Abstract
Specific cleavage of large DNA molecules at few sites, necessary for the analysis of genomic DNA or for targeting individual genes in complex genomes, requires endonucleases of extremely high specificity. Restriction endonucleases (REase) that recognize DNA sequences of 4–8 bp are not sufficiently specific for this purpose. In principle, the specificity of REases can be extended by fusion to sequence recognition modules, e.g. specific DNA-binding domains or triple-helix forming oligonucleotides (TFO). We have chosen to extend the specificity of REases using TFOs, given the combinatorial flexibility this fusion offers in addressing a short, yet precisely recognized restriction site next to a defined triple-helix forming site (TFS). We demonstrate here that the single chain variant of PvuII (scPvuII) covalently coupled via the bifunctional cross-linker N-(γ-maleimidobutryloxy) succinimide ester to a TFO (5′-NH2-[CH2]6 or 12-MPMPMPMPMPPPPPPT-3′, with M being 5-methyl-2′-deoxycytidine and P being 5-[1-propynyl]-2′-deoxyuridine), cleaves DNA specifically at the recognition site of PvuII (CAGCTG) if located in a distance of approximately one helical turn to a TFS (underlined) complementary to the TFO (‘addressed’ site: 5′-TTTTTTTCTCTCTCTCN∼10CAGCTG-3′), leaving ‘unaddressed’ PvuII sites intact. The preference for cleavage of an ‘addressed’ compared to an ‘unaddressed’ site is >1000-fold, if the cleavage reaction is initiated by addition of Mg2+ ions after preincubation of scPvuII-TFO and substrate in the absence of Mg2+ ions to allow triple-helix formation before DNA cleavage. Single base pair substitutions in the TFS prevent addressed DNA cleavage by scPvuII-TFO.
INTRODUCTION
The development of artificial or semisynthetic nucleases capable of highly specific cleavage of any desired sequence would greatly facilitate physical mapping and manipulation of genomes as well as targeting genes in vivo. Strategies for generating such programmable nucleases include the coupling of DNA-binding domains (1–5) or triple-helix forming oligonucleotides (TFOs) (6–8) to non-specific nucleases or ‘chemical’ nucleases. An inherent disadvantage of using non-specific nucleases of natural or chemical origin, however, is that they do not cleave the DNA at a defined phosphodiester bond but rather produce DNA fragments with heterogeneous ends. In this respect, Type II restriction endonucleases (REase), though not yet exploited for this purpose, appear superior to non-specific nucleases for the development of a programmable nuclease, because REases cleave DNA at specific sites, 4–8 bp in length, leaving defined ends [review: (9)]. REases are very accurate enzymes: the ratio of activities at cognate and non-cognate sites is very large; e.g. in pAT153 EcoRV cleaves it canonical site (GATATC) by a factor of 106 faster than the next best site (GTTATC) [review: (10)]. Almost all restriction enzymes require Mg2+ ions for phosphodiester bond cleavage [reviews: (11,12)]. In some cases, e.g. EcoRV, Mg2+ ions or other divalent metal ions, such as Ca2+ ions, are required also for specific DNA-binding (13,14).
A REase–TFO conjugate would have a bipartite recognition sequence, consisting of the recognition site of the REase and the DNA sequence matching to the TFO (Figure 1). DNA triple helices are formed by the binding of synthetic oligonucleotides within the major groove of duplex DNA, where they make specific hydrogen bond contacts with the Hoogsteen faces of the purine bases [reviews: (15–17)]. Polypyrimidine TFOs bind in parallel orientation, polypurine TFOs in an antiparallel with respect to the duplex purine strand. The parallel motif has been most widely studied and is characterized by the formation of C+.GC and T.AT triplets (the notation X.ZY refers to a triplet, in which the third strand base X interacts with the duplex ZY base pair, forming hydrogen bonds to base Z). Recent work has demonstrated that by using a combination of nucleoside analogs it is possible to generate stable triplexes with high sequence specificity at physiological pH even to target sites not consisting of homopurine tracts (18–22).
Figure 1.
General strategy to generate a programmed restriction enzyme. The single chain variant of PvuII is covalently linked via a (CH2)6- or (CH2)12 - linker to a TFO using GMBS, a bifunctional chemical cross-linker specific for amino and sulfhydryl groups. The final constructs, scPvuII-C6-TFO and scPvuII-C12-TFO, have a bipartite recognition site: the PvuII recognition site and the TFS.
We have produced a conjugate of a PvuII variant (23) and a TFO containing modified bases (19) using a bifunctional cross-linker. This REase–TFO conjugate has a bipartite recognition sequence consisting of the PvuII recognition site (CAGCTG) and the double-stranded (ds) DNA sequence (5′-TTTTTTTCTCTCTCTC-3′/5′-GAGAGAGAGAAAAAAA-3′) addressed by the TFO. We demonstrate here that at neutral pH and Mg2+ concentrations of 1–2 mM, this REase-TFO cleaves DNA specifically at the bipartite recognition sequence (addressed PvuII sites), leaving unaddressed PvuII sites intact. This specificity, in principle, suffices to target a unique sequence in a genomic context.
MATERIALS AND METHODS
Proteins and oligonucleotides
The mutant coding for the His6-tagged single cysteine variant scPvuII-H6G4C was generated using pRIZ'-scPvuII (23) as template by a PCR-based site-directed mutagenesis method (24); the protein was expressed and purified similarly as described previously (25). TFOs containing a 5′-C6-spacer or 5′-C12-spacer with a N-terminal group were purchased form Eurogentec: 5′-NH2-[CH2]6/12-MPMPMPMPMPPPPPPT-3′, where M is 5-methyl-2′-deoxycytidine and P is 5-[1-propynyl]-2′-deoxyuridine (19,26).
Fluorescence anisotropy measurements
To demonstrate that the single chain variant of PvuII (scPvuII) variant which we used required divalent cations for strong DNA-binding, fluorescence titrations were carried out with 5′-hexafluorecein-labeled 5′-TCTAGGCAGCTGGGAT-3′ (hybridized to its non-labeled complement) at various concentrations and scPvuII-H6G4C (0–450 nM) in 50 mM Tris–HCl (pH 7.5), 100 mM NaCl in the presence of 10 mM Ca2+ or 3 mM EDTA, respectively, at 23°C in a F-4500 Hitachi fluorimeter), similarly as described by Reid et al. (27).
Electrophoretic mobility shift assay (EMSA)
To demonstrate the high stability of the triple-helix used in our experiments EMSAs were performed with the TFO and a complementary double-stranded oligodeoxyribonucleotide. The TFO (5–500 nM) was incubated at 37°C for 1 h with 32P-labeled double-stranded 5′-TTTTTTTCTCTCTCTC-3′/5′-GAGAGAGAGAAAAAAA-3′ (50 nM) in 10 mM Tris–phosphate (pH 7.2), 1 mM spermine in a volume of 15 µl. Subsequently, 5 µl of 10 mM Tris–phosphate (pH 7.2), 25% (w/v) sucrose were added to each sample and complex formation was analyzed by electrophoresis on 15% polyacrylamide gels in 10 mM Tris–phosphate (pH 7.2). Gels were dried and the radioactive bands visualized using an instant imager (Canberra Packard).
Restriction enzyme cleavage protection assay
Cleavage protection assays were performed with a 150 bp DNA substrate containing a FokI site and an adjacent triple-helix forming site (TFS) overlapping the cleavage site of FokI. Substrate (1 µM) was preincubated in 10 mM Tris–phosphate (pH 7.2) with either 10 µM TFO or a non-specific hexadecadeoxyribonucleotide for 4 h at 37°C in a volume of 70 µl. Forty units FokI (NEB) (in FokI cleavage buffer) were added to give a final volume of 100 µl. At selected time points (0.5, 1, 1.5, 2, 5, 10, 20, 30, 45 and 60 min), 10 µl aliquots were withdrawn and the reaction terminated by adding EDTA (50 mM final concentration). Loading buffer (5 µl) [10% (w/v) Ficoll, 10% (v/v) glycerol, 0.2% (w/v) bromphenol blue and 0.2% (w/v) xylene cyanol] were then added and cleavage protection was analyzed by electrophoresis on 12% polyacrylamide gels in 80 mM Tris–phosphate (pH 8.2) and 2 mM EDTA. Products were visualized by ethidium bromide (0.1 µg/ml) staining and illumination with UV light.
Protein-TFO cross-linking
For activation of the oligonucleotide, 80 µM TFO was incubated with 40 mM bifunctional cross-linker N-(γ-maleimidobutryloxy) succinimide ester (GMBS, Pierce) in 500 µl phosphate-buffered saline (PBS) [140 mM NaCl, 2.7 mM KCl, 4.3 mM Na2HPO4 and 1.4 mM K2HPO4 (pH 7.2)] for 1 h at 23°C. Unreacted cross-linker was removed by passage through NAP™-5 and subsequently NAP™-10 (Amersham Sephadex™ G-25 DNA grade) equilibrated with water. The desalted, activated TFO was dried and dissolved in 80 µl PBS. For cross-linking of oligonucleotide and protein, 80 µM activated TFO was incubated for 1 h at 23°C with 20 µM scPvuII-H6G4C, which had been dialyzed for 4 h against PBS to remove DTT. Cross-linking yield was analyzed by 15% SDS–PAGE.
Protein-TFO purification
Uncross-linked scPvuII-H6G4C was separated from cross-linked scPvuII-C6/C12-TFO by anion exchange chromatography using DE52 in 10 mM Tris–HCl (pH 7.5), 3 mM EDTA using a linear gradient of NaCl (100–400 mM). scPvuII-H6G4C eluted in the flow through whereas scPvuIIC6/C12-TFO eluted at a concentration of 350–390 mM–NaCl. Purified scPvuII-C6/C12-TFO was dialyzed against PBS containing 25% PEG 10 000 and then stored at 4°C.
DNA cleavage assays
For DNA cleavage experiments, radioactively labeled 239 bp substrates (containing a single PvuII site) were generated by PCR using [α-32P]dATP. All substrates share the same sequence except for that replaced by the TFS (only the pyrimidine rich strand is shown, underlined) adjacent to the PvuII site (bold): 3 bp, 5′-…CGGTACTCGACGTTTTTTTCTCTCTCTCAGACAGCTG…-3′; 5 bp, 5′-…CGGTACTCGATTTTTTTCTCTCTCTCGCAGACAGCTG…-3′; 7 bp, 5′-…CGGTACTCTTTTTTTCTCTCTCTCCAGCAGACAGCTG…-3′; 9 bp, 5′-…CGGTACTTTTTTTCTCTCTCTCCACAGCAGACAGCTG…-3′; 11 bp, 5′-…CGGTTTTTTTTCTCTCTCTCCGCACAGCAGACAGCTG…-3′; 13 bp, 5′-…CGTTTTTTTCTCTCTCTCGGCGCACAGCAGACAGCTG…-3′.
For control reaction an unaddressed 478 bp substrate that contains the same central sequence as the 239mer with the exception that it does not carry a TFS was generated by PCR as well. All substrates were purified using the QIAquick PCR Purification Kit (Qiagen) and analyzed by PAGE. Concentrations were calculated assuming 50 ng/µl DNA corresponds to an OD260 of 1. To analyze DNA cleavage, 45 nM of these substrates were cleaved by 5 nM scPvuII-C6/C12-TFO at 37°C in 10 mM Tris–phosphate (pH 7.2), 10 mM MgCl2, 1 mM spermine. Reactions were terminated after 5, 10, 20, 30, 60 or 110 min by addition of EDTA to a final concentration of 50 mM. Reaction products were analyzed by electrophoresis on 12% polyacrylamide gels. Gels were dried and the radioactive bands visualized by autoradiography using an instant imager system (Canberra Packard). To analyze targeted cleavage, 40 nM scPvuII-C6/C12-TFO was preincubated with addressed and unaddressed substrate in competition (each at 45 nM) in 10 mM Tris–phosphate (pH 7.2), 1 mM spermine, 3 mM EGTA at 23°C overnight. EGTA was included in the preincubation buffer to chelate divalent metal ions that even at low concentrations would have stimulated specific binding to the PvuII site of the unaddressed substrate. Cleavage reactions were initiated at 37°C by adding an equal volume of 10 mM Tris–HCl (pH 7.2) with MgCl2 to various final concentrations: 1.25, 2.5 or 5 mM. Cleavage products were analyzed as described above. Similar experiments were done with variants of the 239 bp substrate, which carry a modified TFS, 5′-TTTTTTTYTCTCTCTC-3′ (Y = A, G or T)/5′-GAGAGAGAXAAAAAAA-3′ (X = T, C or A), in a distance of 9 bp to the PvuII site. To analyze targeted cleavage of larger substrates by scPvuII-C12-TFO, a 5556 bp plasmid with 5 PvuII sites, one of which is adjacent to a TFS, was used. Preincubation and cleavage of the supercoiled and HindIII-linearized plasmid were done as described for the 239 and 478 bp substrates. Cleavage products were separated by electrophoresis on 0.8% agarose gels that were subsequently stained with SYBR gold and analyzed using the Biometra software. A similar plasmid cleavage experiment was carried out in competition with a large excess of bacteriophage λ-DNA (0.007 pmol plasmid DNA with a single addressed PvuII site and 3 PvuII sites, 0.68 pmol λ-DNA with 15 PvuII sites, 0.006 pmol scPvuII-C12-TFO, preincubated in the absence of Mg2+ for 72 h at 23°C; cleavage was initiated by addition of MgCl2 and NaCl to give a final concentration of 2.5 mM MgCl2 and 100 mM NaCl).
RESULTS
To generate a programmed restriction enzyme we coupled a single chain variant of PvuII with a TFO (Figure 1). PvuII is a homodimeric Type II REase (9,11) that recognizes the double-stranded DNA sequence CAG/CTG, cleaving it as indicated (28,29). It is one of the best studied restriction enzymes, both in terms of structure (30–32) and function (33). PvuII requires Mg2+ for both specific DNA-binding and DNA cleavage (33,34).
We used the monomeric scPvuII (23) as the fusion partner to generate a programmable restriction enzyme as this allowed us to introduce a unique single cysteine for the coupling of the TFO. To this end, a His6-tag followed by four glycines and one cysteine residue was added to the C-terminus of scPvuII. This variant binds specific DNA in the presence of Ca2+ (as an analogue of Mg2+) with 27 nM affinity, and with 20 µM affinity in its absence, as shown by fluorescence anisotropy measurements (data not shown).
TFOs bind via Hoogsteen hydrogen bonds in the major groove of the DNA, either in a parallel (pyrimidine motif, Y.RY) or antiparallel (purine motif, R.YR) orientation with respect to the purine strand of the Watson–Crick base pairs (35). Repulsion of the negatively charged DNA strands can be overcome by adding spermine (36). Pyrimidine motif triple helices are unstable at physiological pH because of the requirement for cytosine protonation that occurs at relatively acidic pH (pKa = 4.5) (37,38). With 5-methylcytosine (M) replacing cytosine, this pH restriction is reduced via the contribution of the methyl group to base stacking and the exclusion of water from the major groove of the DNA (39). Using propynyl-deoxyuridine (P) instead of thymine also increases triple-helix stability (19,40). For this study, we utilized the TFO 5′-NH2-[CH2]6/12-MPMPMPMPMPPPPPPT-3′. Triple-helix formation between this TFO and a complementary double-stranded hexadecadeoxyribonucleotide or a 150 bp PCR product containing the TFS (TFS: 5′-TTTTTTTCTCTCTCTC-3′/5′-GAGAGAGAGAAAAAAA-3′) was demonstrated by EMSA and restriction cleavage protection assay. A binding constant of KAss = 6.4 × 107 M−1 was determined for the TFO and the double-stranded DNA by electrophoretic mobility shift experiments (data not shown).
The TFO was coupled with the bifunctional cross-linker GMBS (N-[γ-maleimidobutyryloxy]succinimide ester) via amide bond formation between the 5′-NH2-group of the TFO and the succinimide group of GMBS. The NH2-group is connected to the TFO by 6 or 12 methylene groups, resulting in different linker lengths in the final scPvuII-C6-TFO or scPvuII-C12–TFO conjugates, respectively. The maleimide group of the GMBS-modified TFO reacts with the single cysteine at the C-terminal end of the scPvuII-H6G4C variant to create a covalent scPvuII-C6/C12-TFO conjugate. Coupling yields exceeded 70%. Residual amounts of uncross-linked scPvuII-H6G4C were removed by anion exchange chromatography.
Activity and specificity of scPvuII-C6/C12-TFO were tested with DNA substrates of similar sequence generated by PCR: (i) a 239 bp substrate with a TFS adjacent to a PvuII site (‘addressed’ DNA substrate) and; (ii) a 478 bp substrate that incorporates the 239 bp substrate sequence but does not have a TFS adjacent to its PvuII site (‘unaddressed’ DNA substrate). In PvuII cleavage buffer, scPvuII-C6/C12-TFO exhibits the same cleavage activity with both substrates, demonstrating that the presence of the TFS next to the PvuII site does not influence DNA cleavage under conditions where triple-helix formation does not take place (data not shown).
In targeted DNA cleavage experiments, the TFO of scPvuII-C6/C12-TFO should guide the enzyme to the PvuII site adjacent to the TFS. Since the kinetics of triplex formation are slow, these experiments require a preincubation of the DNA with the scPvuII-C6/C12-TFO conjugate. Initial experiments were carried out with scPvuII-C12-TFO. Targeting was investigated by cleaving the 239 bp substrate (with the TFS at a distance of 9 bp from the PvuII site) and the 478 bp substrate (without the TFS) in competition: scPvuII-C12-TFO was preincubated overnight with DNA substrates in a buffer compatible with triple-helix formation and depleted of divalent metal ions to prevent binding to PvuII sites without the adjacent TFS. Preincubation was carried out using near equimolar amounts of scPvuII-C12-TFO and addressed DNA substrate to obtain near stoichiometric binding of the TFS and avoid excess free enzyme in the reaction mixture. Cleavage was initiated by adding PvuII cleavage buffer with MgCl2 at different final concentrations: 1.25, 2.5 or 5 mM. As shown in Figure 2 we observed a very strong cleavage preference of scPvuII-C6/C12-TFO for the addressed substrate. This preference showed a clear MgCl2 concentration dependence: with decreasing concentration of MgCl2 the rate of cleavage of the unaddressed 478 bp substrate was reduced, approaching zero at 1.25 mM MgCl2, whereas the rate of cleavage of the addressed 239 bp substrate was not changed (Figure 2). This dependence of the cleavage rates on the Mg2+ concentration can be understood by considering that DNA-binding and cleavage by PvuII requires Mg2+ (34,41). In the case of addressed cleavage the enzyme is Mg2+ ion-independently bound to its cleavage site via the TFO–TFS interaction, allowing efficient cleavage even at low Mg2+ concentrations that would not readily support unaddressed cleavage. Total addressed substrate was not cleaved to completion since a substoichiometric amount of scPvuII-C12-TFO was used (40 nM enzyme versus 45 nM of each substrate). Experiments in 1.25 mM MgCl2 with different enzyme concentrations confirmed that moles of substrate cleaved correspond exactly to moles of enzyme used, demonstrating that the enzyme does not turnover but remains bound to one of the products via triple-helix formation. With an excess of enzyme over DNA the unaddressed substrate is also cleaved, albeit at a much slower rate (data not shown).
Figure 2.
Unaddressed versus addressed DNA cleavage by scPvuII-C12-TFO at two Mg2+ concentrations. Equimolar (45 nM) amounts of unaddressed 478 bp substrate (diamond; no TFS) and addressed 239 bp substrate (square; having a TFS 9 bp away from the PvuII site) were mixed and preincubated with scPvuII-C12-TFO (40 nM) to allow triple-helix formation. After addition of 1.25 mM (top) or 5 mM (bottom) MgCl2, the kinetics of cleavage were determined. Whereas at 5 mM MgCl2 the unaddressed substrate is cleaved with appreciable rate, it is not cleaved to a significant extent in the presence of 1.25 mM MgCl2. In contrast, the addressed substrate is cleaved with a high rate in the presence of either 1.25 (top) or 5 mM MgCl2 (bottom). P denotes the unaddressed PvuII site. The addressed PvuII site is indicated by an open star.
In the experiment described above, the PvuII site and the TFS on the addressed 239 bp substrate were separated by 9 bp. Since it is likely that the distance between the PvuII site and the TFS influences the efficiency of cleavage, we prepared additional addressed 239 bp substrates having 3, 5, 7, 11 or 13 bp distances. Furthermore, to investigate whether the length of the linker between scPvuII and the TFO has an effect on the addressed cleavage or optimum distance between the PvuII site and the TFS, two conjugates, scPvuII-C12–TFO (12 methylene groups) and scPvuII-C6-TFO (six methylene groups), were prepared and analyzed. Cleavage reactions were carried out using addressed and unaddressed substrate in competition. Cleavage rates of the unaddressed substrates are extremely low in all reactions initiated with 1.25 mM MgCl2: the rate constants for cleavage of the unaddressed substrate by scPvuII-C12-TFO and scPvuII-C6-TFO were kunaddressed(C12) = 0.002 min−1 and kunaddressed(C6) = 0.001 min−1, respectively. Up to 1400-fold higher cleavage rates were observed for the addressed substrate, with the best substrates having a 9 or 11 bp distance between the PvuII site and the TFS. For scPvuII-C12-TFO, the DNA having a 13 bp distance between the PvuII site and the TFS was also a very good substrate. Cleavage rates observed for scPvuII-C12-TFO were higher than for scPvuII-c6-TFO, presumably due to the greater flexibility of the C12-linker. Cleavage rates of substrates with PvuII site and TFS distances shorter than 7 bp decreased, with scPvuII-C6-TFO more so that with the scPvuII-C12-TFO, again indicating a higher flexibility of the C12-linker (Table 1). We assume, that both the C-terminal extension H6G4C of scPvuII and the 5′-NH2-[CH2]6/12-extension of the TFO are needed for the simultaneous binding of the PvuII recognition sequence and the adjacent TFS by the scPvuII–TFO conjugate. Our data suggest that the C12-linker is superior to the C6-linker in this respect.
Table 1.
Rate constants for cleavage by scPvuII-C6/C12-TFO of the 239 bp substrates
| Distance between TFS and PvuII site | kaddressed (scPvuII-C12-TFO) | Cleavage preference (scPvuII-C12-TFO) | kaddressed (scPvuII-C6-TFO) | Cleavage preference (scPvuII-C6-TFO) |
|---|---|---|---|---|
| 3 bp | 1.0 min−1 (±5%) | 440-fold | 0.03 min−1 (±7%) | 20-fold |
| 5 bp | 0.8 min−1 (±6%) | 360-fold | 0.03 min−1 (±17%) | 20-fold |
| 7 bp | 1.2 min−1 (±7%) | 540-fold | 0.30 min−1 (±7%) | 210-fold |
| 9 bp | 3.2 min−1 (±9%) | 1410-fold | 0.50 in−1 (±6%) | 450-fold |
| 11 bp | 3.1 min−1 (±13%) | 1360-fold | 1.00 in−1 (±15%) | 820-fold |
| 13 bp | 2.6 min−1 (±10%) | 1140-fold | 0.20 min−1 (±10%) | 190-fold |
Data are given for the scPvuII-C12–TFO and scPvuII-C6–TFO conjugates that differ in spacer length (12 or 6 methylene groups between the protein and the oligonucleotide). Cleavage rate constants were determined for six different addressed 239 bp substrates that differ in the spacing between the PvuII site and the TFS. SD based on three independent experiments are given in parentheses. Cleavage preference is the ratio of the rate constants for the addressed (239 bp) to unaddressed (478 bp) substrate.
In the presence of 2.5 mM MgCl2 (data not shown) or 5 mM MgCl2 (Figure 2), cleavage preferences are smaller because cleavage of the unaddressed substrate is faster. The decreased cleavage preference at 2.5 mM MgCl2 concentration can be compensated by addition of NaCl at concentrations up to 100 mM (data not shown). Presumably, low Mg2+ concentrations are needed for preferential cleavage at addressed sites, because at higher Mg2+ concentrations DNA-binding by the scPvuII–TFO conjugate is dominated by the protein–DNA interaction and not the TFO–TFS interaction.
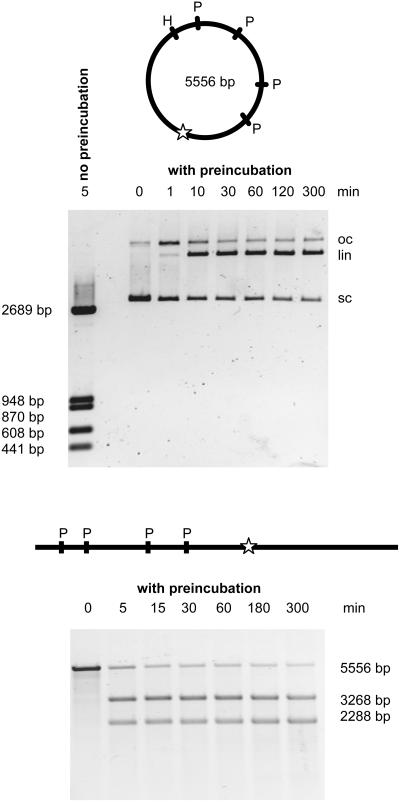
To demonstrate addressed cleavage of larger DNA molecules, a 5556 bp plasmid with five PvuII sites, one having a TFS at a distance of 9 bp, was used (Figure 3). Cleavage of the supercoiled plasmid with PvuII or scPvuII-C12-TFO without preincubation resulted in five cleavage fragments. However, after preincubation of near stoichiometric amounts of scPvuII-C12-TFO and the plasmid, only one cleavage product, the linear plasmid, was obtained and no further cleavage was observed even after incubation for several hours. This result indicates that addressed cleavage of the plasmid is highly specific, because only the addressed PvuII site is cleaved and the four unaddressed sites are unaffected. Highly preferential addressed site cleavage was also observed with the linear form of the plasmid and confirmed by cleavage product analysis (Figure 2). Quantitative intensity analysis of the various DNA bands yielded cleavage rate constants of the addressed site in the supercoiled and the linearized plasmid of 1.4 and 3.9 min−1, respectively. Cleavage of the other PvuII sites was not detectable thus demonstrating a >1000-fold cleavage preference (a conservative estimate considering the detection limit of the intensity analysis).
Figure 3.
Addressed cleavage by scPvuII-C12-TFO of a 5556 bp plasmid. The plasmid contains five PvuII sites, one of which is flanked by a TFS 9 bp away. Top: Plasmid DNA (50 nM) was preincubated with scPvuII-C12-TFO (45 nM) to allow triple-helix formation. After addition of MgCl2 (final concentrations: 2.5 mM MgCl2 and 100 mM NaCl), aliquots of the reaction mixture were taken at the time intervals indicated and reaction products analyzed by agarose gel electrophoresis. The first lane is a cleavage control experiment, where the supercoiled plasmid was incubated for 5 min with scPvuII-C12-TFO in cleavage buffer without prior incubation in the absence of Mg2+: under these conditions the plasmid is cleaved at its 5 PvuII sites. P and H denote the (unaddressed) PvuII and HindIII sites, respectively. The addressed PvuII site is indicated by an open star. Bottom: Addressed cleavage of the same plamid after linearization with HindIII. Now the 5556 bp fagment carries the addressed site at position 3268.
Programmed restriction enzymes are intended to be used for large genome mapping and gene targeting in complex genomes. To demonstrate that our scPvuII–TFO conjugates are able to specifically cleave an addressed PvuII site in a large excess of unaddressed PvuII sites, we have incubated 0.006 pmol scPvuII-C12-TFO with 0.007 pmol plasmid DNA (four PvuII sites, one of which having a TFS at a distance of 9 bp) in the presence of 0.68 pmol bacteriophage λ-DNA (15 PvuII sites), corresponding to a 1460-fold excess of unaddressed PvuII sites over addressed PvuII, and then started the reaction by addition of MgCl2. Only cleavage at the addressed site could be detected (Figure 4).
Figure 4.
Addressed cleavage by scPvuII-C12-TFO in the presence of a 1400-fold excess of unaddressed PvuII sites over addressed PvuII. 0.16 nM scPvuII-C12-TFO, 0.18 nM plasmid DNA with a single addressed PvuII site and 3 PvuII sites and 17 nM λ-DNA with 15 PvuII sites were preincubated in the absence of Mg2+; cleavage was initiated by addition of MgCl2 (final concentrations: 2.5 mM MgCl2 and 100 mM NaCl). The reaction products obtained after the times indicated were analyzed by agarose gel electrophoresis. The gel on the left shows the result of the control experiment (cleavage without preincubation): a typical PvuII cleavage pattern of bacteriophage λ-DNA is seen. The gel on the right shows the result of the cleavage pattern after preincubation: only the cleavage of plasmid DNA is observed, however not cleavage of bacteriophage λ-DNA.
The target site of our programmed restriction enzymes is a composite site consisting of the PvuII site and the TFS. The specificity of the restriction enzyme–TFO conjugate, therefore, depends in part on the specific interaction between the TFO and the TFS. To see how a single ‘Hoogsteen mismatch’ in the triple-helix formed upon interaction of the scPvuII-TFO with the double-stranded DNA substrate affects the rate of cleavage, a variant of the 239 bp substrate with a single addressed PvuII site was used, which had instead of the 5′-TTTTTTTCTCTCTCTC-3′/5′-GAGAGAGAGAAAAAAA-3′ sequence in 9 bp distance to the PvuII site the sequence 5′-TTTTTTTYTCTCTCTC-3′ (Y = A, G or T)/5′-GAGAGAGAXAAAAAAA-3′ (X = T, C or A). The substrates with the M.TA ‘Hoogsteen mismatch’ was not cleaved by the scPvuII-C12-TFO, whereas the substrates with the M.CG and M.AT ‘Hoogsteen mismatch’ were cleaved by a factor of at least 100 more slowly than the reference substrate with no ‘Hoogsteen mismatch’ (M.GC) (Figure 5) indicating a high sequence specificity of the addressing process.
Figure 5.
Addressed cleavage by scPvuII-C12-TFO of single ‘Hoogsteen mismatch’ substrates. Equimolar (45 nM) amounts of unaddressed 478 bp substrate (no TFS) and addressed 239 bp substrate (having a TFS 9 bp away from the PvuII site) were mixed and preincubated with scPvuII-C12-TFO (40 nM) to allow triple-helix formation. After addition of MgCl2 (final concentrations: 2.5 mM MgCl2 and 100 mM NaCl), the kinetics of cleavage was determined. Three different ‘Hoogsteen mismatch’ substrates were used, carrying a mismatch for the Hoogsteen base pairing in the middle of the TFS: instead of a GC bp which would form a Hoogsteen bp with 5-methyl cytosine (M), the ‘Hoogsteen mismatch’ substrates had a TA, CG and AT bp, respectively. Whereas the correct substrate (M.GC) was readily cleaved, the M.TA, M.CG and M.AT ‘Hoogsteen mismatch’ substrates were largely refractory to cleavage.
DISCUSSION
In principle triple helices can be used to target unique DNA sequences for biotechnological and biomedical applications (17,42–55). Of particular interest are conjugates of TFOs with nucleases, which could be used to cleave DNA in vitro for genome analysis and in vivo (or ex vivo) for gene replacement by double-strand break repair involving homologous recombination. We have chosen to extend the specificity of REases using TFOs, given the combinatorial flexibility this fusion offers in addressing a short, yet precisely recognized restriction site next to a defined TFS. Here, we have produced a conjugate of the single chain variant of the type II restriction endonuclease PvuII (scPvuII) (23) with a 16mer oligodeoxyribonucleotide containing 5-methyl-2′-deoxycytidine (for deoxycytidine) and 5-[1-propynyl]-2′-deoxyuridine (for thymidine) and characterized its DNA cleavage properties. The TFO component of this conjugate forms a stable triple-helix at neutral pH, as was shown before (19) and confirmed here. The PvuII component binds to DNA non-specifically in the absence of Mg2+, but specifically in the presence of Mg2+ (34). Thus, in the absence of Mg2+ the conjugate will be directed to the TFS, and only if a PvuII recognition site is in suitable proximity to the TFS (‘addressed PvuII site’) will the DNA be cleaved upon addition of Mg2+. The target site of the scPvuII–TFO conjugate therefore is a composite of the PvuII recognition site and the TFS. We have coupled the TFO to the C-terminus of scPvuII via a (CH2)6 or 12 linker, which together with the C-terminal His6Gly4Cys-extension of our scPvuII variant should provide enough flexibility to allow the scPvuII–TFO conjugate to bind to the PvuII recognition site after addition of Mg2+and to cleave the DNA at addressed PvuII sites in preference over unaddressed sites. These expectations were borne out by the results of DNA cleavage experiments, which showed that
the scPvuII-C6/C12-TFO conjugate is as active as scPvuII in cleaving PvuII recognition sites.
the scPvuII-C6/C12-TFO conjugate shows a >1000-fold preference for addressed over unaddressed PvuII recognition site, provided that the conjugate is preincubated with the DNA in the absence of Mg2+, to allow triple-helix formation to occur before cleavage.
preferential cleavage is only observed if the enzyme concentration is not in excess over addressed PvuII recognition sites, because free scPvuII-C6/C12-TFO conjugate would eventually attack unaddressed PvuII recognition sites.
the preference for cleavage of addressed PvuII recognition site shows a pronounced Mg2+ concentration dependence, little preference at 5 mM (the optimum concentration for PvuII-catalyzed DNA cleavage is around 5 to 10 mM Mg2+) and over 1000-fold preference at 1.25 mM Mg2+ (a sub-optimum concentration for PvuII, but not for the scPvuII-C6/C12-TFO conjugate, because its binding to DNA is supported by the TFO–TFS interaction). This preference is sufficient to cleave DNA at addressed PvuII recognition sites even in the presence of large amounts of unaddressed PvuII recognition sites, as typically encountered in complex genomic DNA.
the optimal distance between the PvuII recognition site and the TFS for addressed cleavage is ∼10 bp (a helical turn), but this distance is less critical for the scPvuII-C12-TFO conjugate compared to the scPvuII-C6-TFO conjugate, presumably because of the greater flexibility provided by the longer linker.
addressed PvuII sites with a single mutation (causing a ‘Hoogsteen mismatch’) in the TFS are refractory to cleavage by the scPvuII-C12-TFO conjugate.
We conclude from these results that the scPvuII-C6/C12-TFO conjugate is able to cleave addressed PvuII sites on high molecular weight DNA with the specificity required for recombinant DNA work and for this purpose could be used instead of homing endonucleases or other ‘rare cutters’ (e.g. REase with 8 bp recognition sites). In principle, this or similar constructs could therefore be used to map chromosomal DNA and to clone very large DNA fragments. Given the fact that TFO can be synthesized that would form triple helices with any sequence that one would like to address (22), the scPvuII variant can be considered a programmable restriction enzyme. In addition, there is no reason why this approach should not work with other restriction enzymes such that the requirement for the presence of a PvuII recognition site is not absolute. Furthermore, this approach could in principle also be used to program other ‘DNA enzymes’, e.g. DNA-methyltransferases.
It has been shown that highly efficient endogenous human gene correction can be achieved using targeted DNA cleavage by a designed nuclease (5,56,57). In this case a designed zinc finger nuclease was used, which contained the non-specific catalytic domain of the Type IIs REase FokI. We believe that restriction endonuclease–TFO conjugates are a useful alternative to designed zinc finger nucleases, because of their simple rules of defining TFS sites. That triple-helix formation can be used to target genes in vivo has been shown recently (58). To target unique sites in complex genomes in vivo by restriction endonuclease–TFO conjugates will require efficient delivery systems (59), stabilization of the TFO by chemical modification (17) [or using PNA (60)] and a procedure to activate the REase [‘caged’ (61) REase] after triple-helix formation has occurred which all is in technological reach.
Acknowledgments
The authors thank Dres. B.A. Connolly, P. Friedhoff, M. Kokkinidis, G. Meiss and G. Silva for fruitful discussions. This work was supported by the European Union (QLKR-CT-2001-00448), the BMBF BioFuture Programme, the Deutsche Forschungsgemeinschaft (Pi 122/12), the Deutscher Akademische Austauschdienst (International Quality Network ‘Biochemistry of Nucleic Acids’) and the Fonds der Chemischen Industrie. A patent application has been filed for which K.E., A.J. and A.P. are co-inventors. Funding to pay the Open Access publication charges for this article was provided by Justus-Liebig-University Giessen.
Conflict of interest statement. None declared.
REFERENCES
- 1.Kim Y.G., Cha J., Chandrasegaran S. Hybrid restriction enzymes: zinc finger fusions to Fok I cleavage domain. Proc. Natl Acad. Sci. USA. 1996;93:1156–1160. doi: 10.1073/pnas.93.3.1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kim Y.G., Chandrasegaran S. Chimeric restriction endonuclease. Proc. Natl Acad. Sci. USA. 1994;91:883–887. doi: 10.1073/pnas.91.3.883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ebright R.H., Ebright Y.W., Pendergrast P.S., Gunasekera A. Conversion of a helix–turn–helix motif sequence-specific DNA binding protein into a site-specific DNA cleavage agent. Proc. Natl Acad. Sci. USA. 1990;87:2882–2886. doi: 10.1073/pnas.87.8.2882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Oakley M.G., Dervan P.B. Structural motif of the GCN4 DNA binding domain characterized by affinity cleaving. Science. 1990;248:847–850. doi: 10.1126/science.2111578. [DOI] [PubMed] [Google Scholar]
- 5.Durai S., Mani M., Kandavelou K., Wu J., Porteus M.H., Chandrasegaran S. Zinc finger nucleases: custom-designed molecular scissors for genome engineering of plant and mammalian cells. Nucleic Acids Res. 2005;33:5978–5990. doi: 10.1093/nar/gki912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Strobel S.A., Dervan P.B. Site-specific cleavage of a yeast chromosome by oligonucleotide-directed triple-helix formation. Science. 1990;249:73–75. doi: 10.1126/science.2195655. [DOI] [PubMed] [Google Scholar]
- 7.Pei D., Corey D.R., Schultz P.G. Site-specific cleavage of duplex DNA by a semisynthetic nuclease via triple-helix formation. Proc. Natl Acad. Sci. USA. 1990;87:9858–9862. doi: 10.1073/pnas.87.24.9858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moser H.E., Dervan P.B. Sequence-specific cleavage of double helical DNA by triple helix formation. Science. 1987;238:645–650. doi: 10.1126/science.3118463. [DOI] [PubMed] [Google Scholar]
- 9.Roberts R.J., Vincze T., Posfai J., Macelis D. REBASE—restriction enzymes and DNA methyltransferases. Nucleic Acids Res. 2005;33:D230–D232. doi: 10.1093/nar/gki029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roberts R.J., Halford S.E. Chapter 2: Type II restriction endonucleases. In: Linn S.M., Lloyd R.S., Roberts R.J., editors. Nucleases. 2nd edition. Cold Spring Harbor NY: Cold Spring Harbor Laboratory Press; 1993. pp. 35–88. [Google Scholar]
- 11.Pingoud A., Fuxreiter M., Pingoud V., Wende W. Type II restriction endonucleases: structure and mechanism. Cell. Mol. Life Sci. 2005;62:685–707. doi: 10.1007/s00018-004-4513-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pingoud A., Jeltsch A. Structure and function of type II restriction endonucleases. Nucleic Acids Res. 2001;29:3705–3727. doi: 10.1093/nar/29.18.3705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thielking V., Selent U., Köhler E., Landgraf A., Wolfes H., Alves J., Pingiud A. Mg2+ confers DNA binding specificity to the EcoRV restriction endonuclease. Biochemistry. 1992;31:3727–3732. doi: 10.1021/bi00130a001. [DOI] [PubMed] [Google Scholar]
- 14.Vipond I.B., Halford S.E. Specific DNA recognition by EcoRV restriction endonuclease induced by calcium ions. Biochemistry. 1995;34:1113–1119. doi: 10.1021/bi00004a002. [DOI] [PubMed] [Google Scholar]
- 15.Thuong N.T., Hélène C. Sequence specific recognition and modification of double helical DNA by oligonucleotides. Angew. Chem. Int. Ed. Engl. 1993;32:666–690. [Google Scholar]
- 16.Soyfer V.N., Potoman V.N. Triple-Helical Nucleic Acids. New York: Springer-Verlag; 1996. [Google Scholar]
- 17.Fox K.R. Targeting DNA with triplexes. Curr. Med. Chem. 2000;7:17–37. doi: 10.2174/0929867003375506. [DOI] [PubMed] [Google Scholar]
- 18.Gowers D.M., Fox K.R. Towards mixed sequence recognition by triple helix formation. Nucleic Acids Res. 1999;27:1569–1577. doi: 10.1093/nar/27.7.1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lacroix L., Lacoste J., Reddoch J.F., Mergny J.L., Levy D.D., Seidman M.M., Matteucci M.D., Glazer P.M. Triplex formation by oligonucleotides containing 5-(1-propynyl)-2′-deoxyuridine: decreased magnesium dependence and improved intracellular gene targeting. Biochemistry. 1999;38:1893–1901. doi: 10.1021/bi982290q. [DOI] [PubMed] [Google Scholar]
- 20.Robles J., Granda A., Pedroso E., Luque F.J., Eritja R., Orozco M. Nucleic acid triple helices: stability effects of nucleobase modifications. Curr. Org. Chem. 2002;6:1333–1368. [Google Scholar]
- 21.Buchini S., Leumann C.J. Recent improvements in antigene technology. Curr. Opin. Chem. Biol. 2003;7:717–726. doi: 10.1016/j.cbpa.2003.10.007. [DOI] [PubMed] [Google Scholar]
- 22.Rusling D.A., Powers V.E., Ranasinghe R.T., Wang Y., Osborne S.D., Brown T., Fox K.R. Four base recognition by triplex-forming oligonucleotides at physiological pH. Nucleic Acids Res. 2005;33:3025–3032. doi: 10.1093/nar/gki625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Simoncsits A., Tjornhammar M.L., Rasko T., Kiss A., Pongor S. Covalent joining of the subunits of a homodimeric type II restriction endonuclease: single-chain PvuII endonuclease. J. Mol. Biol. 2001;309:89–97. doi: 10.1006/jmbi.2001.4651. [DOI] [PubMed] [Google Scholar]
- 24.Kirsch R.D., Joly E. An improved PCR-mutagenesis strategy for two-site mutagenesis or sequence swapping between related genes. Nucleic Acids Res. 1998;26:1848–1850. doi: 10.1093/nar/26.7.1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wende W., Grindl W., Christ F., Pingoud A., Pingoud V. Binding, bending and cleavage of DNA substrates by the homing endonuclease Pl-SceI. Nucleic Acids Res. 1996;24:4123–4132. doi: 10.1093/nar/24.21.4123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Arimondo P.B., Angenault S., Halby L., Boutorine A., Schmidt F., Monneret C., Garestier T., Sun J.S., Bailly C., Helene C. Spatial organization of topoisomerase I-mediated DNA cleavage induced by camptothecin–oligonucleotide conjugates. Nucleic Acids Res. 2003;31:4031–4040. doi: 10.1093/nar/gkg457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reid S.L., Parry D., Liu H.H., Connolly B.A. Binding and recognition of GATATC target sequences by the EcoRV restriction endonuclease: a study using fluorescent oligonucleotides and fluorescence polarization. Biochemistry. 2001;40:2484–2494. doi: 10.1021/bi001956p. [DOI] [PubMed] [Google Scholar]
- 28.Gingeras T.R., Greenough L., Schildkraut I., Roberts R.J. Two new restriction endonucleases from Proteus vulgaris. Nucleic Acids Res. 1981;9:4525–4536. doi: 10.1093/nar/9.18.4525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Blumenthal R.M., Gregory S.A., Cooperider J.S. Cloning of a restriction-modification system from Proteus vulgaris and its use in analyzing a methylase-sensitive phenotype in Escherichia coli. J. Bacteriol. 1985;164:501–509. doi: 10.1128/jb.164.2.501-509.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Horton J.R., Cheng X. PvuII endonuclease contains two calcium ions in active sites. J. Mol. Biol. 2000;300:1049–1056. doi: 10.1006/jmbi.2000.3938. [DOI] [PubMed] [Google Scholar]
- 31.Cheng X., Balendiran K., Schildkraut I., Anderson J.E. Structure of PvuII endonuclease with cognate DNA. EMBO J. 1994;13:3927–3935. doi: 10.1002/j.1460-2075.1994.tb06708.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Athanasiadis A., Vlassi M., Kotsifaki D., Tucker P.A., Wilson K.S., Kokkinidis M. Crystal structure of PvuII endonuclease reveals extensive structural homologies to EcoRV. Nature Struct. Biol. 1994;1:469–475. doi: 10.1038/nsb0794-469. [DOI] [PubMed] [Google Scholar]
- 33.Nastri H.G., Evans P.D., Walker I.H., Riggs P.D. Catalytic and DNA binding properties of PvuII restriction endonuclease mutants. J.Biol. Chem. 1997;272:25761–25767. doi: 10.1074/jbc.272.41.25761. [DOI] [PubMed] [Google Scholar]
- 34.Conlan L.H., Dupureur C.M. Dissecting the metal ion dependence of DNA binding by PvuII endonuclease. Biochemistry. 2002;41:1335–1342. doi: 10.1021/bi015843x. [DOI] [PubMed] [Google Scholar]
- 35.Frank-Kamenetskii M.D., Mirkin S.M. Triplex DNA structures. Annu. Rev. Biochem. 1995;64:65–95. doi: 10.1146/annurev.bi.64.070195.000433. [DOI] [PubMed] [Google Scholar]
- 36.Singleton S.F., Dervan P.B. Equilibrium association constants for oligonucleotide-directed triple helix formation at single DNA sites: linkage to cation valence and concentration. Biochemistry. 1993;32:13171–13179. doi: 10.1021/bi00211a028. [DOI] [PubMed] [Google Scholar]
- 37.Cheng Y.K., Pettitt B.M. Stabilities of double- and triple-strand helical nucleic acids. Prog. Biophys. Mol. Biol. 1992;58:225–257. doi: 10.1016/0079-6107(92)90007-s. [DOI] [PubMed] [Google Scholar]
- 38.Singleton S.F., Dervan P.B. Influence of pH on the equilibrium association constants for oligodeoxyribonucleotide-directed triple helix formation at single DNA sites. Biochemistry. 1992;31:10995–11003. doi: 10.1021/bi00160a008. [DOI] [PubMed] [Google Scholar]
- 39.Xodo L.E., Manzini G., Quadrifoglio F., van der Marel G.A., van Boom J.H. Effect of 5-methylcytosine on the stability of triple-stranded DNA—a thermodynamic study. Nucleic Acids Res. 1991;19:5625–5631. doi: 10.1093/nar/19.20.5625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Phipps A.K., Tarkoy M., Schultze P., Feigon J. Solution structure of an intramolecular DNA triplex containing 5-(1-propynyl)-2′-deoxyuridine residues in the third strand. Biochemistry. 1998;37:5820–5830. doi: 10.1021/bi972811u. [DOI] [PubMed] [Google Scholar]
- 41.Spyridaki A., Matzen C., Lanio T., Jeltsch A., Simoncsits A., Athanasiadis A., Scheuring-Vanamee E., Kokkinidis M., Pingoud A. Structural and biochemical characterization of a new Mg(2+) binding site near Tyr94 in the restriction endonuclease PvuII. J. Mol. Biol. 2003;331:395–406. doi: 10.1016/s0022-2836(03)00692-2. [DOI] [PubMed] [Google Scholar]
- 42.Kalish J.M., Seidman M.M., Weeks D.L., Glazer P.M. Triplex-induced recombination and repair in the pyrimidine motif. Nucleic Acids Res. 2005;33:3492–3502. doi: 10.1093/nar/gki659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Uil T.G., Haisma H.J., Rots M.G. Therapeutic modulation of endogenous gene function by agents with designed DNA-sequence specificities. Nucleic Acids Res. 2003;31:6064–6078. doi: 10.1093/nar/gkg815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Buchini S., Leumann C.J. Recent improvements in antigene technology. Curr. Opin. Chem. Biol. 2003;7:717–726. doi: 10.1016/j.cbpa.2003.10.007. [DOI] [PubMed] [Google Scholar]
- 45.Potaman V.N. Applications of triple-stranded nucleic acid structures to DNA purification, detection and analysis. Expert Rev. Mol. Diagn. 2003;3:481–496. doi: 10.1586/14737159.3.4.481. [DOI] [PubMed] [Google Scholar]
- 46.Knauert M.P., Glazer P.M. Triplex forming oligonucleotides: sequence-specific tools for gene targeting. Hum. Mol. Genet. 2001;10:2243–2251. doi: 10.1093/hmg/10.20.2243. [DOI] [PubMed] [Google Scholar]
- 47.Vasquez K.M., Narayanan L., Glazer P.M. Specific mutations induced by triplex-forming oligonucleotides in mice. Science. 2000;290:530–533. doi: 10.1126/science.290.5491.530. [DOI] [PubMed] [Google Scholar]
- 48.Wang G., Levy D.D., Seidman M.M., Glazer P.M. Targeted mutagenesis in mammalian cells mediated by intracellular triple helix formation. Mol. Cell. Biol. 1995;15:1759–1768. doi: 10.1128/mcb.15.3.1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vasquez K.M., Wilson J.H. Triplex-directed modification of genes and gene activity. Trends Biochem. Sci. 1998;23:4–9. doi: 10.1016/s0968-0004(97)01158-4. [DOI] [PubMed] [Google Scholar]
- 50.Helene C., Toulme J.J. Specific regulation of gene expression by antisense, sense and antigene nucleic acids. Biochim. Biophys. Acta. 1990;1049:99–125. doi: 10.1016/0167-4781(90)90031-v. [DOI] [PubMed] [Google Scholar]
- 51.Gorman L., Glazer P.M. Directed gene modification via triple helix formation. Curr. Mol. Med. 2001;1:391–399. doi: 10.2174/1566524013363771. [DOI] [PubMed] [Google Scholar]
- 52.Faruqi A.F., Seidman M.M., Segal D.J., Carroll D., Glazer P.M. Recombination induced by triple-helix-targeted DNA damage in mammalian cells. Mol. Cell. Biol. 1996;16:6820–6828. doi: 10.1128/mcb.16.12.6820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Faruqi A.F., Datta H.J., Carroll D., Seidman M.M., Glazer P.M. Triple-helix formation induces recombination in mammalian cells via a nucleotide excision repair-dependent pathway. Mol. Cell. Biol. 2000;20:990–1000. doi: 10.1128/mcb.20.3.990-1000.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Culver K.W., Hsieh W.T., Huyen Y., Chen V., Liu J., Khripine Y., Khorlin A. Correction of chromosomal point mutations in human cells with bifunctional oligonucleotides. Nat. Biotechnol. 1999;17:989–993. doi: 10.1038/13684. [DOI] [PubMed] [Google Scholar]
- 55.Chan P.P., Glazer P.M. Triplex DNA: fundamentals, advances, and potential applications for gene therapy. J. Mol. Med. 1997;75:267–282. doi: 10.1007/s001090050112. [DOI] [PubMed] [Google Scholar]
- 56.Kandavelou K., Mani M., Durai S., Chandrasegaran S. ‘Magic’ scissors for genome surgery. Nat. Biotechnol. 2005;23:686–687. doi: 10.1038/nbt0605-686. [DOI] [PubMed] [Google Scholar]
- 57.Urnov F.D., Miller J.C., Lee Y.L., Beausejour C.M., Rock J.M., Augustus S., Jamieson A.C., Porteus M.H., Gregory P.D., Holmes M.C. Highly efficient endogenous human gene correction using designed zinc-finger nucleases. Nature. 2005;435:646–651. doi: 10.1038/nature03556. [DOI] [PubMed] [Google Scholar]
- 58.Majumdar A., Puri N., Cuenoud B., Natt F., Martin P., Khorlin A., Dyatkina N., George A.J., Miller P.S., Seidman M.M. Cell cycle modulation of gene targeting by a triple helix-forming oligonucleotide. J. Biol. Chem. 2003;278:11072–11077. doi: 10.1074/jbc.M211837200. [DOI] [PubMed] [Google Scholar]
- 59.van der Woude I., Wagenaar A., Meekel A.A., ter Beest M.B., Ruiters M.H., Engberts J.B., Hoekstra D. Novel pyridinium surfactants for efficient, nontoxic in vitro gene delivery. Proc. Natl Acad. Sci. USA. 1997;94:1160–1165. doi: 10.1073/pnas.94.4.1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lohse J., Dahl O., Nielsen P.E. Double duplex invasion by peptide nucleic acid: a general principle for sequence-specific targeting of double-stranded DNA. Proc. Natl Acad. Sci. USA. 1999;96:11804–11808. doi: 10.1073/pnas.96.21.11804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Marriott G., Ottl J., Heidecker M., Gabriel D. Light-directed activation of protein activity from caged protein conjugates. Methods Enzymol. 1998;291:95–116. doi: 10.1016/s0076-6879(98)91009-6. [DOI] [PubMed] [Google Scholar]