Abstract
Eukaryotic DNA replication produces sister chromatids that are linked together until anaphase by cohesin, a ring-shaped protein complex that is thought to act by embracing both chromatids. Cohesin is enriched at centromeres, as well as discrete sites along chromosome arms where transcription positions the complex between convergent gene pairs. A relationship between cohesin and Sir-mediated transcriptional silencing has also begun to emerge. Here we used fluorescence microscopy and site-specific recombination to characterize interactions between newly replicated copies of the silent HMR mating-type locus. HMR was tagged with lac-GFP and flanked by binding sites for an inducible site-specific recombinase. Excision of the locus in cells with sister chromatids produced two chromatin circles that remained associated with one another. Pairing of the circles required silent chromatin, cohesin, and the RSC chromatin-remodeling complex. Chromatin immunoprecipitation showed that targeting of cohesin to the locus is Sir-dependent, and functional tests showed that silent chromatin acts in a continuous fashion to maintain cohesion. Remarkably, loss of silencing led to loss of cohesin from linear chromosomal templates but not from excised chromatin circles. The results are consistent with a model in which cohesin binds silent chromatin via topological linkage to individual chromatids.
Keywords: Sir, transcriptional silencing, silent chromatin, sister chromatid cohesion, cohesin, RSC
Faithful segregation of chromosomes between dividing cells relies on sister chromatid cohesion, a process by which newly replicated sister chromatids adhere to one another until they align on the bipolar metaphase spindle (Nasmyth 2001; Uhlmann 2004). When all chromosome pairs attach to microtubules from both spindle poles (biorientation), cohesion of sister chromatids is destroyed and anaphase separation commences. Cohesion is reestablished in the next cell cycle as chromosomes are duplicated during S phase. Sister chromatid cohesion relies on a multisubunit complex, termed cohesin, that consists of a heterodimer of SMC proteins Smc1 and Smc3, as well as two non-SMC proteins, Scc3 and Scc1/Mcd1 (hereafter referred to as Scc1). The proteins are thought to form a ring-shaped complex with a central hole large enough for two nucleosomal fibers. According to one compelling model, cohesin embraces both sister chromatids to counter the opposing force of spindle microtubules (Gruber et al. 2003). In a competing model, sisters are “snapped” together by interacting pairs of cohesin complexes (Milutinovich and Koshland 2003). In either case, destruction of cohesin at anaphase is triggered by separase, a site-specific protease that cleaves Scc1.
Cohesin has been mapped to centromeres and discrete sites along chromosomal arms in both budding and fission yeasts (Blat and Kleckner 1999; Megee et al. 1999; Tanaka et al. 1999; Laloraya et al. 2000; Glynn et al. 2004; Lengronne et al. 2004). Rules governing binding specificity have emerged despite the absence of a clear consensus binding site. A general correlation was found between cohesin enrichment and elevated AT content in Saccharomyces cerevisiae. Regions between convergent genes were also highly enriched among bound sites (Glynn et al. 2004; Lengronne et al. 2004). That cohesin localized to these regions in a transcription-dependent manner led to the hypothesis that the complex is mobile and can be repositioned by passage of the transcriptional machinery (Lengronne et al. 2004). Cohesin is also targeted for function at specific sites. The complex associates with broad regions surrounding double-strand breaks so that post-replicative repair of sister chromatids can proceed by homologous recombination (Strom et al. 2004; Unal et al. 2004). Cohesin also associates with a specific site on the native yeast 2-μm plasmid, where it facilitates segregation of the extrachromosomal element (Yang et al. 2004). In Schizosaccharomyces pombe, cohesin associates with pericentric heterochromatin, where it facilitates the proper segregation of chromosomes (Bernard et al. 2001; Nonaka et al. 2002).
In S. cerevisiae, heterochromatin-like structures known as silent chromatin repress the expression of genes at the silenced mating-type loci and at telomeres (Rusché et al. 2003). Silent chromatin is nucleated by cis-acting silencer sequences that assemble various combinations of localized factors, including ORC (the replication origin recognition complex), Rap1, Abf1, and Sir1. Together, the bound proteins recruit a complex of factors (Sir2–4) that binds throughout the silenced domain. Spreading of the Sir2–4 complex is facilitated by Sir2, a histone deacetylase that creates preferred histone-binding sites for Sir3 and Sir4. Sir2 also acts independently of Sir3 and Sir4 to create domains of transcriptional repression at the rDNA. At the mating-type loci, Sir-mediated repression is confined from spreading into adjacent transcriptionally poised domains of the genome by cis-acting boundary elements, as well as by other mechanisms involving histone modifications, histone variants, and histone-binding proteins (Donze et al. 1999; Ladurner et al. 2003; Meneghini et al. 2003; Kimura and Horikoshi 2004; Tackett et al. 2005).
Early mapping studies of Scc1 on S. cerevisiae chromosomes found binding sites near the silent loci, apparently coinciding with the boundary elements in the case of HMR (Blat and Kleckner 1999; Laloraya et al. 2000). The function of boundary elements was compromised in some cohesin mutants, suggesting that the complex might normally act to delimit silent domains (Donze et al. 1999). Further evidence of a connection between silencing and sister chromatid cohesion arose from studies on de novo transcriptional repression. Bell and coworkers (Lau et al. 2002) found that Scc1 negatively regulates establishment of the silent state. Still further work indicated that sister cohesion plays a positive role (Suter et al. 2004). In mutants lacking CTF4, CTF18, or DCC1, all genes that link DNA replication to the establishment of cohesion, transcriptional repression of reporter genes at HMR and telomeres is compromised.
Despite evidence of a relationship between silencing and cohesin in budding yeast, there has been no demonstration that silenced loci on sister chromatids are held together. In this study, we used fluorescence microscopy and chromatin immunoprecipitation (ChIP) to monitor cohesion of HMR alleles that were uncoupled from chromosomes at mitosis by site-specific recombination. We found that pairs of excised chromatin circles were associated with one another by a process that required both cohesin and Sir-mediated repression at HMR. Inhibition of silencing in cells with sister chromatids disrupted cohesion, prompting loss of the complex from unexcised chromosomal templates but not from excised circles. The results suggest that silent chromatin targets and maintains cohesin function by a mechanism that involves topological linkage of the ring-shaped complex to individual chromatids.
Results
Experimental rationale and design
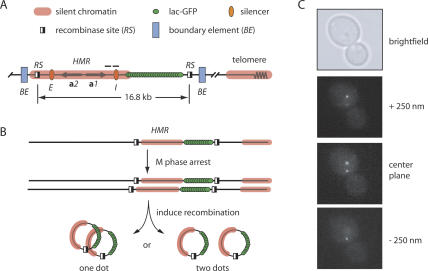
Studies of cohesion often rely on direct visualization of sister chromatids that are illuminated either by GFP-tagged bacterial repressors or by FISH probes. The approaches are well suited for characterization of global regulators of cohesion but not modifiers of cohesion at specific loci. In the latter case, loss of cohesion at a specific site will likely be masked by the persistence of cohesion at neighboring sites (Megee and Koshland 1999). While studying the localization of HMR on newly replicated chromosomes, we were concerned that cohesion along the arm of chromosome III would mask potential interactions between the silenced loci of sister chromatids. To overcome this constraint, we used strains in which the entire HMR locus could be uncoupled from the remainder of the chromosome by site-specific recombination (Gartenberg et al. 2004). Target sites for the R recombinase of Zygosaccharomyces rouxii (RS sites) were placed at positions that bracketed the E and I silencers (Fig. 1A). Flanking chromosomal sequences that possess defined boundary element activity did not lie between the recombinase sites (Donze et al. 1999). In this way, HMR could be uncoupled from the boundary elements and other neighboring sequences that might independently promote cohesion.
Figure 1.
General methodology. (A) A schematic diagram of the GFP-tagged and excisable HMR locus. RS sites were situated to include the a1 and a2 mating-type genes and both silencers in the excised circle yet exclude functionally defined boundary elements. The silent domain at HMR most likely extends beyond the representative translucent red line used to represent it in this figure. Horizontal bars denote regions examined by ChIP in Figure 5. (B) Flow chart of the general experimental procedure. (C) Three successive fluorescent images from a representative z-stack (each separated by 250 nm), as well as a bright-field image. Fixed cells rather than live cells were used to avoid counting highly mobile loci more than once. Microscopy with live cells, nevertheless, yielded similar results (data not shown).
HMR circles from sister chromatids were visualized by the lac-GFP tagging approach (Straight et al. 1996). An array of lac operators (lacop) was inserted between the HMR-I silencer and the adjacent RS site in strains that expressed lac-GFP constitutively (Gartenberg et al. 2004). Cells with sister chromatids were arrested in M phase by treatment with the benzimidazole microtubule inhibitors nocodazole and benomyl (see Materials and Methods for details). Induction of a GAL1-recombinase gene fusion in arrested cells produced a pair of 16.8-kb circles that contained the a mating-type genes, both E and I silencers, and the lacop array (Fig. 1B). Southern blot analysis showed that the reaction proceeded to roughly 80% completion under these experimental conditions (data not shown), and Northern blot analysis showed that the excised locus remained transcriptionally repressed (Gartenberg et al. 2004; see below). After fixation with paraformaldehyde, serial fluorescent images were obtained along the vertical axis (z-stacks) and GFP foci were counted (e.g., see Fig, 1C). Pairs of excised HMR circles that were associated produced one bright fluorescent dot, whereas two dots of slightly weaker intensity were observed if the circles dissociated.
HMR loci on sister chromatids colocalize in a Sir-dependent manner
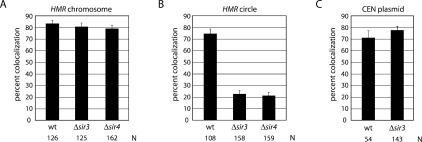
Preliminary experiments were performed without induced recombination to test whether chromosomal HMR loci on sister chromatids were associated during M-phase arrest. Consistent with findings for sites on chromosomal arms (Guacci et al. 1997; Michaelis et al. 1997), the two fluorescent signals coalesced in 83% of the cells, while the remaining 17% of the cell population contained two dots (Fig. 2A). Nearly identical values were obtained with a congenic strain that lacked the SIR3 gene. This result demonstrates that Sir3 is not a general mediator of chromosome arm cohesion, at least near the domain containing HMR. When the experiment was repeated under conditions that excised HMR, the fluorescent signals corresponding to the pair of chromatin circles colocalized in 74% of the cells (Fig. 2B). These results indicate that linkage to the chromosome is not required for association of HMR loci. Strikingly, the percentage of colocalized circles dropped to 22% when the SIR3 gene was deleted. A Δsir4 strain, like the Δsir3 strain, yielded unpaired circles in the vast majority of the cells examined (only 21% remained colocalized) (Fig. 2A). The fluorescent signals were often but not always well separated, presumably a consequence of microtubule-independent diffusion of chromatin circles before fixation (Gartenberg et al. 2004). We conclude that colocalization of HMR is mediated by a Sir-dependent mechanism. Cohesion of neighboring regions of the chromosome, on the other hand, persists in the absence of silencing.
Figure 2.
Sir-mediated repression is required for colocalization of HMR loci from sister chromatids. (A) Colocalization of GFP-tagged chromosomal HMR loci in M-phase-arrested cells. The percentages of colocalized (one dot) loci are reported with error bars representing the standard error of proportion (see Materials and Methods for description of error and statistical analyses). Strains CRC52 (wt), CRC54 (Δsir3), and CSW20 (Δsir4) were used. (N) Number of cells examined. (B) Colocalization of GFP-tagged HMR circles. (C) Colocalization of TetGFP-tagged CEN plasmid pPCM14tetO (Megee and Koshland 1999) in strains PMY127 (wt) and CRC60 (Δsir3). There is no significant difference between values obtained for (1) silent HMR before or after excision, (2) nonsilent circles in Δsir3 and Δsir4 strains, or (3) plasmid pPCM14tetO in wild-type and Δsir3 strains.
Sir1 was recently found to be a functional component of centromeric chromatin (Sharp et al. 2003). This discovery raised the possibility that the protein acts in concert with the Sir2–4 complex to establish cohesion of sister centromeres. Therefore, we measured the influence of SIR3 on the pairing of a TetGFP-tagged centromeric plasmid. Centromere loss from the plasmid was previously shown to cause a fourfold decrease in cohesion (Megee and Koshland 1999). As shown in Figure 2C, sister centromeric plasmids colocalized in roughly 70% of the cells, irrespective of whether SIR3 was deleted or not. We conclude that the Sir2–4 complex does not contribute to centromeric cohesion. The finding is consistent with the inability to detect Sir2–4 at centromeres by ChIP (Sharp et al. 2003).
Silent chromatin is required in cis for HMR colocalization
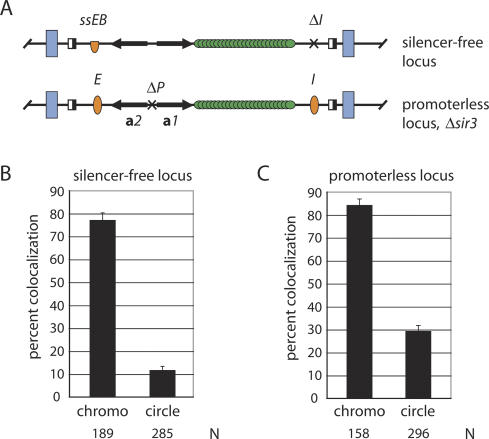
The Sir proteins control transcription at a number of loci, including many genes found at telomeres. To test whether pairing of HMR requires that the proteins act in cis, we generated a silencing competent strain that lacked Sir function at HMR. For this purpose, the I silencer was omitted and the E silencer was replaced with a nonfunctional derivative (Fig. 3A; Li et al. 2001). As shown in Figure 3B, the HMR alleles remained closely associated when embedded in the chromosome (77% of the cells contained single dots). After excision, however, the percentage of cells with paired circles dropped to 11%. Thus, the loss of HMR pairing in sir mutants cannot be attributed to derepression of an unlinked locus. Silent chromatin is required in cis for the colocalization of HMR.
Figure 3.
Colocalization of modified HMR alleles. (A) Schematic diagrams of HMR loci. The ssEB silencer (half oval) contains synthetic Rap1 and Abf1 sites and lexA operators that replace the ORC-binding site. Both the ssEB silencer and the ΔI silencer deletion are identical to ones used in Li et al. (2001). The ΔP deletion of the a gene promoters is identical to the one used in Cheng and Gartenberg (2000). In both constructs, the lacop array was inserted immediately downstream of the a1 gene. (B) Colocalization of an HMR locus lacking functional silencers in strain CSW18. (C) Colocalization of the promoterless locus in strain YH02 (Δsir3). There is no significant difference between values for a promoter-containing CRC81 (Δsir3) strain (data not shown) and the promoterless YH02 (Δsir3) strain.
Inactivation of Sir proteins or elimination of silencers at HMR results in expression of the resident a1 and a2 mating-type genes. In our experiments, dissociation of HMR circles could result from either loss of silent chromatin or gain of transcription of these genes. To distinguish between these scenarios, we deleted the divergent a1 and a2 promoters from HMR in a strain that lacked SIR3 (Fig. 3A). A functional test confirmed that the deletion blocked a1 gene expression (Y. Hom and M.R. Gartenberg, unpubl.). Figure 3C shows that pairing was not restored in HMR circles that lacked the a gene promoters. We infer that silent chromatin, not the absence of transcription, is responsible for colocalization of HMR loci.
HMR colocalization requires cohesin
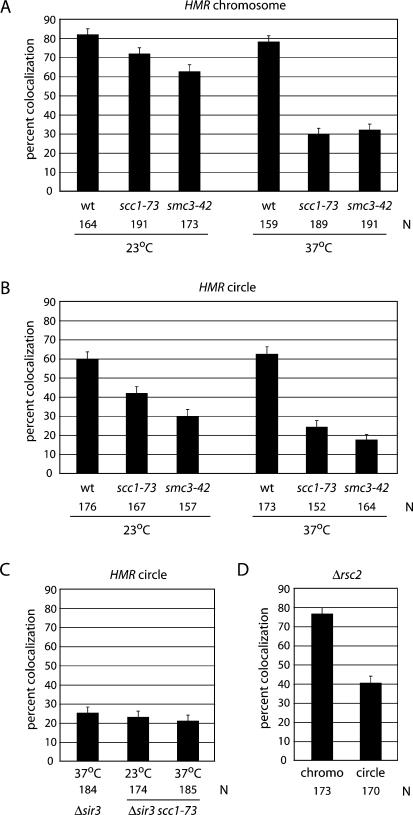
Recent mapping studies using high-resolution oligonucleotide tiled arrays found cohesin within HMR, as well as in adjoining sequences (Lengronne et al. 2004). To determine whether cohesin participates in pairing HMR loci, we introduced to our strain a temperature-sensitive allele of the gene encoding Scc1 (scc1-73). In the absence of excision, incubation at nonpermissive temperature caused a decrease in the percentage of cells with single dots from 71% to 29% (Fig. 4A). In contrast, little change was seen in the wild-type counterpart upon temperature shift. This behavior parallels that of other GFP-tagged domains when essential cohesin subunits are inactivated (Guacci et al. 1997; Michaelis et al. 1997). That HMR loci in the context of unrecombined sister chromatids do not remain associated at the elevated temperature suggests that cohesin pairs the silent domains. To eliminate the influence of neighboring chromosomal sequences, we repeated the experiment with excised HMR circles. In this case, M-phase-arrested cells were incubated in galactose for 2 h at permissive temperature followed by an additional 2 h at nonpermissive temperature. At the end of this extended cell cycle arrest, the majority of circles in the wild-type (wt) strain were still paired (62%), whereas only 24% of the mutant cells contained paired HMR circles. Similar results were obtained when a different cohesin subunit (Smc3) was inactivated conditionally (Fig. 4A,B). Moreover, the defect in pairing HMR circles was not increased in a sir3 scc1 double mutant relative to either single mutant (Fig. 4B,C). This indicates that both SIR3 and SCC1 act in the same pathway. Colocalization of HMR circles was also diminished at the permissive temperature for the cohesin mutants (Fig. 4B). In the background of one SCC1 allele (mcd1-1), pairing of the circles was almost entirely absent (data not shown). Taken together, the results indicate that both Scc1 and Smc3 act together as part of the cohesin complex to cohese silenced copies of HMR, whether the loci remain within sister chromatids or whether they are converted to extrachromosomal circles.
Figure 4.
Pairing of HMR loci requires the cohesin complex. (A) Colocalization of chromosomal HMR requires cohesin. Cultures were pregrown in raffinose at 23°C. Following the addition of dextrose, half of the culture was maintained at 23°C and half was shifted to the nonpermissive temperature (37°C) for 2 h. Strains CRC52 (wt), CRC83 (scc1-73), and CRC85 (smc3-42) were used. (B) Colocalization of HMR circles requires cohesin. Two hours after the addition of galactose, half of the culture was maintained at 23°C and the other half was shifted to 37°C for an additional 2 h. The percentage colocalization of chromosomal and excised loci in wild-type strains was not influenced significantly by temperature. However, temperature shifts caused significant differences in the mutant strains (p < 0.01). Differences between wild-type and mutant strains were also statistically significant at both 23°C (p < 0.05) and 37°C (p < 0.01). (C) Colocalization of HMR circles is not decreased in a Δsir3 scc1-73 double mutant. There is no significant difference between colocalization percentages for CRC54 (Δsir3), CRC95 (Δsir3 scc1-73), and CRC83 (scc1-73) (see B). Cultures were grown as in B. (D) The RSC chromatin-remodeling complex contributes to cohesion of HMR. Strain CRC66 (Δrsc2) exhibited a slight growth defect when grown at 30°C. Colocalization of circles in the wild type (CRC52) (Fig. 2) and rsc2 mutant (CRC66) was significantly different (p < 0.01), whereas colocalization of unexcised loci was not.
The RSC chromatin-remodeling complex has been linked to sister chromatid cohesion (Baetz et al. 2004; Huang et al. 2004; Yang et al. 2004). Genetic interactions between RSC and the cohesion pathway have been identified, RSC associates with cohesin-binding sites, and the complex was shown to support functional cohesion of the 2-μm plasmid and the arms of chromosome V. The exact role of RSC in cohesion, however, is still debated (Riedel et al. 2004). We tested whether RSC was involved in cohesion of HMR circles by repeating our assay in the absence of the nonessential subunit Rsc2. Whereas deletion of the rsc2 gene had little effect on cohesion of HMR in a chromosomal context (76% vs. 83% in wild type) (cf. Figs. 2A and 4D), the percentage of cells with paired HMR circles dropped to 40%, a value that is significantly different from that seen in a wild-type strain (p < 0.01). Based on the selective influence of RSC2 on the excised locus, we conclude the remodeling complex facilitates cohesion of HMR.
Silent chromatin at HMR is anchored to the nuclear periphery via two parallel pathways defined by yKu and Esc1 (Gartenberg et al. 2004). It seemed possible that colocalization of HMR circles by cohesin might also involve attachment of both circles to a common perinuclear docking site. Elimination of both Esc1 and Ku, however, did not alter the pairing of HMR loci, indicating that cohesion and anchorage of silent chromatin are independent events (Supplementary Fig. S1).
Silent chromatin targets cohesin to HMR
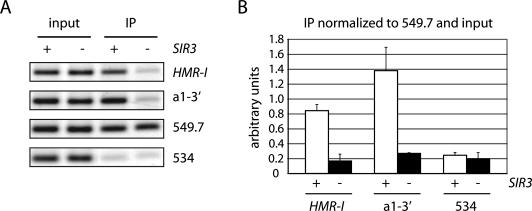
ChIP was used to investigate the binding of cohesin at HMR. A TAP (tandem affinity purification) tag was appended to SCC1, and IgG sepharose was used to precipitate formaldehyde cross-linked chromatin from M-phase-arrested cells. For simplicity, unexcised chromosomal loci were used exclusively in this set of experiments. The PCR analysis in Figure 5A revealed that a fragment containing the HMR-I silencer was present in the immunoprecipitated material but that a known cohesin-free control on chromosome V (designated 534) (Tanaka et al. 1999) was absent. When the experiment was repeated with a silencing-deficient Δsir3 strain, HMR-I was no longer precipitated. Sir-dependent Scc1 recruitment was also detected at a second HMR region between the a1 coding region and the HMR-I silencer (Fig. 5), as well as within the a1 open reading frame (data not shown). These results indicate that cohesin binding is not restricted to silencers. Quantitative comparison of both HMR fragments and the 534 control to a well-characterized cohesin-binding site on chromosome V (designated 549.7) confirmed that binding of Scc1 to HMR required Sir3 (Fig. 5B). We conclude that cohesin is targeted specifically to silenced loci by silent chromatin. Colocalization of HMR in the experiments described above can most easily be explained by the direct action of cohesin at the locus.
Figure 5.
Silent chromatin targets Scc1 to HMR. (A) ChIP of TAP-tagged Scc1 in strains CRC67 (SCC1-TAP) and CRC68 (SCC1-TAP Δsir3). Primer pairs amplified a region containing the I silencer of HMR (HMR-I) and an adjacent region immediately downstream of a1 (a1-3′, refer to Fig. 1). Chromosome V regions 549. 7 and 534 served as controls (Tanaka et al. 1999). Cultures grown in dextrose were exposed to nocodazole for 3 h. (B) Quantitative analysis. HMR-I, a1-3′, and 534 signals in IP samples were measured relative to 549.7 and normalized to the same ratios in the input samples. Plotted data represent the mean and standard deviation of three independent trials.
Cohesion is not required for maintenance of transcriptional silencing
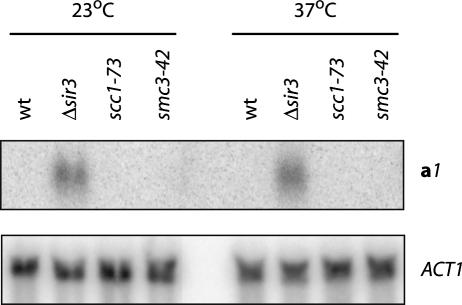
Northern blot analysis was used to measure transcriptional silencing in strains with conditional cohesion defects. Expression of the circle-borne a1 mating-type gene was examined in cultures retained at the permissive temperature or those that had been shifted to the nonpermissive temperature after the addition of galactose. No a1 transcript was found in the scc1-73 or smc3-42 mutants at either temperature, indicating that silencing was maintained in the extrachromosomal templates after cohesion was eliminated (Fig. 6). Silencing was also maintained when HMR cohesion was disrupted by loss of Rsc2 (data not shown). Together, these results show that sister chromatid cohesion is not required for maintenance of transcriptional silencing.
Figure 6.
Maintenance of silencing in HMR circles in the absence of cohesin. Two hours after the addition of galactose to produce chromatin circles, half of each culture was maintained at 23°C, whereas the other half was shifted to 37°C for 2 h. Strains CRC76 (Δmat), CRC81 (Δmat Δsir3), CRC83 (MATα scc1-73), and CSW21 (Δmat smc3-42) were used. The a1 gene and ACT1 control were probed sequentially.
Silencing is required for maintenance of cohesion
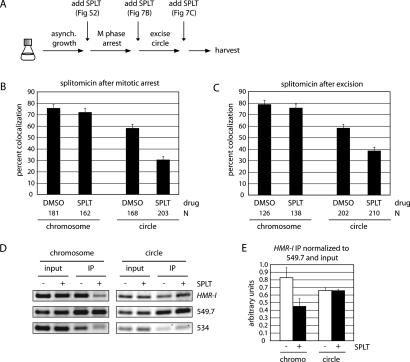
The preceding experiments uncovered a role for silent chromatin in targeting sister chromatid cohesion to HMR. We next asked whether silent chromatin also helps maintain cohesion of the locus. To address this question, we inactivated silencing in cells with cohesed sister chromatids. In the first of two approaches, we antagonized silencing with splitomicin (SPLT), a small molecule inhibitor of the Sir2 histone deacetylase activity (Bedalov et al. 2001). Preliminary experiments showed that the drug inhibits silencing of HMR circles in our strains (Supplementary Fig. S2A). Splitomicin also interferes with cohesion of HMR circles (but not the chromosomal arm), when added to asynchronously growing cultures (Fig. 7A; Supplementary Fig. S2B). Thus, Sir2 deacetylation activity, along with Sir3 and Sir4, promote cohesion of HMR. Splitomicin was next added to cells arrested in M phase with nocodazole. Galactose was added simultaneously in this experiment to evaluate cohesion of HMR (Fig. 7A,B). After 4 h, only 30% of the splitomicin-treated cells contained paired HMR circles versus 57% when treated with solvent (DMSO) alone. This result indicated that inhibition of silencing impairs pre-existing cohesion of HMR loci. Importantly, similar results were obtained when splitomicin was added after HMR had been excised. In Figure 7C, 38% of splitomicin-treated cells contained colocalized HMR circles versus 58% when treated with the solvent alone. Therefore, inhibition of Sir2 activity interferes with cohesion, even after the HMR locus has been converted to a covalently closed circle of chromatin. Taken together, these results demonstrate that silencing acts continuously to maintain cohesion of silent chromatin.
Figure 7.
Loss of HMR cohesion in cultures treated with a Sir2-deacetylase inhibitor. (A) Flow chart of the experimental procedures. (SPLT) Splitomicin. (B) Colocalization of HMR. Splitomicin (or DMSO) was added simultaneously with benomyl and galactose (or dextrose) to nocodazole-arrested cultures of CRC52. Images were collected 4 h later. (C) Colocalization of HMR. Splitomicin (or DMSO) was added 2 h after benomyl and galactose (or dextrose). Images were collected 2 h later. In each experiment, the difference in values for HMR circle colocalization in SPLT- and DMSO-treated cells was significant (p < 0.01), whereas the difference in values for unexcised loci was not. (D) ChIP of Scc1 in strain CRC67 (SCC1-TAP). Cultures were grown as in C. (E) Quantitative analysis is as in Figure 5B.
ChIP was used to determine the impact of splitomicin on cohesin bound at HMR. The behaviors of both chromosomal and excised loci were examined by adding the drug to M-phase-arrested cells that had or had not undergone recombination. In the case of the nonexcised locus, treatment with splitomicin resulted in loss of Scc1 from the HMR-I region (Fig. 7D). The protein was also diminished from the a1-3′ region and the a1 open-reading frame (data not shown). Thus, silencing acts continuously to maintain cohesin at HMR when it is embedded within a chromosome. The finding is consistent with absence of Scc1 at HMR in Δsir3 cells (Fig. 5). Strikingly, different results were obtained when splitomicin was added after excision. In this case, cohesin remained bound to the circle (Fig. 7D). The most recognizable distinction between the excised and unexcised loci is a topological one. Excised circles lack DNA ends and are topologically closed, whereas yeast chromosomes are linear and topologically open. The results suggest that excised circles retain cohesin selectively when silencing is lost because the ring-shaped protein complex is topologically trapped.
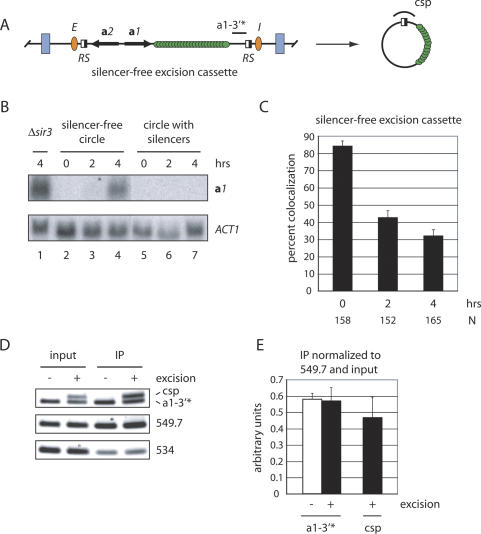
To explore this notion further, we used an alternative approach to inactive silencing. Previously, we showed that silencers act continuously in cis to maintain silencing. When the mating-type genes were uncoupled from neighboring silencers by site-specific recombination, transcription was restored, even in nocodazole-arrested cells (Cheng and Gartenberg 2000). Here, we adapted this approach by repositioning the lacop array and RS sites within HMR such that induced recombination produced a lacGFP-tagged circle that contained the mating-type genes but that lacked E and I (Fig. 8A). This strain is fundamentally different from the one in Figure 3A in which functional silencers were completely absent from the chromosomal locus. Northern blot analysis of cells arrested in mitosis showed that the a1 gene in this construct was repressed when present on the chromosome but that transcripts ultimately reappeared after excision (Fig. 8B).
Figure 8.
Cohesion of HMR is lost when silencers are removed after M-phase arrest. (A) Schematic diagram of the HMR excision cassette that produces silencer-free circles. Note the position of RS sites relative to silencers. a1-3′⋆ and csp designate PCR-amplified regions for ChIP. (B) Northern blot analysis of a1 mRNA. The MATa locus was either deleted or replaced with MATα, leaving the a1 gene at HMR as the sole cellular copy. ACT1 mRNA serves as an internal loading control. The time interval after the addition of galactose is labeled above each lane. (Lanes 1) Strain CRC81 (circle with silencers, Δsir3 Δmat). (Lanes 2–4) Strain CSW22 (silencer-free circle, MATα). (Lanes 5–7) Strain CRC76 (circle with silencers, Δmat). (C) Colocalization of HMR circles that lack silencers. Strain CSW22 was used. The difference between values from the 2-h and 4-h time points is significant (p < 0.05). (D) ChIP of Scc1 in strain CSW34 (SCC1-TAP). Benomyl and galactose (or dextrose) were added to nocodazole-arrested cells for 2 h prior to formaldehyde treatment. a1-3′⋆ primers amplify both excised and unexcised loci, whereas csp primers amplify only the excised circle. (E) Quantitative analysis is as in Figure 5B.
Fluorescence microscopy showed that the unexcised HMR loci in this strain were held together in 84% of the cells (Fig. 8C). Following excision, however, silencer-free circles were paired in only 42% of the cells 2 h after galactose addition and this value dropped to 32% 2 h later. In contrast, 72% of silencer-bearing circles were held together 2 h after galactose addition (Fig. 2B), and roughly 60% remained cohesed 2 h later (see Fig. 4B [at 23°C or 37°C] or Fig. 7B,C [DMSO]). These data show that cohesion is not maintained in circles that cannot maintain silencing.
ChIP showed that cohesin was bound to this excision cassette equally well before and after excision (Fig. 8D). One set of primers amplified a region (a1-3′⋆) immediately upstream of the RS site adjacent to HMR-I (nearly identical to the a1-3′ region tested in Fig. 5). Similar results were obtained with a primer pair spanning the a1 open reading frame (data not shown). These findings rule out the possibility that silencer-free circles fail to cohere because they lack cohesin. To be certain that the positive ChIP signal was not due to a subpopulation of unexcised templates, the chromatin precipitates were probed again with circle-specific PCR (csp) using primers that flank the single RS site of the circle (Fig. 8A). These primers confirmed that Scc1 remained bound after excision (Fig. 8D). Repetition of the experiment in a Δsir4 strain showed that binding of Scc1 was Sir-dependent (Supplementary Fig. S3), indicating that the process of induced recombination was not responsible for loading cohesin to the chromatin circle. We conclude that silencing must be maintained to preserve cohesin function. However, retention of cohesin after circularization of the template is silencing-independent.
Discussion
Silent chromatin-dependent cohesion of HMR
The specificity of most noncentromeric cohesin-binding sites has been attributed to transcription. Passage of RNA polymerase is thought to reposition the cohesin ring to the ends of genes, resulting in accumulation of the complex where genes converge. Nevertheless, cohesin has been found at silent chromatin domains, like HMR, where transcription and other DNA transactions are hindered or absent. Using a functional assay for pairing of discrete chromosomal elements, we show that cohesion of HMR is mediated specifically by silent chromatin. Without Sir3, Sir4, or the deacetylase activity of Sir2, chromatin circles bearing the locus dissociated. The cytological findings were supported by ChIP experiments that showed that cohesin is bound at HMR in a Sir-dependent manner. The role of the Sir proteins in cohesin recruitment is specific to silent chromatin since the silencing factors did not influence the binding or function of cohesin at other well-characterized sites. Indeed, uncoupling HMR from the remainder of the chromosome was necessary to visualize Sir-dependent cohesion of the locus. Together, these results indicate that retention of cohesin by silent chromatin results in cohesion of HMR. It is likely that similar events occur at other Sir-silenced loci.
We postulate that there exists a direct interaction between components of the silencing and cohesion pathways. Such an interaction could facilitate capture of preformed complexes that migrate from other chromosomal positions or promote the de novo loading of cohesin. Accumulation of cohesin at HMR is not due simply to the absence of transcription. Otherwise, deletion of the mating-type gene promoters would have restored cohesion in silencing-deficient strains. However, we have not yet excluded the possibility that undocumented transcription in the region serves this purpose.
In addition to critical roles for Sir proteins and cohesin, our work also found that cohesion of HMR required Rsc2 of the RSC chromatin-remodeling complex. The effect was localized (not global) because pairing of the remainder of the chromosome arm was Rsc2-independent. At present we do not know whether RSC acts directly at silent chromatin or through some other indirect mechanism. However, we note that RSC binds tRNA genes, promotes their expression (Ng et al. 2002), and that tRNA transcription and cohesin contribute to demarcation of the silent chromatin domain at HMR (Donze and Kamakaka 2001). Considering the recent ties between RSC and cohesion (Baetz et al. 2004; Huang et al. 2004; Yang et al. 2004), it is tempting to speculate that RSC acts at the juncture between silent and nonsilent domains to impart cohesion of HMR.
Silencing-mediated cohesion at other yeast loci
Sir2 was recently shown to mediate Scc1 binding to the rDNA by an indirect mechanism that appears to involve transcriptional suppression (Kobayashi et al. 2004; Kobayashi and Ganley 2005). At the rDNA array, cohesin enhances genomic stability by preventing unequal sister chromatid exchange, possibly by maintaining proper register of tandemly repeated rDNA units. Silent chromatin-mediated recruitment of cohesin might serve a similar function at the quiescent mating-type loci where post-replicative recombination between homologous sequences at HMR, HML and MAT could lead to unprogrammed mating-type switching or lethal chromosomal deletions. Such rearrangements have been observed in cohesion mutants (Warren et al. 2004). In S. pombe, impairment of cohesin also leads to genomic rearrangements involving the heterochromatic mating-type loci (Nonaka et al. 2002).
In fission yeast, cohesin is recruited to heterochromatin by an interaction between the Scc3 cohesin subunit (designated Psc3 in S. pombe) and Swi6, a fundamental heterochromatin component homologous to mammalian HP1 (Nonaka et al. 2002). In strains lacking Swi6 (or other factors required for heterochromatin function), chromosomes lag on the elongating anaphase spindle (Bernard et al. 2001; Nonaka et al. 2002; Bailis et al. 2003). Budding yeast does not possess an HP1 homolog, and transcriptional silencing, a hallmark of heterochromatin, has not been found at centromeres. If cohesion of budding yeast centromeres requires distinct chromatin domains (Bloom and Carbon 1982), the typical Sir2–4 pathway does not appear to participate.
Silencing and sister chromatid cohesion
The simplest explanation for Sir-mediated cohesion is that the cohesin complex has been co-opted from its essential role in chromosome segregation for a secondary role in transcriptional silencing. The exact functional relationship between cohesin and silent chromatin, however, is not yet clear. Our own work shows that the silencing persists after cohesin inactivation, indicating that maintenance of the silent state does not require that sister chromatids be held together. Instead, the existing body of evidence indicates that cohesin influences establishment of the silent state. In assays that measure de novo transcriptional repression, Scc1 was shown to hinder silencing until anaphase when the protein is destroyed (Lau et al. 2002). Artificial depletion of Scc1 allowed silencing to occur earlier. These regulatory functions appear to be important because silencing defects emerge when the replication-coupled deposition of cohesion is impaired (Suter et al. 2004). As suggested by Suter et al., uncontrolled establishment and spreading of silent chromatin may create a situation in which the effective concentration of a silencing factor becomes too limiting to form silencing-competent chromatin. Indeed, silencing defects have been seen in the absence of Dot1, a histone methyltransferase, and the H2A variant Htz1, both of which ordinarily create chromatin states that are incompatible with silencing elsewhere in the genome (van Leeuwen et al. 2002; Meneghini et al. 2003).
Silent chromatin, maintenance of cohesion, and the protein ring model
The proposition that cohesin binds sister chromatids topologically by forming a protein ring is attractive, in part, because it provides a basis for how the complex functions in an equivalent manner at different locations throughout the genome. Encircling chromatin rather than binding DNA directly avoids the need for sequence-specific association. A stable ring would be expected to slide along DNA and not dissociate until encountering a DNA end. Indeed, recent biochemical experiments showed that cohesin was released from circular minichromosomes by DNA linearization (Ivanov and Nasmyth 2005). Our site-specific recombination approach combined with conditional silencing tools afforded us the opportunity to test predictions of the protein ring model in vivo, at least in the context of silent chromatin. If cohesin binds DNA topologically, then its persistence on a given DNA segment should depend on whether that DNA is linear or circular. We reasoned that conditional inactivation of silencing in cells with sister chromatids might permit mobilization of the complex from HMR since the locus was found to be cohesin-free in a sir-null mutant. In our experiments, this was achieved by treating M-phase-arrested cells with an inhibitor of Sir2. We found that cohesin was diminished at HMR when it resided within a linear chromosome but that it persisted at the locus when it was excised and converted to a covalently closed circle. The results support a topological model for cohesin binding to DNA. We envision that the ring-shaped complex remains trapped by catenation and cannot slide away when HMR is circularized.
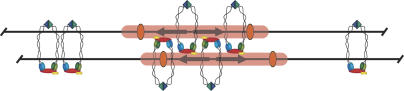
A second prediction of the protein ring model is that circularization of sister chromatid DNA segments should trap cohesin complexes that embrace both DNA duplexes. We expected that cohesion of HMR circles by trapped cohesin rings would persist following the inactivation of silencing. This was not observed. When Sir2 was inhibited or when silencers were omitted, HMR circles retained cohesin but failed to remain cohesed. To accommodate the protein ring model, we stipulate that silenced loci represent exceptions where cohesin does not embrace both sister chromatids. The loading of Sir proteins on nucleosomal templates, as well as the prospect of folding them into higher-order 30-nm fibers, may generate chromatids that are too large for both to fit within a ring. A different mode of cohesin binding, one in which sister chromatids are bridged by complexes that embrace individual chromatids, may have evolved to accommodate these specialized chromatin domains (Fig. 9). Conceptually, silent chromatin could mediate bridges by interacting with cohesin complexes bound to the opposing chromatid. An alternative model contends that cohesin embraces both sister chromatids as a ring but one that dissociates at a slow but measurable rate. In this model, silent chromatin stabilizes cohesin by slowing the dissociation rate. The paucity of evidence for disassembly of functioning cohesin by dissociation, however, makes this alternative seem unlikely (Haering et al. 2004).
Figure 9.
A model for distinct binding modes of cohesin in the chromosomal region containing HMR. Among silent domains, cohesin associates by encircling individual chromatids but not both sisters. In this representation, silent chromatin mediates bridging by interacting with cohesin bound to opposing chromatids. Other modes of Sir-stabilization are possible. Silencing-independent cohesin rings are shown to either side of HMR, binding chromatids together by embracing both DNA duplexes.
Cohesion of heterochromatin in higher eukaryotes
Prolonged pairing of the heterochromatic domains within sister chromatids has been noted in higher eukaryotes, where cohesion of chromosome arms but not centromeres is removed at prophase. In lymphocytes, heritable silencing of several developmentally regulated genes delays their separation into spatially resolved chromatids following DNA replication (Azuara et al. 2003). Similarly, during mitosis, the major satellite repeats of mouse pericentric heterochromatin were found to be the last centromeric sequences to separate (Guenatri et al. 2004). Importantly, this delay was lost in the absence of Suv39h, a histone methyltransferase required for HP1 recruitment to heterochromatin. How heterochromatin and cohesin cooperate to pair domains in higher eukaryotes and how pairing contributes to chromosome function will be of considerable future interest.
Materials and methods
Strain construction
All strains are listed in Table 1. Those used for microscopy contain an integrated copy of pGVH60, a lac-GFP (S65T) expression vector (provided by G. Van Houwe and S.M. Gasser, University of Geneva, Geneva, Switzerland) that lacks the repressor's tetramerization domain (Straight et al. 1996). CRC52 was derived from MRG2201 by first reconstituting the ade2-1 locus and then integrating pGVH60. Growth of CRC52 on 5-FOA yielded CRC54 (Δsir3::HIS3) or CSW10 (SIR3). Null mutants were obtained by PCR-mediated gene replacement using one of the following markers: kanMX, hphMX, natMX, or the Kluyveromyces lactis URA3. TAP-tagged SCC1 (OpenBiosystems, Inc) was transferred to CRC52 by PCR-mediated gene replacement after converting the adjacent HIS3MX marker to natMX. Strain CSW18 was obtained in two steps by first crossing CRC52 with YCL49 to generate segregants that contained an HMR locus that lacked silencers. In the second step, a 256 lacop array was integrated downstream of the a1 gene using pAFS52-lPCR, a derivative of pAFS52 (Straight et al. 1996). The same plasmid was used to modify HMR in strain CSW22, which was derived from THC23 by integrating both pRINT (a GAL1-recombinase R expression vector) (Raghuraman et al. 1997) and pGVH60, as well as crossing with a congenic strain to acquire SIR3 and MATα. Strains CRC83 and CRC85 are segregants from crosses between a CRC52 derivative and strains K5832 and K5824, respectively. To generate strain YH02, the pop-in/pop-out gene replacement technique was used to substitute the divergent a genes of HMR in strain CRC54 with promoterless versions (Cheng et al. 1998). All genomic modifications were confirmed by PCR, Southern blotting, and/or functional tests. Microscopy was used to verify that G1-phase cells of each strain contained only one fluorescent dot.
Table 1.
Yeast strains
| Strain | Genotype | Reference |
|---|---|---|
| W303-1A | MATaade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 | Thomas and Rothstein 1989 |
| K5824 | W303-1A smc3-42 psi+ | Michaelis et al. 1997 |
| K5832 | W303-1A scc1-73 psi+ | Michaelis et al. 1997 |
| MRG2201 | W303-1A RS :: HMRE-a2a1-HMRI-TRP1-256lacop :: RS (LEU2 :: GAL1-R)2 :: leu2-3,112 SIR3 :: URA3 :: Δsir3 :: HIS3 ADE2 :: HIS3P-lacGFP/URA3P-tetGFP :: ade2-1 | Gartenberg et al. 2004 |
| THC23 | W303-1A HMRE :: RS-a2a1-RS :: HMRI Δsir3 :: HIS3 | Cheng et al. 1998 |
| YCL49 | W303-1A MATα RS :: 6lexopssEB-a2a1-ΔHMRI :: RS Δlys2 Δbar1 :: hisG Δsir1 :: loxP-kanMX-loxP | Li et al. 2001 |
| PMY127 | W303-1A his3-11,15 :: (GAL-R :: HIS3)n LEU2 :: tetR-GFP :: leu2-3,112 bar1 | Megee and Koshland 1999 |
| CRC52 | MRG2201 ADE2 :: HIS3P-lacGFP :: ade2-1 | This study |
| CRC54 | CRC52 Δsir3 :: HIS3 | This study |
| CRC60 | PMY127 Δsir3 :: kanMX | This study |
| CRC66 | CRC52 Δrsc2 :: kanMX | This study |
| CRC67 | CRC52 SCC1-TAP :: natMX | This study |
| CRC68 | CRC54 SCC1-TAP :: natMX | This study |
| CRC76 | CRC52 Δmat :: natMX | This study |
| CRC81 | CRC54 Δmat :: natMX | This study |
| CRC83 | CRC52 MATα scc1-73 SIR3 | This study |
| CRC85 | CRC52 smc3-42 SIR3 | This study |
| CRC95 | CRC83 Δsir3 :: kanMX | This study |
| CSW10 | CRC52 SIR3 | This study |
| CSW18 | CSW10 RS :: 6lexopssEB-a2a1-256lacop-TRP1-ΔI :: RS | This study |
| CSW20 | CSW10 Δsir4 :: kanMX | This study |
| CSW21 | CRC85 Δmat :: natMX | This study |
| CSW22 | THC23 MATα HMRE :: RS-a2a1-256lacop-TRP1-RS :: HMRI ADE2 :: HIS3P-lacGFP :: ade2-1 (LEU2 :: GAL1-R)4 :: leu2-3,112 | This study |
| CSW34 | CSW22 SCC1-TAP :: HIS3MX | This study |
| CSW35 | CSW34 Δsir4 :: kanMX | This study |
| YH02 | CRC81 RS :: HMRE-a2a1Δp-HMRI-TRP1-256lacop :: RS | This study |
Cell growth, fixation, and microscopy
Freshly streaked cells were grown in SC-trp media containing 2% dextrose for ∼8 h before diluting 1/200 into YPA media + 2% raffinose for overnight growth. When cultures reached an OD of 0.2, nocodazole (1 mg/mL in DMSO) was added to a final concentration of 10 μg/mL. After 3 h, roughly 90% of the cells had adopted a dumbbell-shaped appearance characteristic of an M-phase arrest. The arrest was extended by benomyl (Cf = 10 μg/mL from a 10 mg/mL stock in DMSO), which was added along with galactose (Cf = 2%) to induce excision. For nonexcision controls, dextrose was added instead of galactose. After two more hours, cells were harvested by microcentrifugation at 3000 × g (see figure legends for exceptions). Splitomicin (20 mM in DMSO) was added to a final concentration of 20 μM when specified. Cell pellets were resuspended in 100 μL of 4% paraformaldehyde at 4°C for 12 min and then washed three times with a solution containing 1.2 M sorbitol and 0.1 M K2PO4 (pH 7.8). Fixation was confirmed by time-lapse microscopy, which showed the cessation of chromatin movement.
Samples were mounted on microscope slides containing 1.4% agarose plugs. Images were collected with a Zeiss Axioplan II fluorescence microscope (100× objective) and Axiocam HR camera. Z-stacks composed of 17 elevations, each separated by 250 nm and a 250-msec acquisition time, were collected on fields of 10–30 cells. Data were analyzed with Zeiss AxioVision software. Ten percent to 20% of the cells were excluded from analysis because they either lacked detectable dots, contained ambiguous dots (optical resolution in the xy plane is ∼200 nm), or contained single dots on either side of the bud neck (indicative of nuclear division between cells). Most data sets were based on three independent trials composed of 100–300 M-phase cells. Individual trials were compared with one another for homogeneity by the χ2 tests of homogeneity of proportions (Fleiss 1981). The trials were then pooled and the proportion of cells containing single spots (reported as the percentage of the total pool) was graphed with error bars indicating the standard error of proportion (Warren et al. 2004). Values of percent colocalization for different experimental conditions and strains were compared with one another for statistical significance by χ2 tests where appropriate. The results of these tests are noted in the figure legends.
ChIP
Cross-linking and immunoprecipitation were performed according to Lengronne et al. (2004) using IgG Sepharose 6 Fast Flow (Amersham). See Supplementary Table S1 for primer sequences. PCR reactions, run either individually or in multiplex, were stained with Vistra Green (Amersham), imaged by digital photography (Alpha Innotech), and found to be within the linear range (data not shown).
Supplementary Material
Acknowledgments
We thank Susan Gasser, Vincent Guacci, Paul Megee, Frank Uhlmann, and Andrew Vershon for reagents, and Nancy Walworth for use of her microscope in early parts of this work. We thank Steve Zheng and Brehon Laurent for comments on the manuscript. This work was funded by a grant from the NIH (GM51402).
Supplemental material is available at http://www.genesdev.org.
Article published online ahead of print. Article and publication date are at http://www.genesdev.org/cgi/doi/10.1101/gad.1356305.
References
- Azuara V., Brown, K.E., Williams, R.R., Webb, N., Dillon, N., Festenstein, R., Buckle, V., Merkenschlager, M., and Fisher, A.G. 2003. Heritable gene silencing in lymphocytes delays chromatid resolution without affecting the timing of DNA replication. Nat. Cell Biol. 5: 668-674. [DOI] [PubMed] [Google Scholar]
- Baetz K.K., Krogan, N.J., Emili, A., Greenblatt, J., and Hieter, P. 2004. The ctf13-30/CTF13 genomic haploinsufficiency modifier screen identifies the yeast chromatin remodeling complex RSC, which is required for the establishment of sister chromatid cohesion. Mol. Cell. Biol. 24: 1232-1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailis J.M., Bernard, P., Antonelli, R., Allshire, R.C., and Forsburg, S.L. 2003. Hsk1-Dfp1 is required for heterochromatin-mediated cohesion at centromeres. Nat. Cell Biol. 5: 1111-1116. [DOI] [PubMed] [Google Scholar]
- Bedalov A., Gatbonton, T., Irvine, W.P., Gottschling, D.E., and Simon, J.A. 2001. Identification of a small molecule inhibitor of Sir2p. Proc. Natl. Acad. Sci. 98: 15113-15118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard P., Maure, J.F., Partridge, J.F., Genier, S., Javerzat, J.P., and Allshire, R.C. 2001. Requirement of heterochromatin for cohesion at centromeres. Science 294: 2539-2542. [DOI] [PubMed] [Google Scholar]
- Blat Y. and Kleckner, N. 1999. Cohesins bind to preferential sites along yeast chromosome III, with differential regulation along arms versus the centric region. Cell 98: 249-259. [DOI] [PubMed] [Google Scholar]
- Bloom K.S. and Carbon, J. 1982. Yeast centromere DNA is in a unique and highly ordered structure in chromosomes and small circular minichromosomes. Cell 29: 305-317. [DOI] [PubMed] [Google Scholar]
- Cheng T.-H. and Gartenberg, M.R. 2000. Yeast heterochromatin is a dynamic structure that requires silencers continuously. Genes & Dev. 14: 452-463. [PMC free article] [PubMed] [Google Scholar]
- Cheng T.-H., Li, Y.-C., and Gartenberg, M.R. 1998. Persistence of an alternate chromatin structure at silenced loci in the absence of silencers. Proc Natl. Acad. Sci. 95: 5521-5526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donze D. and Kamakaka, R.T. 2001. RNA polymerase III and RNA polymerase II promoter complexes are heterochromatin barriers in Saccharomyces cerevisiae. EMBO J. 20: 520-531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donze D., Adams, C.R., Rine, J., and Kamakaka, R.T. 1999. The boundaries of the silenced HMR domain in Saccharomyces cerevisiae. Genes & Dev. 13: 698-708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleiss J.L. 1981. Statistical methods for rates and proportions. Wiley, New York.
- Gartenberg M.R., Neumann, F.N., Laroche, T., Blaszczyk, M., and Gasser, S.M. 2004. Sir-mediated repression can occur independently of chromosomal and subnuclear contexts. Cell 119: 955-967. [DOI] [PubMed] [Google Scholar]
- Glynn E.F., Megee, P.C., Yu, H.G., Mistrot, C., Unal, E., Koshland, D.E., DeRisi, J.L., and Gerton, J.L. 2004. Genome-wide mapping of the cohesin complex in the yeast Saccharomyces cerevisiae. PLoS Biol. 2: E259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber S., Haering, C.H., and Nasmyth, K. 2003. Chromosomal cohesin forms a ring. Cell 112: 765-777. [DOI] [PubMed] [Google Scholar]
- Guacci V., Koshland, D., and Strunnikov, A. 1997. A direct link between sister chromatid cohesion and chromosome condensation revealed through the analysis of MCD1 in S. cerevisiae. Cell 91: 47-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenatri M., Bailly, D., Maison, C., and Almouzni, G. 2004. Mouse centric and pericentric satellite repeats form distinct functional heterochromatin. J. Cell Biol. 166: 493-505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haering C.H., Schoffnegger, D., Nishino, T., Helmhart, W., Nasmyth, K., and Lowe, J. 2004. Structure and stability of cohesin's Smc1–kleisin interaction. Mol. Cell 15: 951-964. [DOI] [PubMed] [Google Scholar]
- Huang J., Hsu, J.M., and Laurent, B.C. 2004. The RSC nucleosome-remodeling complex is required for cohesin's association with chromosome arms. Mol. Cell 13: 739-750. [DOI] [PubMed] [Google Scholar]
- Ivanov D. and Nasmyth, K. 2005. A topological interaction between cohesin rings and a circular minichromosome. Cell 122: 849-860. [DOI] [PubMed] [Google Scholar]
- Kimura A. and Horikoshi, M. 2004. Partition of distinct chromosomal regions: Negotiable border and fixed border. Genes Cells 9: 499-508. [DOI] [PubMed] [Google Scholar]
- Kobayashi T. and Ganley, A.R. 2005. Recombination regulation by transcription-induced cohesin dissociation in rDNA repeats. Science 309: 1581-1584. [DOI] [PubMed] [Google Scholar]
- Kobayashi T., Horiuchi, T., Tongaonkar, P., Vu, L., and Nomura, M. 2004. SIR2 regulates recombination between different rDNA repeats, but not recombination within individual rRNA genes in yeast. Cell 117: 441-453. [DOI] [PubMed] [Google Scholar]
- Ladurner A.G., Inouye, C., Jain, R., and Tjian, R. 2003. Bromodomains mediate an acetyl-histone encoded antisilencing function at heterochromatin boundaries. Mol. Cell 11: 365-376. [DOI] [PubMed] [Google Scholar]
- Laloraya S., Guacci, V., and Koshland, D. 2000. Chromosomal address of the cohesion component Mcd1p. J. Cell Biol. 151: 1047-1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau A., Blitzblau, H., and Bell, S.P. 2002. Cell-cycle control of the establishment of mating-type silencing in S. cerevisiae. Genes & Dev. 16: 2935-2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengronne A., Katou, Y., Mori, S., Yokobayashi, S., Kelly, G.P., Itoh, T., Watanabe, Y., Shirahige, K., and Uhlmann, F. 2004. Cohesin relocation from sites of chromosomal loading to places of convergent transcription. Nature 430: 573-578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y.-C., Cheng, T.-H., and Gartenberg, M.R. 2001. Establishment of transcriptional silencing in the absence of DNA replication. Science 291: 650-653. [DOI] [PubMed] [Google Scholar]
- Megee P.C. and Koshland, D. 1999. A functional assay for centromere-associated sister chromatid cohesion. Science 285: 254-257. [DOI] [PubMed] [Google Scholar]
- Megee P.C., Mistrot, C., Guacci, V., and Koshland, D. 1999. The centromeric sister chromatid cohesion site directs Mcd1p binding to adjacent sequences. Mol. Cell 4: 445-450. [DOI] [PubMed] [Google Scholar]
- Meneghini M.D., Wu, M., and Madhani, H.D. 2003. Conserved histone variant H2A.Z protects euchromatin from the ectopic spread of silent heterochromatin. Cell 112: 725-736. [DOI] [PubMed] [Google Scholar]
- Michaelis C., Ciosk, R., and Nasmyth, K. 1997. Cohesins: Chromosomal proteins that prevent premature separation of sister chromatids. Cell 91: 35-45. [DOI] [PubMed] [Google Scholar]
- Milutinovich M. and Koshland, D.E. 2003. Molecular biology. SMC complexes—Wrapped up in controversy. Science 300: 1101-1102. [DOI] [PubMed] [Google Scholar]
- Nasmyth K. 2001. Disseminating the genome: Joining, resolving, and separating sister chromatids during mitosis and meiosis. Annu. Rev. Genet. 35: 673-745. [DOI] [PubMed] [Google Scholar]
- Ng H.H., Robert, F., Young, R.A., and Struhl, K. 2002. Genome-wide location and regulated recruitment of the RSC nucleosome-remodeling complex. Genes & Dev. 16: 806-819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonaka N., Kitajima, T., Yokobayashi, S., Xiao, G., Yamamoto, M., Grewal, S.I., and Watanabe, Y. 2002. Recruitment of cohesin to heterochromatic regions by Swi6/HP1 in fission yeast. Nat. Cell Biol. 4: 89-93. [DOI] [PubMed] [Google Scholar]
- Raghuraman M.K., Brewer, B.J., and Fangman, W.L. 1997. Cell cycle-dependent establishment of a late replication program. Science 276: 806-809. [DOI] [PubMed] [Google Scholar]
- Riedel C.G., Gregan, J., Gruber, S., and Nasmyth, K. 2004. Is chromatin remodeling required to build sister-chromatid cohesion? Trends Biochem. Sci. 29: 389-392. [DOI] [PubMed] [Google Scholar]
- Rusché L.N., Kirchmaier, A.L., and Rine, J. 2003. The establishment, inheritance, and function of silenced chromatin in Saccharomyces cerevisiae. Annu. Rev. Biochem. 72: 481-516. [DOI] [PubMed] [Google Scholar]
- Sharp J.A., Krawitz, D.C., Gardner, K.A., Fox, C.A., and Kaufman, P.D. 2003. The budding yeast silencing protein Sir1 is a functional component of centromeric chromatin. Genes & Dev. 17: 2356-2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straight A.F., Belmont, A.S., Robinett, C.C., and Murray, A.W. 1996. GFP tagging of budding yeast chromosomes reveals that protein–protein interactions can mediate sister chromatid cohesion. Curr. Biol. 6: 1599-1608. [DOI] [PubMed] [Google Scholar]
- Strom L., Lindroos, H.B., Shirahige, K., and Sjogren, C. 2004. Postreplicative recruitment of cohesin to double-strand breaks is required for DNA repair. Mol. Cell 16: 1003-1015. [DOI] [PubMed] [Google Scholar]
- Suter B., Tong, A., Chang, M., Yu, L., Brown, G.W., Boone, C., and Rine, J. 2004. The origin recognition complex links replication, sister chromatid cohesion and transcriptional silencing in Saccharomyces cerevisiae. Genetics 167: 579-591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tackett A.J., Dilworth, D.J., Davey, M.J., O'Donnell, M., Aitchison, J.D., Rout, M.P., and Chait, B.T. 2005. Proteomic and genomic characterization of chromatin complexes at a boundary. J. Cell Biol. 169: 35-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T., Cosma, M.P., Wirth, K., and Nasmyth, K. 1999. Identification of cohesion sites at centromeres and along chromosome arms. Cell 98: 847-858. [DOI] [PubMed] [Google Scholar]
- Thomas B.J. and Rothstein, R. 1989. Elevated recombination rates in transcriptionally active DNA. Cell 56: 619-630. [DOI] [PubMed] [Google Scholar]
- Uhlmann F. 2004. The mechanism of sister chromatid cohesion. Exp. Cell Res. 296: 80-85. [DOI] [PubMed] [Google Scholar]
- Unal E., Arbel-Eden, A., Sattler, U., Shroff, R., Lichten, M., Haber, J.E., and Koshland, D. 2004. DNA damage response pathway uses histone modification to assemble a double-strand break-specific cohesin domain. Mol. Cell 16: 991-1002. [DOI] [PubMed] [Google Scholar]
- van Leeuwen F., Gafken, P.R., and Gottschling, D.E. 2002. Dot1p modulates silencing in yeast by methylation of the nucleosome core. Cell 109: 745-756. [DOI] [PubMed] [Google Scholar]
- Warren C.D., Eckley, D.M., Lee, M.S., Hanna, J.S., Hughes, A., Peyser, B., Jie, C., Irizarry, R., and Spencer, F.A. 2004. S-Phase checkpoint genes safeguard high-fidelity sister chromatid cohesion. Mol. Biol. Cell 15: 1724-1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X.M., Mehta, S., Uzri, D., Jayaram, M., and Velmurugan, S. 2004. Mutations in a partitioning protein and altered chromatin structure at the partitioning locus prevent cohesin recruitment by the Saccharomyces cerevisiae plasmid and cause plasmid missegregation. Mol. Cell. Biol. 24: 5290-5303. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.