Abstract
The yeast checkpoint kinases Mec1 and Rad53 are required for genomic stability in the presence of replicative stress. When replication forks stall, the stable maintenance of replisome components requires the ATR kinase Mec1/Ddc2 and the RecQ helicase Sgs1. It was unclear whether either Mec1 or Sgs1 action requires the checkpoint effector kinase, Rad53. By combining sgs1Δ with checkpoint-deficient alleles, we can now distinguish the role of Mec1 at stalled forks from that of Rad53. We show that the S-phase-specific mec1-100 allele, like the sgs1Δ mutation, partially destabilizes DNA polymerases at stalled forks, yet combining the mec1-100 and sgs1Δ mutations leads to complete disassociation of the replisome, loss of RPA, irreversible termination of nucleotide incorporation, and compromised recovery from hydroxyurea (HU) arrest. These events coincide with a dramatic increase in both spontaneous and HU-induced chromosomal rearrangements. Importantly, in sgs1Δ cells, RPA levels at stalled forks do not change, although Ddc2 recruitment is compromised, explaining the partial Sgs1 and Mec1 interdependence. Loss of Rad53 kinase, on the other hand, does not affect the levels of DNA polymerases at arrested forks, but leads to MCM protein dissociation. Finally, confirming its unique role during replicative stress, Mec1, and not Tel1, is shown to modify fork-associated histone H2A.
Keywords: Replicative stress, checkpoint, DNA polymerases, Mec1, Sgs1, chromosome instability
Intact S-phase checkpoint mechanisms are essential for cell survival and proliferation in the presence of DNA replicative stress, which can be caused by the stalling of replication forks at DNA lesions, at DNA-bound protein complexes (Ivessa et al. 2003), or as a result of reduced nucleotide levels induced by the addition of hydroxyurea (HU). Importantly, DNA replication defects and genomic instability are both hallmarks of oncogenic transformation. Indeed, cancer cells appear to persist in a state of perpetual replicative stress, which correlates with low but continuous signs of an activated DNA damage response, such as histone H2AX and CHK2 phosphorylation (Halazonetis 2004). In budding yeast, the ATR kinase homolog Mec1 and its downstream effector kinase Rad53, the hCHK2 homolog, are both central to the DNA damage checkpoint signaling cascade.
A role for ATM-related kinases in the cellular response to replication fork stalling is conserved in all eukaryotes. The affinity of the mammalian ATRIP for replication protein A (RPA) suggests a model in which ATR-ATRIP is recruited to sites of damage or to abnormal structures generated at stalled replication forks that contain extended regions of RPA-bound single-stranded DNA (ssDNA) (Zou and Elledge 2003). Mec1 requires a cofactor Ddc2, the counterpart to mammalian ATRIP, and loss of either subunit abrogates the checkpoint-dependent phosphorylation of Rad53 and Pds1 proteins, precluding a checkpoint response (Paciotti et al. 2000). Once recruited, Mec1 may act by phosphorylating fork-associated targets such as RPA (Brush et al. 1996; Kim and Brill 2003; Bartrand et al. 2004) or the replication/checkpoint adaptor protein Mrc1 (Alcasabas et al. 2001; Osborn and Elledge 2003).
In mammals, the ATR kinase was also shown to bind and phosphorylate the RecQ helicase BLM (Davies et al. 2004; Li et al. 2004). RecQ helicases are a family of 3′-5′ DNA-unwinding enzymes conserved from bacteria to man, which includes a single budding yeast homolog called Sgs1. Mutations in three of five human RecQ helicases are responsible for genetic disorders that correlate with chromosomal loss, increase rates of translocation, and cause premature aging or cancer (for review, see Mohaghegh and Hickson 2001). BLM helicase, like the yeast Sgs1 protein, associates with DNA repair foci in S-phase cells, and was recently shown to be an intermediary in the response to stalled replication forks, physically interacting with 53BP1 and γ-H2AX in human cells (Sengupta et al. 2004).
In budding yeast, elimination of Sgs1 helicase leads to elevated rates of meiotic and mitotic recombination (Watt et al. 1996), increased frequencies of spontaneous gross chromosomal rearrangements (GCR) (Myung and Kolodner 2002), as well as aberrant DNA replication phenotypes (Versini et al. 2003; Liberi et al. 2005). When replication forks are stalled by the addition of HU, sgs1-deficient cells suffer a partial loss of fork-associated DNA polymerases (Cobb et al. 2003). It was proposed but not proven that the chromosome instability arises from loss of polymerases at stalled forks.
One way to categorize the various phenotypes associated with a loss of Sgs1 is to determine whether or not they require its helicase activity, and/or the associated type I topoisomerase, Top3. For instance, Sgs1 contributes to the activation of Rad53 in response to HU, on a pathway that is redundant with break-induced signaling pathways (Frei and Gasser 2000). This activity requires intact Sgs1, but neither its helicase function nor the activity of Top3 (Bjergbaek et al. 2005). In contrast, the contribution of Sgs1 to replication fork stability on HU requires both the helicase activity and Top3 interaction (Cobb et al. 2003; Bjergbaek et al. 2005). Moreover, loss of Sgs1's polymerase stabilizing function appears to be epistatic with loss of the strand-exchange factor Rad51, consistent with the observation that Rad51-dependent cruciform structures accumulate at stalled forks in sgs1 cells (Liberi et al. 2005).
By monitoring cells as they synchronously enter S phase, we have shown that both Mec1/Ddc2 and Mrc1 are required to stabilize DNA polymerase ε (pol ε) and α (pol α) at stalled replication forks during the first hour of HU-induced arrest (Cobb et al. 2003; Katou et al. 2003; Bjergbaek et al. 2005). This occurs prior to Rad53 kinase activation. Consistently, fork-bound polymerases remain bound at stalled forks in cells that carry a complete rad53 deletion (Cobb et al. 2003). Inexplicably, however, an active-site mutation, rad53-K227A, appears to provoke a partial loss of both DNA pol ε and pol α on HU (Lucca et al. 2004). Other differences in the response to replicative stress have been reported for different checkpoint mutants. For instance, a complete deletion of mec1 increased the rate of spontaneous GCR far more significantly than the loss of the G2 damage checkpoint in rad9 or rad53 cells (Kolodner et al. 2002). Nonetheless, the survival rate of a rad53 mutant after exposure to HU was just as compromised as a mec1Δ strain (Weinert et al. 1994), and strains lacking Rad53 are unable to resume replication after fork stalling (Lopes et al. 2001; Tercero et al. 2003). While these studies suggest that the functions of Mec1/Ddc2 and Rad53 kinase at stalled forks are distinct, they do not reveal how their modes of action differ.
Past results supported the argument that Sgs1, Mrc1, Mec1/Ddc2, and Rad53 all contribute to cellular recovery after replication fork arrest, yet the relationship between the maintenance of engaged replicative polymerases and prevention of irreversible fork collapse remained unclear, because these proteins act on overlapping pathways. Here we dissect the roles of the Mec1/Ddc2 complex and Rad53 kinase in preserving replication fork integrity, by combining an S-phase-specific allele of mec1 with a complete deletion of sgs1. We detect a dramatic synergism between sgs1Δ and mec1-100 mutations in promoting fork collapse and in destabilizing replication polymerases at stalled forks, a defect that cannot be attributed to impaired activation of the downstream kinase Rad53. The sgs1 and mec1-100 mutations affect the binding of RPA and Mec1/Ddc2 at stalled forks differentially, and collectively lead to complete polymerase loss. This is not the case in cells lacking Rad53, although other replisome components, like the MCM helicase, are found displaced from stalled forks in this mutant. Finally, we recover phosphorylated H2A at stalled replication forks and show that its modification depends exclusively on Mec1. These data directly link the loss of polymerases and RPA from forks and an inability to recover from replicative stress, with dramatic increases in both spontaneous and HU-induced chromosomal rearrangements. This suggests mechanisms through which ATR and BLM maintain genomic stability.
Results
Sgs1p and Mec1p contribute independently to genomic stability and cell viability
Previous studies have implicated the Saccharomyces cerevisiae replication checkpoint in the suppression of spontaneous genomic instability (for review, see Kolodner et al. 2002). Cells with deletions for Mec1 were shown to be highly synergistic with the loss of Sgs1 for GCR (Myung and Kolodner 2002). Surprisingly, the synergism with sgs1 was much less pronounced for mutants that lose the DNA damage-induced checkpoint response, such as rad24, rad53,or tel1 (Myung and Kolodner 2002). While this suggested a special relationship between the S-phase functions of Sgs1 and Mec1, there were no data to link this instability to their roles at stalled replication forks.
Given that the complete deletion of MEC1 compromises both the intra-S and the G2/M checkpoint responses, we made use of the mec1-100 allele, which is deficient for the replication checkpoint but which maintains a functional G2/M arrest in response to strand breaks (Paciotti et al. 2001). The mutation reflects two amino acid substitutions (F1179S and N1700S), upstream of the C-terminal PI3-kinase domain in a region shared with the fission yeast and mammalian ATM/ATR enzymes. We introduced the appropriate markers to monitor GCR and backcrossed to generate isogenic strains bearing either the mec1-100 allele, an sgs1 deletion, or both. Spontaneous and HU-induced GCR, and viability during chronic exposure to HU, were then monitored. Finally, we scored the strains for their ability to recover from nucleotide depletion and resume DNA replication.
The rate of spontaneous GCR monitored in the mec1-100 allele is 187-fold above that in wild-type cells, while that of sgs1Δ increases by 67-fold (Table 1). By deleting sgs1 in the mec1-100 background, we see the rate of GCRs rise synergistically to a value 573-fold above the wild-type rate. This phenotype is unique to the mec1-sgs1 combination; in rad53-11 sgs1Δ cells, GCR rates are 177-fold above wild type, which is not even additive (177 < 67 + 123-fold). Therefore, with respect to chromosome instability, the mec1-100 allele shows synergistic effects with sgs1Δ much like mec1Δ (Myung and Kolodner 2002). This genetic interaction becomes even more severe when cells are treated with 0.2 M HU. Under these conditions, mec1-100 cells showed a 6 × 103-fold increase in GCR rate over wild type, and the double mutant reaches 1.62 × 105 times the wild-type GCR rate (Table 1). This elevated GCR rate in mec1-100 sgs1Δ cells is dramatically exacerbated by HU, increasing by another 667-fold (+HU/-HU), while the same ratio is 2.4-fold in wild-type cells (Table 1).
Table 1.
Effect of sgs1Δ and mec1-100 mutations on spontaneous and HU-induced GCR rates
| Genotype | GCR | GCR after 0.2M HU | Fold increase (+HU/-HU) |
|---|---|---|---|
| Wild-type (S288c) | 2.4 [0.7–4.1] × 10-10 (1) | 5.6 [0.7–10.0] × 10-10 (1) | 2.4 |
| sgs1Δ | 1.6 [0.3–28.4] × 10-8 (67) | 6.9 [3.2–10.6] × 10-8 (123) | 4.4 |
| mec1-100 | 4.5 [2.4–6.6] × 10-8 (187) | 3.4 [3.0–3.8] × 10-6 (6000) | 75.3 |
| rad53-11 | 3.0 [2.0–3.9] × 10-8 (123) | a | N/D |
| sgs1Δ mec1-100 | 1.4 [1.2–1.6] × 10-7 (573) | 9.1 [4.8–13.4] × 10-5 (162,000) | 667 |
| sgs1Δ rad53-11 | 4.2 [3.7–4.7] × 10-8 (177) | a | N/D |
Too few survivors were recovered under these conditions to determine rates of GCR. [] indicates the highest and lowest rates observed in the fluctuation tests. The numbers in parentheses are the fold increases in the rate relative to that of the wild-type strain.
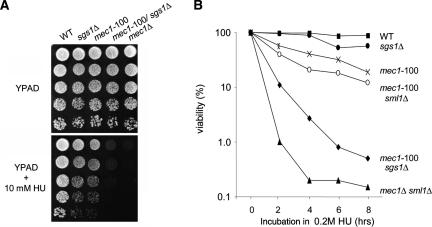
Coincident with this extreme chromosome instability, we monitor a severe loss of cell viability both when the double mutant is plated on low levels of HU (Fig. 1A), or after cells have been arrested for increasing periods of time on HU, and released for growth in the absence of drug (Fig. 1B). Although neither sgs1Δ nor mec1-100 mutations alone are highly sensitive, the mec1-100 sgs1Δ cells are nearly as compromised as the mec1Δ strain. This is not suppressed by up-regulating dNTP levels (i.e., by sml1 deletion) (Fig. 1B), which is necessary for viability in the mec1Δ background. Observing the S-phase-specific defects of the mec1-100 strain and its high rates of GCR, we speculated that this mutation might be sufficient to irreversibly destabilize replication polymerases and cause fork collapse, as reported for the more pleiotropic mec1Δ mutation (Tercero and Diffley 2001; Cobb et al. 2003).
Figure 1.
Highly synergistic effects of mec1-100 and sgs1Δ mutations on chromosome stability and recovery from HU. (A) Drop assays on YPD ± 10 mM HU were performed with exponentially growing cultures of the indicated W303-1a (GA-180) derivates, using a 1:5 serial dilution series: sgs1Δ (GA-1761), mec1-100 (GA-2474), mec1-100 sml1Δ (GA-2478), mec1-100 sgslΔ (GA-2514), or mec1Δ sml1Δ (GA-2895). (B) Cell viability was monitored as colony outgrowth from cultures synchronized by α-factor and held in YPD + 0.2 M HU for indicated times.
Combining mec1-100 and sgs1Δ mutations synergistically promotes fork collapse
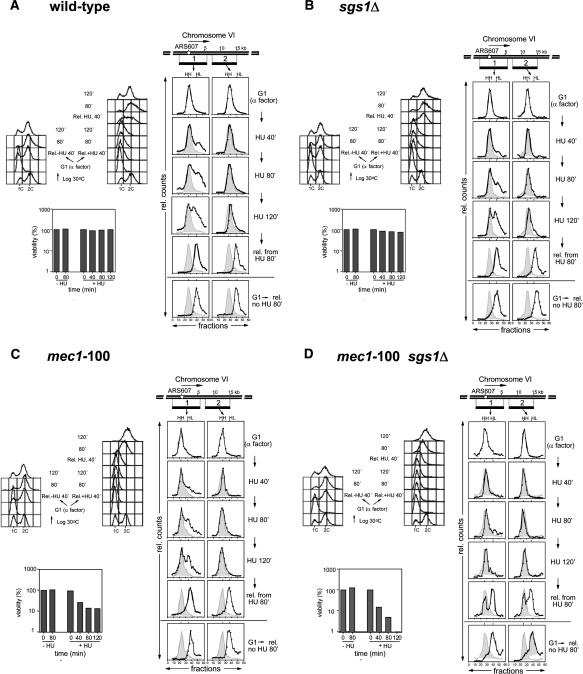
When yeast cells enter S phase in media containing HU, early origins fire normally, yet the rate of replication is severely reduced due to low dNTP levels. In wild-type cells, DNA polymerases remain fork-associated or progress very slowly along the chromosome, allowing efficient recovery when nucleotide levels are restored. In mec1Δ cells, on the other hand, forks that encounter damage collapse (Tercero and Diffley 2001). To see if fork collapse correlates with the synergistic effects on GCR rates scored for the mec1-100 sgs1Δ double mutant, we monitored replication fork progression in HU with a density isotope substitution method (Fig. 2; Tercero et al. 2000), using probes that recognize DNA fragments at the origin (fragment 1) or at a site ∼15 kb away (fragment 2). This monitors nucleotide incorporation genome-wide, as well as locally.
Figure 2.
Replication fork collapse in mec1-100 sgs1Δ mutants. For all panels, cells were grown in minimal medium with heavy (H) isotopes and blocked in G1 with α-factor. The cultures were held in α-factor for an additional 30 min in light (L) isotope before dividing and releasing into fresh medium ± 0.2 M HU. Samples were taken at 40, 80, and 120 min, when cells were released from HU and their recovery was monitored. DNA content as determined by flow cytometry and cell viability for all strains was scored at the indicated time points. A time course of DNA replication at ARS607 was analyzed by density transfer after release from α-factor arrest into medium with 0.2 M HU, using specific probes recognizing the ClaI/SalI fragments 1 and 2. The relative amounts of radioactivity in the hybridized DNA are plotted against the gradient fraction number. The positions of unreplicated (heavy-heavy, HH) and fully replicated heavy-light (HL) are indicated. At later time points, the position of the initial HH peak is shown for comparison (gray area). Corresponding FACS analysis and survival assays are shown for each isogenic strain bearing the following mutations: wild-type (YJT110) (A); sgs1Δ (YVR1) (B); mec1-100 (GA-2931) (C); and mec1-100 sgs1Δ (GA-2930) (D).
In wild-type, sgs1Δ and mec1-100 single mutant cells, we clearly detect the replication of fragment 1 by 120 min in HU, although between 30% and 35% of the forks stall within this zone. Little of fragment 2 becomes fully replicated (Fig. 2A,C), consistent with data from Santocanale and Diffley (1998), who found that most forks stall within 10 kb of an origin in cells exposed to high concentrations of HU. In mec1-100 sgs1Δ cells, on the other hand, no replication of fragment 1 can be detected under identical conditions (Fig. 2D). Given that there are no differences for the timing of S-phase onset, budding index (Supplementary Fig. 1), and bubble arc appearance (Fig. 3), nor in the level of Orc2 recovered at origins by chromatin immunoprecipitation (ChIP), we conclude that the mec1-100 sgs1Δ strain, unlike either single mutant, suffers severe attenuation of fork progression on HU.
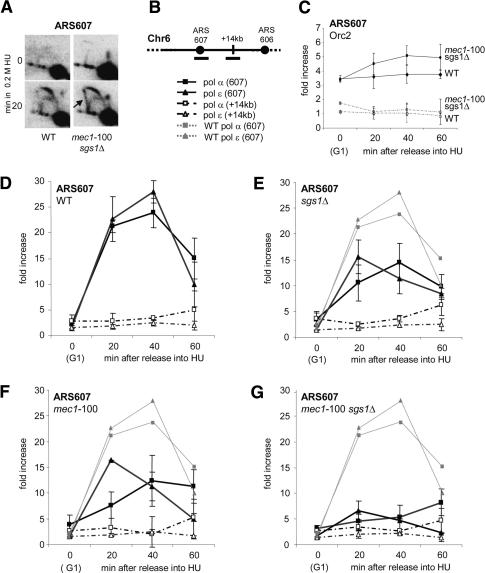
Figure 3.
Loss of DNA polymerases at stalled replication forks in mec1-100 sgs1Δ cells. (A) Neutral/neutral 2D gel analysis was performed at ARS607 in wild-type (GA-1020) and mec1-100 sgs1Δ (GA-2514) cells released from α-factor arrest into YPD + 0.2 M HU. Genomic DNA was prepared from cells collected at 0 (G1) or 20 min after release, and the Southern transfer was probed with a 2.6-kb fragment spanning ARS607. (B) Primers were designed to amplify genomic regions on Chr 6 corresponding to early-firing origin ARS607 (filled symbols) and a nonorigin site, +14 kb (open symbols). ChIP was performed on cultures synchronized in G1 by α-factor arrest, and released into pre-warmed YPD + 0.2 M HU, prior to fixation with 1% formaldehyde at the indicated time points. (C) ChIP with anti-Myc (9E10) is used to quantify Myc-Orc2 presence at ARS607 in isogenic wild-type (GA-2897, diamonds) and mec1-100 sgs1Δ (GA-2896, ovals) cells. (D-G) ChIP with anti-Myc (9E10) or anti-HA (12CA5) precipitated HA-tagged DNA pol α (squares) or Myc-tagged DNA pol ε (diamonds). The strains used were wild-type strains GA-2238 and GA-2448 in D; sgs1Δ strains GA-2256 and GA-2450 in E; mec1-100 strains GA-2567 and GA-2515 in F; and mec1-100 sgs1Δ strains GA-2578 and GA-2516 in G. In E-G, wild-type signals are shown as light-gray dashed lines for comparison. Controls and quantitation are described in Materials and Methods. Standard deviation is calculated from duplicate runs and multiple independent experiments.
To monitor the reversibility of fork stalling in these cultures, cells were released from HU arrest by placing them in fresh, drug-free media. Under these conditions, wild-type, mec1-100, and sgs1Δ cells all resume DNA replication satisfactorily (Fig. 2A-C). By 80 min, both fragments 1 and 2 are fully replicated, indicating that a large fraction of replication forks recover and continue DNA synthesis after HU removal. In contrast, the mec1-100 sgs1Δ double mutant shows significant amounts of unreplicated DNA even after release into fresh media (Fig. 2D). We estimate that significantly fewer than 50% of the replication forks resume DNA synthesis in the double mutant, since the replication of fragments 1 and 2 could initiate from any origin on the chromosomal arm to complete replication by 80 min. We conclude that a high fraction of DNA replication forks collapse irreversibly in the mec1-100 sgs1Δ strain on HU. This is reminiscent of the fork collapse reported for the mec1Δ strain on MMS (Tercero and Diffley 2001), and is likely to account for the loss of viability observed for these cells (Figs. 1B, 2D).
Sgs1 and Mec1 cooperate to stabilize DNA polymerases at stalled replication forks
To see if fork collapse and high GCR rates are due to a loss of replicative polymerases at forks, we performed ChIP for both DNA pol α and pol ε, comparing wild-type and mutant strains as they synchronously enter S phase in the presence of 0.2 M HU (Cobb et al. 2003; Bjergbaek et al. 2005). During the first hour in HU, the abundance of Myc-tagged DNA pol ε and HA-tagged DNA pol α was analyzed at the early-firing origin ARS607 (filled symbols) by real-time PCR (rtPCR). As a negative control, we probed for a site +14 kb away from the origin (Fig. 3B, open symbols). The values plotted are direct ratios of the mean rates of fragment accumulation monitored by rt-PCR in immunoprecipitates over control precipitates.
In Figure 3D, we show that both DNA pol ε and pol α are efficiently associated with ARS607 by 20 min after release from a pheromone arrest. In the absence of HU, the polymerases progress rapidly through both the origin and distal sites, and genomic replication is completed by ∼30 min (Cobb et al. 2003). However, in HU-containing medium, both polymerases remain associated with the stalled fork for ∼60 min (Fig. 3D, filled symbols), and migrate slowly into the fragment at +14 kb by 60 min (Fig. 3D, open symbols and stippled lines). When the same assay is performed in either sgs1Δ or mec1-100 cells, we see a partial loss of DNA pol ε and pol α at ARS607 (2- to 2.5-fold reduction) as compared with the isogenic wild-type strain (Fig. 3D-F).
A much more striking loss of polymerases occurs in the mec1-100 sgs1Δ cells. We see that both DNA pol ε and pol α levels drop to near background levels at ARS607 (Fig. 3G), as occurs in mec1Δ and mec1Δ sgs1Δ cells (Supplementary Fig. 2). We also observe a transient enrichment of DNA pol α and pol ε at the late-firing origin ARS501 in mec1-100 cells (Supplementary Fig. 3), confirming that late origins fire precociously in these mutants (Santocanale and Diffley 1998). ARS501 serves as a positive control both for the assay and the mec1-100 defect for Rad53 activation (Fig. 4A,B; Paciotti et al. 2001; Tercero et al. 2003).
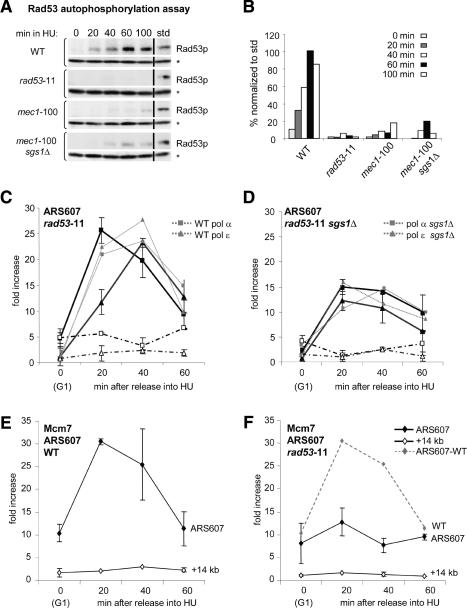
Figure 4.
Rad53 is needed to stabilize MCM proteins but not DNA polymerases at stalled forks. (A) ISA analysis of Rad53 autophosphorylation was performed on wild-type (GA-1020), rad53-11 (GA-2240), mec1-100 (GA-2474), and mec1-100 sgs1Δ (GA-2514) cells. For each strain, the upper box shows the incorporation of γ32-ATP into Rad53, and the bottom panel shows the same blot probed with anti-RNaseH42 to normalize loading (*). Time (in minutes) after α-factor release is indicated above each panel, and “std” is 5 μL of a standard containing a known amount of a HU-activated Rad53. For every sample, protein concentration was determined by Coomassie blue staining prior to equally loading gels. Dried filters were exposed for equal times on a Bio-Rad PhosphorImager, before reprobing for RNaseH42 to normalize signals. (B) Quantification of Rad53 autophosphorylation displayed as a normalized percentage of std. Shown is an average of two experiments with standard deviations between 5% and 15%. (C,D) ChIP was performed as described in Figure 3 for HA-tagged DNA pol α (squares) and Myc-tagged DNA pol ε (diamonds) in rad53-11 strain GA-2574 and rad53-11 sgs1Δ strain GA-2576. (E,F) ChIP was performed for Myc-tagged Mcm7 (diamonds) in cultures released from α-factor into 0.2 M HU as in Figure 3 using wild type (GA-1003) and rad53-11 (GA-3054). rtPCR was performed as described for ARS607 (filled symbols) and +14 kb (open symbols). Wild-type and sgs1Δ signals are shown in light-gray dashed lines for comparison.
The drop in polymerase levels in mec1-100 sgs1Δ cells is not due to aberrant initiation timing as demonstrated by 2D gel analysis of replication intermediates (Fig. 3A). Furthermore, it is presumed that prereplication complexes are not disrupted, since Orc2 recovery at ARS607 is similar in wild-type and mutant cells (Fig. 3C). Finally, the budding index is not significantly altered in any of these mutants, either in the presence or absence of HU (Supplementary Fig. 1), and progression through S phase in the absence of HU occurs normally (see ChIP for DNA pol ε and FACS analysis) (Supplementary Fig. 4). Thus, there must be a true reduction in the level of replicative enzymes bound to stalled forks in mec1-100 sgs1Δ cells. This correlates with an accumulation of aberrant X-shaped structures in neutral-neutral 2D gels of mec1-100 sgs1Δ mutants treated ∼20 min with HU (Fig. 3A, see arrow). These may reflect nonproductive fork-associated recombination events.
Polymerase stability at stalled forks is independent of Rad53 checkpoint activation
We next asked whether the defects on HU reflect the double mutant's inability to activate Rad53 kinase and thereby delay progression into mitosis. Indeed, rad53Δ cells, like both the mec1Δ and mec-100 sgs1Δ double mutant, are known to lose viability after exposure to high HU concentrations (Desany et al. 1998; Lopes et al. 2001), and irreversible fork collapse was reported to occur in both rad53Δ and mec1Δ strains on MMS (Tercero and Diffley 2001). Our previous work indicated that DNA polymerases remained efficiently bound at stalled forks in a rad53Δ strain (Cobb et al. 2003), yet in these experiments the rad53Δ mutation was coupled with sml1Δ, to prevent cell death (Zhao et al. 1998). The sml1 mutation up-regulates ribonucleotide reductase genes (RNR1-4), which might conceivably influence replisome stability indirectly. Thus, to test whether the loss of Rad53 activity contributes to the synergism between the mec1-100 and sgs1Δ mutations, we tested a recessive, activity-dead allele called rad53-11, which fails to become phosphorylated and to activate the checkpoint, yet which does not require sml1 deletion for survival (Weinert et al. 1994; Pellicioli et al. 1999).
An in-gel Rad53 autophosphorylation assay confirms that on HU, Rad53 is activated by 60 min in wild-type cells, but is inactive in a rad53-11 mutant, and is strongly reduced in the mec1-100 allele (Paciotti et al. 2001). In the mec1-100 sgs1Δ strain, we see slightly more Rad53 activity, perhaps reflecting the higher rates of DNA breakage and activation of the G2 checkpoint through Rad9 (Fig. 4A,B). Since impaired Rad53 activation might accelerate progression into mitosis, we tested whether we could enhance the viability of the double mutant by providing time for recovery from HU. Delaying the G2/M transition by placing the HU-arrested cells transiently in nocodazole-containing media, did not, however, increase survival (Supplementary Fig. 5), arguing that the loss of viability in mec1-100 or mec1-100 sgs1Δ strains is not due simply to premature entry into mitosis or uncontrolled spindle elongation.
We next analyzed the effect of the rad53-11 allele on DNA polymerase stability at HU-arrested forks by monitoring whether a loss of Rad53 activity is synergistic with the deletion of sgs1. ChIP experiments performed with an isogenic rad53-11 mutant show nearly identical levels of DNA pol ε and pol α at ARS607 as the wild-type and the rad53Δ sml1Δ control strains (Fig. 4C; Cobb et al. 2003). In these mutant samples, we also detect the recruitment of DNA polymerases to the late-firing origin ARS501, confirming that Rad53 failed to activate the checkpoint response that suppresses late origin firing (Supplementary Fig. 3; Santocanale and Diffley 1998). Importantly, when rad53-11 is combined with a deletion of sgs1, we detect no synergism whatsoever, and the levels of fork-associated DNA pol ε and pol α are identical to the levels scored in sgs1Δ cells (Fig. 4D). Thus, the loss of DNA polymerases at stalled forks in mec1 cells, and in the mec1-100 sgs1Δ double mutant, does not reflect Mec1's role as an activator of Rad53 kinase and its downstream checkpoint response. This result supports the hypothesis that both Mec1 and Sgs1 have a Rad53-independent function at replication forks (Desany et al. 1998; Tercero and Diffley 2001; Bjergbaek et al. 2005).
Why then are stalled replication forks unable to recover in the absence of Rad53 activity? One of the phenotypes of rad53-deficient cells arrested on HU is the accumulation of long stretches of ssDNA at stalled forks (Sogo et al. 2002), which could result from an uncoupling of polymerases from the MCM helicase (Byun et al. 2005). Similarly, cells lacking the histone chaperone Asf1, which interacts with Rad53, show aberrant replisome stability with the MCM helicase being displaced along the template (Franco et al. 2005). To see if the rad53-11 strain would have a similar phenotype, we monitored MCM proteins at replication forks stalled on HU, as described above. Indeed, fork-associated Mcm7 levels drop significantly in the rad53-11 mutant, and it does not move into the +14-kb fragment (Fig. 4E,F). Similar results were obtained for a tagged Mcm4 subunit, where we see that Rad53 checkpoint activation, but not Sgs1, is necessary for Mcm4 maintenance (Supplementary Fig. 6). Thus, the primary defect at stalled forks in rad53-deficient cells appears to be the displacement of MCM proteins, which does not necessarily lead to polymerase loss. We do not know whether MCM proteins become displaced along the DNA fiber, or are completely lost from stalled forks.
RPA recovery at replication forks is diminished in the mec1-100 sgs1Δ double mutant
To identify the mechanisms through which mec1-100 and sgs1Δ cells lose functional replication forks, we looked at factors that might be differently regulated by Mec1 and Rad53, yet which also interact with Sgs1. One likely candidate was the single-strand binding complex, RPA, which interacts strongly with Sgs1 both in the presence and absence of HU (Cobb et al. 2003). This interaction is conserved, as BLM and WRN helicases both bind human RPA tightly (Brosh et al. 2000; Doherty et al. 2005). Importantly, functional RPA is known to be necessary for the recruitment of pol α-primase (Tanaka and Nasmyth 1998) and pol ε to active forks (Lucca et al. 2004), and the phosphorylation of Rpa2 in response to DNA damage requires Mec1, but not Rad53 (Brush et al. 1996; Kim and Brill 2003). Indeed, in response to HU, Rpa2 is fully phosphorylated in the rad53-11 mutant, yet lacks damage-specific modifications in the mec1-deficient strain. To see if the presence of RPA was affected by either the mec1-100 or sgs1Δ mutation, we assayed for Rpa1 at stalled forks, as described in Figure 3.
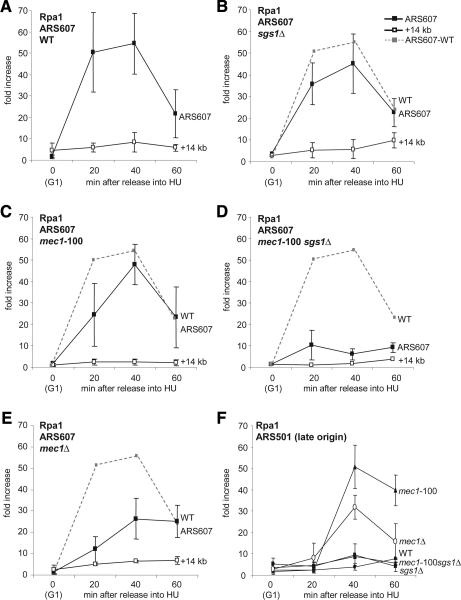
Rpa1 was immunoprecipitated from wild-type and mutant cells synchronously released into S phase in the presence of 0.2 M HU (Fig. 5A). In wild-type, sgs1Δ, and mec1-100 cells, there is no significant change in the level of Rpa1 present at the early firing origin ARS607 in HU-arrested cells (Fig. 5A-C). On the other hand, there is a striking and complete loss of Rpa1 at stalled forks in the mec1-100 sgs1Δ double mutant (Fig. 5D). This effect is even more severe than that observed for mec1Δ cells (Fig. 5E). These results indicate that Mec1 activity is necessary to maintain RPA at stalled forks, which was not the case for the Rad53 kinase (Tanaka and Nasmyth 1998). Given that Rpa1 remains bound in the mec1-100 mutant, but is lost when this mutation is coupled with sgs1Δ (Fig. 5D), we conclude that Sgs1 activity must contribute to Rpa1 binding when Mec1 activity is compromised. Loss of Rpa1 correlates with irreversible fork collapse and high GCR rates in the mec1-100 sgs1Δ double mutant.
Figure 5.
Rpa1 is displaced from stalled replication forks in mec1-100 sgs1Δ cells. ChIP was performed on Myc-tagged Rpa1 (squares) in cultures release from α-factor into 0.2 M HU as described in Figure 3 using the following strains: wild-type (GA-1113) (A), sgs1Δ (GA-2439) (B), mec1-100 (GA-2571) (C), mec1-100 sgs1Δ (GA-2581) (D), and mec1Δ sml1Δ (GA-2582) (E). rtPCR-amplified regions correspond to ARS607 (filled symbols) and +14 kb (open symbols), with the wild-type signal for Rpa1 shown as a dashed line. From the same experiment, the level of Myc-Rpa1 at the late origin ARS501 is shown for indicated wild-type and mutant strains.
We monitored Rpa1 binding at the late-firing origin ARS501 in the same set of strains under identical conditions. Consistent with a lack of activated Rad53 and the precocious firing of late origins, Rpa1 is present at ARS501 in mec1-100 and mec1Δ cells, yet it is absent in the mec1-100 sgs1Δ double mutant (Fig. 5F; see also Tanaka and Nasmyth 1998). This suggests that RPA binding is destabilized at both early- and late-firing origins in the double mutant.
Mec1-Ddc2 recruitment to forks is compromised in sgs1Δ, but not in mec1-100, cells
The Mec1/Ddc2 complex has been shown to be recruited to stalled forks (Katou et al. 2003; Osborn and Elledge 2003), apparently through the affinity of Ddc2 for RPA (Zou and Elledge 2003). Given that Sgs1 binds Rpa1, it was possible that the RecQ helicase might influence the association of Mec1/Ddc2 near stalled forks. To test whether Mec1/Ddc2 recruitment is altered in mec1-100 or sgs1Δ mutants, we monitored the recruitment of the Ddc2 protein to ARS607 by ChIP (Fig. 6). The presence of Ddc2 is assumed to reflect the binding of the Mec1/Ddc2 heterodimer, since in both yeast and human cells, the vast majority of the Mec1/ATR kinase is recovered in a complex with Ddc2/ATRIP (Rouse and Jackson 2002; Zou and Elledge 2003) and DDC2 disruption completely abrogates the checkpoint response (Paciotti et al. 2000).
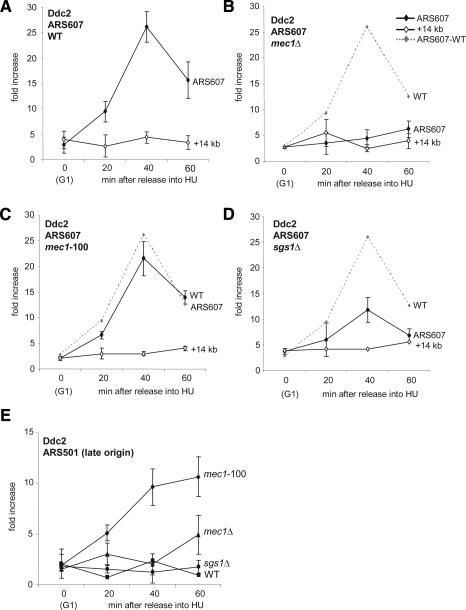
Figure 6.
Ddc2 recruitment drops in sgs1Δ but not mec1-100 strains. ChIP was performed on HA-tagged Ddc2 (diamonds) in cultures released from α-factor into 0.2 M HU exactly as described in Figure 3 using the following strains: wild-type (GA-2462) (A), mec1Δ (GA-2463) (B), mec1-100 (GA-2475) (C), and sgs1Δ (GA-2519) (D). rtPCR-amplified regions correspond to ARS607 (filled symbols) and +14 kb (open symbols), with the wild-type signal for Ddc2 shown as a dashed line. From the same experiment, the level of Ddc2 at the late origin ARS501 is shown for indicated wild-type and mutant strains (E).
For Ddc2 localization we use an HA epitope-tagged version of the protein that is fully functional, based on the cellular response and viability under DNA-damaging conditions (data not shown). Consistent with previous reports (Katou et al. 2003; Osborn and Elledge 2003), we see that HA-Ddc2 is recruited to ARS607 in wild-type cells during an HU arrest, peaking at ∼40 min after release from pheromone arrest (Fig. 6A). This is 20 min later than the first appearance of DNA polymerases or Sgs1 helicase at early-firing origins (Cobb et al. 2003), but coincides with Mec1 appearance by ChIP (data not shown). We find that in mec1Δ cells, Ddc2 recruitment to stalled forks is completely abolished (Fig. 6B). In the mec1-100 background, on the other hand, we see no significant drop in the efficiency of Ddc2 binding to stalled forks (Fig. 6C). These data support previous immunofluorescence studies that showed the proper association of Ddc2 with S-phase-specific repair foci in the mec1-100 allele in response to MMS (Tercero et al. 2003). We argue that the instability of polymerases does not stem from an absence of mec1-100/Ddc2 complex recruitment, but rather from altered activity of the complex, supporting the hypothesis that Mec1/Ddc2 targets fork-associated proteins to stabilize the replisome (Cha and Kleckner 2002; Osborn and Elledge 2003).
We next examined Ddc2 recruitment in sgs1Δ cells. We see a partial, but reproducible twofold decrease in the amount of Ddc2 recovered at stalled forks (Fig. 6D). This is all the more noteworthy because we do not detect a significant drop in Rpa1 levels in this strain (Fig. 5B). This may mean that sgs1-deficient cells accumulate inappropriate strand exchanges (Liberi et al. 2005) that preclude efficient Mec1 binding. Alternatively, Sgs1 may be needed for a conformational change in RPA that favors either Mec1/Ddc2 and/or DNA polymerase interaction (see Discussion).
Finally, we monitored whether the Mec1/Ddc2 complex is recruited to late-firing origins, or whether it only binds those that fire early and then stall. Indeed, Ddc2 is recovered at the late origin ARS501 when it is inappropriately activated in the mec1-100 mutant, but not in wild-type or sgs1Δ cells (Fig. 6E). This shows that Mec1/Ddc2 can be recruited to any active fork arrested by HU, and argues that the unscheduled firing of late origins is a further source of damage that requires Mec1 action.
Mec1-dependent H2A phosphorylation at stalled replication forks
Histone H2A or its variant H2AX is a critical target of ATR and ATM kinases at sites of double-strand breaks (DSB), and it also becomes modified in response to HU in mammalian cells (for review, see Liu et al. 2003; Thiriet and Hayes 2005). This modification helps recruit downstream kinases as well as chromatin-modifying enzymes, to maintain the checkpoint arrest. In budding yeast, the two major H2A isoforms both carry the serine at position 129, typical of H2AX, which becomes phosphorylated in response to damage by either Tel1 or Mec1 kinase, but not by Dun1 or Rad53 (Downs et al. 2000; Shroff et al. 2004). Similarly, both ATR and ATM kinases modify H2AX in fission yeast and vertebrates (Nakamura et al. 2004). Given that loss of the C-terminal phospho-acceptor serine increases sensitivity to S-phase damage (MMS), we examined whether or not H2A-P is directly associated with stalled forks.
Phospho-specific antibodies to H2A-P (a gift from W. Bonner, NIH, Bethesda, MD) were used to monitor the presence of the modified histone near HU-arrested replication forks. In parallel, we precipitated the Myc-tagged DNA pol ε to confirm fork position (data not shown). Using the indicated primers, we detect a strong enrichment of H2A-P at stalled forks in wild-type cells treated with HU, while in its absence we detect no significant phosphorylation of H2A-P (Fig. 7B,C). Thus H2A-P modification is specific to stalled forks and not to replication per se.
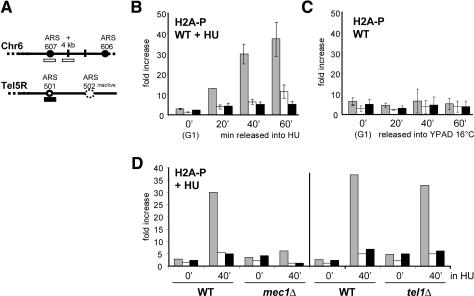
Figure 7.
Modification of H2A at replication forks is Mec1-specific. (A) Primers as previously described in Figure 3 were used to amplify the early-firing origin ARS607 (stippled) and an origin-proximal site (+4 kb; white), or late-firing origin ARS501 (black). (B) ChIP was performed on a wild-type (GA-2448) culture as described in Figure 3, released from α-factor into 0.2 M HU, using a phospho-specific rabbit polyclonal antibody recognizing the Ser 129-P H2A epitope (a gift from W. Bonner). Myc-tagged DNA pol ε was precipitated in parallel (data not shown). (C) ChIP for H2A-P as in B was performed on a wild-type culture following synchronous release from pheromone arrest into YPD at 16°C, in the absence of HU. (D) ChIP as described in B except that the strains used were mec1Δ (GA-2588) and tel1Δ (GA-2002). Here the ratio of absolute fold enrichments is reported after the rtPCR signals are normalized to a wild-type control in duplicate independent experiments: The scaling factor is 1.00 for mec1Δ and 0.268 for tel1Δ. Error bars were similar for both wild-type and mutant strains.
To see if this phosphorylation event is mediated by both Tel1 and Mec1, as shown for DSBs, we performed the HU arrest and quantitative H2A ChIP experiment in appropriate mutants. The amount of H2A-P at stalled forks in the mec1Δ strain drops to background levels, but there is no significant change in the tel1Δ strain (Fig. 7D). Given that there is no DNA PK homolog in yeast, this suggests that Mec1 alone modifies yeast H2A at stalled replication forks. This unique function underscores the singular importance of Mec1 during replication fork stalling and recovery (Cha and Kleckner 2002), quite apart from its ability to activate the downstream checkpoint kinase Rad53. The cross-talk between Mec1 and Sgs1 may be further reflected in the ability of RecQ helicases to be bound and potentially regulated by H2A-P (Nakamura et al. 2004; Sengupta et al. 2004).
Discussion
Chromosomal breaks and rearrangements are not only correlated with neoplastic transformation but also can cause malignancy. Previous work showed that in yeast an increase in both spontaneous and induced rearrangements is strikingly elevated in cells mutated for ATM, and even more so for cells lacking ATR, or its ortholog Mec1 (Kolodner et al. 2002). It was recently proposed that gross chromosomal rearrangements of this type are likely to arise from stalled forks (Lambert et al. 2005). We now use the synthetic behavior of a double mutant to show that this increase in chromosomal breaks and rearrangements correlates strictly with replication fork collapse, which entails a rapid displacement of DNA polymerases and RPA. This is due, in turn, to the loss of Mec1(ATR) kinase activity on substrates at sites of replication stalling. A partial loss-of-function mutant, mec1-100, which has very minor phenotypes on its own, has highly synergistic rates of GCR and replication fork collapse when coupled with deletion of the gene encoding the RecQ helicase Sgs1.
The simplest interpretation of our findings is as follows: In a strain that bears a mutated but catalytically active Mec1 kinase, mec1-100, we find a partial displacement of polymerases, although RPA remains bound and the Mec1-Ddc2 complex is recruited to stalled forks at near wild-type levels. The mec1-100 mutation allows a fairly efficient resumption of replication after removal of HU, although the strain shows a slight sensitivity to HU. We conclude that in this background there is a second pathway that stabilizes the replisome, or allows its reestablishment, enabling recovery from HU arrest. This second pathway depends almost entirely on the activity of the RecQ helicase, Sgs1, because in the mec1-100 sgs1Δ double mutant we observe a complete collapse of replication forks. This coincides with the displacement not only of replicative polymerases, but also of RPA. Coincident with these events, there is a synergistic increase in gross chromosomal rearrangements, presumably reflecting strand breakage, and the abolition of fork recovery potential. Enhanced strand breakage was also reported to occur on replicating DNA in Xenopus extracts depleted for XBLM helicase, although the mechanisms leading to such instability were not addressed (Li et al. 2004).
There are three important conclusions from our observations: First, based on the strong synergism observed coordinately for gross chromosomal rearrangements, polymerase displacement, and loss of fork recovery potential, we argue that these events are mechanistically linked. Second, we have identified fork-associated targets that are dependent on Rad53 checkpoint activation (MCM proteins) or that are unique to Mec1, being Rad53-independent (DNA polymerases and H2A-P at forks). Third, we elucidate the role played by Sgs1 helicase in this process and find that Sgs1 becomes essential to promote polymerase stability in the mec1-100 background. This suggests that Mec1/Ddc2 and Sgs1 contribute independently to polymerase stabilization, and that either Sgs1 or the partial mec1-100 activity is sufficient to ensure fork recovery (Fig. 8). Given that both Sgs1 and Mec1/Ddc2 bind RPA, and that RPA in turn promotes DNA pol α/primase initiation, it was not unexpected that both pathways for replisome stability converge on RPA, which itself is a target of checkpoint kinase modification (for review, see Binz et al. 2004).
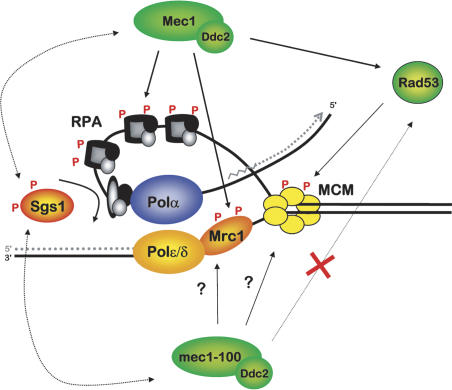
Figure 8.
Mec1/Ddc2 and Sgs1 stabilize RPA and DNA polymerases at stalled forks. This model summarizes the pathways that stabilize the replisome in cells exposed to HU. Sgs1, like large T antigen, is proposed to provoke a conformational change in RPA that promotes stable binding of DNA pol α as a primosome. Mec1/Ddc2 kinase also acts on Mrc1 to stabilize polymerases, while Rad53 either uncouples or displaces the MCM complex.
How can Sgs1 directly modulate RPA function, if the level of RPA bound at stalled forks does not change in an sgs1Δ strain? It is well-established that RPA can bind ssDNA in two modes, a high-affinity footprint that covers 29-30 nucleotides (nt), and a less tightly bound “primosome” complex that associates with DNA pol α/primase, leaving an RPA-DNA contact of ∼10 nt (for review, see Binz et al. 2004; Arunkumar et al. 2005). RPA is also known to bind the virally encoded helicase, large T antigen (Tag) through its 70-kDa and 32-kDa subunits. It was recently shown that interaction with Tag provokes a conformational change in RPA that strongly favors formation of a primosome complex with DNA pol α/primase, switching RPA's DNA-binding mode. Given that RecQ helicases, notably, BLM, WRN, and Sgs1, all bind the large RPA subunits with high affinity (Brosh et al. 2000; Cobb et al. 2003; Doherty et al. 2005), we propose that Sgs1, like Tag, may induce a conformational change in RPA that promotes its interaction with DNA pol α. This may, in turn, promote primosome formation at stalled forks (see Fig. 8). While the Sgs1 function is not absolutely essential in the presence of fully functional Mec1 kinase, it becomes critical for maintenance of replicative polymerases in the mec1-100 background. We propose that the maintenance of RPA at stalled forks in the sgs1Δ strain reflects the binding of RPA in its non-primosome, high-affinity form (Arunkumar et al. 2005). This may be influenced by checkpoint kinase-induced phosphorylation. Because Sgs1 is also necessary for maximal Mec1/Ddc2 levels at stalled forks, a tertiary complex of RPA, Sgs1, and Mec1/Ddc2 may also exist. Intriguingly, BLM and Sgs1 are targets of ATR-family kinases that are activated in response to fork-associated damage (Brush and Kelly 2000; Davies et al. 2004; Li et al. 2004).
What other targets of Mec1/Ddc2 are essential to stabilize the replisome? This pathway involves proteins other than the Sgs1 helicase, and is undoubtedly tightly regulated. With respect to DNA pol α and pol ε, it has been proposed that hyperphosphorylation of RPA by PI3-related kinases alters the interaction of RPA with several ligands, reducing its affinity for Tag, DNA pol α, and ATR, while increasing affinity for p53 (for review, see Binz et al. 2004). A partially modified form of RPA may change its mode of DNA binding such that it is able to maintain or re-establish contact with DNA pol α/primase, to allow resumption of DNA replication recovery once conditions improve. Mrc1 is also an important target of Mec1 kinase at stalled forks, and in mrc1Δ mutants, DNA pol ε is also partially destabilized on HU (Katou et al. 2003; Bjergbaek et al. 2005). Given that this effect is synergistic with loss of Sgs1, while loss of Rad53 is not (Fig. 4), we propose that Mrc1 also contributes to replisome stability at stalled forks on a pathway separate from Sgs1 (Fig. 8). Finally, we find that H2A phosphorylation at stalled forks depends on Mec1, but not Tel1. This observation clearly distinguishes the “stalled fork” response from the “DNA damage” response, where Mec11/Tel1 kinase redundancy has been established (Nakamura et al. 2004; Shroff et al. 2004). The modification of H2A at stalled forks may regulate the accessibility of DNA to enzymes involved in repair and fork restart.
A critical target of Mec1 kinase is, of course, Rad53, and we show here that a loss of Rad53 kinase activity leads to a drop in MCM levels at stalled forks, although fork-associated levels of DNA pol α and pol ε, and RPA, remain stable (Fig. 4; Tanaka and Nasmyth 1998; Cobb et al. 2003 for rad53Δ sml1Δ). We propose that some of the MCM modifications attributed to ATR-like kinases are actually due to the effector kinase Rad53 (Cortez et al. 2004; Yoo et al. 2004; Byun et al. 2005). The inefficient maintenance of MCM helicase could lead indirectly to fork collapse through an uncoupling of DNA unwinding from DNA synthesis. This is consistent with the high levels of ssDNA that accumulate at stalled forks in HU-arrested rad53-deficient cells (Sogo et al. 2002). Also consistent with our results, we note that DNA pol α levels actually increase at stalled forks in strains lacking Asf1, a histone chaperone and Rad53-binding protein, while MCM helicase becomes mislocalized from the replisome (Franco et al. 2005).
It is thus also possible that some of the defects in rad53 cells that lead to fork collapse are linked to the role of Rad53 in regulating histone levels during a checkpoint response. Rad53 down-regulates histones to release the histone chaperone Asf1 (Emili et al. 2001). Consistently, overexpression of Asf1 can partially suppress the lethality of a rad53 mutation on HU. Importantly, the ability of a cell to survive histone overexpression and degrade histones is independent of Mec1, and requires an intact Rad53 kinase (Gunjan and Verreault 2003). This further distinguishes the functions of Mec1 and Sgs1 during replicative stress from those of Rad53.
What are the implications of the dramatic synergy detected between a partial defect in the ATR kinase and RecQ helicase mutation? Since many cancer therapies still rely on DNA-damaging agents that create irreparable damage in S phase, our results support the argument that cell death might be significantly increased if ATR kinase and BLM helicase activities were coordinately compromised during treatment with HU or DNA intercalating drugs. To test this, it will be important to see if the ATR/RecQ synergy observed in yeast similarly enhances HU sensitivity in higher eukaryotic cells.
Materials and methods
All strains used are listed in Table 2 and are derived from either S288C for the GCR assays or W303-1a (MATa ade2-1 ura3-1 his3-11,15 trp1-1 leu2-3,112 can1-100) for all other experiments. Viability was calculated by plating ∼500 cells in triplicate onto YPD plates and scoring after 3 d at 30°C. Drop assays were a 1:5 dilution series of uniformly diluted cultures on YPD plates ± 10 mM HU.
Table 2.
S. cerevisiae strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| GA-180 | MATa,ade2-1, trp1-1, his3-11, -15, ura3-1, leu2-3,-112, can1-100 | R. Rothstein (W303-1A) |
| GA-1003 | MATa,ade2-1, trp1-1, his3-11, -15, ura3-1, leu2-3,-112, can1-100; CDC47-6Myc::URA | Tanaka and Nasmyth 1998 |
| GA-1020 | MATa,ade2-1, trp1-1, his3-11, -15, ura3-1, leu2-3,-112, can1-100, pep4::LEU2 | R. Rothstein (W303-1A) |
| GA-1113 | MATa,ade2-1, trp1-1, his3-11, -15, ura3-1, leu2-3,-112, can1-100; RFA1-18Myc::TRP1 from K7141 | This study |
| Tanaka and Nasmyth 1998 | ||
| GA-1761 | GA-1020 with sgs1::TRP1 | Bjergbaek et al. 2005 |
| GA-2002 | GA-1020 with tel1::URA3; Rad53-13Myc::KanMX6 | This study |
| GA-2238 | MATa,ade2-1, trp1-1, his3-11, -15, ura3-1, leu2-3,-112, can1-100, CDC17-3HA::TRP1 | Aparicio et al. 1999 |
| GA-2239 | MATa,ade2-1, trp1-1, his3-11, -15, ura3-1, leu2-3,-112, can1-100, CDC17-3HA::TRP1; rad53-11 | Aparicio et al. 1999 |
| GA-2240 | MATa,ade2-1, trp1-1, his3-11, -15, ura3-1, leu2-3,-112, can1-100 rad53-11 | Aparicio et al. 1999 |
| GA-2256 | GA-2238 with sgs1::LEU2 | This study |
| GA-2439 | GA-1113 with sgs1::LEU2 | This study |
| GA-2448 | GA-1020 with POL2-13Myc::KanMX6 | Bjergbaek et al. 2005 |
| GA-2450 | GA-1020 with sgs11-3::TRP1, POL2-13Myc::KanMX6 | Bjergbaek et al. 2005 |
| GA-2462 | MATa,ade2-1, trp1-1, his3-11, -15, ura3-1, leu2-3,-112, can1-100, DDC2-3HA::URA3, same as YLL683.8/4A | Paciotti et al. 2001 |
| GA-2463 | MATa,ade2-1, trp1-1, his3-11, -15, ura3-1, leu2-3,-112, can1-100, DDC2-3HA::URA3, mec1::HIS3, sml1::KanMX6, same as DMP3048 | Paciotti et al. 2001 |
| GA-2474 | MATa,ade2-1, trp1-1, his3-11, -15, ura3-1, leu2-3,-112, can1-100, mec1-100::LEU2(HIS3), derived from DMP3343/6C | This study |
| Paciotti et al. 2001 | ||
| GA-2475 | GA-2474 with DDC2-3HA::URA3 | This study |
| GA-2478 | MATa,ade2-1, trp1-1, his3-11, -15, ura3-1, leu2-3,-112, can1-100, mec1-100::LEU2(HIS3), sml1::KanMX6 derived from DMP3343/6C | This study |
| Paciotti et al. 2001 | ||
| GA-2514 | GA-2474 with sgs11-3::TRP1 | This study |
| GA-2515 | GA-2474 with POL2-13Myc::KanMX6 | This study |
| GA-2516 | GA-2474 with sgs11-3::TRP1, POL2-13Myc::KanMX6 | This study |
| GA-2519 | GA-2462 with sgs11-3::TRP1 | This study |
| GA-2567 | GA-2474 with CDC17-3HA::TRP1 | This study |
| GA-2571 | GA-2474 with RFA1-18Myc::TRP1 | This study |
| GA-2574 | GA-2239 with POL2-13Myc::KanMX6 | This study |
| GA-2576 | GA-2239 with sgs1::LEU2, POL2-13Myc::KanMX6 | This study |
| GA-2578 | GA-2474 with sgs11-3::LEU2; CDC17-3HA::TRP1 | This study |
| GA-2581 | GA-2474 with sgs11-3::LEU2; RFA1-18Myc::TRP1 | This study |
| GA-2582 | GA-1113 with mec1::HIS3, sml1::KanMX6 | This study |
| GA-2588 | GA-180 with POL2-13Myc::KanMX6; mec1::HIS3, sml1::KanMX6 | This study |
| GA-2895 | GA-180 with mec1::HIS3, sml1::KanMX6 | This study |
| GA-2896 | MATa,ade2-1, trp1-1, his3-11, -15, ura3-1, leu2-3,-112, can1-100, ORC2-9Myc::LEU2, sgs11-3::TRP1; mec1-100:: LEU2 (HIS3) | This study |
| GA-2897 | MATa,ade2-1, trp1-1, his3-11, -15, ura3-1, leu2-3,-112, can1-100, ORC2-9Myc::LEU2 | This study |
| GA-2930 | YJT110 with sgs1::KanMX6, mec1-100::LEU2(HIS3) | This study |
| GA-2931 | YJT110 with mec1-100::LEU2(HIS3) | This study |
| GA-3050 | MATa,CAN1, hxt13::URA3 same as E1557 | Lengronne and Schwob 2002 |
| GA-3053 | GA-3050 with mec1-100::LEU2(HIS3), sgs1::TRP | This study |
| GA-3054 | GA-1003 with rad53-11 | This study |
| GA-3056 | GA-3050 with sgs11-3::TRP | This study |
| GA-3057 | GA-3050 with mec1-100::LEU2(HIS3) | This study |
| GA-3062 | GA-3050 with rad53-11 | This study |
| GA-3063 | GA-3050 with rad53-11, sgs11-3::TRP | This study |
| YJT110 | W303 MATa with ARS608::HIS3;ARS609::TRP1, ADE+ | Tercero et al. 2003 |
| YVR1 | YJT110 with sgs1::KanMX6 | Tercero et al. 2003 |
GCR rates were calculated as in Myung and Kolodner (2002) for the following strains derived from the wild-type strain GA-3050 (same as E1557 in S288c background) (Lengronne and Schwob 2002): sgs1Δ (GA-3056), mec1-100 (GA-3057), and mec1-100 sgs1Δ (GA-3053), rad53-11 (GA-3062), rad53-11 sgs1Δ (GA-3063) cells were grown in YPD overnight to a density of 0.5 × 106 cells/mL, and incubated ± 0.2 M HU for 2 h, washed, and grown in YPD overnight. GCR rates were determined by scoring Canr-FOAr colonies due to loss of URA3 and CAN1 genes on Chr 5L. Values reported are from two to three different experiments using five colonies per strain, and mutation rates were calculated by fluctuation analysis (Lea and Coulson 1948).
ChIP was performed using either monoclonal antibodies against HA (12CA5) to precipitate HA-tagged Ddc2 and HA-tagged pol α, Myc (9E10) to precipitate Myc-tagged DNA pol ε, Myc-tagged Mcm7, and Myc-tagged Orc2, or phospho-specific rabbit polyclonal antibody against an epitope containing H2AS129P (a gift from W. Bonner) as described (Cobb et al. 2003), with IP washes at 0.5 M NaCl. In all cases cells were synchronized in G1 with α-factor at 30°C and then released into S phase in either the presence of 0.2 M HU at 30°C or YPD alone at 16°C. BSA-saturated Dynabeads incubated with the same cell extracts served as the background control for each time point. rtPCR quantifies DNA that was amplified with a Perkin-Elmer ABI Prism 7700 or 7000 Sequence Detector System. Sequences of the primers/probes used are available upon request. The data for each strain are averaged over two or three independent ChIP experiments with rtPCR performed in triplicate or duplicate where indicated (standard deviation is shown by error bars). The fold increase represents the ratio of the signal accumulation rates obtained from the antibody-coupled Dynabeads (IP) divided by the signal obtained from BSA-coated Dynabeads (background) after both signals were first normalized to the signal from the input fraction (Cobb et al. 2003). rtPCR monitors T½ within the exponential curve of product accumulation, and the replicate samples ensure a highly quantitative evaluation of product accumulation.
Neutral 2D gel analysis was performed as described (Huberman et al. 1987) with yeast genomic DNA isolated from 7 × 108 cells using a G-20 column (QIAGEN) followed by digestion with PstI and ClaI. Density transfer assays were performed and analyzed as described (Tercero et al. 2000). Rad53 in situ autophosphorylation assay (ISA) is described in Bjergbaek et al. (2005) and Pellicioli et al. (1999). Rat anti-RnaseH42 was kindly provided by U. Wintersberger (University of Vienna, Vienna, Austria) was used to normalize ISA signals.
Supplementary Material
Acknowledgments
We thank M.P. Longhese, W. Bonner and U. Wintersberger for reagents and P. Pasero, K. Shimada, H. Van Attikum, E. Fanning, and J. Diffley for helpful discussions. The Gasser laboratory thanks the Swiss Cancer League, Swiss National Science Foundation, European RTN Checkpoints and Cancer, and fellowships from the American Cancer Society to J.A.C. (PF-01-142-01-CCG) and the Danish Cancer Society to L.B. (DP00060). J.A.T.'s work is supported by grants BMC2003-00699 from Ministerio de Educación y Ciencia and GR/SAL/0144/2004 from Comunidad de Madrid and by an institutional grant from Fundación Ramón Areces to the CBMSO.
Supplemental material is available at http://www.genesdev.org.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.361805.
References
- Alcasabas A.A., Osborn, A.J., Bachant, J., Hu, F., Werler, P.J., Bousset, K., Furuya, K., Diffley, J.F., Carr, A.M., and Elledge, S.J. 2001. Mrc1 transduces signals of DNA replication stress to activate Rad53. Nat. Cell Biol. 3: 958-965. [DOI] [PubMed] [Google Scholar]
- Aparicio O.M., Stout, A.M., and Bell, S.P. 1999. Differential assembly of Cdc45p and DNA polymerases at early and late origins of DNA replication. Proc. Natl. Acad. Sci. 96: 9130-9135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arunkumar A.I., Klimovich, V., Jiang, X., Ott, R.D., Mizoue, L., Fanning, E., and Chazin, W.J. 2005. Insights into hRPA32 C-terminal domain-mediated assembly of the simian virus 40 replisome. Nat. Struct. Mol. Biol. 12: 332-339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartrand A.J., Iyasu, D., and Brush, G.S. 2004. DNA stimulates Mec1-mediated phosphorylation of replication protein A. J. Biol. Chem. 279: 26762-26767. [DOI] [PubMed] [Google Scholar]
- Binz S.K., Sheehan, A.M., and Wold, M.S. 2004. RPA phosphorylation and the cellular response to DNA damage. DNA repair 3: 1015-1024. [DOI] [PubMed] [Google Scholar]
- Bjergbaek L., Cobb, J.A., Tsai-Pflugfelder, M., and Gasser, S.M. 2005. Mechanistically distinct roles for the RecQ helicase Sgs1p at stalled replication forks. EMBO J. 24: 405-417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosh Jr R.M., Li, J.L., Kenny, M.K., Karow, J.K., Cooper, M.P., Kureekattil, R.P., Hickson, I.D., and Bohr, V.A. 2000. Replication protein A physically interacts with the Bloom's syndrome protein and stimulates its helicase activity. J. Biol. Chem. 275: 23500-23508. [DOI] [PubMed] [Google Scholar]
- Brush G.S. and Kelly, T.J. 2000. Phosphorylation of the RPA large subunit in the S. cerevisiae checkpoint response. Nucleic Acids Res. 28: 3725-3732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brush G.S., Morrow, D.M., Hieter, P., and Kelly, T.J. 1996. The ATM homologue MEC1 is required for phosphorylation of RPA in yeast. Proc. Natl. Acad. Sci. 93: 15075-15080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byun T.S., Pacek, M., Yee, M.C., Walter, J.C., and Cimprich, K.A. 2005. Functional uncoupling of MCM helicase and DNA polymerase activities activates the ATR-dependent checkpoint. Genes & Dev. 19: 1040-1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha R.S. and Kleckner, N. 2002. ATR homolog Mec1 promotes fork progression, thus averting breaks in replication slow zones. Science 297: 602-606. [DOI] [PubMed] [Google Scholar]
- Cobb J.A., Bjergbaek, L., Shimada, K., Frei, C., and Gasser, S.M. 2003. DNA polymerase stabilization at stalled replication forks requires Mec1 and the RecQ helicase Sgs1. EMBO J. 22: 4325-4336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortez D., Glick, G., and Elledge, S.J. 2004. Minichromosome maintenance proteins are direct targets of the ATM and ATR checkpoint kinases. Proc. Natl. Acad. Sci. 101: 10078-10083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies S.L., North, P.S., Dart, A., Lakin, N.D., and Hickson, I.D. 2004. Phosphorylation of the Bloom's syndrome helicase and its role in recovery from S-phase arrest. Mol. Cell. Biol. 24: 1279-1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desany B.A., Alcasabas, A.A., Bachant, J.B., and Elledge, S.J. 1998. Recovery from DNA replicational stress is the essential function of the S-phase checkpoint pathway. Genes & Dev. 12: 2956-2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doherty K.M., Sommers, J.A., Gray, M.D., Lee, J.W., von Kobbe, C., Thoma, N.H., Kureekattil, R.P., Kenny, M.K., and Brosh Jr., R.M. 2005. Physical and functional mapping of the RPA interaction domain of the Werner and Bloom syndrome helicases. J. Biol. Chem. 280: 29494-29505. [DOI] [PubMed] [Google Scholar]
- Downs J.A., Lowndes, N.F., and Jackson, S.P. 2000. A role for S. cerevisiae histone H2A in DNA repair. Nature 408: 1001-1004. [DOI] [PubMed] [Google Scholar]
- Emili A., Schieltz, D.M., Yates III, J.R., and Hartwell, L.H. 2001. Dynamic interaction of DNA damage checkpoint protein Rad53 with chromatin assembly factor Asf1. Mol. Cell 7: 13-20. [DOI] [PubMed] [Google Scholar]
- Franco A.A., Lam, W.M., Burgers, P.M., and Kaufman, P.D. 2005. Histone deposition protein Asf1 maintains DNA replisome integrity and interacts with replication factor C. Genes & Dev. 19: 1365-1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frei C. and Gasser, S.M. 2000. The yeast Sgs1 helicase acts upstream of Rad53 in the DNA replication checkpoint and colocalizes with Rad53 in S-phase-specific foci. Genes & Dev. 14: 81-96. [PMC free article] [PubMed] [Google Scholar]
- Gunjan A. and Verreault, A. 2003. Rad53 kinase-dependent surveillance mechanism that regulates histone protein levels in S. cerevisiae. Cell 115: 537-549. [DOI] [PubMed] [Google Scholar]
- Halazonetis T.D. 2004. Constitutively active DNA damage checkpoint pathways as the driving force for the high frequency of p53 mutations in human cancer. DNA Repair 3: 1057-1062. [DOI] [PubMed] [Google Scholar]
- Huberman J.A., Spotila, L.D., Nawotka, K.A., el-Assouli, S.M., and Davis, L.R. 1987. The in vivo origin of the yeast 2 micron plasmid. Cell 51: 473-481. [DOI] [PubMed] [Google Scholar]
- Ivessa A.S., Lenzmeier, B.A., Bessler, J.B., Goudsouzian, L.K., Schnakenberg, S.L., and Zakian, V.A. 2003. The S. cerevisiae helicase Rrm3p facilitates replication past nonhistone protein-DNA complexes. Mol. Cell 12: 1525-1536. [DOI] [PubMed] [Google Scholar]
- Katou Y., Kanoh, Y., Bando, M., Noguchi, H., Tanaka, H., Ashikari, T., Sugimoto, K., and Shirahige, K. 2003. S-phase checkpoint proteins Tof1 and Mrc1 form a stable replication-pausing complex. Nature 424: 1078-1083. [DOI] [PubMed] [Google Scholar]
- Kim H.S. and Brill, S.J. 2003. MEC1-dependent phosphorylation of yeast RPA1 in vitro. DNA Repair 2: 1321-1335. [DOI] [PubMed] [Google Scholar]
- Kolodner R.D., Putnam, C.D., and Myung, K. 2002. Maintenance of genome stability in Saccharomyces cerevisiae. Science 297: 552-557. [DOI] [PubMed] [Google Scholar]
- Lambert S., Watson, A., Sheedy, D.M., Martin, B., and Carr, A.M. 2005. Gross chromosomal rearrangements and elevated recombination at an inducible site-specific replication fork barrier. Cell 121: 689-702. [DOI] [PubMed] [Google Scholar]
- Lea D.E. and Coulson, C.A. 1948. The distribution of the numbers of mutants in bacterial populations. J. Genet. 49: 264-285. [DOI] [PubMed] [Google Scholar]
- Lengronne A. and Schwob, E. 2002. The yeast CDK inhibitor Sic1 prevents genomic instability by promoting replication origin licensing in late G1. Mol. Cell 9: 1067-1078. [DOI] [PubMed] [Google Scholar]
- Li W., Kim, S.M., Lee, J., and Dunphy, W.G. 2004. Absence of BLM leads to accumulation of chromosomal DNA breaks during both unperturbed and disrupted S phases. J. Cell Biol. 165: 801-812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberi G., Maffioletti, G., Lucca, C., Chiolo, I., Baryshnikova, A., Cotta-Ramusino, C., Lopes, M., Pellicioli, A., Haber, J.E., and Foiani, M. 2005. Rad51-dependent DNA structures accumulate at damaged replication forks in sgs1 mutants defective in the yeast ortholog of BLM RecQ helicase. Genes & Dev. 19: 339-350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J.S., Kuo, S.R., and Melendy, T. 2003. Comparison of checkpoint responses triggered by DNA polymerase inhibition versus DNA damaging agents. Mutat. Res. 532: 215-226. [DOI] [PubMed] [Google Scholar]
- Lopes M., Cotta-Ramusino, C., Pellicioli, A., Liberi, G., Plevani, P., Muzi-Falconi, M., Newlon, C.S., and Foiani, M. 2001. The DNA replication checkpoint response stabilizes stalled replication forks. Nature 412: 557-561. [DOI] [PubMed] [Google Scholar]
- Lucca C., Vanoli, F., Cotta-Ramusino, C., Pellicioli, A., Liberi, G., Haber, J., and Foiani, M. 2004. Checkpoint-mediated control of replisome-fork association and signalling in response to replication pausing. Oncogene 23: 1205-1213. [DOI] [PubMed] [Google Scholar]
- Mohaghegh P. and Hickson, I.D. 2001. DNA helicase deficiencies associated with cancer predisposition and premature ageing disorders. Hum. Mol. Genet. 10: 741-746. [DOI] [PubMed] [Google Scholar]
- Myung K. and Kolodner, R.D. 2002. Suppression of genome instability by redundant S-phase checkpoint pathways in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. 99: 4500-4507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T.M., Du, L.L., Redon, C., and Russell, P. 2004. Histone H2A phosphorylation controls Crb2 recruitment at DNA breaks, maintains checkpoint arrest, and influences DNA repair in fission yeast. Mol. Cell. Biol. 24: 6215-6230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborn A.J. and Elledge, S.J. 2003. Mrc1 is a replication fork component whose phosphorylation in response to DNA replication stress activates Rad53. Genes & Dev. 17: 1755-1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paciotti V., Clerici, M., Lucchini, G., and Longhese, M.P. 2000. The checkpoint protein Ddc2, functionally related to S. pombe Rad26, interacts with Mec1 and is regulated by Mec1-dependent phosphorylation in budding yeast. Genes & Dev. 14: 2046-2059. [PMC free article] [PubMed] [Google Scholar]
- Paciotti V., Clerici, M., Scotti, M., Lucchini, G., and Longhese, M.P. 2001. Characterization of mec1 kinase-deficient mutants and of new hypomorphic mec1 alleles impairing subsets of the DNA damage response pathway. Mol. Cell. Biol. 21: 3913-3925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellicioli A., Lucca, C., Liberi, G., Marini, F., Lopes, M., Plevani, P., Romano, A., Di Fiore, P.P., and Foiani, M. 1999. Activation of Rad53 kinase in response to DNA damage and its effect in modulating phosphorylation of the lagging strand DNA polymerase. EMBO J. 18: 6561-6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouse J. and Jackson, S.P. 2002. Lcd1p recruits Mec1p to DNA lesions in vitro and in vivo. Mol. Cell 9: 857-869. [DOI] [PubMed] [Google Scholar]
- Santocanale C. and Diffley, J.F. 1998. A Mec1- and Rad53-dependent checkpoint controls late-firing origins of DNA replication. Nature 395: 615-618. [DOI] [PubMed] [Google Scholar]
- Sengupta S., Robles, A.I., Linke, S.P., Sinogeeva, N.I., Zhang, R., Pedeux, R., Ward, I.M., Celeste, A., Nussenzweig, A., Chen, J., et al. 2004. Functional interaction between BLM heli-case and 53BP1 in a Chk1-mediated pathway during S-phase arrest. J. Cell Biol. 166: 801-813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shroff R., Arbel-Eden, A., Pilch, D., Ira, G., Bonner, W.M., Petrini, J.H., Haber, J.E., and Lichten, M. 2004. Distribution and dynamics of chromatin modification induced by a defined DNA -double-strand break. Curr. Biol. 14: 1703-1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sogo J.M., Lopes, M., and Foiani, M. 2002. Fork reversal and ssDNA accumulation at stalled replication forks owing to checkpoint defects. Science 297: 599-602. [DOI] [PubMed] [Google Scholar]
- Tanaka T. and Nasmyth, K. 1998. Association of RPA with chromosomal replication origins requires an Mcm protein, and is regulated by Rad53, and cyclin- and Dbf4-dependent kinases. EMBO J. 17: 5182-5191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tercero J.A. and Diffley, J.F. 2001. Regulation of DNA replication fork progression through damaged DNA by the Mec1/Rad53 checkpoint. Nature 412: 553-557. [DOI] [PubMed] [Google Scholar]
- Tercero J.A., Labib, K., and Diffley, J.F.X. 2000. DNA synthesis at individual replication forks requires the essential initiation factor Cdc45p. EMBO J. 19: 2082-2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tercero J.A., Longhese, M.P., and Diffley, J.F. 2003. A central role for DNA replication forks in checkpoint activation and response. Mol. Cell 11: 1323-1336. [DOI] [PubMed] [Google Scholar]
- Thiriet C. and Hayes, J.J. 2005. Chromatin in need of a fix: Phosphorylation of H2AX connects chromatin to DNA repair. Mol. Cell 18: 617-622. [DOI] [PubMed] [Google Scholar]
- Versini G., Comet, I., Wu, M., Hoopes, L., Schwob, E., and Pasero, P. 2003. The yeast Sgs1 helicase is differentially required for genomic and ribosomal DNA replication. EMBO J. 22: 1939-1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watt P.M., Hickson, I.D., Borts, R.H., and Louis, E.J. 1996. SGS1, a homologue of the Bloom's and Werner's syndrome genes, is required for maintenance of genome stability in S. cerevisiae. Genetics 144: 935-945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinert T.A., Kiser, G.L., and Hartwell, L.H. 1994. Mitotic checkpoint genes in budding yeast and the dependence of mitosis on DNA replication and repair. Genes & Dev. 8: 652-665. [DOI] [PubMed] [Google Scholar]
- Yoo H.Y., Shevchenko, A., Shevchenko, A., and Dunphy, W.G. 2004. Mcm2 is a direct substrate of ATM and ATR during DNA damage and DNA replication checkpoint responses. J. Biol. Chem. 279: 53353-53364. [DOI] [PubMed] [Google Scholar]
- Zhao X., Muller, E.G., and Rothstein, R. 1998. A suppressor of two essential checkpoint genes identifies a novel protein that negatively affects dNTP pools. Mol. Cell 2: 329-340. [DOI] [PubMed] [Google Scholar]
- Zou L. and Elledge, S.J. 2003. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science 300: 1542-1548. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.