bp, and they were discarded from the data set. Collectively, of the 127 genes surveyed, 83 were transcriptionally active and 41 were inactive, with three genes being discarded (Table 1).
Figure 2.
Examples of RT-PCR Verification of Expression for Cen8 Genes.
In the top panel, hypothetical genes Cen8.t00047 and Cen8.t01279 are expressed in all four tissues, while hypothetical gene Cen8.t01782 is only expressed in calli; product size matches the prediction for genes Cen8.t00047 and Cen8.t01782 but is 210 bp larger than predicted for Cen8.t01279. In the middle panel, gene Cen8.t01639 matches CB639805, an EST with no start codon, and RT-PCR confirmed that the nearest start codon is 131 bp upstream; for gene Cen8.t01884, the primers amplified both its cognate transcript (the 399-bp fragment) and another transcript of ∼1100 bp (arrow) from its paralog; the predicted 394-bp fragment was not observed in gene Cen8.t00761, but a smaller fragment derived from its paralog was detected in all four tissues (arrow). In the bottom panel, gene Cen8.t01401 matched two nonoverlapping ESTs CB649599 and AU067849 that showed strong homology to an Arabidopsis cation-chloride cotransporter NP_174333, and RT-PCR confirmed that the two ESTs represent partial transcripts of this gene; for gene Cen8.t01706, its fl-cDNA AK119925 and EST CB646495 both show similarity to fl-cDNA AK121112 mapped to chromosome 9, and RT-PCR using two pairs of primers revealed an ∼2300-bp (1200 + 1100) gap between these two transcripts, one primer pair failed to amplify the expected 11,680-bp fragment from the genomic DNA. M, molecular marker; G, genomic DNA; L, cDNA from leaves; R, cDNA from roots; E, cDNA from etiolated leaves/shoots; C, cDNA from calli; N, negative control (without adding reverse transcriptase).
Table 1.
RT-PCR Analysis of Genes Located in the 3.5-Mb Cen8 Virtual Contig
| Genes with Complete Transcripts | Genes with Incomplete Transcripts | Genes with Protein Homologs | Hypothetical Genes | |
|---|---|---|---|---|
| Total no. of genes | 79 | 27 | 40 | 79 |
| No. of genes tested | 71 | 27 | 22 | 78 |
| RT-PCR negative | 0 | 1 | 8 | 35 |
| RT-PCR positive | 71 (100%) | 26 (96.3%) | 14 (63.6%) | 43 (55.1%) |
| Four tissues | 63 | 19 | 10 | 16 |
| Three tissues | 4 | 1 | 2 | 5 |
| Two tissues | 1 | 3 | 0 | 13 |
| One tissue | 3 | 3 | 2 | 9 |
We also conducted RT-PCR analysis on 71 of the 79 genes with a complete transcript match because these genes were targets for ChIP analysis (see below). We wanted to test how many of these genes are expressed in the four cDNA samples, especially in etiolated seedlings that were also used as tissue for ChIP analysis. This analysis revealed that 63 genes were expressed in all four cDNA samples, four in three samples, and another four in one or two samples (gene category A5 in Supplemental Table 3 online). We confirmed that 65 genes were expressed in 14-d-old etiolated seedlings. Altogether, we estimated the existence of 222 (225 − 3) genes in this 3.5-Mb region. Among them, 162 genes were confirmed to be transcriptionally active based on both RT-PCR analysis and sequence similarity to rice transcripts, 41 genes whose expression was not confirmed by RT-PCR were labeled as inactive genes, and the remaining 19 genes were not tested by RT-PCR (Table 2).
Table 2.
Distribution of Genes and Repeats along the 3.5-Mb Cen8 Virtual Contig
| Feature | Domain C1 | Domain B1 | Domain A | Domain B2 | Domain C2 |
|---|---|---|---|---|---|
| Genetic position (cM) | 54.0–54.3 | 54.3 | 54.3 | 54.3 | 54.3–55.4 |
| Coordinate (kb) | 1–437 | 437–1350 | 1350–2100 | 2100–2749 | 2749–3490 |
| Size (kb) | 437 | 913 | 750 | 649 | 741 |
| Active genes | 28 | 32 | 16 | 38 | 48 |
| Protein homology | 12 | 20 | 7 | 24 | 17 |
| No protein homology | 16 | 12 | 9 | 14 | 31 |
| Gene density (kb per active gene) | 15.6 | 28.5 | 46.9 | 17.1 | 15.4 |
| Active gene coverage | 24.0% | 15.9% | 12.8% | 19.8% | 21.2% |
| Inactive or unanalyzed genes | 15 | 14 | 13 | 7 | 11 |
| Protein homology | 6 | 6 | 10 | 2 | 2 |
| Ab initio prediction | 9 | 8 | 3 | 5 | 9 |
| Total genes | 43 | 46 | 29 | 45 | 59 |
| Retrotransposon (kb) | 135.9 | 390.0 | 277.8 | 214.8 | 209.4 |
| Transposon (kb) | 10.1 | 55.0 | 32.9 | 33.1 | 51.3 |
| MITE (kb) | 10.2 | 15.4 | 8.4 | 20.1 | 17.6 |
| Others or unknown (kb) | 8.1 | 10.9 | 62.2 | 8.4 | 7.1 |
| Total repeats (kb) | 164.2 (37.6%) | 471.3 (51.6%) | 381.3 (50.8%) | 276.3 (42.6%) | 285.4 (38.5%) |
Active genes have matches with rice transcripts and/or are RT-PCR positive. Inactive genes have no matches with rice transcripts and/or are RT-PCR negative. Unanalyzed genes have no matches with rice transcripts and/or RT-PCR was not done. MITE, miniature inverted-repeat transposable element.
We previously reported four active genes in the ∼750-kb CENH3 binding domain (Nagaki et al., 2004). Our new annotation and RT-PCR data provide compelling evidence for the presence of 16 active genes in this domain (see Supplemental Table 4 online). Of these 16 active genes, 12 were expressed in all four cDNA samples, with gene Cen8.t00969 being the only one that was expressed in a single tissue (calli) (see Supplemental Figure 1 online). The putative function can be assigned for seven of the 16 active genes. Nine genes have significant homologs in Arabidopsis (E ≤ 2.1 × 10−43) based on BLASTp search against annotated Arabidopsis proteins (The Institute for Genomic Research [TIGR] V5, http://www.tigr.org/tdb/e2k1/ath1/).
Active Genes Located in the Recombination-Free Region of Cen8
The recombination-free region of Cen8 spans 2312 kb. The 750-kb CENH3 binding domain is located approximately in the middle (Figure 1). The two outer domains (C1 and C2) in the Cen8 virtual contig show low recombination rates, 1457 kb/cM and 674 kb/cM, respectively, compared with an average of 283 kb/cM across the rice genome (Harushima et al., 1998). An in-depth annotation of the Cen8 virtual contig provides a unique opportunity for us to examine how genes, especially active genes, are organized and distributed in a large chromosomal domain with severe or complete recombination suppression.
The 750-kb CENH3 binding domain (A) shows the lowest density of active genes (47 kb per gene) and lowest active gene coverage (12.8% of total DNA sequence) compared with the surrounding domains (Table 2). The active genes within this domain are generally scattered over its entire length. Two subdomains were found to be extremely poor in active genes, one between gene numbers 98 and 100 and the other between gene numbers 109 and 117, together covering more than half (402 kb) of the 750-kb domain (see Supplemental Figure 2 online). There is only one active gene within each of these two subdomains (numbers 99 and 113, respectively).
Domains C1 and C2, which flank the recombination-free region, show relatively higher densities of active genes (∼16 kb per gene or 21.2 to 24.0% of total sequence) than domain A but differ little from the recombination-free domain B2, which has a gene density of 17 kb per active gene (19.8% of total sequence) (Table 2). Transcribed sequences are not evenly distributed within the virtual contig. This is particularly prominent in the 913-kb recombination-free domain B1 (Figure 3; gene numbers 44 to 89 in Supplemental Figure 2 online). Specifically, a 475-kb region within domain B1 (from gene number 44 to 71) has an active gene coverage of 19.6%; by contrast, the second region of 251 kb (from gene number 72 to 76) harbors only a single active gene, and the third region of 187 kb (from gene number 77 to 89) adjacent to domain A has a much higher coverage of active genes (27.1% of total sequence) (see Supplemental Figure 2 online).
Figure 3.
Sequence Features along the ∼3.5-Mb Cen8 Virtual Contig.
Genes were annotated using a combination of transcript/protein homolog search and ab initio prediction and then tested for expression using RT-PCR. Repetitive DNA was identified using RepeatMasker with the TIGR Oryza repeat database (Ouyang and Buell, 2004) and centromere retrotransposon of rice (Nagaki et al., 2005) as the filters. The cutoff is a minimum score of 225 and ≤35% of divergence. All genes and repeats are plotted against their coordinates on the virtual contig.
Repetitive DNA in Cen8
We used RepeatMasker to reveal the abundance and distribution of repetitive DNA across the five domains in Cen8. We found that 1,578,492 bp DNA, or 45.23% of the ∼3.5-Mb sequence, is repetitive DNA (Table 2); the majority of this consists of retrotransposons (77.79%), followed by DNA transposons (11.56%), miniature inverted-repeat transposable elements (4.54%), unknown repeats (3.52%), and the centromeric CentO satellite repeat (2.42%) (Dong et al., 1998; Cheng et al., 2002) or other satellite DNA (0.18%). Up to 97.2% of the repetitive DNA is located within intergenic regions (Figure 3). We also detected 44,210 bp of repetitive DNA (2.8% of total repeats) within 75 of the 222 annotated genes. Most of this (34,626 bp) is intronic sequences, with 7557 bp being protein coding sequences and 2027 bp being untranslated sequences. Regions of low gene density mostly correspond to regions of higher retrotransposon density. Despite such local variations in density, the overall distribution of retrotransposons and other repetitive elements appears to be similar among the five domains throughout the 3.5-Mb virtual contig, with only a modest enrichment in domains A and B1 of the recombination-free region (Figure 3, Table 2).
Histone Modifications Associated with Cen8 Genes
It has been recognized in numerous organisms that histone H4 acetylation (H4Ac; at Lys 5, 8, 12, and 16) and histone H3 dimethylation at Lys 4 (H3K4me2) are preferentially associated with transcriptionally active euchromatin, while histone H3 dimethylation at Lys 9 (H3K9me2) is a hallmark of repressive heterochromatin (Jenuwein and Allis, 2001). We asked if histone modification patterns are identical for genes located in the CENH3 binding domain, in the recombination-free zone, and in the flanking regions that are recombinationally competent. We conducted ChIP analysis on 150 active and 26 inactive genes using antibodies against three modified histones: H4Ac, H3K4me2, and H3K9me2. We observed that 77% of the active genes, versus 42% of the inactive genes, showed enrichment for modifications of H4Ac, H3K4me2, or both (Table 3). Of the 127 genes showing H4Ac or H3K4me2, 71% of them are associated with both modifications, reflecting their concerted role in regulating transcription of Cen8 genes. By contrast, we detected a much smaller proportion of genes (only 6%), including four of the active genes (3%) and seven of the inactive genes (27%), that were selectively enriched for H3K9me2 only. The enrichment of both H3K9me2/H3K4me2 or H4Ac on 13 genes may reflect differences in the transcription or histone modification status of these genes between different cells.
Table 3.
Histone Methylation and Acetylation Associated with Cen8 Genes
| Category | H4Ac | H3K4me2 | H4Ac and H3K4me2 | H4Ac and H3K9me2 H3K4me2 and H3K9me2 | H3K9me2 | None |
|---|---|---|---|---|---|---|
| Active genes | 14 (10) | 19 (17) | 83 (72) | 10 (7) | 4 (3) | 21 (16) |
| Inactive genes | 2 | 2 | 7 | 3 | 7 | 4 |
| Total | 16 | 21 | 90 | 13 | 11 | 25 |
Numbers in parentheses indicate genes expressed in etiolated seedlings that were used for ChIP analysis.
We observed a maximum relative fold enrichment (RFE) up to 101 for H4Ac and 237 for H3K4me2, a striking contrast to the maximum RFE of just 11 for H3K9me2 (Figure 4). To better describe the fluctuation of H3K4me2 and H4Ac levels of adjacent genes, we arbitrarily chose a cutoff of RFE ≥16 for both H3K4me and H4Ac and cataloged the 140 H3K4me2- or H4Ac-associated genes into two main groups. Group I includes 33 genes with RFE ≥16 for both H3K4me2 and H4Ac and another 33 genes for either H3K4me2 or H4Ac, while group II includes the remaining 74 genes that have a RFE <16. The 66 genes with RFE of H3K4me2 and/or H4Ac ≥16 were mapped as 53 H3K4me2 and/or H4Ac hot spots in the virtual contig. Most of these hot spots (42) are represented by a single gene, with the neighboring genes displaying either decreased levels of H3K4me2/H4Ac, no H3K4me2/H4Ac, or the presence of H3K9me2. Each of the other 11 hot spots contains two to three physically linked genes with each hot spot spanning ∼2 to 16 kb. Overall, these hot spots do not show a tendency to cluster and scatter across all five domains of the virtual contig.
Figure 4.
ChIP-PCR Analysis of Histone Methylation and Acetylation Associated with Cen8 Genes.
Primers from 150 active and 26 inactive genes were used in quantitative real-time PCR, using gene model Cen8.t00238 (putative NB-ARC domain) as the reference that shows none of H4Ac, H3K4me2, and H3K9me2. For each primer pair, significance of enrichment difference between antibody binding fraction and mock treatment was tested using Student's t test (α = 0.05). RFE is then calculated as 2−ΔΔCT (see Methods) and indicated by the height of color-coded lines.
ChIP data from all 16 active and three out of the four inactive genes from the ∼750-kb CENH3 binding domain A showed that only three genes (one active and two inactive) were not enriched for H3K4me2 or H4Ac. Furthermore, these genes displayed the highest level of H4Ac compared with genes in the flanking domains (see Supplemental Figure 3 online). Approximately 40% of the ChIP-positive genes are highly methylated at H3K4 and/or highly acetylated at H4, as defined by their RFE exceeding the cutoff of 16 (see Supplemental Figure 3 online).
The ChIP data on H3K4me2/H4Ac contrast with our earlier inference that the CENH3 binding region contains a high level of H3K9me2 and a low level of H3K4me2 (Nagaki et al., 2004). To understand the reason for this apparent difference between the old and new data sets, we repeated the analysis of the 22 sequences from this ∼750-kb CENH3 binding domain previously used by Nagaki et al. (2004). Ten of the 11 sequences previously shown to be enriched for H3K9me2 by gel-based ChIP were found to be enriched for H3K9me2 by real-time PCR ChIP. However, four of these 11 sequences also showed some levels of enrichment for H3K4me2, and an additional six out of the other 11 sequences for which no association with H3K4me2 or H3K9me2 was previously detected were found to be enriched for H3K4me2. The difference between the old and new ChIP results for some of the primer sets is likely to have resulted from the different techniques used in the studies. The real-time PCR method used in this study is more sensitive, allowing us to detect low levels of H3K4me2 that were missed using the gel-based PCR method employed by Nagaki et al. (2004).
In addition to differences in sensitivity between the two sets of experiments, there are also systematic differences in the locations of the PCR primer pairs. Whereas the new data set uses almost exclusively primers designed from the coding sequences of the predicted genes, we found that of the 22 primer pairs from domain A in the previous data set, 11 pairs were from intergenic regions, 10 pairs were derived from introns and intron-exon junctions of the predicted genes, and the remaining pair was derived from an exon-intergenic junction. Notably, 9 of the 11 sequences enriched for CENH3 and 7 of the 10 sequences enriched for H3K9me2 in the previous study are from intergenic regions. Active genes are known to undergo histone replacement (Schwartz and Ahmad, 2005), and replacement forms of histone H3 are enriched for active histone modifications in both plants and animals (Waterborg, 1990; Johnson et al., 2004; McKittrick et al., 2004). Our results are consistent with the view that CENH3 and H3K9me2 are found primarily over nontranscribed regions of domain A because transcription results in replacement by histones that are enriched for active modifications.
DISCUSSION
Our complete sequence and annotation of the 3.5-Mb virtual contig provides a detailed characterization of an entire recombination-free region encompassing a centromere in a multicellular eukaryote. This has important implications for understanding the basis for suppression of meiotic recombination. The near universality of dense tandemly repetitive sequences in such regions had led to the assumption that these repeats, or the heterochromatin that assembles on them, are responsible for recombination suppression (Roberts, 1965; Khush and Rick, 1967, 1968). We demonstrate this assumption to be inadequate because rice Cen8 is just as recombination-free as other plant and animal centromeres, yet dense tandem repeats occupy only ∼60 kb of the 2312-kb recombination-free region.
It has been hypothesized that meiotic recombination is largely restricted to genes (Thuriaux, 1977). This hypothesis is supported by the fact that several recombination hot spots identified in the maize genome are associated with genes (Patterson et al., 1995; Fu et al., 2001, 2002; Yao et al., 2002). In addition, the retrotransposon-rich regions surrounding these hot spots lack recombination (Fu et al., 2002; Yao et al., 2002). Fu et al. (2002) proposed that a gene or a gene cluster may adopt a less condensed chromatin configuration and be more accessible to the recombination machinery during meiosis. This association between recombination and genic regions, however, cannot explain why the region associated with Cen8, which includes numerous active genes, is free of recombination (Figure 3).
The Cen8 recombination-free region is moderately enriched in transposable elements relative to the arms. Transposable elements appear to suppress meiotic recombination near the bronze locus in maize (Dooner and Martinez-Ferez, 1997), raising the possibility that similar transposon-based suppression accounts for the recombination-free region at rice Cen8. However, transposon enrichment is not especially striking at Cen8. Distally, there is no clear distinction between the recombination-free region and the surrounding region that recombines in the overall density of transposable elements. Transposable element density is inversely proportional to gene density, suggesting that the expansions in the physical map that result from insertions underlie reduced gene density. Insertions into recombination-free regions are expected to resist elimination, and when they occur between genes, they are unlikely to be culled out by purifying selection. The fact that Cen8 is still only moderately enriched in transposons indicates that the enrichment process, and thus the recombination-free region, is relatively young.
Our evidence that recombination-free regions around centromeres do not correspond to either genes or repetitive elements raises the question as to whether sequence features are involved at all in suppression of recombination at centromeres. This question was previously raised by Haupt et al. (2001), who found a fivefold difference in the recombination suppression of the apparently similar left and right pericentromeres of Arabidopsis CEN1, although the presence of megabases of satellite DNA in between complicated interpretation of this difference. In budding yeast, ectopically located centromeres are known to suppress recombination in their new flanking regions (Lambie and Roeder, 1986); but whether this is because of the sequence itself or the centromeric chromatin that forms there cannot be determined given that yeast centromeres are defined by sequence. In the case of rice Cen8, we cannot rigorously rule out the presence of some as-yet-unidentified sequence feature that distinguishes the Cen8 region from recombination-competent regions of the genome; however, we consider this unlikely. Functionally normal centromeres are known to form de novo in ordinary regions of the human genome, which strongly argues that centromeres are maintained epigenetically (Choo, 2001). Since recombination-free regions appear to be general features of centromeres, it seems most likely that they are important for normal centromere function during meiosis. This suggests that recombination-free regions around centromeres are themselves epigenetic features.
What might these epigenetic features be? Our inability to detect consistent histone modification differences between the recombination-free and flanking regions around Cen8 suggests that gross modification differences are not essential for recombination differences around centromeres. The same situation appears to hold for budding yeast centromeres, which lack pericentric heterochromatin and do not appear to alter adjacent nucleosome structure when they are ectopically located (Bloom and Carbon, 1982), even though they are able to suppress recombination in flanking regions (Lambie and Roeder, 1986). There is also no evidence to suggest that histone modifications around Cen8 are radically different between the etiolated seedlings that we assayed and meiotic cells where recombination occurs. In Arabidopsis, the distributions of H3K4me2 and H3K9me2 on chromosomes appear to be essentially the same in interphase, mitosis, and meiosis within the limits of immunofluorescent cytology (Jasencakova et al., 2003). In nematodes where histone modifications can be easily assayed from premeiosis to postmeiosis, the only obvious changes in the patterns of eight different histone modifications take place after recombination is initiated and are primarily restricted to the X chromosome (Kelly et al., 2002). Although we cannot exclude the possibility of an altered histone modification pattern in rice meiocytes, taken together, these observations suggest that CENH3-containing chromatin itself is the primary feature that underlies suppression of recombination around centromeres.
It is difficult to envision the direct action of CENH3-containing chromatin on recombination over the ∼1-Mb distances between domain A and domains C1/C2 of rice Cen8. Rather, we suggest that the effect is indirect. Specifically, organisms might deposit proteins that inhibit recombination over long stretches on either side of centromeric chromatin. For example, ectopically located human CENP-A recruits the cohesin subunit hSMC1 (Van Hooser et al., 2001), and ectopic kinetochores in budding yeast locally enhance cohesin binding (Weber et al., 2004). Although cohesins are necessary for repair of meiotic double-strand breaks in budding yeast (Klein et al., 1999), those breaks do not occur in regions of high cohesin density (Blat et al., 2002; Glynn et al., 2004). This suggests that cohesins inhibit interactions between homologs that are necessary for meiotic recombination, which has been hypothesized to occur in chromatin loops between cohesin binding sites (Blat et al., 2002). Consistent with this possibility, mutations in the cohesin subunit Psc3 lead to increased rearrangement at the cohesin-enriched mating type locus of fission yeast, even though the underlying heterochromatin remains unchanged (Nonaka et al., 2002). In support of the possibility that cohesins inhibit recombination around centromeres, we note that ectopic yeast centromeres enhance binding of cohesins and reduce meiotic recombination over 20 to 50 kb, a length that is >100-fold larger than the region occupied by the single CENH3-containing nucleosome of budding yeast (Lambie and Roeder, 1986; Weber et al., 2004). Thus, high concentrations of cohesins might inhibit recombination in extensive regions around centromeric DNA packaged into CENH3-containing nucleosomes. Recruitment of cohesins by pericentric heterochromatin (Bernard et al., 2001; Nonaka et al., 2002; Fukagawa et al., 2004) may be an independent means of ensuring cohesion around centromeres, resulting in more extensive recombination suppression as a consequence.
In animals, CENH3-containing nucleosomes are present in long arrays that are separated by arrays of H3-containing nucleosomes (Blower et al., 2002; Chueh et al., 2005). Remarkably, patterns of covalent modifications on H3 and H4 in centromeric arrays are very different from those found over the rest of the genome. Specifically, H3K4me2 is detected, whereas trimethylated H3K4 (H3K4me3), methylated H3K9, and acetylated H3 and H4 are strikingly depleted at centromeres, in contrast with the situation for other parts of the genome (Sullivan and Karpen, 2004). This observation has lead to the concept of centrochromatin, which is functionally distinguished from euchromatin and heterochromatin by both CENH3 and histone modifications maintained through interphase and mitosis. However, these findings appear to conflict with the observation of striking hyperacetylation of H4 at interphase barley centromeres (Jasencakova et al., 2001; Wako et al., 2002, 2003), suggesting that plants and animals have very different centrochromatin compositions. Our high-resolution mapping of rice Cen8 centrochromatin provides a more satisfying explanation for this apparent discrepancy between centromeres in different organisms that are functionally the same. We find that H3K4me2 and H4 acetylation are primarily features of active genes at rice centromeres, which otherwise are enriched in H3K9me2. Our previous finding that H3K4me2 is reduced in Cen8 relative to euchromatic control loci (Nagaki et al., 2004) appears to reflect both technical limitations of gel-based PCR and the fact that many of the primers chosen for that study were in intergenic regions. The cytological characterization of modifications at animal centromeres detected an overall reduction of H3K4me2 at centromeres relative to euchromatic arms, although not nearly as much as for the other modifications (Sullivan and Karpen, 2004), and similar differences in H3K4me2 levels were found in fission yeast centromeres (Cam et al., 2005). Therefore, the results for plants, fungi, and animals with respect to centromeric H3K4me2 levels may be similar.
Other differences between rice and animal centromeres might simply reflect differences in sequence composition. Unlike animal centromeres, rice Cen8 essentially lacks satellite DNA sequences but is highly enriched in active genes and transposons that are absent from the centromeres of flies and humans. The histone modifications that we find enriched at genes and near transposons of rice Cen8 are characteristic of modifications found for these sequence elements genome wide. By contrast, animal centromeric satellites have no counterparts outside of centromeric regions for comparison of histone modifications. We propose that the remarkable H3 and H4 modification features seen at animal centromeres (Sullivan and Karpen, 2004) are not special for centromeres but instead are characteristic of satellite DNA arrays. Tighter packaging of phased nucleosomes at satellite DNA arrays would inhibit accessibility of the machinery that is responsible for chromatin modification, resulting in reduction of most H3 and H4 tail modifications, including acetylation and H3K9 methylation. The presence of moderate levels of H3K4me2 and absence of H3K4me3 would result from occlusion of the Set1 H3K4 methyltransferase, which would be reduced in its ability to complete the conversion of H3K4me2 to H3K4me3 in satellite DNA nucleosome arrays. In support of this idea, we note that H3K4me3 was observed to be depleted in the large pericentric satellite regions found on some human chromosomes but not in other pericentric heterochromatin (Sullivan and Karpen, 2004). Thus, centrochromatin would be epigenetically maintained by assembly of CENH3-containing nucleosomes, but the distribution of histone modifications would reflect the composition and abundance of sequence elements that inhabit centromeric DNA.
METHODS
Construction of the Cen8 Virtual Contig
We previously sequenced 11 rice (Oryza sativa) BAC clones that span the centromere of rice chromosome 8 (Nagaki et al., 2004). A virtual contig of 1,654,753 bp had been constructed by assembling sequences of these 11 clones and four additional flanking clones from GenBank. We have extended this virtual contig by adding sequences of seven BAC/PAC clones to the north and 12 clones to the south side. We retrieved clone sequences from GenBank between February 12 and April 10, 2004, trimmed overlapping sequences, and linked the unique sequences to form a contiguous sequence of 3,489,829 bp. This Cen8 virtual contig corresponds to the genetic region from 54.0 to 55.4 cM on the linkage map (Figure 1).
Computational Gene Annotation
We annotated the ∼3.5-Mb Cen8 sequence for gene models using both DNA and protein homology searches as well as ab initio predictions. We first searched the Cen8 sequence against the rice transcript sequences, which consist of 32,127 fl-cDNAs from rice variety Nipponbare (downloaded from the KOME website, http:/cdna01.dna.affrc.go.jp/cDNA/, version October 24, 2003), the same rice variety that was used for genome sequencing, and 286,163 EST sequences (March 15, 2004, GenBank EST database, rice) using Blast2seq available in the NCBI Blastall program. ESTs and fl-cDNAs were parsed out at a stringency of at least 90% identity over 80% the length of a given entry sequence. We then aligned these EST and fl-cDNA sequences with the Cen8 virtual contig using Spidey (Wheelan et al., 2001) to delineate the gene intron-exon structure. We considered an EST or fl-cDNA as the corresponding transcript for Cen8 when (1) it shows at least 95% identity with the genome sequence over at least 95% of its length, and (2) it has a poly(A) tail or spans a minimum of two exons with clear splicing sites. RT-PCR was then used to confirm the authenticity of ambiguous ESTs and fl-cDNAs (see below for RT-PCR procedures). For each gene model, we assembled multiple EST and fl-cDNA sequences into a consensus sequence using CAP3 (Huang and Madan, 1999) and corrected sequencing errors whenever possible. Alternative splicing variants for the same gene were differentially labeled by adding the suffix .1, .2, .3, and so on to the gene name, with suffix .1 standing for gene model with the longest protein coding sequence. ESTs and fl-cDNAs that encode <50 amino acids were considered as noncoding RNAs and were excluded from further analysis.
We then searched Cen8 sequence against the NCBI nonredundant protein database (downloaded on March 18, 2004) using BLASTx. In this case, Cen8 sequence was first masked for repetitive elements using RepeatMasker (http://www.repeatmasker.org/, version 20040306) at the cutoff score of ≥225 and divergence ≤35%, with TIGR Oryza repeat database (Ouyang and Buell, 2004; ftp://ftp.tigr.org/pub/data/TIGR_Plant_Repeats/) and centromere retrotransposon of rice (Nagaki et al., 2005) as the filters. Protein matches with an expect value of ≤10−20 are regarded as being significant, excluding matches to transposon-related proteins or to entries labeled as hypothetical protein from rice. We used the DPS/NAP program (Huang et al., 1997) to infer tentative gene structure.
We performed ab initio gene predictions using five gene finders, including FGENESH, Genemark.hmm (rice matrix), Genscan (maize matrix), Genscan+ (Arabidopsis matrix), and GlimmerR. Only gene models that were predicted by two gene finders with similar exon-intron structure were included.
We classified gene models based on the sources of supporting evidence. Gene models having less perfect, but significant, protein homologs were labeled as “putative XXX protein” (E ≤ 10−20) or “XXX-like protein” (E ≤ 10−10); gene models whose existence was supported by cognate rice transcripts or by positive RT-PCR, but without significant protein homologs, were labeled as “unknown protein”; and finally, gene models predicted by gene prediction programs whose expression was not confirmed by RT-PCR were labeled as “hypothetical protein” (see Supplemental Table 1 online). In all cases, we excluded genes that showed similarity to transposons or had many copies in the rice genome.
RT-PCR Analysis
We used RT-PCR to test the expression of gene models for which we did not identify the corresponding transcript sequences, including hypothetical genes only predicted by gene finders and some of the genes with protein homologs. We also used RT-PCR to verify the expression spectrum of all the gene models for which only incomplete transcripts are available and most of the gene models with complete transcripts. We defined an EST or fl-cDNA sequence as an incomplete transcript when (1) it covers only a single exon and contains no poly(A) tail (for which the possibility of genomic DNA contamination cannot be excluded), or (2) it represents only partial coding sequence that either lacks start/stop codon surrounding sequence or contains internal sequence gap. For an EST or fl-cDNA to be considered as a complete transcript, it should contain complete coding sequence (at least 153 bp or 50 amino acids), covering at least two exons or containing a poly(A) tail when it contains only one exon. The amplified products were directly sequenced when necessary.
We designed primers using Primer3 (http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi) in conjunction with Netprimer (http://www.premierbiosoft.com/netprimer/). To increase the specificity of amplification, we designed primers from part of a gene that showed divergence from its paralogs, if any, by aligning the predicted coding sequence against the virtual contig of the entire rice genome (http://www.tigr.org/tdb/e2k1/osa1/, version 2). Only in a few cases were primers designed from the predicted untranslated regions or intronic sequences. Each primer was between 24 and 32 bp long with an annealing temperature from 65 to 82°C (see Supplemental Table 2 online).
We isolated Nipponbare mRNA from four different tissues or treatments for RT-PCR, including (1) leaves and (2) roots collected from 14-d-old plants growing in a Biotron greenhouse set at 32°C for 16 h and 30°C for 8 h; (3) etiolated leaves/shoots collected from 14-d-old plants growing in the darkness in the same Biotron greenhouse; and (4) calli that were induced by keeping seeds on Murashige and Skoog medium with supplement of 2 mg/L 2,4-D for 22 d and collected after another 12 d of growing on new Murashige and Skoog medium.
We extracted total mRNA using the Qiagen RNeasy plant mini kit (catalog number 74904), which is supplemented with an on-column DNA digestion for 20 min (Qiagen RNase-Free DNase set, catalog number 79254). Before reverse transcription, we treated the mRNA with DNA-free (Ambion; catalog number 1906). The absence of detectable contamination with genomic DNA was confirmed using RT (−) control, without adding Superscript II reverse transcriptase to the reverse transcription reaction. Primers were first tested on rice genomic DNA to get specific amplification before use in RT-PCR. The RT-PCR program was 94°C for 3 min, followed by 37 cycles of 94°C for 30 s, 62 to 67°C for 40 s, and 72°C for 40 s and ended by a 4-min extension at 72°C.
ChIP
We conducted ChIP using 2-week-old etiolated seedlings. We immunoprecipitated DNA associated with modified histones using antidimethyl-histone H3 (Lys 4) (H3K4me2), antidimethyl-histone H3 (Lys 9) (H3K9me2), and antiacetyl-histone H4 (Lys 5, 8, 12, and 16) (H4Ac) antibodies (Upstate Biotechnology); mock-precipitated extract with normal rabbit serum was used as negative control. We designed primers using the criteria as described above to amplify genomic regions sized between 148 and 332 bp. We conducted quantitative real-time PCR analysis on a DNA Engine Option System (MJ Research) to determine the relative enrichment of modified histone-associated sequences in the bound fractions over the mock treatment. PCR reactions were performed in triplicate using the DyNAmo HS SYBR Green qPCR kit (MJ Research) and run at 95°C for 15 min, 45 cycles of 95°C for 10 s, 62 to 67°C for 30 s, and 72°C for 30 s. The PCR cycle threshold (CT) was manually set to let the signals be slightly above the fluorescent intensity baseline (0.05 for most of the experiments). For each primer pair, we calculated the RFE of the amplified product with the comparative CT method according to Saffery et al. (2003), using gene Cen8.t00238 as the reference that shows no enrichment of H4Ac, H3K4me2, and H3K9me2. We calculated CT difference for mock DNA as ΔCT (mock DNA) = CT (gene of interest) − CT (Cen8.t00238) and that for bound fraction as ΔCT (bound DNA) = CT (gene of interest) − CT (Cen8.t00238). We then calculated the RFE as 2−ΔΔCT, where ΔΔCT = ΔCT (bound DNA) − ΔCT (mock DNA). We performed one-tailed Student's t test of ΔΔCT values at the significant level of α = 0.05.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. RT-PCR Verification of Expression for 16 Active Genes in the ∼750-kb CENH3 Binding Domain.
Supplemental Figure 2. Distribution of Genes and Repetitive Elements within the ∼3.5-Mb Cen8 Virtual Contig.
Supplemental Figure 3. Comparison of H3 Methylation at Lys 4 (H3K4me2) and H4 Acetylation (H4Ac) Levels among the Five Cen8 Domains.
Supplemental Table 1. List of Genes Predicted in the 3.5-Mb Cen8 Virtual Contig.
Supplemental Table 2. Primers Used in RT-PCR and ChIP Analyses.
Supplemental Table 3. RT-PCR Analysis of Cen8 Genes.
Supplemental Table 4. Active Genes Located in the CENH3 Binding Domain.
Supplementary Material
Acknowledgments
We thank John Crow at the University of Minnesota (Twin Cities, MN) for help with computational analysis. This research was supported by Grant FG02-01ER15266 from the Department of Energy and partly by Grant 9975827 from the National Science Foundation to J.J.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Jiming Jiang (jjiang@wisc.edu).
Online version contains Web-only data.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.105.037945.
References
- Amor, D.J., Kalitsis, P., Sumer, H., and Choo, K.H.A. (2004). Building the centromere: From foundation proteins to 3D organization. Trends Cell Biol. 14, 359–368. [DOI] [PubMed] [Google Scholar]
- Anderson, L.K., Doyle, G.G., Brigham, B., Carter, J., Hooker, K.D., Lai, A., Rice, M., and Stack, S.M. (2003). High-resolution crossover maps for each bivalent of Zea mays using recombination nodules. Genetics 165, 849–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beadle, G.W. (1932). A possible influence of the spindle fibre on crossing-over in Drosophila. Proc. Natl. Acad. Sci. USA 18, 160–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard, P., Maure, J.F., Partridge, J.F., Genier, S., Javerzat, J.P., and Allshire, R.C. (2001). Requirement of heterochromatin for cohesion at centromeres. Science 294, 2539–2542. [DOI] [PubMed] [Google Scholar]
- Blat, Y., Protacio, R.U., Hunter, N., and Kleckner, N. (2002). Physical and functional interactions among basic chromosome organizational features govern early steps of meiotic chiasma formation. Cell 111, 791–802. [DOI] [PubMed] [Google Scholar]
- Bloom, K.S., and Carbon, J. (1982). Yeast centromere DNA is in a unique and highly ordered structure in chromosomes and small circular minichromosomes. Cell 29, 305–317. [DOI] [PubMed] [Google Scholar]
- Blower, M.D., Sullivan, B.A., and Karpen, G.H. (2002). Conserved organization of centromeric chromatin in flies and humans. Dev. Cell 2, 319–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cam, H.P., Sugiyama, T., Chen, E.S., Chen, X., FitzGerald, P.C., and Grewal, S.I.S. (2005). Comprehensive analysis of heterochromatin- and RNAi-mediated epigenetic control of the fission yeast genome. Nat. Genet. 37, 809–819. [DOI] [PubMed] [Google Scholar]
- Cheng, Z.K., Dong, F., Langdon, T., Ouyang, S., Buell, C.B., Gu, M.H., Blattner, F.R., and Jiang, J. (2002). Functional rice centromeres are marked by a satellite repeat and a centromere-specific retrotransposon. Plant Cell 14, 1691–1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choo, K.H.A. (2001). Domain organization at the centromere and neocentromere. Dev. Cell 1, 165–177. [DOI] [PubMed] [Google Scholar]
- Chueh, A.C., Wong, L.H., Wong, N., and Choo, K.H.A. (2005). Variable and hierarchical size distribution of L1-retroelement-enriched CENP-A clusters within a functional human neocentromere. Hum. Mol. Genet. 14, 85–93. [DOI] [PubMed] [Google Scholar]
- Davis, C.R., Kempainen, R.R., Srodes, M.S., and McClung, C.R. (1994). Correlation of the physical and genetic maps of the centromeric region of the right arm of linkage group III of Neurospora crassa. Genetics 136, 1297–1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong, F., Miller, J.T., Jackson, S.A., Wang, G.L., Ronald, P.C., and Jiang, J. (1998). Rice (Oryza sativa) centromeric regions consist of complex DNA. Proc. Natl. Acad. Sci. USA 95, 8135–8140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dooner, H.K., and Martinez-Ferez, I.M. (1997). Recombination occurs uniformly within the bronze gene, a meiotic recombination hotspot in the maize genome. Plant Cell 9, 1633–1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu, H., Park, W., Yan, X., Zheng, Z., Shen, B., and Dooner, H.K. (2001). The highly recombinogenic bz locus lies in an unusually gene-rich region of the maize genome. Proc. Natl. Acad. Sci. USA 98, 8903–8908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu, H., Zheng, Z., and Dooner, H.K. (2002). Recombination rates between adjacent genic and retrotransposon regions in maize vary by 2 orders of magnitude. Proc. Natl. Acad. Sci. USA 99, 1082–1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukagawa, T., Nogami, M., Yoshikawa, M., Ikeno, M., Okazaki, T., Takami, Y., Nakayama, T., and Oshimura, M. (2004). Dicer is essential for formation of the heterochromatin structure in vertebrate cells. Nat. Cell Biol. 6, 784–791. [DOI] [PubMed] [Google Scholar]
- Glynn, E.F., Megee, P.C., Yu, H.G., Mistrot, C., Unal, E., Koshland, D.E., DeRisi, J.L., and Gerton, J.L. (2004). Genome-wide mapping of the cohesin complex in the yeast Saccharomyces cerevisiae. PLoS Biol. 2, 1325–1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harushima, Y., et al. (1998). A high-density rice genetic linkage map with 2275 markers using a single F2 population. Genetics 148, 479–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haupt, W., Fischer, T.C., Winderl, S., Fransz, P., and Torres-Ruiz, R.A. (2001). The CENTROMERE1 (CEN1) region of Arabidopsis thaliana: Architecture and functional impact of chromatin. Plant J. 27, 285–296. [DOI] [PubMed] [Google Scholar]
- Huang, X., Adams, M.D., Zhou, H., and Kerlavage, A.R. (1997). A tool for analyzing and annotating genomic sequence. Genomics 46, 37–45. [DOI] [PubMed] [Google Scholar]
- Huang, X., and Madan, A. (1999). CAP3: A DNA sequence assembly program. Genome Res. 9, 868–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson, M.S., See, C.G., Mulligan, L.M., and Lauffart, B.F. (1996). A 9.75-Mb map across the centromere of human chromosome 10. Genomics 33, 258–270. [DOI] [PubMed] [Google Scholar]
- Jasencakova, Z., Meister, A., and Schubert, I. (2001). Chromatin organization and its relation to replication and histone acetylation during the cell cycle in barley. Chromosoma 110, 83–92. [DOI] [PubMed] [Google Scholar]
- Jasencakova, Z., Soppe, W.J.J., Meister, A., Gernand, D., Turner, B.M., and Schubert, I. (2003). Histone modifications in Arabidopsis—High methylation of H3 lysine 9 is dispensible for constituitive heterochromatin. Plant J. 33, 471–480. [DOI] [PubMed] [Google Scholar]
- Jenuwein, T., and Allis, C.D. (2001). Translating the histone code. Science 293, 1074–1080. [DOI] [PubMed] [Google Scholar]
- Jin, W.W., Lamb, J.C., Vega, J.M., Dawe, R.K., Birchler, J.A., and Jiang, J. (2005). Molecular and functional dissection of the maize B centromere. Plant Cell 17, 1412–1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, L., Mollah, S., Garcia, B.A., Mouratore, T.L., Shabanowitz, J., Hunt, D.F., and Jacobsen, S.E. (2004). Mass spectrometry analysis of Arabidopsis histone H3 reveals distinct combinations of post-translational modifications. Nucleic Acids Res. 32, 6511–6518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly, W.G., Schaner, C.E., Dernburg, A.F., Lee, M.-H., Kim, S.K., Villeneuve, A.M., and Reinke, V. (2002). X-chromosome silencing in the germline of C. elegans. Development 129, 479–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khush, G.S., and Rick, C.M. (1967). Studies on the linkage map of chromosome 4 of the tomato and on the transmission of induced deficiencies. Genetica 38, 74–94. [Google Scholar]
- Khush, G.S., and Rick, C.M. (1968). Cytogenetic analysis of the tomato genome by means of induced deficiencies. Chromosoma 23, 452–484. [Google Scholar]
- Klein, F., Mahr, P., Galova, M., Buonomo, S.B.C., Michaelis, C., Nairz, K., and Nasmyth, K. (1999). A central role for cohesions in sister chromatid cohesion, formation of axial elements, and recombination during yeast meiosis. Cell 98, 91–103. [DOI] [PubMed] [Google Scholar]
- Künzel, G., Korzun, L., and Meister, A. (2000). Cytologically integrated physical restriction fragment length polymorphism maps for the barley genome based on translocation breakpoints. Genetics 154, 397–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambie, E.J., and Roeder, G.S. (1986). Repression of meiotic crossing over by a centromere (CEN3) in Saccharomyces cerevisiae. Genetics 114, 769–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahtani, M.M., and Willard, H.F. (1998). Physical and genetic mapping of the human X chromosome centromere: Repression of recombination. Genome Res. 8, 100–110. [DOI] [PubMed] [Google Scholar]
- Mather, K. (1939). Crossing over and heterochromatin in the X chromosome of Drosophila melanogaster. Genetics 24, 413–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKittrick, E., Gaften, P.R., Ahmad, K., and Henikoff, S. (2004). Histone H3.3 is enriched in covalent modifications associated with active chromatin. Proc. Natl. Acad. Sci. USA 101, 1525–1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaki, K., Cheng, Z.K., Ouyang, S., Talbert, P.B., Kim, M., Jones, K.M., Henikoff, S., Buell, C.R., and Jiang, J. (2004). Sequencing of a rice centromere uncovers active genes. Nat. Genet. 36, 138–145. [DOI] [PubMed] [Google Scholar]
- Nagaki, K., Neumann, P., Zhang, D., Ouyang, S., Buell, C.R., Cheng, Z., and Jiang, J. (2005). Structure, divergence, and distribution of the CRR centromeric retrotransposon family in rice. Mol. Biol. Evol. 22, 845–855. [DOI] [PubMed] [Google Scholar]
- Nakaseko, Y., Adachi, Y., Funahashi, S., Niwa, O., and Yanagida, M. (1986). Chromosome walking shows a highly homologous repetitive sequence present in all the centromere regions of fission yeast. EMBO J. 5, 1011–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonaka, N., Kitajima, T., Yokobayashi, S., Xiao, G., Yamamoto, M., Grewal, S.I.S., and Watanabe, Y. (2002). Recruitment of cohesin to heterochromatic regions by Swi6/HP1 in fission yeast. Nat. Cell Biol. 4, 89–93. [DOI] [PubMed] [Google Scholar]
- Ouyang, S., and Buell, C.R. (2004). The TIGR Plant Repeat Databases: A collective resource for the identification of repetitive sequences in plants. Nucleic Acids Res. 32, 360–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson, G.I., Kubo, K.M., Shroyer, T., and Chandler, V.L. (1995). Sequences required for paramutation of the maize b gene map to a region containing the promoter and upstream sequences. Genetics 140, 1389–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts, P.A. (1965). Difference in the behaviour of eu- and heterochromatin: Crossing over. Nature 205, 725–726. [DOI] [PubMed] [Google Scholar]
- Saffery, R., Sumer, H., Hassan, S., Wong, L.H., Craig, J.M., Todokoro, K., Anderson, M., Stafford, A., and Choo, K.H.A. (2003). Transcription within a functional human centromere. Mol. Cell 12, 509–516. [DOI] [PubMed] [Google Scholar]
- Schueler, M.G., Higgins, A.W., Rudd, M.K., Gustashaw, K., and Willard, H.F. (2001). Genomic and genetic definition of a functional human centromere. Science 294, 109–115. [DOI] [PubMed] [Google Scholar]
- Schwartz, B.E., and Ahmad, K. (2005). Transcriptional activation triggers deposition and removal of the histone variant H3.3. Genes Dev. 19, 804–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman, J.D., and Stack, S.M. (1995). Two-dimensional spreads of synaptonemal complexes from Solanaceous plants. VI. High-resolution recombination nodule map for tomato (Lycopersicon esculentum). Genetics 141, 683–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh, K., Ishii, T., Parco, A., Huang, N., Brar, D.S., and Khush, G.S. (1996). Centromere mapping and orientation of the molecular linkage map of rice (Oryza sativa L.). Proc. Natl. Acad. Sci. USA 93, 6163–6168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan, B.A., and Karpen, G.H. (2004). Centromeric chromatin exhibits a histone modification pattern that is distinct from both euchromatin and heterochromatin. Nat. Struct. Mol. Biol. 11, 1076–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanksley, S.D., et al. (1992). High-density molecular linkage maps of the tomato and potato genomes. Genetics 132, 1141–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thuriaux, P. (1977). Is recombination confined to structural genes on the eukaryotic genome? Nature 268, 460–462. [DOI] [PubMed] [Google Scholar]
- Van Hooser, A.A., Ouspenski, I.I., Gregson, H.C., Starr, D.A., Yen, T.J., Goldberg, M.L., Yokomori, K., Earnshaw, W.C., Sullivan, K.F., and Brinkley, B.R. (2001). Specification of kinetochore-forming chromatin by the histone H3 variant CENP-A. J. Cell Sci. 114, 3529–3542. [DOI] [PubMed] [Google Scholar]
- Wako, T., Fukuda, M., Furushima-Shimogawara, R., Belyaev, N.D., and Fukui, K. (2002). Cell cycle-dependent and lysine residue-specific dynamic changes of histone H4 acetylation in barley. Plant Mol. Biol. 49, 645–653. [DOI] [PubMed] [Google Scholar]
- Wako, T., Houben, A., Furushima-Shimogawara, R., Belyaev, N.D., and Fukui, K. (2003). Centromere-specific acetylation of histone H4 in barley detected through three-dimensional microscopy. Plant Mol. Biol. 51, 533–541. [DOI] [PubMed] [Google Scholar]
- Warburton, P.E. (2004). Chromosomal dynamics of human neocentromere formation. Chromosome Res. 12, 617–626. [DOI] [PubMed] [Google Scholar]
- Waterborg, J.H. (1990). Sequence analysis of acetylation and methylation in 2 histone H3 variants of alfalfa. J. Biol. Chem. 265, 17157–17161. [PubMed] [Google Scholar]
- Weber, S.A., Gerton, J.L., Polancic, J.E., DeRisi, J.L., Koshland, D., and Megee, P.C. (2004). The kinetochore is an enhancer of pericentric cohesin binding. PLoS Biol. 2, 1340–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner, J.E., Endo, T.R., and Gill, B.S. (1992). Toward a cytogenetically based physical map of the wheat genome. Proc. Natl. Acad. Sci. USA 89, 11307–11311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheelan, S.J., Church, D.M., and Ostell, J.M. (2001). Spidey: A tool for mRNA-to-genomic alignments. Genome Res. 11, 1952–1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, J.Z., et al. (2004). Composition and structure of the centromeric region of rice chromosome 8. Plant Cell 16, 967–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao, H., Zhou, Q., Li, J., Smith, H., Yandeau, M., Nikolau, B.J., and Schnable, P.S. (2002). Molecular characterization of meiotic recombination across the 140-kb multigenic al-sh2 interval of maize. Proc. Natl. Acad. Sci. USA 99, 6157–6162. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.