Abstract
Genome-wide 70-mer oligonucleotide microarrays of rice (Oryza sativa) and Arabidopsis thaliana were used to profile genome expression changes during light-regulated seedling development. We estimate that the expression of ∼20% of the genome in both rice and Arabidopsis seedlings is regulated by white light. Qualitatively similar expression profiles from seedlings grown under different light qualities were observed in both species; however, a quantitatively weaker effect on genome expression was observed in rice. Most metabolic pathways exhibited qualitatively similar light regulation in both species with a few species-specific differences. Global comparison of expression profiles between rice and Arabidopsis reciprocal best-matched gene pairs revealed a higher correlation of genome expression patterns in constant light than in darkness, suggesting that the genome expression profile of photomorphogenesis is more conserved. Transcription factor gene expression under constant light exposure was poorly conserved between the two species, implying a faster-evolving rate of transcription factor gene expression in light-grown plants. Organ-specific expression profiles during seedling photomorphogenesis provide genome-level evidence for divergent light effects in different higher plant organs. Finally, overrepresentation of specific promoter motifs in root- and leaf-specific light-regulated genes in both species suggests that these cis-elements are important for gene expression responses to light.
INTRODUCTION
Because plants are both photosynthetic and sessile, plant development is dramatically regulated by environmental light signals. The transition of plant growth from skotomorphogenesis to photomorphogenesis is an example of a typical light-regulated process and has been intensively studied at the physiological, genetic, and biochemical levels (Yeh and Lagarias, 1998; Neff et al., 2000), and more recently, also at the genomic level (Ma et al., 2001, 2005b; Tepperman et al., 2001; Ohgishi et al., 2004).
Studies in the eudicot model plant Arabidopsis thaliana have systematically dissected light signal pathways and discovered many of the molecular components involved. Three major types of photoreceptors exist: phytochromes, cryptochromes, and phototropins. The photoreversible phytochromes are a small family that mediates responses to far-red (FR) and red (R) regions of the spectrum. The functions of phytochromes include modulation of germination, deetiolation, leaf expansion, stem and petiole elongation, entrainment of the circadian clock, and controlling the time to flowering (Nagy and Schäfer, 2002; Quail, 2002). Arabidopsis has five phytochromes (Sharrock and Quail, 1989), and rice (Oryza sativa) has three (Goff et al., 2002). The cryptochromes perceive blue light (B) and UV-A light to mediate deetiolation and to regulate circadian rhythms and time of flowering (Cashmore et al., 1999; Lin, 2002). Finally, phototropins, a separate blue light receptor system, mediate transient physiological changes, such as phototropic curving, regulation of stomatal aperture, and chloroplast movement responses (Liscum et al., 2003; Wada et al., 2003).
To understand the light signaling downstream of photoreceptors, a number of light-signaling deficient mutants have been identified in Arabidopsis beginning with the pioneering work of Maarten Koornneef (Koornneef et al., 1980). Analysis of some of these mutants suggested the presence of specific signaling pathways triggered by different photoreceptors, specifically phytochrome A and B and cryptochromes during seedling deetiolation (Quail, 2002). While some studies have indicated an integration of upstream pathways during light-regulated seedling development (Quail, 2002), genome profiling analyses are consistent with a model where differences also exist in specific photoreceptor-triggered signaling pathways (Ma et al., 2001; Tepperman et al., 2001; Wang et al., 2002). In addition, a recent survey of light-regulated transcriptional profiles indicated significant differences among Arabidopsis seedling organ types (Ma et al., 2005b). Nevertheless, all studies support a massive reprogramming of genome expression by light. Amid the present swirl of uncertainty about the mechanisms underlying genome-level control of transcription, chromatin-level regulation stands out as one route (Hsieh and Fischer, 2005). Chromatin remodeling plays an important role in regulating chromatin states that control transcription. Alternatively, cis-regulation through transcriptional cascades has been found to be crucial for the regulation of many signaling pathways in plants.
Despite the fact that monocot plants feed the majority of the world's population, relatively little is known about their photomorphogenic processes at the molecular and genomic levels. The seedling photomorphogenesis of monocots, including rice, is promoted by FR, R, and B light, as is true for Arabidopsis (Mohr, 1962; Pjon and Furuya, 1967; Wilkins, 1977). At the morphological level, light irradiation inhibits the elongation of coleoptiles and the gravitropism of roots in monocots (Feldman and Briggs, 1987; Kelly and Leopold, 1992; Kiss et al., 2003). Additionally, light regulation of plastid development and function has been reported in several grass species (Mullet, 1993). In rice, three phytochrome genes (Kay et al., 1989; Dehesh et al., 1991; Basu et al., 2000), two cryptochrome genes (Matsumoto et al., 2003), and two phototropin genes (Kasahara et al., 2002) have been reported. Various genes are regulated by light in monocots (Lissemore and Quail, 1988). Several genetic analyses of photoreceptors in rice have recently been conducted (Izawa et al., 2000; Takano et al., 2001; Haga et al., 2005), while relatively less is known in other monocots (Markelz et al., 2003).
A genomic study of rice gene expression in response to light is important as a foundation for understanding this process in all monocots. Approximately 150 million years of evolutionary divergence has provided ample opportunity for the evolution of distinct features in light signal transduction of monocots and eudicots, limiting the direct transfer of knowledge gained in well-characterized eudicots, such as Arabidopsis, to monocots (Chaw et al., 2004). In addition, as one of the most important crops in the world, rice has been domesticated for ∼9000 years (Khush, 1997), potentially further altering its genomic light responses. Furthermore, manipulation of light signal transduction may be a possible route of modifying cereal plant agronomic traits (Liu et al., 2004; Sawers et al., 2005). With an established synteny with other cereal crops, findings in rice can be easily adopted to other cereals (Gale and Devos, 1998; Shimamoto and Kyozuka, 2002).
Advances in microarray technology have made possible large-scale transcriptional profiling experiments in plants (Meyers et al., 2004). Sequencing of the rice genome (Sasaki and Burr, 2000; Goff et al., 2002; Yu et al., 2002, 2005) enabled us to apply this functional genomics tool to study expression profiles of the entire rice genome as a complement to genetic studies. Here, we report an effort to analyze systematically the genome expression properties of rice seedling development during photomorphogenesis in comparison with Arabidopsis. In this analysis, 70-mer oligonucleotide microarrays that represent 36,926 rice and 25,676 Arabidopsis known and predicted genes were employed. Light-regulated massive reprogramming of the transcriptome was found for both species. Similar patterns of light regulation were identified between rice and Arabidopsis. Statistical analysis revealed that photomorphogenesis-associated genome expression profiles are relatively more conserved than those of skotomorphogenesis. Organ-specific expression profiles during seedling photomorphogenesis provide genome-level evidence for the divergent light effects in discrete organ types. Finally, our data imply that transcriptomes underlying light regulation in plants are characterized mainly by cis-acting elements rather than chromatin-level mechanisms.
RESULTS
Experimental Conditions for the Rice Seedling Photomorphogenesis Study
Our laboratory growth conditions have been optimized for Arabidopsis seedlings to provide sufficient light for photomorphogenesis but to be moderate enough so that light stress responses are minimized (Osterlund and Deng, 1998; Ma et al., 2001). To define the optimal light intensity range for our rice seedling photomorphogenesis study, we grew indica rice seedlings at a series of constant white (W) light intensities. Coleoptile length and root gravitropic growth were used as indicators of photomorphogenesis (Pjon and Furuya, 1967). We noted that both the inhibition of coleoptile elongation and the gravitropic response of crown roots during early seedling development were saturated at light intensities above 200 μmol·m−2·s−1 (Figure 1). Ten-day-old seedlings at light intensities ranging from 150 to 400 μmol·m−2·s−1 did not show obvious morphology differences or light stress responses, which include anthocyanin accumulation, leaf petiole inhibition, and photosynthesis inhibition. We therefore selected a white light intensity of 220 μmol·m−2·s−1 for further experiments. At this light intensity, rice seedlings have typical photomorphogenic responses but do not have observable stress effects.
Figure 1.
Relationship between White Light Irradiation Intensities and the Growth Responses of Rice Seedlings.
(A) Effect of light intensities on coleoptile inhibition.
(B) Effect of light intensities on crown root gravitropic growth. Each point represents the mean of 20 measurements.
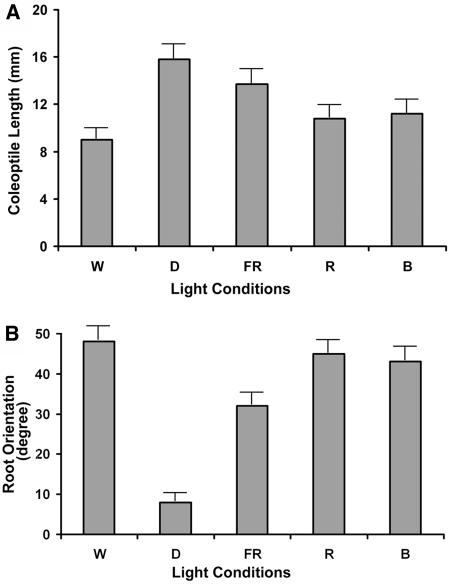
As is the case for Arabidopsis, rice photomorphogenesis involves several morphological changes. Light signals inhibit the elongation of shoots while accelerating the expansion of leaves. In constant darkness (D), rice seedlings exhibit a typical elongation of coleoptiles and mesocotyls. By contrast, continuous W light or monochromatic FR, R, or B light inhibits shoot growth. Mesocotyls were barely seen, therefore not measurable, under any of these light conditions. We used the average length of coleoptiles as a measure of the extent of the light responses (Figure 2A). A comparison indicates that the R and B light treatments we used were about twice as strong as the FR light to induce light responses in rice coleoptile growth inhibition.
Figure 2.
Morphological Phenotype Comparison of Rice Seedlings under Different Light Qualities and in Darkness.
Ten-day-old rice seedlings were grown under continuous W, FR, R, and B light and in darkness. Effect of distinct light qualities on coleoptile inhibition (A) and crown root gravitropic growth (B) were measured based on 15 seedlings or more. Error bars represent se.
Rice seedling roots exhibit dramatic morphological changes in response to light signals. Rice seedlings have five crown roots that emerge sequentially from the nodal portion of shoots, whereas Arabidopsis seedlings have only one main root. The growth orientation of crown roots is subject to light irradiation (Takano et al., 2001). The light-enhanced gravitropic response of rice crown roots was measured by the growth angle between a crown root and the surface (Figure 2B). Seedling roots under R or B light had ∼80% of the W light–enhanced gravitropic response, whereas the FR light effect was ∼60% of the W light response. In darkness, secondary roots emerged from the node between the mesocotyl and coleoptile, which grew horizontally. Irradiance by continuous FR, R, B, or W light inhibited secondary roots in a similar manner to light-modulated mesocotyl growth inhibition.
Strategy for Profiling Light-Regulated Genome Expression
To reveal genome expression profiles specific to photomorphogenesis, we compared transcriptomes of photomorphogenic seedlings grown under W light, as well as FR, R, and B light, with dark-grown rice and Arabidopsis seedlings. Spotted 70-mer long oligonucleotide microarrays for rice (Ma et al., 2005a) and Arabidopsis (Ma et al., 2005b) were utilized for this purpose. Ten-day-old rice seedlings grown under each light condition were compared with the same age seedlings in darkness (Figure 3A, top). Labeled RNA was probed to a microarray representing 36,926 unique known and predicted genes (Ma et al., 2005a). Independent sample preparations were performed for probe labeling for three hybridizations. Quantified microarray hybridization signals went through an automatic processing pipeline with manual inspection to correct the background, to normalize experimental variations, to filter problematic spots, and to check the data quality (see Methods).
Figure 3.
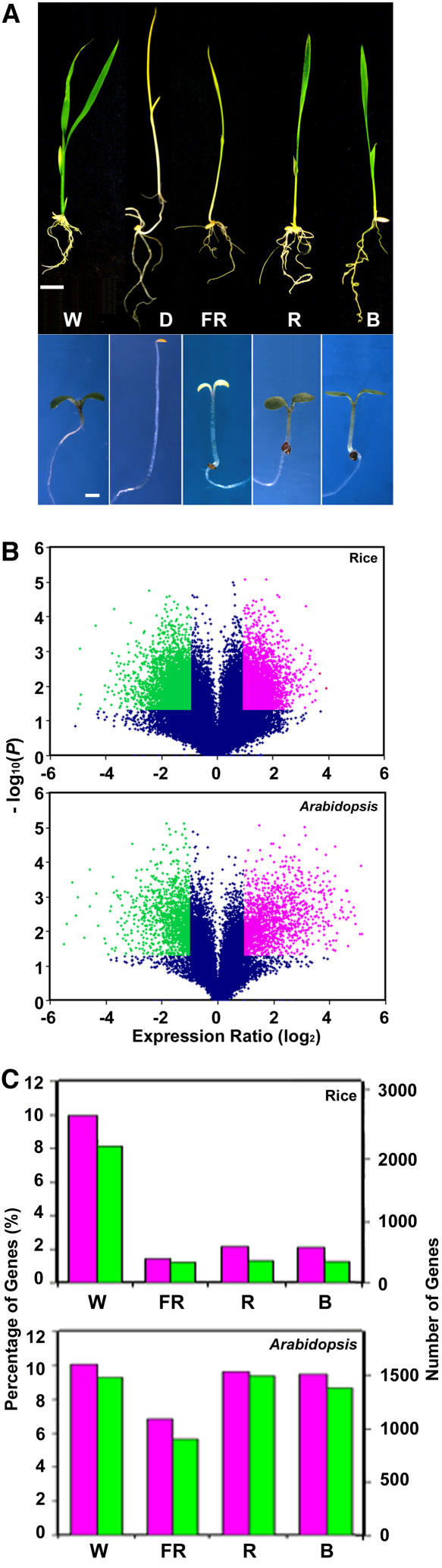
Light-Regulated Morphological and Genome Expression Changes between Rice and Arabidopsis.
(A) Top: 10-d-old rice seedlings grown under continuous W, FR, R, and B light and in darkness. Bar = 10 mm. Bottom: 6-d-old Arabidopsis seedlings. Bar = 1 mm.
(B) Volcano plot where log2-transformed gene expression intensity ratios are plotted against the negative log10-transformed P value from a Student's t test. Genes with statistically different expression (P value < 0.05) and fold changes above 2 are considered to be induced genes and are shown in red. Genes with statistically different expression and fold changes below −2 are considered to be repressed genes and are shown in green.
(C) Summary of expressed genes induced (red) and repressed (blue) beyond twofold by light in each light quality. Both percentages and numbers of genes are shown.
Similar to the strategy for rice, the genome expression profiles of 6-d-old Arabidopsis seedlings grown under distinct light qualities were compared with dark-grown seedlings (Figure 3A, bottom) by means of a similar 70-mer oligonucleotide microarray for Arabidopsis that represents 25,676 genes (Wellmer et al., 2004; Ma et al., 2005b). Exactly the same data processing standards and methods were used for Arabidopsis as in rice.
To gauge the reliability of our data, we compared Arabidopsis expression data with a similar, previously published study using EST-based microarrays covering a quarter of the genome (Ma et al., 2001). A general consistency was found between these independent studies. The sensitivity of detection of the 70-mer oligonucleotide array reported here was found to be similar to the EST-based array (Wang et al., 2003; Lee et al., 2004), and the dynamic range of ratios was ∼1.5 times that of the EST-based array. Whereas EST collections have up to a 30% error rate from frequent mix-ups during collection and preparation (Ma et al., 2001), 70-mer oligonucleotides are entirely handled by automated robots and, thus, likely are less prone to error in identification. Cross-hybridization among gene family members, which may mask changes in transcription in EST-based arrays (Girke et al., 2000; Finkelstein et al., 2002), was effectively addressed during the design of the 70-mer oligonucleotides (Wang et al., 2003; Lee et al., 2004; Ma et al., 2005a). Additionally, our microarray data were confirmed by RT-PCR results in a number of selected rice and Arabidopsis representative genes (see Supplemental Figure 1 online) and are consistent with various reports in the literature.
Light Regulates the Expression of a Significant Portion of the Genome
Examination of the expression ratios of genes between light-grown and dark-grown rice seedlings indicated that 18% of the rice genome was regulated by W light at the seedling stage. We found that 2774 genes were induced and 2232 genes were repressed by at least twofold, with a P value below 0.05. Together, 5006 genes, out of 27,406 genes with detected expression at the seedling stage, were regulated by W light (see Supplemental Table 1 online). A volcano plot provides a clear view of the definition of differentially expressed genes (Figure 3B). Under the light quality conditions, smaller portions of the genome were regulated (Figure 3C) compared with W light, with FR light regulating a slightly smaller portion of the genome than R or B light. It is important to note that FR light is intermediate between darkness and R and B light in comparison to W light in phenotype, just as in the genome-wide expression analysis (Figure 3A).
In Arabidopsis, under W light conditions, we identified 1557 induced genes and 1443 repressed genes with at least a twofold induction or repression level and a P value below 0.05 (see Supplemental Table 2 online). These numbers correspond to 19% of all expression-detectable genes (15,547). Arabidopsis has a more dramatic regulation of genes by light than rice (Figure 3B). Most light-regulated rice genes only had a modest fold change in each direction, whereas a significant fraction of light-regulated Arabidopsis genes fell into the high fold-change end of the plot. For light qualities, the percentages of FR, R, and B light-regulated genes, both induced and repressed, are shown in Figure 3C. Similar to rice, in Arabidopsis, FR light regulated a smaller portion of the genome than either R or B light. However, unlike in rice, in Arabidopsis, the number of monochromatic light-regulated genes was more close to the number of W light–regulated genes.
Qualitatively Similar Effects of Distinct Light Qualities on Genome Expression
To compare the effect of light qualities on genome expression, we further analyzed detailed expression profiles of seedlings under W light and monochromatic light. Using a hierarchical clustering algorithm (Figure 4A), we found that most W light–regulated genes also responded to FR, R, or B light in the same trend. On the other hand, a few genes were specifically regulated by monochromatic light, as reported previously in Arabidopsis by Ma et al. (2001). Unlike W light, each monochromatic light was only able to regulate genome expression weakly; most failed to reach the twofold and P value threshold in rice and are illustrated by the fainter color in the FR, R, and B light lanes. Importantly, the vast majority of these light-regulated genes were turned up or down by monochromatic lights in the same direction as W light. This finding still holds true when we extend our analysis to genes with 1.5-fold or higher light regulation and a P value < 0.05. Therefore, we concluded that W light and the monochromatic light conditions we explored had qualitatively similar effects in regulating genome expression but that W light had a quantitatively stronger effect than monochromatic light in genome expression regulation.
Figure 4.
Light Regulation of Genome Expression.
(A) Overview of light-regulated genome expression by cluster display. W, continuous W light versus dark; FR, continuous far red light versus dark; R, continuous red light versus dark; B, continuous blue light versus dark. The color scale is shown at the bottom. Positive numbers represent fold of induction, and negative numbers represent fold of repression. All rice seedlings were grown at 28°C for 10 d, while Arabidopsis seedlings were grown at 20°C for 6 d. All of those genes that exhibited a twofold or higher differential expression in at least one time point were included.
(B) to (I) Expression profiles of eight representative genes with reciprocal best-matched genes from rice and Arabidopsis, respectively. Biosynthesis pathways ([B] and [C]), utilization/assimilation/degradation pathways ([D], [E], [H], and [I]), and pathways that generate precursor metabolites and energy ([F] and [G]) were represented. (B) O-acetylserine thiollyase (At3g03630 and OsJRFA065652); (C) chlorophyll synthetase (At3g51820 and OsJRFA068855); (D) glyceraldehyde-3-phosphate dehydrogenase (At1g42970 and OsIFCC017765); (E) galactose (galactoside/glucose catabolism) (At5g51820 and OsJRFA068502); (F) ribose 5-phosphate isomerase (At5g44520 and OsJRFA060861); (G) light-harvesting chlorophyll a/b binding protein (At3g54890 and OsIFCC010436); (H) Phe ammonia-lyase (At3g10340 and OsIFCC040013); (I) CTP oxidase (pyrimidine ribonucleotide metabolism) (At4g20320 and OsJRFA070411). Bars in each graph of (B) to (I) correspond to the log2-transformed expression ratios of W, FR, R, and B light compared with dark. Arabidopsis data are shown in patterned bars and rice in filled bars.
In Arabidopsis, gene expression profiles regulated by each monochromatic light also correlated well with W light (Figure 4A). Consistent with the number of light-regulated genes, cluster lanes for monochromatic lights were quantitatively more similar to the W light lane in Arabidopsis than in rice.
Light Regulation of Metabolic Pathways
To examine light effects on different functional groups of genes, we explored the light regulation of various gene functional categories. By functional assignment using Gene Ontology (GO) terms (Gene Ontology Consortium, 2000), we found that light regulated almost all major gene functional categories in both rice and Arabidopsis (see Supplemental Figure 2 online). For the majority of these categories, induced and repressed gene members were similarly distributed both in rice and in Arabidopsis.
The annotation of metabolic pathway genes is more definitive, in general, than other gene groups at this point. Therefore, we initially focused on metabolic pathways to compare the effect of light on rice and Arabidopsis genome expression. Moreover, metabolic pathway genes are often the targets of signaling cascades. We followed the standardized AraCyc-defined metabolic pathways (Mueller et al., 2003), which currently include 1568 Arabidopsis enzyme genes, to identify genes in each pathway. In this analysis, rice genes were grouped into pathways based on their best homologs (see Methods) in the Arabidopsis genome. A total of 898 rice enzyme genes were integrated into pathways.
Light induces expression of genes in most major pathways both in rice and in Arabidopsis. We found examples of light-induced expression of sugar synthesis genes (Figures 4D to 4F), amino acid and protein biosynthesis genes (Figure 4B), and photosynthetic genes (Figures 4C and 4G) in both species. A small number of genes was repressed in expression, such as the suberin biosynthesis gene (Figure 4H) and a gene encoding pyrimidine metabolism enzymes (Figure 4I).
To further examine differences between the species in light regulation of metabolic pathways, entire biosynthetic pathways, rather than selected enzymes from those pathways, were analyzed (Figure 5). By comparing the light regulation of biosynthetic pathways for carbohydrates (Figure 5A), nucleotides (Figure 5B), amino acids (Figure 5C), secondary metabolites (Figure 5D), fatty acids (Figure 5F), and cofactors (Figure 5G), we found a general light induction in both species. Only a few pathways are repressed by light, such as the polyamine biosynthesis pathway (Figure 5E). Despite the general similarity, the highly regulated steps in the pathway may be different in rice and Arabidopsis. For example, in the Leu biosynthesis pathway (Figure 5D), the genes encoding the 2-keto-isovalerate to 3-carboxy-3-hydroxy-isocaproate step and the 2-d-threo-hydroxy-3-carboxy-isocaproate to 2-keto-4-methyl-pentanoate step were only weakly induced in rice but highly induced in Arabidopsis, whereas enzymes catalyzing 2-keto-4-methy-pentanoate to Leu synthesis were highly induced in rice but weakly induced in Arabidopsis.
Figure 5.
Diagram of Representative Biosynthesis Pathways for Rice (Left) and Arabidopsis (Right).
Each pathway is shown as glyphs consisting of nodes, which represent the metabolites, and lines, which represent the reactions. Expression-level change of each reaction is shown in a color relative to the expression level. Missing gene expression data, which may come from lack of annotated enzyme, lack of microarray probe, or lack of expression, are represented by gray lines.
To compare comprehensively the light regulation of major metabolic pathways between rice and Arabidopsis, we aligned all identified homologous gene pairs in related pathways for gene expression comparison. Pathways for the biosynthesis of most primary and secondary metabolites (Figure 6A), utilization pathways (Figure 6B), and energy pathways (Figure 6C) show variable degrees of similarity in light-regulated expression between the two species. Interestingly, photosynthetic pathways and sugar metabolism pathways had more homologs induced by light in both species than other pathways. Fewer homologous pairs were identified as having expression suppressed by light in both species. On the other hand, more than half of the homologous pairs in most categories were not regulated by light in either plant. In addition, cases exist where light regulation of homologous genes occurs in one species but not in the other. Descriptions of these metabolic pathways, along with two homologous gene pair examples for each category, are summarized in Supplemental Table 3 online. A number of hormone biosynthesis genes were significantly light regulated in both rice and Arabidopsis, suggesting their important function in light regulation of development.
Figure 6.
Shared Transcriptional Signature of Light Regulation in Major Metabolic Pathways.
(A) Five representative biosynthesis pathways.
(B) Six representative utilization/assimilation/degradation pathways.
(C) Three representative precursor metabolites and energy pathways.
Conserved patterns in the gene expression data sets from rice and Arabidopsis that corresponded to five biosynthesis (A), six utilization/assimilation/degradation (B), and three precursor metabolites and energy pathways (C) (http://www.arabidopsis.org/tools/aracyc/) were identified. For each pathway, rectangular blocks represented the measured changes in expression of each gene. All homolog pairs in each pathway are shown. In each pathway block, rice genes are in the top row, while Arabidopsis is in the bottom row.
Limited Chromatin-Scale Transcriptional Regulation by Light
Chromatin-scale regulation is one mechanism that controls genome expression (He and Amasino, 2005). Evidence of chromatin-scale regulation has been found in eukaryotes (Hurst et al., 2004), where genes physically near to each other on the chromosome are coordinately expressed. Small domains (∼10 genes) of similarly expressed genes have been observed in plants by exploring assorted huge data sets (Williams and Bowles, 2004) or by focused study of organ-specific expression data (Ma et al., 2005a, 2005b; Schmid et al., 2005). Alternatively, chromatin structure may affect transcription at the cytologically observable scale, with domains as large as several megabases (Jiao et al., 2005).
To reveal possible chromatin-level regulation during photomorphogenesis, our first attempt was to examine the possible presence of small domains of similarly light-regulated genes. In order to identify groups of neighboring and similarly expressed genes systematically, we calculated the average pair-wise Pearson correlation of light-to-dark expression ratios for adjacent genes in sliding widows from 2 to 25 genes. By randomly shuffling the order of genes, we also created random data sets of the same scale to estimate the significance of observation in the ordered data set. The results up to a window size of 15 are presented in Table 1. In rice, the ordered data set is not significantly different from the randomized data set in any tested window size, although slightly more clusters in the ordered data set were observed at some window sizes. In Arabidopsis, somewhat more coexpressed neighboring genes were found in the ordered data set than in the randomized data set, most evident at window sizes below five (Table 1). In previous studies of organ-specific expression data, window sizes of 10 exhibited coregulation of thousands of genes rather than the few hundred coregulated here (Ma et al., 2005a, 2005b).
Table 1.
Coregulation of Neighboring Genes along the Chromosome
| Window Size
|
2
|
5
|
10
|
15
|
|||||
|---|---|---|---|---|---|---|---|---|---|
| P Value | Ordered | Randomized | Ordered | Randomized | Ordered | Randomized | Ordered | Randomized | |
| Rice | 10−3 | 114 | 181 | 281 | 359 | 484 | 413 | 608 | 433 |
| 10−2 | 508 | 596 | 1025 | 1167 | 1397 | 1408 | 1850 | 1501 | |
| Arabidopsis | 10−3 | 477 | 112 | 536 | 242 | 657 | 365 | 685 | 434 |
| 10−2 | 560 | 371 | 937 | 762 | 1388 | 1084 | 1030 | 1205 | |
The number of genes that are physically linked as similarly light-regulated genes at different window sizes in ordered and randomized data sets are shown.
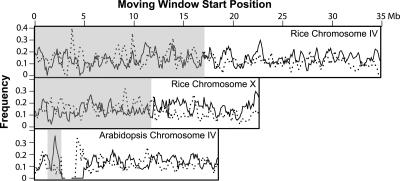
To test whether a relationship exists between light-regulated gene expression and cytologically defined chromosome structures, the frequency of occurrence of differentially regulated genes was estimated along entire chromosomes (Figure 7). Using a 100-kb moving window that advances 50 kb each step, the percentage of W light–induced and W light–repressed genes was calculated and depicted. Significantly, no obvious difference was observed between heterochromatin and euchromatin in rice chromosomes in terms of light-regulated gene expression. Rice chromosomes 4 and 10, which have the most distinct cytological differences (Cheng et al., 2001), are presented in Figure 7. With the exception of centromeric heterochromatin, whose sequences are largely unavailable, Arabidopsis has only limited heterochromatic features, with one located in the short arm of chromosome 4 (Fransz et al., 2000). A peak of light-induced genes was found in this region (Figure 7); however, the peak is narrow and does not cover a significant portion of the heterochromatin region. This sharp peak may be the result of a few induced genes within a region of the genome with low gene density biasing the overall sample. Light-repressed genes were not abundant in this same region.
Figure 7.
Distribution of Light-Regulated Genes along Representative Chromosomes.
For each chromosome, the frequency (percentage) of light-induced genes (solid line) or light-repressed genes (dotted line) is shown in a series of 100-kb windows with moving steps of 50 kb. The position of the window start point along the chromosome is given at the top. Cytologically defined heterochromatic regions are highlighted with a gray background.
Cross-Species Similarity of Transcriptional Profiles
To extend the assumption that important biological mechanisms are likely to be conserved during evolution (McCarroll et al., 2004; Murray et al., 2004), we compared shared patterns of expression across the best-matched homologous gene pairs between rice and Arabidopsis. A genome-wide survey of the rice and Arabidopsis genomes identified 7196 reciprocal best-matched homologous gene pairs based on sequence. These pairs have a simple one-to-one relationship and are likely enriched for potential orthologs (Ma et al., 2005a; Yu et al., 2005). The majority of these reciprocal best-matched gene pairs in both rice (72%) and Arabidopsis (71%) were expressed at the seedling stage. Significantly, a slightly higher percentage of these potential orthologs were induced by light than is the case for the entire genome (Figure 8). By contrast, a smaller percentage of potentially orthologous genes were repressed compared with the total genome.
Figure 8.
Proportion of Light-Regulated Genes in the Entire Genome and among the Rice–Arabidopsis Reciprocal Best-Matched Genes.
Numbers in each bar are percentages of light-induced and light-repressed genes in all expressed genes or in all expressed best-matched genes.
In order to judge the significance of the light regulation of potential orthologs across species, we compared the correlation of gene expression intensities between species under each light condition and in darkness (Figure 9). The cross-species Pearson correlation of gene expression intensities under W light is 0.225, which is significant with P value of 10−51. The correlation of dark-grown seedlings is 0.158 (P value < 10−32). Correlations for B and R light–grown seedlings are more similar to W light. Seedlings under FR light gave a correlation of 0.185 (P value < 10−32) and were closest to dark-grown seedlings. Although taken individually, many reciprocal best-matched pairs lack a clear correlation; as a group, reciprocal best-matched pairs do show conserved light-regulated expression patterns between rice and Arabidopsis. As a control, randomized pairings of genes did not show such high correlations (Figure 9).
Figure 9.
Correlated Regulation of Reciprocal Best-Matched Gene Expression during Photomorphogenesis in Rice and Arabidopsis.
Microarray measurements of gene expression at each light condition were paired for 7196 best-matched genes from rice and Arabidopsis. Pearson correlations (r) with their significance (P values) for best-matched gene pairs at different light conditions are shown by arrows on the right. A distribution of Pearson correlations of 100,000 random pairings of rice and Arabidopsis gene expression data is shown on the left.
Evolutionary Mode of Regulatory Gene Expression
To reveal further the variation of expression shaped by evolutionary forces in different functional groups, we classified reciprocal best-matched gene pairs based on their biological function. We focused on genes with regulatory functions, since they are key factors controlling cell fate or plant development. Based on GO annotation and a literature review, 455 best-matched gene pairs were identified as transcription factors, 353 pairs as signal transducers, and 311 pairs as ubiquitin-proteasome pathway genes (Ma et al., 2005a). For control purposes, we also identified 231 pairs as protein biosynthesis pathway genes.
The cross-species Pearson correlations of best-matched gene pair expression intensities were examined for each functional category. As shown in Table 2, transcription factors had the highest divergence in expression between the two species. Signal transducers and ubiquitin-proteasome pathway genes had a slightly higher correlation, which suggested weak conservation in expression. On the other hand, protein synthesis pathway genes had a much higher conservation of expression. These trends were consistent among W light, darkness, and monochromatic light conditions.
Table 2.
Correlation of Expression of Four Major Functional Categories at Each Light Quality in Rice and Arabidopsis
| D | W | FR | R | B | |
|---|---|---|---|---|---|
| Protein synthesis (231) | 0.316 10−5 | 0.353 10−6 | 0.375 10−7 | 0.373 10−7 | 0.336 10−6 |
| Protein degradation (311) | 0.203 10−3 | 0.190 10−2 | 0.152 10−2 | 0.142 10−2 | 0.195 10−2 |
| Signaling (353) | 0.129 10−2 | 0.164 10−2 | 0.161 10−2 | 0.135 10−2 | 0.167 10−2 |
| Transcription factor (455) | 0.088 10−1 | 0.057 10−1 | 0.101 10−1 | 0.161 10−2 | 0.107 10−1 |
Conserved genome expression was measured by the Pearson correlation (r) of the normalized expression of reciprocal best-matched genes. Statistical significance (P value) of correlation is shown after each Pearson correlation value. The total number of genes analyzed for each group is shown in parenthesis.
Light-Regulated Genome Expression Profiles Exhibit Clear Organ Specificity
Seedling development of both rice and Arabidopsis is dramatically regulated by environmental light in several aspects. Each organ type, such as the shoot or root of rice seedlings, exhibits distinct developmental responses to light, although they appeared to share common light perception and signaling systems (Cashmore et al., 1999; Quail, 2002).
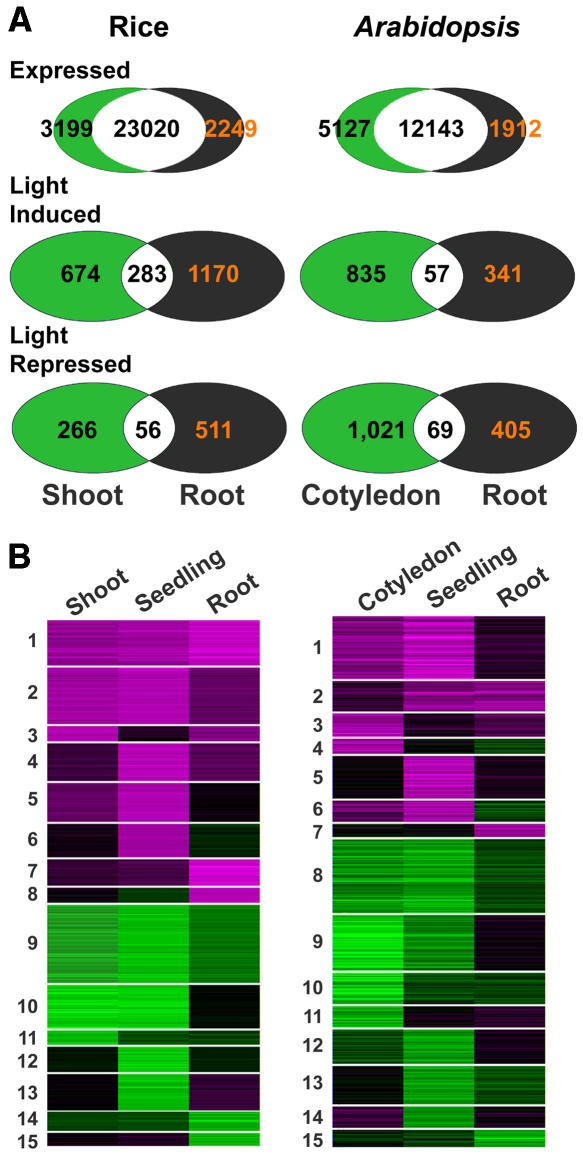
As a first step toward understanding why light triggers distinct developmental responses in different organs, we compared genome expression profiles of representative W light–grown rice or Arabidopsis organs with their dark-grown counterparts. Rice shoots and roots, and Arabidopsis cotyledons and roots were selected for this comparison, together with whole seedlings as a control. As shown in Figure 10A, a significant overlap of genome expression occurs in those light- and dark-grown organ pairs in both species: ∼90% for rice and ∼70% for Arabidopsis. In both rice and Arabidopsis, light-regulated expression of a large number of genes occurred in all organs examined. It is interesting to note that Arabidopsis roots appear to have a smaller fraction of expressed genes that are subject to light regulation than cotyledons, whereas rice roots have even more light-regulated genes than shoots. Only a limited overlap exists for light-regulated genes between shoots or cotyledons and roots (Figure 10A).
Figure 10.
Light-Regulated Genome Expression in Separate Organs.
(A) Venn diagrams of expressed genes, light-induced genes, and light-repressed genes in each organ.
(B) All genes with differential expression in shoots, roots, or whole seedlings were divided into 15 clusters using K-means algorithm. The color scale is the same as in Figure 4A.
To further examine organ-specific light-regulated gene expression, we clustered all W light–regulated genes in shoots (or cotyledons), roots, and whole seedlings in both rice and Arabidopsis. The K-means clustering method was used to group all light-regulated genes into 15 clusters whose pattern of light regulation is similar across organs (Figure 10B). The cluster results revealed that in rice, 50% of the shoot-specific light-regulated genes and 36% of the root-specific light-regulated genes had similar regulation in the other organ but with a weaker magnitude (with a 1.5-fold change and P value of 0.05 cutoff). The percentage of genes weakly regulated in the other organ went down to ∼20% in Arabidopsis cotyledons or roots. The rest of the organ-specific light-regulated genes did not show even weak light responses in the other organ sample examined and, thus, likely represent true organ-specific light regulation. We also noted that a small number (18 in rice and 38 in Arabidopsis) of genes exhibited organ-specific and opposite light regulation.
Light-Regulated Expression of Genes with Diverse Functions
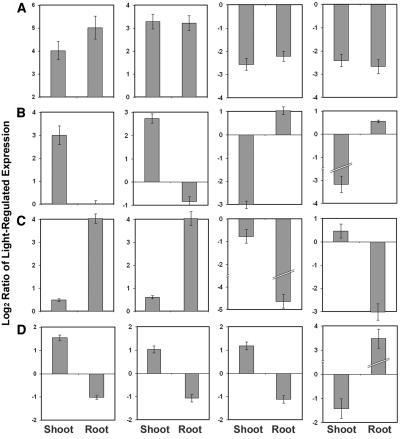
In each species, both seedling-wide light-regulated genes and organ-specific light-regulated genes belong to diverse functional groups. Similar groups of signal transducers, including kinases and transcription factors, were regulated by light in both shoots and roots. For example, the transcript level of HY5, a well-studied transcription factor in Arabidopsis, was induced in roots in a similar manner to what was previously reported in leaves. We also found that certain photosynthesis pathway genes were similarly regulated in both shoots and roots. Several representative genes from rice (Figure 11A) and Arabidopsis (see Supplemental Figure 3A online) are illustrated.
Figure 11.
Organ-Specific Light-Regulated Gene Expression.
(A) Expression profiles of four rice genes with similar light regulation in shoots and in roots. From left to right: OsIFCC043471, putative photosystem I reaction center subunit II precursor; OsIFCC036192, AP2 domain transcription factor; OsIFCC002877, ketol-acid reductoisomerase; OsIFCC042813, unknown function protein.
(B) Expression profiles of four rice genes with light regulation only in shoots. From left to right: OsIFCC011070, Leu-rich repeat transmembrane protein kinase; OsJRFA103597, aldehyde oxidase; OsIFSC048192, extensin; OsIFCC028968, 1-aminocyclopropane-1-carboxylate oxidase (ethylene biosynthesis).
(C) Expression profiles of four rice genes with light regulation only in roots. From left to right: OsIFCC004995, ribosomal protein L11; OsIFCC019376, actin binding protein; OsIFCC032722, sugar porter; OsIFSC045999, G-protein coupled receptor.
(D) Expression profiles of four rice genes with opposite light regulation in shoots and roots. From left to right: OsIFCC004650, putative glutathione transferase; OsIFCC002739, putative cytochrome P450; OsIFCC033515, putative SCARECROW-like transcription factor; OsIFCC010934, putative thionin.
Bars in each graph of (A) to (D) correspond to the log2-transformed expression ratios of W light compared with dark.
Some genes with regulatory and metabolic functions regulated by light are found only in shoots or cotyledons but not in roots. Many metabolic genes, such as energy pathway genes, and cell structure genes, were specifically regulated in shoots but not in roots (Figure 11B; see Supplemental Figure 3B online). Similarly, many genes with diverse metabolic and regulatory functions were specifically regulated by light in roots (Figure 11C; see Supplemental Figure 3C online). Interestingly, ethylene biosynthesis and signaling pathway genes were specifically repressed in shoots, both in rice (Figure 11B) and in Arabidopsis (see Supplemental Figure 3B online). Auxin signaling pathway genes were specifically induced in Arabidopsis shoots but not in roots (see Supplemental Figure 3B online). These results suggest an organ-specific interplay of auxin and ethylene hormones with light. A few genes show opposite light regulation between the two organs, but the functional implications of this are not clear (Figure 11D; see Supplemental Figure 3D online).
It is interesting to note that the light-regulated expression profiles among shoots, roots, and seedlings revealed several clusters of genes that were light regulated in whole seedlings but not in either shoots or roots. We speculate that the parts of the seedling missing from the shoot (cotyledon) or root, such as Arabidopsis hypocotyls, rice mesocotyls, and shoot apical meristems, also make a significant contribution toward the overall light regulation of gene expression in seedlings.
Organ-Specific Light-Regulated Genes Share Common Regulatory Elements in Their Promoters
It is well established that transcription of genes is regulated by promoter sequences. Coexpressed genes are likely to share common regulatory elements or motifs in their promoters. In some cases, the binding promoter motifs underlying light control have been characterized experimentally (Terzaghi and Cashmore, 1995). Computational approaches have also been developed and applied to light regulation of gene expression (Hudson and Quail, 2003). Our light regulation data sets provide unique coexpressed gene lists for common regulatory motif searches by separating different organ types.
Currently used computational approaches can be divided into alignment methods that are based on sequence alignment and enumerative methods that statistically analyze the frequency of exactly matched overrepresented motifs (Ohler and Niemann, 2001). To identify common regulatory motifs reliably, an enumerative algorithm specifically tailored for Arabidopsis, Sift (Hudson and Quail, 2003), and a Gibbs sampling alignment algorithm, AlignACE (Hughes et al., 2000), were both used to search for promoter motifs in light-regulated genes in Arabidopsis cotyledons and roots, in cotyledons only, or in roots only. Only motifs identified using both strategies were counted. A list of the promoter motifs identified using this two-strategy approach, along with their significance in the enumerative search, is provided in Figure 12.
Figure 12.
Motifs Discovered from Light-Regulated Arabidopsis Gene Promoters.
Sequence logo, which represents a motif matrix, a common name, if known, the significance from Sift, and the enrichment in rice and Arabidopsis are provided. Enrichment was acquired by subtracting the presence of a motif in all gene promoters from the presence in target gene promoters.
(A) Motifs were overrepresented in light-induced genes in common between cotyledons and root.
(B) Motifs were overrepresented in light-induced genes specific for cotyledons.
(C) Motifs were overrepresented in light-induced genes specific for root.
(D) Motifs were overrepresented in light-repressed genes common between cotyledons and root.
(E) Motifs were overrepresented in light-repressed genes specific for cotyledons.
The most well-studied promoter element, the G-box, CACGTG (Menkens et al., 1995; Chattopadhyay et al., 1998; Martinez-Garcia et al., 2000), was clearly recognized in light-induced genes in all three organ-specific categories of genes. The flanking sequences identified in different organs, however, do have differences. In light-induced genes common for both cotyledons and roots, a motif of TRAAACACGTKT is identified (Figure 12A). In cotyledon-specific genes, a more standardized G-box, AACACGTGTT, is overrepresented (Figure 12B). Roots have a longer motif, CGACCACGTTATTA, containing the core G-box (Figure 12C). These data suggest versatile possible functions for the core G-box through the use of different flanking sequences to enhance its organ specificity.
Two computationally identified phyA-induced motifs (Hudson and Quail, 2003), SORLIP 1, GCCAC, and SORLIP 5, GAGTGAG, were overrepresented in light-induced genes. SORLIP 1 was found in both cotyledon and root common genes and root-specific genes. Root-specific genes employ an extra flanking sequence together with the core SORLIP 1 motif (Figure 12C). SORLIP 5 was overrepresented in both cotyledon-specific and root-specific genes. Our data suggest that the motif GTGAG is the core of SORLIP 5. One novel cotyledon-specific and one novel root-specific motif were overrepresented in our analysis (Figures 12B and 12C).
Part of a previously identified cis-regulatory element conferring light repression in peas (Pisum sativum), DE1 (TACTA) (Inaba et al., 2000), was overrepresented in cotyledon-specific light-repressed genes together with a flanking sequence in the 5′ end (Figure 12E). This sequence identified by us is also close to a computationally identified phyA-repressed motif (Hudson and Quail, 2003), SORLREP 1 (TACTAGT). Two other computationally identified phyA-repressed motifs, SORLREP 3 and SORLREP 4, were overrepresented in repressed genes. Three novel motifs were overrepresented in light-repressed genes in cotyledons (Figure 12E). One of them, CATGCA, has similarity to a variant of the G-box, CACATG (Blecken et al., 1994). No element is overrepresented in root-specific light-repressed genes in the results of both enumerative and alignment searches. Three other phyA-induced motifs and three other phyA-repressed motifs are reported by searching FR light downstream genes (Hudson and Quail, 2003). We were unable to observe overrepresentation of these in our analysis by combining both searching approaches.
By searching for the existence of these motifs in corresponding light-regulated rice gene promoters, we found that all these motifs were similarly enriched in rice as they were in Arabidopsis (Figure 12). The enrichment was scored by comparing the existence of these motifs in light-regulated gene promoters with their existence in the promoters of all the genes covered by microarrays. However, de novo identification of consensus promoter elements from light-regulated gene collections similar to what we did with Arabidopsis was not successful in rice, possibly due to the unusually high GC content and the preliminary stage of rice gene annotation.
DISCUSSION
Light as a Key Regulator for Rice Seedling Development Mainly through Transcriptional Cascades
Genome profiling revealed that 18% of the rice genome is regulated by light in seedlings, providing genomic support for the notion that light is a key regulator for seedling development of monocots (Markelz et al., 2003). In Arabidopsis, a similar portion of the genome is regulated by light during seedling development (Figure 3). Comparisons between W light–regulated gene expression profiles and each distinct monochromatic light-regulated gene expression profile in rice revealed a qualitative similarity, which is analogous to what was found in Arabidopsis (Figure 4A). A quantitative difference was found between W light and monochromatic light in the degree of genome expression regulation in rice but not in Arabidopsis. The quantitatively weaker regulation of gene expression by monochromatic light in rice may imply that either higher monochromatic light intensities or the coactivation of multiple photoreceptor systems that occurs under white light are required to achieve full photomorphogenesis. An important observation is that rice habitats have a much higher light intensity than do those for Arabidopsis. The weaker effects of monochromatic light in rice under our laboratory conditions may reflect the fact that our monochromatic light intensities are not sufficiently high to achieve the optimal effect on photomorphogenesis. However, due to equipment limitations, the current monochromatic light intensities are the highest ones we can achieve.
A survey of gene functional categories suggests that most of them are regulated by light during seedling development (see Supplemental Figure 2 online). Metabolic pathways, in particular, are significantly controlled by light. Moreover, many metabolic pathway genes exhibited similar signatures of light-regulated expression in both rice and Arabidopsis. In the major metabolic pathways, conserved light-regulated expression patterns were found for the best-matched gene pairs between rice and Arabidopsis (Figure 5), although differences were also found in each pathway between the two species.
The profound effect of light on transcriptomes prompted us to explore the underlying controlling mechanism. A signaling cascade that includes mainly a transcriptional regulatory cascade has been proposed for light regulation (Quail, 2002). Alternatively, chromatin-based regulation of transcription may be employed to control large-scale gene expression changes (Reyes et al., 2002; Williams and Bowles, 2004). In this study, we identified only a limited number of genes that exist in small chromosomal domains with similar light-regulated expression (Table 1). Previous similar analyses of organ-specific expression were able to identify several thousand genes in coregulated domains of ∼10 genes each (Ma et al., 2005a, 2005b). In addition to limited chromosomal domains that are light-regulated domains, no cytological-scale patterns of light-regulated transcriptional regulation were found (Figure 7). Cytological-scale patterns of expression previously had been reported in a developmental expression study of a rice chromosome (Jiao et al., 2005). On the other hand, we were able to identify more than a dozen cis-acting motifs overrepresented in light-induced and light-repressed gene promoter regions. Many of these also exhibit organ specificity (Figure 12). The combination of these data suggests that photomorphogenesis, as a response to the environment, is more directly regulated by a complex transcriptional cascade, with only limited, if any, regulation by chromatin organization.
Photomorphogenesis Is More Conserved Than Skotomorphogenesis
To extend our observation of conservation in metabolic pathways between distinct species to the entire genome, we employed a recently developed cross-species gene expression comparison strategy (McCarroll et al., 2004; Murray et al., 2004). Based on reciprocal best-matched gene pair comparisons (Ma et al., 2005a; Yu et al., 2005), we found the existence of analogous genome expression programs comprising shared patterns of expression under the same light conditions in eudicots and monocots (Figure 9).
Photomorphogenesis has been proposed to be a default pathway of plants (Wei et al., 1994) as dark-grown gymnosperms and some algae form chloroplasts. According to this view, skotomorphogenesis evolved later on the evolutionary time scale when photomorphogenesis became repressed in darkness. Skotomorphogenesis is plausibly a more recently established mechanism in higher plants and, therefore, is more prone to change than the ancient photomorphogenesis system established earlier during evolution. The lower correlation for gene expression in dark-grown seedlings compared with light-grown seedlings suggests that the gene expression program during skotomorphogenesis evolves faster than the gene expression program during photomorphogenesis (Figure 9). Consistent with this fact, a smaller portion of the reciprocal best-matched genes between rice and Arabidopsis were repressed by light than are repressed in the entire genome (Figure 8). These light-repressed genes have higher expression during skotomorphogenesis than during photomorphogenesis. On the other hand, light-induced genes were slightly enriched for the best-matched genes compared with the entire genome both in rice and in Arabidopsis (Figure 8). Not surprisingly, a greater degree of conservation exists among the induced genes expressed during photomorphogenesis than among the repressed genes.
Unlike the clear conservation in expression in metabolic pathway genes (Figures 5 and 6), regulatory genes evolve much faster in expression (Table 2). Transcription factors had limited conservation in expression. Signaling genes and ubiquitin-proteasome pathway genes, on the other hand, evolved slightly slower in expression than transcription factors in general. However, their conservation in expression was still clearly weaker than metabolic pathway genes, such as protein synthesis genes. It should be noted that the effect of light on transcription factor expression is dramatically different depending on the period length of light exposure (Jiao et al., 2003). Our data reported here only examined long-time light effects on transcription factor expression, which may be distinct from the effect of short-time light exposure.
It is interesting to note that in comparing genomic expression patterns between fruit fly and nematode in aging, genes involved in several biochemical processes showed higher conservation in expression (McCarroll et al., 2004). These authors did not report high conservation for regulatory genes. One the other hand, while comparing closely related fruit fly subgroups, Rifkin et al. (2003) found that transcription factors have relatively stable expression profiles compared with their downstream targets. The reason behind these seemingly contrasting results is not clear. It is worth mentioning that a recent study reports that sequence divergence may affect the estimation of gene expression levels and cause spurious results when a single-species microarray is used to gauge closely related species (Gilad et al., 2005).
Although we used reciprocal best-match gene pairs for analysis (Ma et al., 2005a; Yu et al., 2005), it is difficult to assert that these best-match pairs are functional orthologs. Rice has been through at least one whole genome duplication-diploidization cycle, and Arabidopsis has probably been through two such independent cycles since the divergence of these lineages from a common ancestor (Blanc and Wolfe, 2004; Paterson et al., 2004). Tandemly arrayed genes also shaped the genomes of Arabidopsis and rice significantly (Zhang and Gaut, 2003; Yu et al., 2005). These considerations may substantially blur the correlation in expression.
Divergence of Light Effect in Different Organs
Evidence for the existence of photoreceptors in various organs, including roots, had been reported decades ago (Briggs and Siegelman, 1965). Recent studies in Arabidopsis have demonstrated the presence of phytochromes in all organ types (Somers and Quail, 1995; Goosey et al., 1997; Sharrock and Clack, 2002). The presence of phytochromes in various organs has also been reported in rice (Kay et al., 1989).
Although different organs may share the same set of photoreceptors, light effects in distinct organs are obviously different. Light triggers cotyledon expansion and leaf development but inhibits stem growth in Arabidopsis (Neff et al., 2000). In roots, negative phototropic growth is induced by light (Parks and Poff, 1986; Feldman and Briggs, 1987; Blancaflor and Masson, 2003). Light also plays a possible role in the initiation of lateral roots (Furuya and Torrey, 1964; Ohno and Fujiwara, 1967). Light regulation of gene expression in roots has been discussed (Hemm et al., 2004; Sato-Nara et al., 2004). Strong evidence exists that organ-specific effects on transcription occur during the process of light-regulated seedling development (Leu et al., 1995; Huq et al., 2004; Monte et al., 2004; Ma et al., 2005b).
In this study, we globally explored the light-regulated transcriptional profiles of the entire genome at the organ level in rice and compared this with Arabidopsis. By examining the expression profiles, we found a strong impact of light on genome expression in roots that is similar to shoots or cotyledons (Figure 10). Moreover, the limited overlap between light-regulated genes in shoots (or cotyledons) and in roots (Figure 10) suggests that light signaling pathways diverge significantly in separate organs. A suite of transcription factors and regulatory genes may function upstream of light signaling pathways with similar light regulation in shoots and in roots. Parts of the shared signaling pathways result in similar regulation of a suite of metabolic pathway genes. On the other hand, shoots (or cotyledons) and roots also employ distinct downstream signal transducers. These result in organ-specific effecter genes performing distinct developmental responses to light in discrete organs. Consistent with this hypothesis, common and organ-specific cis-acting promoter elements were identified (Figure 12). Specifically, ethylene- and auxin-related genes exhibit organ specificity, suggesting not only the involvement of phytohormones in photomorphogenesis, but also organ-specific usage of phytohormones to regulate spatially distinct photomorphogenic processes.
METHODS
Plant Materials
The rice strain used in this study was Oryza sativa subsp indica cv 93-11. Caryopses were dehusked and surface sterilized (Garg et al., 2002). The caryopses were sown on Murashige and Skoog growth medium agar with 0.8% sucrose. Caryopses were treated at 37°C for 2 d immediately after plating. Plants were grown in environmentally controlled chambers at 28°C for 10 d. Continuous W light was fluorescent light with an intensity of 220 μmol·m−2·s−1. Low W light intensity was obtained by putting variable layers of Kimwipe tissue papers or white printing papers on top of plant culture vessels. The monochromatic light growth chambers (E-30LED2/3; Percival Scientific; described in Osterlund and Deng, 1998) have intensities of 161 μmol·m−2·s−1 for FR light (730 nm), 109 μmol·m−2·s−1 for R light (660 nm), and 16 μmol·m−2·s−1 for B light (470 nm). Shoots were excised from 10-d-old seedlings above the upper node of the mesocotyl. Roots were excised below the shoot–root joint node. Therefore, the node, which includes the shoot apical meristem, and the mesocotyl of dark-grown seedlings were not included as either shoot or root.
Arabidopsis thaliana (ecotype Columbia-0) was surface sterilized, and stratification treatment of seeds was performed as previously described (Ma et al., 2001). Seedlings were grown on GM agar plates containing 1% sucrose. The seedlings were grown under continuous light or in darkness for 6 d at 20°C. The W light intensity used was provided by fluorescent light tubes with an intensity of 160 μmol·m−2·s−1. Monochromatic light chambers were the same as for rice, only with different temperature settings (described in Osterlund and Deng, 1998). Cotyledons were isolated from seedlings from the branch point of the two cotyledons without including the shoot apical meristem, while roots were excised below the junction of shoot and root (Ma et al., 2005b).
Oligonucleotide Microarray
The rice 70-mer oligonucleotide set was based on a combination of FGENESH predicted gene models from an improved indica rice genome sequence and the available full-length cDNAs and ESTs (Ma et al., 2005a). A total of 58,404 70-mer oligonucleotides were designed and custom synthesized by Operon. This oligonucleotide set covers 36,926 rice genes from the recent release of the improved indica rice genome sequence and annotation (Yu et al., 2005) after oligos with potential cross-hybridization were removed (Ma et al., 2005a). The genes covered by our oligonucleotide set include 15,059 full-length cDNA confirmed genes, 5435 predicted gene models with EST support, and 16,638 FGENESH predicted gene models without EST support. Approximately 92% of the nonredundant, full-length cDNA-confirmed genes are covered. Oligonucleotide annotation information is available at the Beijing Genomics Institute–Rice Information System (BGI-RIS) databases (http://rise.genomics.org.cn; Zhao et al., 2004). In this study, we only analyzed oligos representing the 36,926 known and predicted genes of the most current version of indica genome annotation (Ma et al., 2005a).
The Arabidopsis 70-mer oligonucleotide microarray was based on 26,090 unique 70-mer oligonucleotides of the Arabidopsis Genome Oligo Set version 1.0 (Operon). Oligonucleotides of this set correspond to 25,676 protein-coding genes annotated by The Institute for Genomic Research or Munich Information Center for Protein Sequences (Ma et al., 2005b). Information about oligonucleotide annotation is hosted on the oligonucleotide microarray database of Operon (http://www.operon.com/arrays/omad.php).
Rice and Arabidopsis oligonucleotides were printed onto poly-l-Lys–coated microscope slides using contact microarrayers (Ma et al., 2005a, 2005b). Both rice and Arabidopsis microarray slides included the same recommended set of 12 unique, negative-control 70-mer oligonucleotides based on heterologous genes (http://omad.operon.com/arabidopsis/index.php). There are 240 negative control spots on each rice slide and 192 negative control spots on each Arabidopsis microarray slide.
RNA Isolation, Labeling, and Microarray Hybridization
Whole seedlings and excised organs were frozen in liquid nitrogen. Rice total RNA was isolated and purified using the RNeasy kit (Qiagen). Arabidopsis total RNA was extracted using the RNeasy plant kit. For each treatment, three independent biological samples were used for RNA isolation and probe synthesis, with a dye swap to give two repeats labeled in the same direction and a third repeat labeled in the other direction. For light and dark comparison of Arabidopsis cotyledons and roots, the three replicates included two from a prior published work (Ma et al., 2005b) and one new data set with reverse dye labeling. All other samples in this study were new and not previously reported. For each sample, 100 μg of total RNA was labeled with aminoallyl-dUTP (Sigma-Aldrich) by reverse transcription as described previously (Ma et al., 2005b). After reverse transcription for 3 to 4 h, template RNA was degraded. The aminoallyl-dUTP–labeled cDNAs were purified using a Microcon YM-30 filter (Millipore) and resuspended in 0.1 M NaHCO3. The cDNA probe was further fluorescently labeled by conjugating monofunctional Cy3 or Cy5 dye (Amersham) to the aminoallyl functional groups. After coupling at room temperature for 1 h, the labeling reaction was stopped by ethanolamine. The fluorescent dye–labeled probe was separated from unincorporated dye using the QIAquick PCR purification kit (Qiagen) and concentrated for hybridization using a Microcon YM-30 filter.
We followed recently described protocols for microarray hybridization, microarray slide washing, and array scanning (Ma et al., 2005b). Hybridized microarray slides were scanned with a GenePix 4000B scanner (Axon), and independent TIFF images for both Cy3 and Cy5 channels were used for subsequent analysis.
Microarray Data Processing
After manual removal of spots with aberrant morphology, microarray spot intensity signals were acquired using the Axon GenePix Pro 3.0 software package. To identify and remove systematic sources of variation, including dye and spatial effects, spot intensities from the GenePix Pro output files of all repeats of a given sample pair were normalized using the web-based EXPRESSYOURSELF platform (http://bioinfo.mbb.yale.edu/ExpressYourself) with default parameters (Luscombe et al., 2003). This normalization process identified and ameliorated spatial, intensity-based, and dye-specific artifacts using multiple-step corrections, including loess normalization among print-tip groups. To check the quality of the repeats, we set a threshold of 0.80 or better for the overall correlation coefficient between any two repeats. Any data set that did not meet this threshold was not used and further repeated with the same biological sample until three quality repeats were obtained and normalized. The actual correlation coefficients among our quality-checked repeats typically ranged between 0.85 and 0.95.
To determine objectively whether a gene had significant expression in a given sample, we followed a method based on negative control spots described before (Ma et al., 2005b). This method considered both the signal intensity and the reproducibility of each spot among independent biological repeats. To estimate nonspecific hybridization, a distribution of normalized intensities was obtained from the subset of negative control spots present on each array slide. From this distribution, we chose an intensity cutoff at which <10% of the distribution was greater than or equal to this threshold. Then we considered the expression of a gene detectable only if it was above the threshold in two or more repeats out of the three. These criteria had been demonstrated suitable for oligonucleotide arrays with an error rate range of 1 to 3% false negatives (Ma et al., 2005b). We only included genes with expression in at least one channel in each experiment for analysis.
To identify genes differentially expressed between the light- and dark-grown samples, a Student's t test was conducted by comparing log2-transformed light versus dark expression values with two-sample hypothesis and equal variations assumptions. To address the issue of multiple testing errors, we estimated the false discovery rate using a method specifically developed for genome-wide studies (Storey and Tibshirani, 2003). We found this false discovery rate ranged from 2 to 9% with a P value of 0.05 in different sample sets. To reduce further the occurrence of false positives, we added a ratio above a twofold cutoff filter to genes with P < 0.05 (Figure 3B). We used averages of the three individual light versus dark expression ratios from the repeats here and in subsequence analysis. Genes selected by these criteria, as suggested previously (Reymond et al., 2004), were considered to be differentially expressed genes regulated by light in subsequent analyses.
Cluster Analysis
Cluster analysis was applied to all genes showing light versus dark differential expression in at least one comparison in each group of experiments subject to cluster. Differential expression was determined as stated above. Genes missing data in any experiment were removed. Average normalized log2-transformed ratios were subjected to cluster analysis. For hierarchical clustering, a Pearson correlation was used to compute similarities, and the complete linkage clustering algorithm was used. For K-means clustering, we estimated the number of clusters (K) as 15 for both rice and Arabidopsis. Cluster analysis was performed using the software Cluster and visualized using TreeView (Eisen et al., 1998).
Functional Classification
GO annotation was used for gene functional classification. Rice gene annotations using GO terms were downloaded from the BGI-RIS database at http://rise.genomics.org.cn (Zhao et al., 2004). For Arabidopsis genes, we followed the functional annotations using GO terms at The Arabidopsis Information Resource (TAIR) (http://www.arabidopsis.org/info/ontologies/go/; Berardini et al., 2004).
For biochemical pathway genes, we did further classification following the AraCyc database (http://www.arabidopsis.org/tools/aracyc) for Arabidopsis, which is based on MetaCyc pathway collections (Mueller et al., 2003). A rice gene was considered to be associated with a biochemical pathway if it has an Arabidopsis homolog (see below) in that pathway.
Search for Similarly Regulated Chromosomal Domains
Coregulated adjacent genes were identified using the method reported by Spellman and Rubin (2002). Specifically, the average of all possible Pearson correlations was calculated at each given window size. This calculation was performed for both ordered and randomized data sets to reveal the significance of observed coregulated domains.
To reveal any potential correlation between cytological scale domains and light regulation, the percentages of light-regulated genes were calculated in a series of moving windows along each chromosome. More specifically, for a window size W and moving step K, we created a series of M = N/K moving windows with width W, where N denotes the number of genes in this chromosome. For each window, the frequency of light-induced and light-repressed genes were calculated and plotted along the chromosome.
Transcription Correlation Analysis between Rice and Arabidopsis
To compare the expression profiles of conserved genes between rice and Arabidopsis, we selected reciprocal best-matched gene pairs between rice and Arabidopsis, which are based on a TBLASTN search of both rice and Arabidopsis gene sequences (Ma et al., 2005a; Yu et al., 2005). We calculated the Pearson correlation (r) of the log10-transformed normalized transcriptional intensity measurements for reciprocal best-matched genes to measure global correlation between two species.
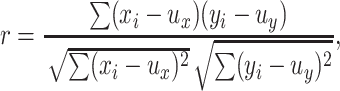 |
where X = (x1, x2,…xn) and Y = (y1, y2,…yn) are log-transformed transcriptional intensity measurements for rice and for Arabidopsis, the mean of these measurements are represented by ux and uy, respectively, and n is the number of best-matched pairs used for calculation. The statistical significance (P value) was assessed by the Student's t test with (n − 2) degrees of freedom. Normal quantile plots and residual plots suggest that log10-transformed normalized transcriptional intensities have a close to normal distribution without patterns affecting the correlation calculation (see Supplemental Figure 3 online).
Motif Search
The genome sequences 2 kb upstream of annotated translation start sites were retrieved from the BGI-RIS and TAIR databases for rice and Arabidopsis, respectively. Both DNA strands were searched using Sift, an enumerative algorithm (Hudson and Quail, 2003), and AlignACE, a Gibbs sampling-based alignment approach (Hughes et al., 2000). For the enumerative search, only elements meeting the critical P value smaller than 10−5 were selected. A MAP score above 10.0 was expected for motif groups identified by AlignACE. Identified elements were aligned, and common motifs found by both search approaches were identified manually. Comparison of detected motifs with known motifs was performed using the PLACE database (Higo et al., 1998), the PlantCARE database (Rombauts et al., 1999), and literature searches.
Accession Number and Data Deposition
Microarray data from this article are deposited with the National Center for Biotechnology Information Gene Expression Omnibus data repository (http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE2360. The processed data can be found in the supplemental data online as well as at http://plantgenomics.biology.yale.edu/.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Table 1. Expression of 36,926 Rice Genes in Response to Light Exposure.
Supplemental Table 2. Expression of 25,676 Arabidopsis Genes in Response to Light Exposure.
Supplemental Table 3. Biochemical Pathways Regulated by Light in Both Rice and Arabidopsis.
Supplemental Figure 1. RT-PCR Verification of Microarray Data.
Supplemental Figure 2. Functional Classification of Expressed, Light-Induced, and Light-Repressed Genes by Gene Ontology Terms.
Supplemental Figure 3. Organ-Specific Light-Regulated Expression in Arabidopsis.
Supplemental Figure 4. Residual Check for the Calculation of Pearson Correlation between Reciprocal Best-Matched Gene Pairs.
Supplementary Material
Acknowledgments
We gratefully acknowledge Xiuqing Zhang (Beijing Genome Institute, Beijing, China) and the DNA microarray laboratory of the Keck Biotechnology Resource Center at Yale for the printing of microarray slides used in this work. We thank Thomas Royce for advice on microarray data processing, Xiangfeng Wang, Chen Chen, and Zhong Guan for advice on computation and algorithms, and Erica Che and Jamila Searchwell for technical assistance. Our research was supported by National Institutes of Health Grant GM-47850 (to X.-W.D.), the 863 Rice Functional Genomics Program from the Ministry of Science and Technology of China, a strategic international cooperation project grant (30221120261) from the National Science Foundation of China, and, in part, by a National Science Foundation Plant Genome Program grant (DBI-0325821). Y.J. was the recipient of a Yale University Joseph F. Cullman, Jr. Fellowship, L.M. was a long-term postdoctoral fellow of the Human Frontier Science Program, and E.S. is a recipient of a Ruth L. Kirschstein National Research Service Award from the National Institutes of Health.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Xing Wang Deng (xingwang.deng@yale.edu).
Online version contains Web-only data.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.105.035840.
References
- Basu, D., Dehesh, K., Schneider-Poetsch, H.J., Harrington, S.E., McCouch, S.R., and Quail, P.H. (2000). Rice PHYC gene: Structure, expression, map position and evolution. Plant Mol. Biol. 44, 27–42. [DOI] [PubMed] [Google Scholar]
- Berardini, T.Z., et al. (2004). Functional annotation of the Arabidopsis genome using controlled vocabularies. Plant Physiol. 135, 745–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc, G., and Wolfe, K.H. (2004). Widespread paleopolyploidy in model plant species inferred from age distributions of duplicate genes. Plant Cell 16, 1667–1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blancaflor, E.B., and Masson, P.H. (2003). Plant gravitropism. Unraveling the ups and downs of a complex process. Plant Physiol. 133, 1677–1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blecken, J., Weisshaar, B., and Herzfeld, F. (1994). Two distinct cis-acting elements are involved in light-dependent activation of the pea elip promoter. Mol. Gen. Genet. 245, 371–379. [DOI] [PubMed] [Google Scholar]
- Briggs, W.R., and Siegelman, H.W. (1965). Distribution of phytochrome in etiolated seedlings. Plant Physiol. 40, 934–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cashmore, A.R., Jarillo, J.A., Wu, Y.J., and Liu, D. (1999). Cryptochromes: Blue light receptors for plants and animals. Science 284, 760–765. [DOI] [PubMed] [Google Scholar]
- Chattopadhyay, S., Ang, L.-H., Puente, P., Deng, X.-W., and Wei, N. (1998). Arabidopsis bZIP protein HY5 directly interacts with light-responsive promoters in mediating light control of gene expression. Plant Cell 10, 673–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaw, S.-M., Chang, C.-C., Chen, H.-L., and Li, W.-H. (2004). Dating the monocot-dicot divergence and the origin of core eudicots using whole chloroplast genomes. J. Mol. Evol. 58, 424–441. [DOI] [PubMed] [Google Scholar]
- Cheng, Z., Buell, C.R., Wing, R.A., Gu, M., and Jiang, J. (2001). Toward a cytological characterization of the rice genome. Genome Res. 11, 2133–2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehesh, K., Tepperman, J., Christensen, A.H., and Quail, P.H. (1991). phyB is evolutionarily conserved and constitutively expressed in rice seedling shoots. Mol. Gen. Genet. 225, 305–313. [DOI] [PubMed] [Google Scholar]
- Eisen, M.B., Spellman, P.T., Brown, P.O., and Botstein, D. (1998). Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA 95, 14863–14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman, L.J., and Briggs, W.R. (1987). Light-regulated gravitropism in seedling roots of maize. Plant Physiol. 83, 241–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein, D., Ewing, R., Gollub, J., Sterky, F., Cherry, J.M., and Somerville, S. (2002). Microarray data quality analysis: Lessons from the AFGC project. Plant Mol. Biol. 48, 119–131. [DOI] [PubMed] [Google Scholar]
- Fransz, P.F., Armstrong, S., de Jong, J.H., Parnell, L.D., van Drunen, C., Dean, C., Zabel, P., Bisseling, T., and Jones, G.H. (2000). Integrated cytogenetic map of chromosome arm 4S of A. thaliana: Structural organization of heterochromatic knob and centromere region. Cell 100, 367–376. [DOI] [PubMed] [Google Scholar]
- Furuya, M., and Torrey, J.G. (1964). The reversible inhibition by red and far-red light of auxin-induced lateral root initiation in isolated pea roots. Plant Physiol. 39, 987–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gale, M.D., and Devos, K.M. (1998). Comparative genetics in the grasses. Proc. Natl. Acad. Sci. USA 95, 1971–1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg, A.K., Kim, J.K., Owens, T.G., Ranwala, A.P., Choi, Y.D., Kochian, L.V., and Wu, R.J. (2002). Trehalose accumulation in rice plants confers high tolerance levels to different abiotic stresses. Proc. Natl. Acad. Sci. USA 99, 15898–15903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gene Ontology Consortium (2000). Gene ontology: Tool for the unification of biology. Nat. Genet. 25, 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilad, Y., Rifkin, S.A., Bertone, P., Gerstein, M., and White, K.P. (2005). Multi-species microarrays reveal the effect of sequence divergence on gene expression profiles. Genome Res. 15, 674–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girke, T., Todd, J., Ruuska, S., White, J., Benning, C., and Ohlrogge, J. (2000). Microarray analysis of developing Arabidopsis seeds. Plant Physiol. 124, 1570–1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goff, S.A., et al. (2002). A draft sequence of the rice genome (Oryza sativa L. ssp. japonica). Science 296, 92–100. [DOI] [PubMed] [Google Scholar]
- Goosey, L., Palecanda, L., and Sharrock, R.A. (1997). Differential patterns of expression of the Arabidopsis PHYB, PHYD, and PHYE phytochrome genes. Plant Physiol. 115, 959–969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haga, K., Takano, M., Neumann, R., and Iino, M. (2005). The rice COLEOPTILE PHOTOTROPISM1 gene encoding an ortholog of Arabidopsis NPH3 is required for phototropism of coleoptiles and lateral translocation of auxin. Plant Cell 17, 103–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, Y., and Amasino, R.M. (2005). Role of chromatin modification in flowering-time control. Trends Plant Sci. 10, 30–35. [DOI] [PubMed] [Google Scholar]
- Hemm, M.R., Rider, S.D., Ogas, J., Murry, D.J., and Chapple, C. (2004). Light induces phenylpropanoid metabolism in Arabidopsis roots. Plant J. 38, 765–778. [DOI] [PubMed] [Google Scholar]
- Higo, K., Ugawa, Y., Iwamoto, M., and Higo, H. (1998). PLACE: A database of plant cis-acting regulatory DNA elements. Nucleic Acids Res. 26, 358–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh, T.-F., and Fischer, R.L. (2005). Biology of chromatin dynamics. Annu. Rev. Plant Biol. 56, 327–351. [DOI] [PubMed] [Google Scholar]
- Hudson, M.E., and Quail, P.H. (2003). Identification of promoter motifs involved in the network of phytochrome A-regulated gene expression by combined analysis of genomic sequence and microarray data. Plant Physiol. 133, 1605–1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes, J.D., Estep, P.W., Tavazoie, S., and Church, G.M. (2000). Computational identification of cis-regulatory elements associated with groups of functionally related genes in Saccharomyces cerevisiae. J. Mol. Biol. 296, 1205–1214. [DOI] [PubMed] [Google Scholar]
- Huq, E., Al-Sady, B., Hudson, M., Kim, C., Apel, K., and Quail, P.H. (2004). Phytochrome-interacting factor 1 is a critical bHLH regulator of chlorophyll biosynthesis. Science 305, 1937–1941. [DOI] [PubMed] [Google Scholar]
- Hurst, L.D., Pal, C., and Lercher, M.J. (2004). The evolutionary dynamics of eukaryotic gene order. Nat. Rev. Genet. 5, 299–310. [DOI] [PubMed] [Google Scholar]
- Inaba, T., Nagano, Y., Reid, J.B., and Sasaki, Y. (2000). DE1, a 12-base pair cis-regulatory element sufficient to confer dark-inducible and light down-regulated expression to a minimal promoter in pea. J. Biol. Chem. 275, 19723–19727. [DOI] [PubMed] [Google Scholar]
- Izawa, T., Oikawa, T., Tokutomi, S., Okuno, K., and Shimamoto, K. (2000). Phytochromes confer the photoperiodic control of flowering in rice (a short-day plant). Plant J. 22, 391–399. [DOI] [PubMed] [Google Scholar]
- Jiao, Y., et al. (2003). A genome-wide analysis of blue-light regulation of Arabidopsis transcription factor gene expression during seedling development. Plant Physiol. 133, 1480–1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao, Y., et al. (2005). A tiling microarray expression analysis of rice chromosome 4 suggests a chromosome-level regulation of transcription. Plant Cell 17, 1641–1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasahara, M., et al. (2002). Photochemical properties of the flavin mononucleotide-binding domains of the phototropins from Arabidopsis, rice, and Chlamydomonas reinhardtii. Plant Physiol. 129, 762–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kay, S.A., Keith, B., Shinozaki, K., Chye, M.L., and Chua, N.-H. (1989). The rice phytochrome gene: Structure, autoregulated expression and binding of GT-1 to a conserved sit in the 5′ upstream region. Plant Cell 1, 351–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khush, G.S. (1997). Origin, dispersal, cultivation and variation of rice. Plant Mol. Biol. 35, 25–34. [PubMed] [Google Scholar]
- Kiss, J.Z., Mullen, J.L., Correll, M.J., and Hangarter, R.P. (2003). Phytochrome A and B mediate red-light-induced positive phototropism in roots. Plant Physiol. 131, 1411–1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef, M., Rolff, E., and Spruit, C.J.P. (1980). Genetic control of light inhibited hypocotyl elongation in Arabidopsis thaliana (L.). Heynh. Z. Pflanzenphysiol. 100, 147–160. [Google Scholar]
- Lee, H.-S., et al. (2004). Sensitivity of 70-mer oligonucleotides and cDNAs for microarray analysis of gene expression in Arabidopsis and its related species. Plant Biotechnol. J. 2, 45–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leu, W.-M., Cao, X.-L., Wilson, T.J., Snustad, D.P., and Chua, N.-H. (1995). Phytochrome A and phytochrome B mediate the hypocotyl-specific downregulation of TUB1 by light in Arabidopsis. Plant Cell 7, 2187–2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, C. (2002). Blue light receptors and signal transduction. Plant Cell 14, S207–S225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liscum, E., Hodgson, D.W., and Campbell, T.J. (2003). Blue light signaling through the cryptochromes and phototropins. So that's what the blues is all about. Plant Physiol. 133, 1429–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lissemore, J.L., and Quail, P.H. (1988). Rapid transcriptional regulation by phytochrome of the genes for phytochrome and chlorophyll a/b-binding protein in Avena sativa. Mol. Cell. Biol. 8, 4840–4850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y., Roof, S., Ye, Z., Barry, C., van Tuinen, A., Vrebalov, J., Bowler, C., and Giovannoni, J. (2004). Manipulation of light signal transduction as a means of modifying fruit nutritional quality in tomato. Proc. Natl. Acad. Sci. USA 101, 9897–9902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luscombe, N.M., Royce, T.E., Bertone, P., Echols, N., Horak, C.E., Chang, J.T., Snyder, M., and Gerstein, M. (2003). ExpressYourself: A modular platform for processing and visualizing microarray data. Nucleic Acids Res. 31, 3477–3482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, L., et al. (2005. a). A microarray analysis of the rice transcriptome and its comparison to Arabidopsis. Genome Res. 15, 1274–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, L., Li, J., Qu, L., Hager, J., Chen, Z., Zhao, H., and Deng, X.W. (2001). Light control of Arabidopsis development entails coordinated regulation of genome expression and cellular pathways. Plant Cell 13, 2589–2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, L., Sun, N., Liu, X., Jiao, Y., Zhao, H., and Deng, X.W. (2005. b). Organ-specific regulation of Arabidopsis genome expression during development. Plant Physiol. 138, 80–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markelz, N.H., Costich, D.E., and Brutnell, T.P. (2003). Photomorphogenic responses in maize seedling development. Plant Physiol. 133, 1578–1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Garcia, J.F., Huq, E., and Quail, P.H. (2000). Direct targeting of light signals to a promoter element-bound transcription factor. Science 288, 859–863. [DOI] [PubMed] [Google Scholar]
- Matsumoto, N., Hirano, T., Iwasaki, T., and Yamamoto, N. (2003). Functional analysis and intracellular localization of rice cryptochromes. Plant Physiol. 133, 1494–1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarroll, S.A., Turphy, C.T., Zuo, S., Pletcher, S.D., Chin, C.-S., Jan, Y.N., Kenyon, C., Bargmann, C.I., and Li, H. (2004). Comparing genome expression patterns across species identifies shared transcriptional profile in aging. Nat. Genet. 36, 197–204. [DOI] [PubMed] [Google Scholar]
- Menkens, A.E., Schindler, U., and Cashmore, A.R. (1995). The G-box: A ubiquitous regulatory DNA element in plants bound by the GBF family of bZIP proteins. Trends Biochem. Sci. 20, 506–510. [DOI] [PubMed] [Google Scholar]
- Meyers, B.C., Galbraith, D.W., Nelson, T., and Agrawal, V. (2004). Methods for transcriptional profiling in plants. Be fruitful and replicate. Plant Physiol. 135, 637–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohr, H. (1962). Primary effects of light on growth. Annu. Rev. Plant Physiol. 13, 465–488. [Google Scholar]
- Monte, E., Tepperman, J.M., Al-Sady, B., Kaczorowski, K.A., Alonso, J.M., Ecker, J.R., Li, X., Zhang, Y., and Quail, P.H. (2004). The phytochrome-interacting transcription factor, PIF3, acts early, selectively, and positively in light-induced chloroplast development. Proc. Natl. Acad. Sci. USA 101, 16091–16098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller, L.A., Zhang, P., and Rhee, S.Y. (2003). AraCyc: A biochemical pathway database for Arabidopsis. Plant Physiol. 132, 453–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullet, J.E. (1993). Dynamic regulation of chloroplast transcription. Plant Physiol. 103, 309–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray, J.I., Whitfield, M.L., Trinklein, N.D., Myers, R.M., Brown, P.O., and Botstein, D. (2004). Diverse and specific gene expression responses to stresses in cultured human cells. Mol. Biol. Cell 15, 2361–2374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy, F., and Schäfer, E. (2002). Phytochromes control photomorphogenesis by differentially regulated, interacting signaling pathways in higher plants. Annu. Rev. Plant Biol. 53, 329–355. [DOI] [PubMed] [Google Scholar]
- Neff, M.M., Fankhauser, C., and Chory, J. (2000). Light: An indicator of time and place. Genes Dev. 14, 257–271. [PubMed] [Google Scholar]
- Ohgishi, M., Saji, K., Okada, K., and Sakai, T. (2004). Functional analysis of each blue light receptor, cry1, cry2, phot1, and phot2, by using combinatorial multiple mutants in Arabidopsis. Proc. Natl. Acad. Sci. USA 101, 2223–2228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohler, U., and Niemann, H. (2001). Identification and analysis of eukaryotic promoters: Recent computational approaches. Trends Genet. 17, 56–60. [DOI] [PubMed] [Google Scholar]
- Ohno, Y., and Fujiwara, A. (1967). Photoinhibition of elongation growth of roots in rice seedlings. Plant Cell Physiol. 8, 141–150. [Google Scholar]
- Osterlund, M.T., and Deng, X.-W. (1998). Multiple photoreceptors mediate the light-induced reduction of GUS-COP1 from Arabidopsis hypocotyl nuclei. Plant J. 16, 201–208. [DOI] [PubMed] [Google Scholar]
- Parks, B.M., and Poff, K.L. (1986). Altering the axial light gradient affects photomorphogenesis in emerging seedlings of Zea mays L. Plant Physiol. 81, 75–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson, A.H., Bowers, J.E., and Chapman, B.A. (2004). Ancient polyploidization predating divergence of the cereals, and its consequences for comparative genomics. Proc. Natl. Acad. Sci. USA 101, 9903–9908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pjon, C.-J., and Furuya, M. (1967). Phytochrome action in Oryza sativa L. I. Growth responses of etiolated coleoptiles to red, far-red and blue light. Plant Cell Physiol. 8, 709–718. [Google Scholar]
- Quail, P.H. (2002). Photosensory perception and signalling in plant cells: New paradigms? Curr. Opin. Cell Biol. 14, 180–188. [DOI] [PubMed] [Google Scholar]
- Reyes, J.C., Hennig, L., and Gruissem, W. (2002). Chromatin-remodeling and memory factors. New regulators of plant development. Plant Physiol. 130, 1090–1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reymond, P., Bodenhausen, N., Van Poecke, R.M.P., Krishnamurthy, V., Dicke, M., and Farmer, E.E. (2004). A conserved transcript pattern in response to a specialist and a generalist herbivore. Plant Cell 16, 3132–3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rifkin, S.A., Kim, J., and White, K.P. (2003). Evolution of gene expression in the Drosophila melanogaster subgroup. Nat. Genet. 33, 138–144. [DOI] [PubMed] [Google Scholar]
- Rombauts, S., Déhais, P., Van Montagu, M., and Rouzé, P. (1999). PlantCARE, a plant cis-acting regulatory element database. Nucleic Acids Res. 27, 295–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki, T., and Burr, B. (2000). International rice genome sequencing project: The effort to completely sequence the rice genome. Curr. Opin. Plant Biol. 3, 138–141. [DOI] [PubMed] [Google Scholar]
- Sato-Nara, K., Nagasaka, K., Yamashita, H., Ishida, J., Enju, A., Seki, M., Shinozaki, K., and Suzuki, H. (2004). Identification of genes regulated by dark adaptation and far-red light illumination in roots of Arabidopsis thaliana. Plant Cell Environ. 27, 1387–1394. [Google Scholar]
- Sawers, R.J., Sheehan, M.J., and Brutnell, T.P. (2005). Cereal phytochromes: Targets of selection, targets for manipulation? Trends Plant Sci. 10, 138–143. [DOI] [PubMed] [Google Scholar]
- Schmid, M., Davison, T.S., Henz, S.R., Pape, U.J., Demar, M., Vingron, M., Scholkopf, B., Weigel, D., and Lohmann, J.U. (2005). A gene expression map of Arabidopsis thaliana development. Nat. Genet. 37, 501–506. [DOI] [PubMed] [Google Scholar]
- Sharrock, R.A., and Clack, T. (2002). Patterns of expression and normalized levels of the five Arabidopsis phytochromes. Plant Physiol. 130, 442–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharrock, R.A., and Quail, P.H. (1989). Novel phytochrome sequences in Arabidopsis thaliana: Structure, evolution and differential expression of a plant regulatory photoreceptor family. Genes Dev. 3, 1745–1757. [DOI] [PubMed] [Google Scholar]
- Shimamoto, K., and Kyozuka, J. (2002). Rice as a model for comparative genomics of plants. Annu. Rev. Plant Biol. 53, 399–419. [DOI] [PubMed] [Google Scholar]
- Somers, D.E., and Quail, P.H. (1995). Temporal and spatial expression patterns of PHYA and PHYB genes in Arabidopsis. Plant J. 7, 413–427. [DOI] [PubMed] [Google Scholar]
- Spellman, P.T., and Rubin, G.M. (2002). Evidence for large domains of similarly expressed genes in the Drosophila genome. J. Biol. 1, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey, J.D., and Tibshirani, R. (2003). Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. USA 100, 9440–9445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takano, M., Kanegae, H., Shinomura, T., Miyao, A., Hirochika, H., and Furuya, M. (2001). Isolation and characterization of rice phytochrome A mutants. Plant Cell 13, 521–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tepperman, J.M., Zhu, T., Chang, H.-S., Wang, X., and Quail, P.H. (2001). Multiple transcription-factor genes are early targets of phytochrome A signaling. Proc. Natl. Acad. Sci. USA 98, 9437–9442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terzaghi, W.B., and Cashmore, A.R. (1995). Light-regulated transcription. Annu. Rev. Plant Physiol. Plant Mol. Biol. 46, 445–474. [Google Scholar]
- Wada, M., Kagawa, T., and Sato, Y. (2003). Chloroplast movement. Annu. Rev. Plant Biol. 54, 455–468. [DOI] [PubMed] [Google Scholar]
- Wang, H., Ma, L., Habashi, J., Li, J., Zhao, H., and Deng, X.W. (2002). Analysis of far-red light-regulated genome expression profiles of phytochrome A pathway mutants in Arabidopsis. Plant J. 32, 723–733. [DOI] [PubMed] [Google Scholar]
- Wang, H.-Y., Malek, R.L., Kwitek, A.E., Greene, A.S., Luu, T.V., Behbahani, B., Frank, B., Quackenbush, J., and Lee, N.H. (2003). Assessing unmodified 70-mer oligonucleotide probe performance on glass-slide microarrays. Genome Biol. 4, R5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, N., Kwok, S.F., von Arnim, A.G., Lee, A., McNellis, T.W., Piekos, B., and Deng, X.-W. (1994). Arabidopsis COP8, COP10, and COP11 genes are involved in repression of photomorphogenic development in darkness. Plant Cell 6, 629–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellmer, F., Riechmann, J.L., Alves-Ferreira, M., and Meyerowitz, E.M. (2004). Genome-wide analysis of spatial gene expression in Arabidopsis flowers. Plant Cell 16, 1314–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkins, M.B. (1977). Light- and gravity-sensing guidance systems in plants. Proc. R. Soc. Lond. B Biol. Sci. 199, 513–524. [DOI] [PubMed] [Google Scholar]
- Williams, E.J.B., and Bowles, D.J. (2004). Coexpression of neighboring genes in the genome of Arabidopsis thaliana. Genome Res. 14, 1060–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh, K.-C., and Lagarias, J.C. (1998). Eukaryotic phytochromes: Light-regulated serine/threonine protein kinases with histidine kinase ancestry. Proc. Natl. Acad. Sci. USA 95, 13976–13981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, J., et al. (2002). A draft sequence of the rice genome (Oryza sativa L. ssp. indica). Science 296, 79–92. [DOI] [PubMed] [Google Scholar]
- Yu, J., et al. (2005). The genomes of Oryza sativa: A history of duplications. PLoS Biol. 3, e38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, L., and Gaut, B.S. (2003). Does recombination shape the distribution and evolution of tandemly arrayed genes (TAGs) in the Arabidopsis thaliana genome? Genome Res. 13, 2533–2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, W., et al. (2004). BGI-RIS: An integrated information resource and comparative analysis workbench for rice genomics. Nucleic Acids Res. 32, D377–D382. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.