Abstract
Our studies in yeast show that there is an essential requirement for either an active thioredoxin or an active glutathione (GSH)–glutaredoxin system for cell viability. Glutathione reductase (Glr1) and thioredoxin reductase (Trr1) are key regulatory enzymes that determine the redox state of the GSH–glutaredoxin and thioredoxin systems, respectively. Here we show that Trr1 is required during normal cell growth, whereas there is no apparent requirement for Glr1. Analysis of the redox state of thioredoxins and glutaredoxins in glr1 and trr1 mutants reveals that thioredoxins are maintained independently of the glutathione system. In contrast, there is a strong correlation between the redox state of glutaredoxins and the oxidation state of the GSSG/2GSH redox couple. We suggest that independent redox regulation of thioredoxins enables cells to survive in conditions under which the GSH–glutaredoxin system is oxidized.
Introduction
Recent reports highlight the key roles played by thiol groups, and in particular the glutathione (GSH)–glutaredoxin and thioredoxin systems in maintaining redox homeostasis in the cell (see, for example, Carmel-Harel & Storz, 2000; Grant, 2001). Glutaredoxins and thioredoxins are small oxidoreductases with two conserved cysteine residues in their active sites (Holmgren, 1989). They were originally identified as hydrogen donors for ribonucleotide reductase, but they also regulate many metabolic enzymes that form disulphides during their catalytic cycle (Rietsch & Beckwith, 1998). Roles have been proposed for them in many cellular processes including protein folding and regulation, reduction of dehydroascorbate, repair of oxidatively damaged proteins, and sulphur metabolism (Holmgren, 1989; Rietsch & Beckwith, 1998). Glutaredoxins and thioredoxins are structurally similar and have been conserved throughout evolution, particularly in the region of their active site (Holmgren & Aslund, 1995). They are thought to be differentially regulated, although both systems obtain reducing power from NADPH. The oxidized form of thioredoxin is reduced directly by NADPH and thioredoxin reductase, whereas glutaredoxin is reduced by GSH using electrons donated by NADPH via glutathione reductase.
The yeast Saccharomyces cerevisiae contains two genes encoding glutaredoxins (GRX1 and GRX2) that share extensive homology with glutaredoxins from bacterial and mammalian species (Luikenhuis et al., 1997). Strains deleted for both genes are viable, but lack heatstable oxidoreductase activity with β-hydroxyethylene disulphide as a model disulphide substrate. The yeast glutaredoxins act as antioxidants and have activity as glutathione peroxidases (Luikenhuis et al., 1997; Collinson et al., 2002). Yeast also contains two genes encoding cytoplasmic thioredoxins (TRX1 and TRX2) which are dispensable during normal growth conditions (Gan, 1991; Muller, 1991). Deletion of both TRX1 and TRX2 affects the cell cycle, prolonging S phase and shortening the G1 interval (Muller, 1991). Thioredoxin function is required for protection against reactive oxygen species (ROS) and provides reducing power for various thioredoxin peroxidase isoenzymes (Park et al., 2000). Genetic screens have identified the yeast thioredoxin reductase (TRR1) (Machado et al., 1997; Pearson & Merril, 1998) but little is known about its role in maintaining the cellular redox environment.
The first evidence that the thioredoxin and GSH–glutaredoxin systems have an overlapping function in redox regulation came from the identification of GLR1, encoding glutathione reductase, in a genetic screen for mutations that confer a requirement for thioredoxins (Muller, 1996). Loss of both TRX1 and TRX2 also results in elevated concentrations of glutathione in both the reduced (GSH) and oxidized (GSSG) forms, indicating a link between the thioredoxin system and glutathione concentrations in the cell (Muller, 1996; Garrido & Grant, 2002). Furthermore, similar to bacteria (Prinz et al., 1997), a quadruple trx1 trx2 grx1 grx2 yeast mutant is non-viable and a single functional disulphide reductase system is necessary for viability (Draculic et al., 2000). Here we investigate further the functional overlap between the GSH–glutaredoxin and thioredoxin systems by focusing on thioredoxin reductase and glutathione reductase. We provide the first evidence in vivo that, unlike glutaredoxins, the redox state of the thioredoxin system is maintained independently of the glutathione system.
Results and Discussion
Viability depends on functional redoxin systems
Previous studies have shown that glr1 trx1 trx2 and trx1 trx2 grx1 grx2 mutants are non-viable (Muller, 1996; Draculic et al., 2000). To test further the requirement for components of the thioredoxin and GSH–glutaredoxin systems, we attempted to construct mutants lacking components of each system. Strains completely lacking the thioredoxin (trx1 trx2 trr1) or glutathione (gsh1 glr1) systems are viable, but strains simultaneously deleted for components of both systems (gsh1 trx1 trx2) and (glr1 trr1) are non-viable (Table 1 and Fig. 1). These results confirm that a single functional disulphide reduction system is essential for viability. The lethality of the glr1 trr1 mutant is somewhat surprising given that this strain should still be able to synthesize GSH and thioredoxins, which are both made in the reduced form. Furthermore, the lethality of the glr1 trr1 mutant could not be rescued by anaerobic growth or by the addition of exogenous GSH, indicating that it is not due to an accumulation of toxic disulphide bonds (data not shown). The basis of the lethality must therefore be the loss of activity of an essential metabolic enzyme such as ribonucleotide reductase. Because glutathione reductase and thioredoxin reductase are key enzymes determining the redox state of the GSH–glutaredoxin and thioredoxin systems respectively, we further compared the glr1 and trr1 mutants to try to understand the overlapping nature of the two systems.
Table 1.
Requirement for components of the GSH–glutaredoxin and thioredoxin systems
| Genotype | Viability |
|---|---|
| glr1 trx1 trx2 | Non-viable1 |
| trx1 trx2 grx1 grx2 | Non-viable2 |
| gsh1 trx1 | Viable |
| gsh1 trx2 | Viable |
| gsh1 trx1 trx2 | Non-viable |
| gsh1 glr1 | Viable |
| trr1 glr1 | Non-viable |
| trr1 trx1 | Viable |
| trr1 trx2 | Viable |
| trr1 trx1 trx2 | Viable |
Yeast genetic analysis was used to determine the viability of mutants lacking multiple components of the GSH–glutaredoxin and thioredoxin systems.
1From Muller (1996).
2From Draculic et al. (2000).
Figure 1.
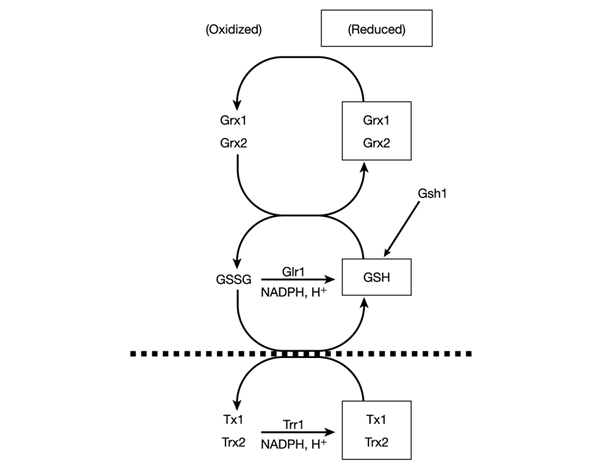
Requirement for components of the GSH–glutaredoxin and thioredoxin systems. Overlapping nature of the GSH–glutaredoxin and thioredoxin systems. The oxidized disulphide form of thioredoxins is reduced directly by NADPH and thioredoxin reductase, whereas oxidized glutaredoxins are reduced by GSH, and oxidized GSSG is in turn reduced in an NADPH-dependent reaction catalysed by glutathione reductase. The redox status of GSH might provide a functional link between the GSH–glutaredoxin and thioredoxin systems because thioredoxins function together with Glr1 to maintain the high intracellular GSH:GSSG ratio. Mutants lacking components of the GSH–glutaredoxin system (above the dotted line) or the thioredoxin system (below the dotted line) are viable. In contrast, mutants that inactivate both the GSH–glutaredoxin and thioredoxin systems are non-viable (see Table 1).
Growth phenotypes of reductase mutants
Previous studies have examined the growth kinetics of glr1 (Grant et al., 1996a) and trr1 (Trotter & Grant, 2002) mutant strains. During fermentative growth, the kinetics of the glr1 mutant was identical to that of the wild-type strain, whereas the trr1 mutant displayed a more prolonged lag phase and a slow growth rate. We reasoned that a growth requirement for Glr1 might become apparent during respiratory growth conditions that would be expected to generate intracellular ROS. The growth of strains was therefore compared in minimal medium containing glycerol and ethanol as respiratory carbon sources. Cells were inoculated to the same initial starting density in minimal SGE medium (see Methods) and monitored throughout the various phases of growth (Fig. 2). However, similarly to fermentative growth, the strain deleted for GLR1 grew at the same rate and reached a similar final cell yield to that of the wild-type strain. In contrast, the strain deleted for trr1 showed a longer lag phase and slower growth rate before reaching a similar cell yield. These findings indicate that Trr1 is required during both fermentative and respiratory cell growth, whereas there is no apparent requirement for Glr1.
Figure 2.
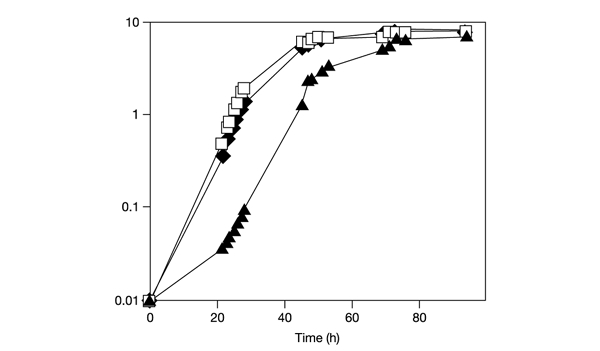
Growth of glutathione reductase and thioredoxin reductase mutants. Wild-type cells (filled diamonds), trr1 mutant cells (filled triangles) and glr1 mutant cells (open squares) were inoculated to the same initial cell density in minimal medium containing glycerol plus ethanol (initial optical density at 600 nm (OD600) = 0.01) and growth was monitored by measuring OD600.
Concentrations of redoxin proteins in trr1 and glr1 mutants
To begin to address the reasons behind the growth differences in the trr1 and glr1 mutants we examined the concentrations of glutaredoxin and thioredoxin proteins by western blot analysis with the use of specific antibodies (Fig. 3). Antibodies were generated against recombinant yeast Trx1 and Grx1, but recognize both Trx1 and Trx2, and Grx1 and Grx2, respectively (data not shown). The amounts of glutaredoxin and thioredoxin proteins were unaffected by the loss of GLR1 compared with the wild-type strain. Glutaredoxin concentrations were also unaffected by the loss of TRR1, whereas there was an elevation of thioredoxins in the trr1 mutant. This is consistent with the increased yAP1-dependent TRX2 transcription previously described for a trr1 mutant (Carmel-Harel & Storz, 2000). Thus, the growth defect of the trr1 mutant is not due to a deficiency in the concentrations of glutaredoxin or thioredoxin proteins.
Figure 3.
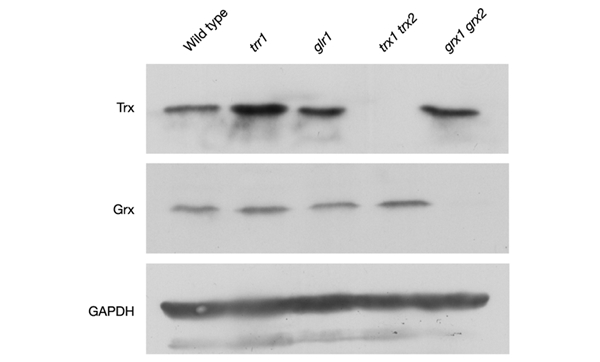
Western blot analysis of glutaredoxin and thioredoxin protein concentrations. Glutaredoxin and thioredoxin protein concentrations were measured in wild-type and glr1, trr1, trx1 trx2 and grx1 grx2 mutant strains by western blot analysis with antibodies specific for thioredoxins (Trx), glutaredoxins (Grx) and glyceraldehyde-3-phosphate dehydrogenase (GAPDH).
Redox status of redoxins in trr1 and glr1 mutants
We next examined the oxidation state of thioredoxins and glutaredoxins in vivo. The cellular oxidation state was preserved by rapidly treating cells with trichloroacetic acid, which protonates free thiol groups. Extracts were reacted with the thiol-specific probe 4-acetamido-4′-maleimidyldystilbene-2,2′-disulphonic acid (AMS), followed by separation by SDS–polyacrylamide gel electrophoresis (SDS– PAGE) and western blot analysis. The migration of all Trx protein from the wild-type strain was decreased after treatment with AMS, indicating that the vast majority is present in the reduced form (Fig. 4A). In contrast, the reduced and oxidized forms of glutaredoxin were present in the wild-type strain in approximately equal amounts.
Figure 4.
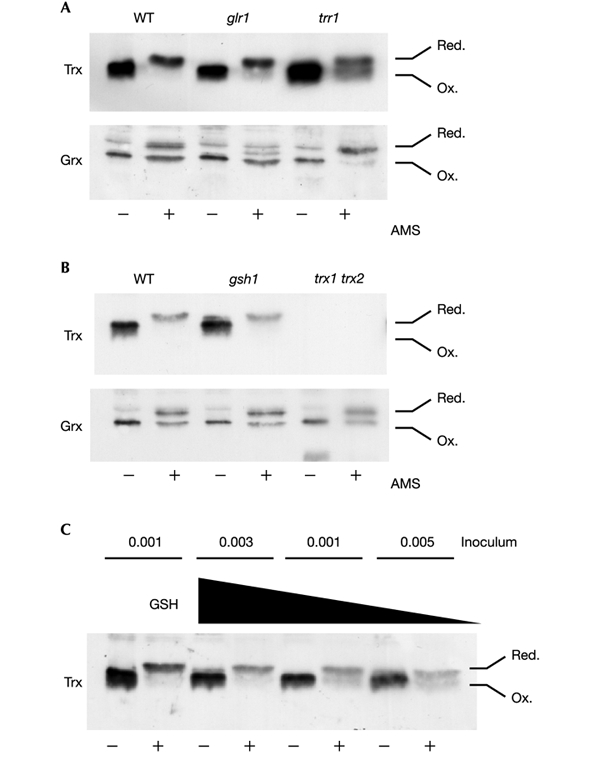
Redox state of glutaredoxins and thioredoxins. Proteins were precipitated with trichloroacetic acid and free thiols were modified by reaction with AMS. Samples were separated by SDS–18% PAGE, and thioredoxins (Trx) and glutaredoxins (Grx) were detected by western blot analysis. Fully oxidized and fully reduced proteins are indicated. (A) Wild-type, glr1 and trr1 mutants; (B) wild-type, gsh1 and trx1 trx2 mutants grown to exponential phase in YEPD medium. The middle glutaredoxin band in the glr1 mutant might have arisen from partial oxidation. (C) Oxidation of thioredoxins in response to GSH depletion. The gsh1 mutant was grown overnight in YEPD medium and washed with distilled water to remove any exogenous GSH. It was then inoculated into SD medium without GSH at different initial cellular concentrations; growth continued for 19 h (OD600 = 0.03, 0.01 and 0.005; lanes 3–8). As a control, the gsh1 mutant was also inoculated at a starting OD600 of 0.001 in the presence of 0.1 mM GSH (lanes 1 and 2).
As might be expected, a loss of TRR1 shifted the thioredoxin redox balance to a more oxidized state, with approximately equal amounts of reduced and oxidized forms present (Fig. 4A). The presence of high concentrations of reduced thioredoxin in a trr1 mutant is unexpected. This finding might indicate that the synthesis of thioredoxins, which are made in the reduced form, is sufficient to maintain a pool of reduced thioredoxin; alternatively, an additional thioredoxin reduction system might exist that can recycle oxidized thioredoxin to the reduced form. In contrast, a loss of GLR1 did not effect the redox state of thioredoxins. This is somewhat surprising given that loss of GLR1 is known to shift the cellular redox balance to a more oxidized state, when the GSH:GSSG ratio is assessed (Grant et al., 1996a; Muller, 1996) (Table 2). These results indicate that the redox state of thioredoxins is maintained largely independently of the glutathione system. In contrast, in the glr1 mutant, glutaredoxins are present in a predominantly oxidized form consistent with the GSSG redox state (Fig. 4A and Table 2). Surprisingly, glutaredoxin was present in a more reduced form in the trr1 mutant. This was further investigated by examining the redox state of glutaredoxins in a strain lacking thioredoxins (Fig. 4B, trx1 trx2). The glutaredoxin redox balance was unaffected by the loss of thioredoxins.
Table 2.
Glutathione concentrations and redox state
| Strain | 1GSH (nmol A−1 ml−1) | 1GSSG (nmol A−1 ml−1) | GSH:GSSG | 2Ehc (mV) |
|---|---|---|---|---|
| Wild type | 3.80 ± 0.25 | 0.059 ± 0.004 | 64 | −222 |
| trx1 trx2 | 12.20 ± 0.62 | 0.86 ± 0.083 | 14 | −218 |
| trr1 | 12.35 ± 0.81 | 0.17 ± 0.044 | 72 | −239 |
| glr1 | 3.78 ± 0.16 | 1.26 ± 0.16 | 3 | −183 |
| grx1 grx2 | 3.56 ± 0.12 | 0.064 ± 0.011 | 56 | −219 |
1Data shown are taken from Garrido & Grant (2002) and Trotter & Grant (2002).
2Redox potential was calculated from the Nernst equation as described in Methods.
Redox potential and glutaredoxin redox state
In the light of the glutaredoxin and thioredoxin redox state data above, we examined the redox state of the glutathione system. Our previous studies measured the cellular concentrations of GSSG and GSH in glutathione/glutaredoxin and thioredoxin mutants (Garrido & Grant, 2002; Trotter & Grant, 2002). Mutants deleted for GLR1, TRR1 and TRX1 TRX2 contain elevated concentrations of GSSG, with a threefold increase in the trr1 mutant, a 15-fold increase in the trx1 trx2 mutant and a 21-fold increase in the glr1 mutant (Table 2). In contrast, loss of glutaredoxins does not affect glutathione concentration or redox state. Additionally, a loss of thioredoxins or thioredoxin reductase results in a threefold increase in the concentration of reduced GSH. This is consistent with the increase in GSH1 and GLR1 expression in thioredoxin mutants which is mediated by the transcription factor yAP-1 (reviewed in Carmel-Harel & Storz, 2000). Thus, the redox state of glutathione, expressed simply as the ratio of GSH to GSSG, is shifted towards a more oxidized form in the glr1 and trx1 trx2 mutants, whereas in the trr1 mutant it is similar to that of the wild type (Table 2).
The redox potential of the glutathione system is better expressed as the redox state of the GSSG/2GSH redox couple, calculated from the Nernst equation, which takes into account the reducing capacity of the redox couple, with the stoichiometry of two GSH molecules per GSSG molecule (Schafer & Buettner, 2001). This analysis showed that the redox potential (Table 2) was unaffected in the trx1 trx2 and grx1 grx2 mutants compared with the wild-type strain. In contrast, a loss of GLR1 caused a 39 mV oxidation, whereas a loss of TRR1 caused a 17 mV reduction. The measured changes in redox state of thioredoxins and glutaredoxins in the trr1 and glr1 mutants are therefore consistent with the known redox potentials of thioredoxins and glutaredoxins, because thioredoxins are generally much more negative (reducing) than glutaredoxins (Aslund et al., 1997; Schafer & Buettner, 2001). Thus, there is a strong correlation between the redox states of glutaredoxins (Fig. 4) and the glutathione pool (Table 2), indicating that the glutaredoxin redox state is determined by the GSSG/2GSH redox couple, whereas the thioredoxin redox state is primarily maintained by thioredoxin reductase.
Thioredoxins are oxidized in the absence of GSH
Strains lacking GSH1 are viable but require a source of exogenous GSH for growth. For example, the gsh1 mutant grows normally on YEPD medium (see Methods), which contains about 0.5 mM GSH (Wu & Moye-Rowley, 1994; Lee et al., 2000). The gsh1 mutant grown on YEPD medium also maintains the wild-type redox state of glutaredoxins and thioredoxins (Fig. 4B). We have previously shown that the gsh1 mutant accumulates intracellular GSH from an external medium such as YEPD. This means that the growth of a gsh1 mutant on medium lacking GSH is entirely dependent on the size of the inoculum used, owing to the intracellular GSH accumulated during pregrowth on YEPD medium (Lee et al., 2000). We could therefore test directly whether the depletion of GSH affects the redox state of thioredoxins by pregrowing the gsh1 mutant in YEPD medium, washing to remove exogenous GSH, and inoculating at different initial cell densities in minimal medium. In the absence of GSH, progressively more oxidized thioredoxin was detected in the gsh1 mutant as the size of the inoculum was decreased (Fig. 4C). In comparison, the gsh1 mutant grown in minimal medium containing GSH maintained thioredoxins in the reduced form (Fig. 4C). These data indicate that as GSH is depleted, thioredoxins become increasingly oxidized. Thus, although shifting the glutathione pool to a more oxidized form in the glr1 mutant does not affect the redox state of thioredoxins, the complete absence of glutathione does result in thioredoxin oxidation.
Summary and conclusions
Glutathione is ubiquitous in eukaryotic cells and has long been considered as the major cellular redox buffer, with the redox state of the glutathione system taken as an indicator of the cellular redox environment (Meister & Anderson, 1983). Our studies show that there is an essential cellular requirement for either an active thioredoxin or GSH–glutaredoxin system. In a glr1 mutant, glutaredoxins exist predominantly in an oxidized, and hence inactivated, form. Independent redox regulation of the thioredoxin system ensures that it can be maintained in conditions under which the GSH–glutaredoxin system is inactive. In contrast with the glr1 mutant, loss of TRR1 not only results in the oxidation of thioredoxins but also shifts the redox state of glutaredoxins to a more reduced form. Simultaneous defects in both disulphide reduction systems might therefore account for the growth defect observed in the trr1 mutant.
Methods
Yeast strains and growth conditions.
Saccharomyces cerevisiae strains used in this study were all isogenic derivatives of CY4 (MATa ura3-52 leu2-3 leu2-112 trp1-1 ade2-1 his3-11 can1-100) (Grant et al., 1996b). Strains deleted for glutaredoxins (grx1::LEU2 grx2::HIS3), thioredoxins (trx1::TRP1 trx2::URA3), glutathione reductase (glr1::TRP1), thioredoxin reductase (trr1::HIS3) and γ-glutamylcysteine synthetase (gsh1::LEU2) have all been described previously (Trotter & Grant, 2002). Standard yeast genetic techniques were used to test the requirements for different components of the GSH–glutaredoxin and thioredoxin systems. Strains were grown in rich YEPD medium (2% (w/v) glucose, 2% (w/v) bactopeptone and 1% (w/v) yeast extract) or minimal SD medium (0.17% (w/v) yeast nitrogen base without amino acids, 5% (w/v) ammonium sulphate and 2% (w/v) glucose, supplemented with appropriate amino acids and bases) (Sherman et al., 1974). For growth on non-fermentable carbon sources, SGE contained 3% (v/v) glycerol and 1% (v/v) ethanol. For anaerobic growth conditions, SD medium was supplemented with 0.1% (v/v) Tween-80 and 30 mg l−1 ergosterol, and plates were maintained in an anaerobic jar containing a gas-generating kit (Oxoid).
Determination of redox states.
Concentrations of GSH and GSSG were determined as described previously (Grant et al., 1998). The redox potential (Eh) of the GSSG/2GSH redox couple was calculated from the Nernst equation (Schafer & Buettner, 2001). The redox state of thioredoxins and glutaredoxins was measured by covalent modification with the thiol-reactive probe AMS (Molecular Probes) as described previously (Frand & Kaiser, 1999).
Western blot analysis.
Plasmid pBAD-YTRX1 contains a His6-tagged version of TRX1 (Mukhopadhyay & Rosen, 2000). Trx1 was purified, by means of the His6 tag, on Ni2+-agarose columns; purified fusion protein was used to generate rabbit anti-yeast thioredoxin antibody (Biogenesis Ltd). The rabbit anti-yeast glutaredoxin antibody has been described previously (Collinson et al., 2002). Protein extracts were subjected to electrophoresis on SDS–18% PAGE minigels and electroblotted to poly(vinylidene difluoride) membrane (Amersham Pharmacia Biotech). Blots were incubated in anti-Grx1 or anti-Trx1 antibody (1:1000 dilution). Bound antibody was detected by chemiluminescence (ECL; Amersham Pharmacia Biotech). Anti-(glyceraldehyde-3-phosphate dehydrogenase) (Chemicon International Inc.) was used as a loading control.
Acknowledgments
We thank B. Rosen for donating plasmids pBAD-YGRX1 and pBAD-YTRX1, and G. Wheeler and K. Quinn for discussions. This work was supported by project grant number 36/G13234 from the Biotechnology and Biological Sciences Research Council (UK).
References
- Aslund F., Berndt K.D. & Holmgren A. (1997) Redox potentials of glutaredoxins and other thiol–disulfide oxidoreductases of the thioredoxin superfamily determined by direct protein–protein redox equilibria. J. Biol. Chem., 272, 30780–30786. [DOI] [PubMed] [Google Scholar]
- Carmel-Harel O. & Storz G. (2000) Roles of the glutathione- and thioredoxin-dependent reduction systems in the Escherichia coli and Saccharomyces cerevisiae responses to oxidative stress. Annu. Rev. Microbiol., 54, 439–461. [DOI] [PubMed] [Google Scholar]
- Collinson E.J. et al. (2002) The yeast glutaredoxins are active as glutathione peroxidases. J. Biol. Chem. 277, 16712–16717. [DOI] [PubMed] [Google Scholar]
- Draculic T., Dawes I.W. & Grant C.M. (2000) A single glutaredoxin or thioredoxin is essential for viability in the yeast Saccharomyces cerevisiae. Mol. Microbiol., 36, 1167–1174. [DOI] [PubMed] [Google Scholar]
- Frand A.R. & Kaiser C.A. (1999) Ero1p oxidizes protein disulfide isomerase in a pathway for disulfide bond formation in the endoplasmic reticulum. Mol. Cell., 4, 469–477. [DOI] [PubMed] [Google Scholar]
- Gan Z.-R. (1991) Yeast thioredoxin genes. J. Biol. Chem., 266, 1692–1696. [PubMed] [Google Scholar]
- Garrido E.O. & Grant C.M. (2002) Role of thioredoxins in the response of Saccharomyces cerevisiae to oxidative stress induced by hydroperoxides. Mol. Microbiol., 43, 993–1003. [DOI] [PubMed] [Google Scholar]
- Grant C.M. (2001) Role of the glutathione/glutaredoxin and thioredoxin systems in yeast growth and response to stress conditions. Mol. Microbiol., 39, 533–541. [DOI] [PubMed] [Google Scholar]
- Grant C.M., Collinson L.P., Roe J.-H. & Dawes I.W. (1996) Yeast glutathione reductase is required for protection against oxidative stress and is a target gene for yAP-1 transcriptional regulation. Mol. Microbiol., 21, 171–179. [DOI] [PubMed] [Google Scholar]
- Grant C.M., MacIver F.H. & Dawes I.W. (1996b) Glutathione is an essential metabolite required for resistance to oxidative stress in the yeast Saccharomyces cerevisiae. Curr. Genet., 29, 511–515. [DOI] [PubMed] [Google Scholar]
- Grant C.M., Perrone G. & Dawes I.W. (1998) Glutathione and catalase provide overlapping defenses for protection against hydrogen peroxide in the yeast Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun., 253, 893–898. [DOI] [PubMed] [Google Scholar]
- Holmgren A. (1989) Thioredoxin and glutaredoxin systems. J. Biol. Chem., 264, 13963–13966. [PubMed] [Google Scholar]
- Holmgren A. & Aslund F. (1995) Glutaredoxin. Methods Enzymol., 252, 283–292. [DOI] [PubMed] [Google Scholar]
- Lee J.-C., Straffon M.J., Jang T.-Y., Grant C.M. & Dawes I.W. (2000) The essential and ancillary role of glutathione in Saccharomyces cerevisiae: studies with a grande gsh1 disruptant strain. FEMS Yeast Res., 1, 57–65. [DOI] [PubMed] [Google Scholar]
- Luikenhuis S., Dawes I.W. & Grant C.M. (1997) The yeast Saccharomyces cerevisiae contains two glutaredoxin genes that are required for protection against reactive oxygen species. Mol. Biol. Cell, 9, 1081–1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machado A.K., Morgan B.A. & Merril G.F. (1997) Thioredoxin reductase-dependent inhibition of MCB cell cycle box activity in Saccharomyces cerevisiae. J. Biol. Chem., 272, 17045–17054. [DOI] [PubMed] [Google Scholar]
- Meister A. & Anderson M.E. (1983) Glutathione. Annu. Rev. Biochem., 52, 711–760. [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay R. & Rosen B.P. (2000) The phosphatase C(X)5R motif is required for catalytic activity of the Saccharomyces cerevisiae Acr2p arsenate reductase. J. Biol. Chem., 276, 34738–34742. [DOI] [PubMed] [Google Scholar]
- Muller E.G.D. (1991) Thioredoxin deficiency in yeast prolongs S phase and shortens the G1 interval of the cell cycle. J. Biol. Chem., 266, 9194–9202. [PubMed] [Google Scholar]
- Muller E.G.D. (1996) A glutathione reductase mutant of yeast accumulates high levels of oxidized glutathione and requires thioredoxin for growth. Mol. Biol. Cell, 7, 1805–1813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S.G., Cha M.-K., Jeong W. & Kim I.-H. (2000) Distinct physiological functions of thiol peroxidase isoenzymes in Saccharomyces cerevisiae. J. Biol. Chem., 275, 5723–5732. [DOI] [PubMed] [Google Scholar]
- Pearson G.D. & Merril G.F. (1998) Deletion of the Saccharomyces cerevisiae TRR1 gene encoding thioredoxin reductase inhibits p53-dependent reporter gene expression. J. Biol. Chem., 273, 5431–5434. [DOI] [PubMed] [Google Scholar]
- Prinz W.A., Aslund F., Holmgren A. & Beckwith J. (1997) The role of the thioredoxin and glutaredoxin pathways in reducing protein disulphide bonds in Escherichia coli. J. Biol. Chem., 272, 15661–15667. [DOI] [PubMed] [Google Scholar]
- Rietsch A. & Beckwith J. (1998) The genetics of disulfide bond metabolism. Annu. Rev. Genet., 32, 163–184. [DOI] [PubMed] [Google Scholar]
- Schafer F.Q. & Buettner G.R. (2001) Redox environment of the cell as viewed through the redox state of the glutathione disulfide/glutathione couple. Free Radical Biol. Med., 30, 1191–1212. [DOI] [PubMed] [Google Scholar]
- Sherman F., Fink G.R. & Lawrence C.W. (1974) Methods in Yeast Genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, New York. [Google Scholar]
- Trotter E.W. & Grant C.M. (2002) Thioredoxins are required for protection against a reductive stress in the yeast Saccharomyces cerevisiae. Mol. Microbiol., 46, 869–878. [DOI] [PubMed] [Google Scholar]
- Wu A. & Moye-Rowley W.S. (1994) GSH1, which encodes γ-glutamylcysteine synthetase, is a target gene for yAP-1 transcriptional regulation. Mol. Cell. Biol., 14, 5832–5839. [DOI] [PMC free article] [PubMed] [Google Scholar]


