Abstract
NDR protein kinases are involved in the regulation of cell cycle progression and morphology. NDR1/NDR2 protein kinase is activated by phosphorylation on the activation loop phosphorylation site Ser281/Ser282 and the hydrophobic motif phosphorylation site Thr444/Thr442. Autophosphorylation of NDR is responsible for phosphorylation on Ser281/Ser282, whereas Thr444/Thr442 is targeted by an upstream kinase. Here we show that MST3, a mammalian Ste20-like protein kinase, is able to phosphorylate NDR protein kinase at Thr444/Thr442. In vitro, MST3 selectively phosphorylated Thr442 of NDR2, resulting in a 10-fold stimulation of NDR activity. MOB1A (Mps one binder 1A) protein further increased the activity, leading to a fully active kinase. In vivo, Thr442 phosphorylation after okadaic acid stimulation was potently inhibited by MST3KR, a kinase-dead mutant of MST3. Knockdown of MST3 using short hairpin constructs abolished Thr442 hydrophobic motif phosphorylation of NDR in HEK293F cells. We conclude that activation of NDR is a multistep process involving phosphorylation of the hydrophobic motif site Thr444/2 by MST3, autophosphorylation of Ser281/2, and binding of MOB1A.
The NDR and LATS family of serine/threonine protein kinases participates in the regulation of cell cycle progression and cell morphology (14, 43, 53). These kinases share a conserved N-terminal regulatory domain that interacts with MOB (3, 6, 18, 33, 50) and S100B (31) proteins. The conserved catalytic (kinase) domain has an insertion of 30 amino acids between subdomains VII and VIII containing the activation segment phosphorylation site Ser281/Ser282 and, in the case of mammalian NDR1 and NDR2, an autoinhibitory sequence (3). The C-terminal regulatory domain encompasses the regulatory hydrophobic motif phosphorylation site Thr444 (32). Recent data show that NDR1 and NDR2 protein kinase activities are stimulated by Ca2+/S100B- and MOB1-binding-induced autophosphorylation on S281/S282 in vitro and in vivo (3, 31). Phosphorylation of the hydrophobic motif phosphorylation site T444/T442 is required for maximal activation and involves an upstream kinase (42, 44).
Genetic evidence suggests that Ste20 (Sterile)-like protein kinases function as upstream kinases of the NDR family. For example, one of the Saccharomyces cerevisiae Ste20-like kinases, Cdc15p, phosphorylates Dbf2p (28), and the Schizosaccharomyces pombe Ste20-like kinase Pak1p/Shk1p genetically interacts with Orb6p (49). Furthermore, the Ste20-like kinase Kic1p functionally interacts with Cbk1p, the closest relative of NDR from Saccharomyces cerevisiae (33), and the Drosophila melanogaster Ste20-like kinase HIPPO phosphorylates WARTS/LATS kinase (16, 20, 34, 45, 51). The closest mammalian homologues of Kic1p and Hippo are the mammalian Ste20-like protein kinases MST1/KRS1, MST2/KRS2, MST3, MST4/MASK, and SOK/YSK (29), which are involved in the regulation of cell morphology, proliferation, and apoptosis (7, 9, 22, 25, 38). This group of kinases contains an N-terminal kinase domain as well as an autoinhibitory C-terminal regulatory domain. Their activities are regulated by phosphorylation and proteolytic cleavage by caspases in response to stress and apoptotic stimuli (8, 19, 23, 37). Phosphorylation of MST1, for example, increases its activity severalfold and influences its subcellular localization (23). Further, the kinase-dead mutants of MST kinases are known to be inhibitors of MST kinase function in vivo (46).
There is evidence that several MST kinases promote apoptosis in response to stress stimuli and caspase activation. MST1, for example, can induce apoptosis and nuclear condensation in BJAB, 293T, and COS-1 cells (15, 46, 47), and MST1 activity correlates with eosinophil apoptosis (10). Expression of MST3 in HEK293 cells leads to DNA fragmentation and apoptosis (19). YSK1 is activated at initial stages of necrotic cell death (36). In contrast, MST4 regulates cell growth and proliferation in HeLa and Phoenix cells (25). MST4 and YSK1 are localized to the Golgi apparatus, and the latter kinase has been shown to be involved in processes such as cell migration, presumably by linking protein transport events with cellular processes such as cell adhesion or polarization of the cytoskeleton (38).
Our recent work revealed that recruitment of the MOB/NDR complex to the plasma membrane results in phosphorylation on Ser281 and Thr444 and activation of the kinase (17). To clarify and elucidate the role of Ste20-like kinases in regulating the activity of the NDR kinases, we performed in vitro and in vivo experiments with MST3, the closest relative of Kic1p from humans. We provide evidence that MST3 can function as an NDR upstream kinase by phosphorylating the hydrophobic motif and, thus, stimulating kinase activity.
MATERIALS AND METHODS
Cell culture.
COS-7 and HEK293F cells were maintained in Dulbecco's modified Eagle's medium containing 10% fetal calf serum, 100 units/ml penicillin, and 100 μg/ml streptomycin and were manipulated as described previously (17).
Antibodies.
Anti-P-Ser-282 and anti-P-Thr-442 antibodies were described previously (42). 12CA5 (hemagglutinin [HA]) and 9E10 (myc) monoclonal antibody hybridoma supernatants were used for detection of HA-NDR, HA-MST3, and myc-MST3 variants. Anti-glutathione S-transferase (anti-GST) antibodies (G7781) were purchased from Sigma. Polyclonal anti-HA Y11 antibodies were purchased from Santa Cruz. Anti-MST3 antibodies were purchased from BD Biosciences. Anti-NDR-NT peptide antibody was raised against the synthetic peptide DEEKRLRRSAHARKETEFLRLKRTRLGL, corresponding to amino acids 60 to 86 of NDR2, as described previously (17), and a rat monoclonal anti-α-tubulin (YL1/2)-producing hybridoma cell line was obtained from ATCC.
Plasmids.
DNA constructs used for transfection were purified from Escherichia coli XL1 Blue using a Qiafilter Maxiprep kit (QIAGEN) according to the manufacturer's protocol. All DNA constructs were verified by DNA sequencing using an ABI PRISM 3700 DNA analyzer (Applied Biosystems) and custom-synthesized primers. Mammalian expression vector pCMV5-encoding HA-tagged NDR2, HA-MST3, and HA-MST3KR were described previously (19, 42). myc-C1-MOB1A was described previously (17). Bacterial expression constructs for pSHP-NDR2 wild type and mutants and pGEX-2T-MOB1A were described previously (3). BamHI-flanked MST3 cDNA constructs were obtained by deleting the internal BamHI site using the QuikChange site-directed mutagenesis protocol (Stratagene) and the appropriate primers (primer sequences are available upon request) and subsequent amplification using primers 5′-CGCGGATCCATGGCTCAC-TCCCCGGTG-3′ and 5′-CGCGGATCCAAAGGAATTTCAGTGGGATG-3′. The resulting PCR product was subcloned with BamHI-BamHI into pcDNA3.0-myc (KpnI-MEQKLISEEDL-BamHI), pEGFP-C1, and pEBG2T.
A 16-amino-acid copolymer of glutamic acid and glycine was added to the N terminus of MST3 using PCR. The linker-MST3 fragment was amplified using primers 5′-AACTGCAGGAAGGTGAGGGCGAAGGTGAGGGCGAAGGTGAGGGCGAAGGTGAGGGCATGGCTCACTCCCCG-3′ and 5′-GCTCTAGATCAGTGGGATGAAGTTCC-3′. The PstI-XbaI fragment was subcloned into pCMV5. To create a pCMV5-HA-NDR2-MST3 fusion construct, HA-tagged NDR2 was amplified using primers 5′-CTTCCAAGCGCTTAGTCGACATGGCTTACCCATACGATGTTCCAGATTACGCTTCGGCAATGACGGCAGGGACTACAACAACC-3′ and 5′-AACTGCAGTAACTTCCCAGCTTTCATGTAGG-3′ and subcloned as a HindIII-PstI fragment into pCMV5 (linker-MST3). The myc-C1-MOB1A was described previously (17).
The pTER-shMST3 vectors were cloned using the HindIII-BglII-digested pTER vector (48) and the oligonucleotides 5′-GATCCCGGCATTGACAATCGGACTCTTCAAGAGAGAGTCCGATTGTCAATGCCTTTTTGGAAA-3′ and 5′-AGCTTTTCCAAAAAGGCATTGACAATCGGACTCTCTCTTGAAGAGTCCGATTGTCAATGCCGG-3′ for shMST3.
Western blotting and immunoprecipitation.
Cell lysis buffer (immunoprecipitation [IP] buffer) contained 20 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1% Nonidet P-40, 10% glycerol, 1 mM Na3VO4, 20 mM β-glycerol phosphate, 1 μM microcystin, 50 mM NaF, 0.5 mM phenylmethylsulfonyl fluoride, 4 μM leupeptin, and 1 mM benzamidine. To detect HA-NDR, SHP-NDR, HA-MST3, and myc-MST3, samples were resolved by 10 or 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to polyvinylidene difluoride membranes. Membranes were blocked in TBST (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, and 0.05% Tween 20) containing 5% skimmed milk powder and then probed overnight at 4°C with the following antibodies: anti-GST, 12CA5 anti-HA, 9E10 anti-myc, and anti-Thr-442P; or anti-Ser-282-P, anti-NDR-NT, and anti-MST3; and anti-α-tubulin YL1/2. Bound antibodies were detected with horseradish peroxidase-linked secondary antibodies and enhanced chemiluminescence. For immunoprecipitations, HEK293F or COS-7 cells transfected with HA-MST3 wild type, HA-MST3-KR, or HA-NDR2 variants were harvested as described above. Cell lysates (0.5 mg protein) were precleared with protein A- or G-Sepharose and mixed subsequently for 3 h at 4°C with anti-HA 12CA5 antibody prebound to protein A-Sepharose. The beads were then washed twice with IP buffer, once with IP buffer containing 1 M NaCl, once again with IP buffer, and twice with 20 mM Tris-HCl, pH 7.5, containing 4 μM leupeptin and 1 mM benzamidine. Samples were then subjected to kinase assays and/or were resolved by 10% SDS-PAGE. HA-MST3, HA-MST3KR, and HA-NDR2 variants were detected by Western blotting using anti-HA 12CA5 monoclonal antibodies.
Bacterial expression of human GST-fused MOB1A and human SHP-fused NDR2.
XL-1 Blue E. coli was transformed with the pGEX-2T-MOB1A plasmid. Mid-logarithmic-phase cells were induced with 0.1 mM isopropyl β-d-thiogalactopyranoside overnight at 20°C. Bacterial lysis buffers contained 20 mM Tris-HCl, pH 8.5, 10 mM 2-mercaptoethanol, 1% NP-40, 0.5 M NaCl, and Complete proteinase inhibitor cocktail (Roche). Bacteria were disrupted using a French press in the presence of 1 mg/ml lysozyme and protease inhibitors, and the fusion proteins were purified on glutathione-Sepharose. SHP-NDR2 wild-type and mutant plasmids were transformed into XL-1 Blue E. coli cells as described for GST-hMOB1A, and protein was purified on Ni-nitrilotriacetic acid-Sepharose.
NDR protein kinase assays.
Transfected COS-7 cells were washed once with ice-cold phosphate-buffered saline (PBS) and harvested 24 or 48 h after transfection in 1 ml of ice-cold phosphate-buffered saline containing 1 mM Na3VO4 and 20 mM β-glycerol phosphate before lysis in 500 μl IP buffer (20 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1% Nonidet P-40, 10% glycerol, 1 mM Na3VO4, 20 mM β-glycerol phosphate, 1 μM microcystin, 50 mM NaF, 0.5 mM phenylmethylsulfonyl fluoride, 4 μM leupeptin, and 1 mM benzamidine). Lysates were centrifuged at 20,000 × g for 20 min, and duplicate aliquots (250 μg) of the supernatant were precleared with protein A-Sepharose for 60 min and mixed for 3 h at 4°C with anti-HA 12CA5 antibody prebound to protein A-Sepharose. The beads were then washed twice with IP buffer, once for 10 min with IP buffer containing 1 M NaCl, again for 10 min with IP buffer, and twice with 20 mM Tris-HCl, pH 7.5, containing 4 μM leupeptin and 1 mM benzamidine. Thereafter, the beads were resuspended in 30 μl of buffer containing 20 mM Tris-HCl, pH 7.5, 10 mM MgCl2, 1 mM dithiothreitol, 100 μM [γ-32P]ATP (∼1,000 cpm/pmol), 1 μM cyclic AMP (cAMP)-dependent protein kinase inhibitor peptide, 4 μM leupeptin, 1 mM benzamidine, 1 μM microcystin, and 1 mM NDR substrate peptide (KKRNRRLSVA). After a 60-min incubation at 30°C, the reaction mixtures were processed as previously described (44).
Purified recombinant SHP-NDR2 wild type and mutants (1 μg) were preautophosphorylated or prephosphorylated for 60 min in the presence of HA-MST3 or HA-MST3KR (immunoprecipitated from 500 μg of lysates of untreated and okadaic acid [OA]-treated HEK293F cells) in 30 μl of buffer containing 20 mM Tris-HCl, pH 7.5, 10 mM MgCl2, 1 mM dithiothreitol, 100 μM ATP, 1 μM cAMP-dependent protein kinase inhibitor peptide, 4 μM leupeptin, 1 mM benzamidine, and 1 μM microcystin. Aliquots of 10 μl of the supernatant containing SHP-NDR or mutants were removed. One aliquot was assayed for activity using our standard conditions (reaction mixture of 30 μl containing 20 mM Tris-HCl, pH 7.5, 10 mM MgCl2, 1 mM dithiothreitol, 100 μM [γ-32P]ATP [∼1,000 cpm/pmol], 1 μM cAMP-dependent protein kinase inhibitor peptide, 4 μM leupeptin, 1 mM benzamidine, 1 μM microcystin, and 1 mM NDR substrate peptide). After incubation for 30 min at 30°C, the reaction mixtures were processed and kinase activity was determined as described for the HA-NDR kinase assay. The second aliquot was resolved by 10% SDS-PAGE and analyzed for phosphorylation and amount of protein using anti-T-442-P and anti-S-282-P antibodies by Western blotting and Coomassie staining, respectively.
Time course analyses of hydrophobic motif phosphorylation and activity of SHP-NDR2 variants (1 μg) in the presence or absence of GST-MOB1A and in the presence or absence of GST-MST3 from untreated or OA-treated HEK293F cells were performed under our kinase assay standard conditions, as described above. Kinase assays were stopped after 0, 15, 30, 60, 90, and 120 min by adding 3 μl of 0.5 M EDTA. The reaction mixtures were processed, and kinase activity was determined as described for the HA-NDR kinase assay. Phosphorylation of NDR was detected by Western blotting, as described above.
Localization of MST3 and NDR2.
Exponentially growing cells were plated on coverslips and transfected the next day with the indicated constructs using Fugene 6 (Roche) as described by the manufacturer. After the 24-h transfection, cells were washed with PBS and fixed in 3% paraformaldehyde-2% sucrose in PBS at pH 7.4 for 10 min at 37°C. They were then permeabilized using 0.2% Triton X-100 in PBS for 2 min at room temperature. All subsequent steps were carried out at room temperature. Coverslips were rinsed twice with PBS and incubated for 1 h with anti-HA Y11 (Santa Cruz) and anti-myc 9E10 diluted in PBS containing 1% bovine serum albumin-1% goat serum. After three 1-min washes in PBS, goat anti-rabbit antibody-fluorescein isothiocyanate (FITC; Sigma) and goat anti-mouse antibody-Texas Red (Sigma) were used as secondary antibodies. DNA was counterstained with 4 μg/ml Hoechst (Sigma). Coverslips were then inverted into 5 μl Vectashield medium (Vector Lab). Images were obtained with an Eclipse E800 microscope using a CoolPix950 digital camera (Nikon) and processed using Adobe Photoshop 6.0 (Adobe Systems Inc.). Only cells with intact nuclei were included in the statistical evaluation. Cells expressing GFP-MST3 were fixed and then stained for DNA without permeabilization and antibody incubation steps.
Cell fractionation.
To separate cytosolic and membrane-associated proteins, cells were subjected to S100/P100 fractionation as follows: cells were collected in PBS and incubated for 20 min at 4°C in S100/P100 buffer (20 mM Tris, 150 mM NaCl, 2.5 mM EDTA, 1 mM EGTA, 1 mM benzamidine, 4 μM leupeptin, 0.5 mM PMSF, 1 μM microcystin, and 1 mM dithiothreitol at pH 7.5) and homogenized by passage through a 26-gauge needle (Beckton Dickinson), and nuclei were removed by centrifugation for 2 min at 1,000 × g at 4°C. The supernatant was then centrifuged at 100,000 × g for 60 min at 4°C. Equal amounts of protein from the supernatant (S100; cytoplasmic fraction) and the pellet (P100; membrane fraction) were analyzed by SDS-PAGE followed by immunoblotting.
RESULTS
Catalytic domain conservation of Ste20-like kinases.
We compared the substrate-binding pockets (27) of the human MST kinases to the budding yeast relatives Kic1p and Cdc15p, which were reported previously to be upstream regulators of Cbk1p and Dbf2p (28, 33). All of the kinases showed very high conservation of their catalytic domain. MST3, YSK1, and MST4 as well as MST1 and -2 have identical residues in their substrate-binding pockets, suggesting similar substrate specificities (see Fig. S1A in the supplemental material). We also compared the hydrophobic motifs of the budding yeast Cbk1p and Dbf2p with human NDR kinases. The hydrophobic motifs of hNDR1 and -2 as well as LATS1 and -2 show a significant similarity to corresponding regions in the yeast kinases (see Fig. S1B in the supplemental material). To test our hypothesis that a mammalian Ste20-like (MST) kinase is the hydrophobic motif kinase of NDR protein kinases, we performed in vitro kinase assays with MST3, the closest relative of the yeast Kic1p-Cbk1p pathway. For this study we used NDR2, because it was easier to produce recombinant full-length protein.
Activation and hydrophobic motif phosphorylation of NDR protein kinase by MST3.
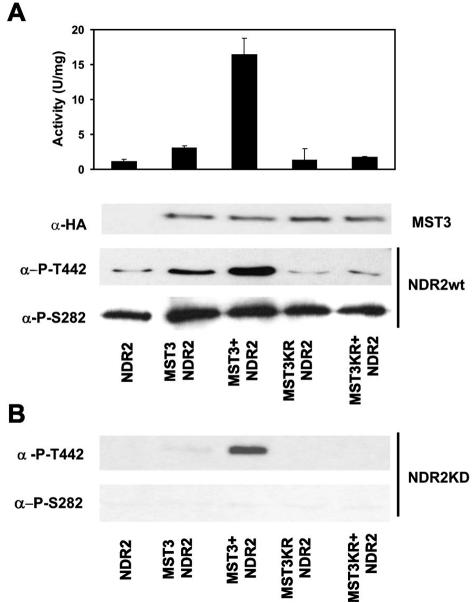
We expressed HA-MST3wt and the kinase-dead HA-MST3KR in HEK293F cells and treated them with 1 μM OA for 1 h (MST3+; MST3KR+) or solvent control (MST3; MST3KR). The HA-tagged MST3 kinase variants were immunoprecipitated with the anti-HA 12CA5 monoclonal antibody and incubated with SHP-NDR2 and catalytically inactive SHP-NDR2KD for 1 h at 30°C. Subsequently, the beads with bound MST3 were removed and the supernatants containing the SHP-NDR proteins were analyzed for activity and phosphorylation status using anti-Ser282-P and anti-Thr442-P specific phospho-antibodies. Kinase assays were carried out for 30 min with the NDR substrate peptide (30). (We previously established that activated MST3 does not significantly phosphorylate the NDR substrate peptide.)
Significantly, preincubation with nonstimulated MST3 only led to a twofold activation of NDR2, while preincubation with OA-activated MST3 resulted in a 10-fold increase in NDR2 activity, suggesting that a certain population of MST3 is active in nonstimulated cells. This result reflected an increase in phosphorylation of both the activation segment phosphorylation site Ser282 and the hydrophobic motif phosphorylation site Thr442 (Fig. 1A). Analysis of the phospho-status of the kinase-dead NDR2 revealed that MST3 phosphorylates NDR2 on Thr442, but not on Ser282 (Fig. 1B). Results from the same experiments performed with the kinase-dead MST3KR confirmed that phosphorylation as well as activation of NDR2 depend on MST3 activity. Significantly, our results also show that OA treatment of HEK293F cells promotes the activation of MST3 similar to that previously found for NDR (32). To have a more robust effect on NDR activation, we used activated MST3 (isolated from OA-treated cells) in the following in vitro experiments.
FIG. 1.
Activation and hydrophobic motif phosphorylation of NDR2 by MST3. (A) Wild-type recombinant SHP-NDR2 was preincubated for 60 min with or without immunoprecipitated HA-MST3 or HA-MST3KR from untreated and OA-treated (+) HEK293F cells. The activity of SHP-NDR2 was determined using the NDR kinase substrate peptide. The results shown are means ± standard deviations of assays carried out in duplicate and are representative of two independent experiments. Samples from each preincubation were also analyzed for phosphorylation by Western blotting using anti-P-T442 and anti-P-S282 antibodies. HA-MST3wt and HA-MST3KR were quantified by Western blot analysis using the α-HA 12CA5 monoclonal. (B) Phosphorylation of kinase-dead NDR2 by MST3. Kinase-dead NDR2K119A was incubated for 60 min with or without immunoprecipitated HA-MST3 or HA-MST3KR from untreated and OA-treated (+) HEK293F cells. Samples were analyzed by Western blotting as for panel A.
Mechanism of NDR activation by MST3 and MOB1.
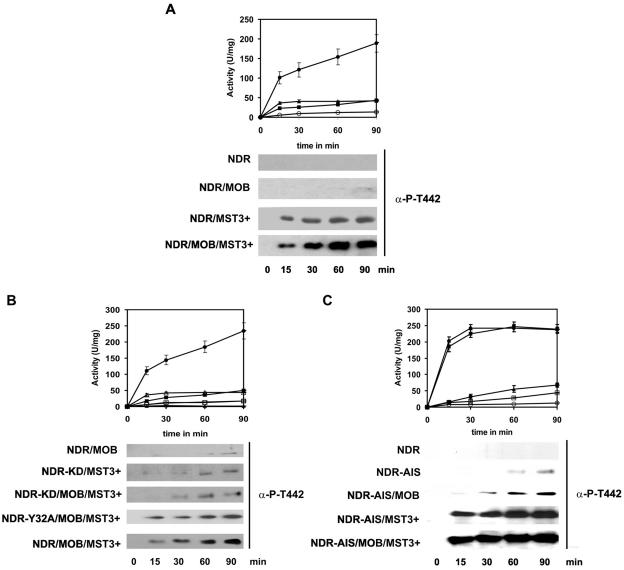
Next, we sought to define the role of the MOB proteins in regulating NDR activity. Previously, we and others demonstrated that NDR1 and -2 are activated by MOB proteins (3, 11). We performed NDR kinase assays in the presence and absence of MOB1A and MST3 using GST-MOB1A and SHP-NDR2 expressed in E. coli, as well as OA-stimulated (1 μM for 1 h) and nonstimulated GST-MST3 expressed in HEK293F cells. Proteins were affinity purified on glutathione-Sepharose (GST-MOB1A and GST-MST3) and Ni-nitrilotriacetic acid-Sepharose (SHP-NDR2), respectively. We performed time course kinase assays (0 to 90 min) with various combinations of the proteins, employing the NDR substrate peptide. NDR2 alone showed only a moderate increase in kinase activity over the time course (Fig. 2A). Addition of MOB1A resulted in a three- to fourfold increase in kinase activity compared with NDR2 alone during the time course (Fig. 2A). Addition of activated MST3 led to a rapid initial increase in kinase activity that reached a maximum after 30 min and remained approximately constant for the remainder of the time course (Fig. 2A). Addition of activated MST3 together with MOB1A resulted in a 20-fold-higher NDR activity (Fig. 2A), suggesting that both MOB1A and MST3 kinase are required for full kinase activation. Nonstimulated MST3 kinase had no significant effect on NDR2 kinase activity either in the presence or absence of MOB1A (data not shown), indicating that activation of MST3 is crucial for NDR activation. However, the Thr442 phosphorylation obtained with wild-type NDR2/MOB1A suggests a minor contribution of hydrophobic motif phosphorylation by NDR2 itself in vitro, as previously reported (42). This activity might also explain the minor increase in Thr442 phosphorylation in NDR2/MOB1A/MST3+ compared to the NDR2/MST3+.
FIG. 2.
Analysis of NDR2 activation and hydrophobic motif phosphorylation by MST3 and MOB1A. (A) Activation and phosphorylation of wild-type NDR2. NDR2 SHP-NDR2 was incubated with GST-MOB1A (▪), GST-MST3 from OA-stimulated HEK293F cells (▴), both (•) or alone (○) under our standard NDR kinase assay conditions. (B) Activation and phosphorylation of kinase-dead and MOB-binding-deficient NDR2. SHP-NDR2 (○), SHP-NDR2/GST-MOB1A (▪), SHP-NDR2/MOB1A/MST3+ (•), SHP-NDR2K119A/MOB1A/MST3+ (▴), SHP-NDR2Y32A (□), and SHP-NDR2Y32A/MOB1A/MST3+ (Δ) were incubated under standard NDR kinase assay conditions. (C) Activation and phosphorylation of the NDR2-AIS mutant. SHP-NDR2 (○), SHP-NDR2-AIS (□), SHP-NDR2-AIS/GST-MOB1A (▴), SHP-NDR2-AIS/MST3+ (▪), and SHP-NDR2-AIS/GST-MOB1A/MST3+ (•) were incubated under standard NDR kinase assay conditions. At the time points indicated, NDR kinase activity was assayed with the NDR substrate peptide; results are expressed as specific activity. Results shown are means ± standard deviations of assays carried out in duplicate and are representative of two independent experiments. Error bars are only shown when larger than the size of the symbols. NDR hydrophobic motif phosphorylation was determined at each time point by Western blotting using the anti-P-T442 antibody.
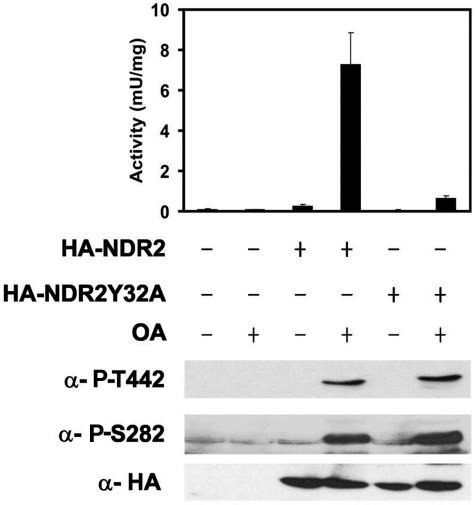
To obtain more detailed information about NDR2 activation by MST3 and MOB1A, we performed in vitro activation and phosphorylation time courses with kinase-dead and MOB-binding-deficient NDR2 mutants (3). The SHP-NDR2 mutants were expressed and purified as described above for wild-type SHP-NDR2. As expected, the MOB1-binding-deficient mutant NDR2Y32A was not activated by MOB1A and showed only a basal activity similar to wild-type NDR2 (Fig. 2B). Addition of activated MST3 resulted in a five- to sixfold increase in NDR activity to levels similar to those observed with wild-type NDR2 (Fig. 2A and B). The activation profiles and phospho-blots of the NDR2Y32A mutant in the presence of MOB1A showed patterns similar to the corresponding profiles and blots of wild-type NDR2 in the absence of MOB1A (Fig. 2A and B). The kinase-dead mutant showed the same degree of Thr442 phosphorylation in the presence and absence of MOB1A, indicating that MOB1A does not affect phosphorylation of NDR by MST3. The Thr442 phosphorylation obtained with the wild-type NDR2/MOB1A/MST3+ and NDR2Y32A/MOB1A/MST3+ assays were similar, indicating that MOB1A promotes an active conformation and release of autoinhibition of the kinase. In agreement with these data, the NDR2-AIS (autoinhibitory sequence) mutant (3) that is released from autoinhibition showed an increase in basal activity; addition of MOB1A resulted in a further twofold increase in NDR-AIS activity (Fig. 2C). Incubation of NDR2-AIS with MST3+ and of NDR2-AIS with MOB1A and MST3+ resulted in very similar activation profiles, indicating that release of autoinhibition and hydrophobic motif phosphorylation are sufficient for full kinase activation (Fig. 2C). We conclude that activation of NDR2 is a multistep process involving phosphorylation of the hydrophobic motif site Thr444/2 by MST3, autophosphorylation of Ser281/2, and binding of MOB1A.
NDR hydrophobic motif site phosphorylation by MST3 in vivo.
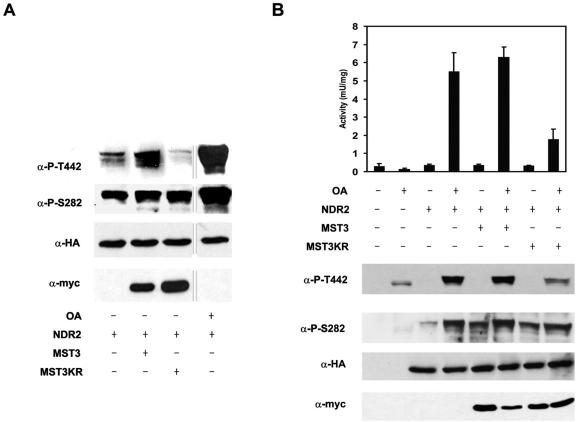
To examine the role of MST3 in NDR activation in vivo, we performed cotransfection experiments with HA-NDR2, myc-MST3, and myc-MST3KR using COS-7 cells. Analysis of the phosphorylation of NDR revealed a small but significant increase in Thr442 phosphorylation in cells expressing MST3 and NDR2 compared with cells expressing NDR2 alone (Fig. 3A). The extent of NDR2 phosphorylation was less than that obtained after OA stimulation, as shown with anti-P-Thr442 (Fig. 3A). Cells expressing the kinase-dead variant MST3KR and NDR2 showed a reduction in Thr442 phosphorylation (Fig. 3A). Inhibition of Thr442 phosphorylation by the kinase-dead MST3KR suggests that the mutant protein acts as an inhibitor of Thr442 phosphorylation mediated by endogenous MST3 (see below). Evidence in support of this hypothesis comes from the inhibition of OA-induced phosphorylation of the NDR2 hydrophobic motif by cotransfection with MST3KR, which resulted in 70% lower NDR activity and Thr442 phosphorylation (Fig 3B). Ser282 phosphorylation was largely unchanged (Fig. 3B). This suggests that MST3KR acts as an inhibitor of Thr442 phosphorylation. The reduced phosphorylation was also reflected by decreased NDR activity.
FIG. 3.
NDR activation and phosphorylation by MST3 in vivo. (A) Effect of cotransfection of MST3 on NDR2 phosphorylation. COS-7 cells were cotransfected with myc-MST3, kinase-dead myc-MST3KR, and HA-NDR2. OA-treated cells transfected with HA-NDR2 were used as a control for NDR2 phosphorylation. All proteins migrated at the expected molecular mass. Cell lysates were immunoblotted with anti-P-T442 (very long exposure), anti-P-S282, anti-HA, and anti-myc. (B) Inhibition of NDR2 activation and hydrophobic motif phosphorylation by MST3KR. COS-7 cells expressing HA-NDR2, wild-type myc-MST3, and myc-MST3KR in various combinations were treated for 1 h with 1 μM OA or with solvent alone. HA-tagged NDR kinase variants were then immunoprecipitated (100 μg of cell-free protein extracts) with anti-HA 12CA5 monoclonal antibody and assayed for kinase activity using the NDR peptide substrate. Bars represent the means ± standard deviations of triplicate immunoprecipitates and are representative of two independent experiments. All cell lysates were immunoblotted with anti-P-T442 (normal exposure times), anti-P-S282, anti-HA 12CA5, and anti-myc.
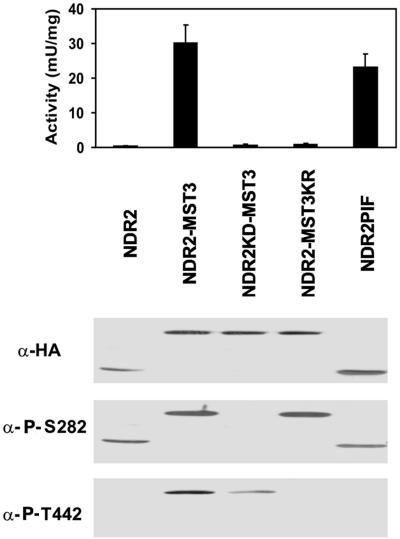
Taken together, our data indicate that MST3 can act in vivo as an NDR hydrophobic motif kinase. We tested this hypothesis further using NDR2-MST3 fusion proteins. To create the NDR2-MST3 fusion protein, a linker of 16 amino acids composed of an alternating copolymer of glutamate and glycine was linked to the NDR2 C terminus and fused to MST3 in frame, as described for ERK2-MEK1 fusions (40). HA-tagged NDR2-MST3, NDR2KD-MST3, and NDR2-MST3KR fusion proteins were expressed in HEK293F cells and analyzed for kinase activity and phosphorylation (Fig. 4). The immunoprecipitated NDR2-MST3 fusion protein was 50- to 100-fold more active than NDR2 from nonstimulated HEK293F cells. The activity was even higher than the previously described constitutively active NDR2-PIF variant (Fig. 4) (42). The NDR2-PIF variant contains the PRK2 hydrophobic motif (EEQEMFRDFDYIADW fused at amino acid 434), which has an Asp residue instead of a Thr or Ser residue and is able to stabilize the kinase in an active conformation, as described for PKB (52). Control fusions with kinase-dead NDR2 were inactive in the kinase assay, indicating that MST3 did not affect the activity measurements. Analysis of the NDR variants with phospho-specific anti-P-T442 antibodies showed that NDR2-MST3 and NDR2KD-MST3, but not NDR2-MST3KR, are phosphorylated on the hydrophobic motif phosphorylation site without OA stimulation, suggesting that the interaction is direct. Similar results were obtained with NDR1-MST3 fusion proteins (data not shown).
FIG. 4.
Activity of NDR2-MST3 fusion proteins. HA-tagged variants of NDR2, NDR2-MST3, NDR2KD-MST3, NDR2-MST3KR, and NDR2-PIF in the pcDNA3.1 vector were expressed in HEK293F cells. The NDR kinase variants were then immunoprecipitated (100 μg of cell-free protein extracts) with anti-HA 12CA5 monoclonal antibody, and kinase activity was assayed using the NDR peptide substrate. Bars represent the means ± standard deviations of triplicate immunoprecipitates and are representative of two independent experiments. Additionally, all immunoprecipitates were subjected to SDS-PAGE and Western blotting using anti-HA 12CA5, anti-P-S282, and anti-P-T442 antibodies. All NDR variants migrated at the expected molecular weight.
Activation and phosphorylation of NDR by endogenous MST3.
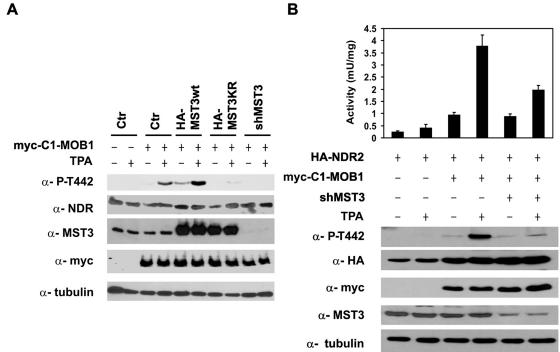
Membrane targeting of NDR leads to phosphorylation and activation of the kinase (17). Constitutive membrane targeting of NDR or MOB using the Lck myristoylation/palmitoylation motif (MGCVCSSN) or inducible membrane targeting using the C1-domain derived from PKCα (amino acids 26 to 126) results in an increase of hydrophobic motif phosphorylation and NDR activity (17). To further establish the in vivo role of MST3 in regulating endogenous NDR, we transfected HEK293F cells with a myc-C1-MOB1A chimera and different MST3 constructs designed to manipulate the levels of the upstream kinase. The cells were stimulated, after 24 h of starvation, with 100 ng/ml TPA or solvent control for 15 min, and hydrophobic motif phosphorylation of endogenous NDR was determined using anti-P-Thr442 antibodies (Fig. 5A). TPA treatment promoted recruitment of endogenous NDR to the membrane and promoted phosphorylation. Overexpression of HA-MST3 increased the phosphorylation of Thr442 in unstimulated and stimulated cells. Knockdown of MST3 protein (Fig. 5A) led to almost complete inhibition of NDR2 phosphorylation. Furthermore, our results show that MST3KR also inhibits phosphorylation of endogenous MST3. The results show a clear correlation between MST3 levels and hydrophobic motif phosphorylation of NDR following TPA-induced membrane recruitment of C1-tagged-MOB1A, indicating that MST3 is indeed a physiological regulator of NDR in vivo.
FIG. 5.
Phosphorylation of endogenous NDR by endogenous MST3. (A) Myc-C1-MOB1A was expressed in HEK293F cells transfected with either pcDNA3.1-HA-MST3, pcDNA3.1-HA-MST3KR, or pTER-shMST3. Two days after transfection, the cells were starved for 24 h and then stimulated for 15 min with TPA (100 ng/ml) prior to harvesting. All cell lysates were subjected to SDS-PAGE and immunoblotted with anti-P-T442, anti-NDR-NT, anti-MST3, anti-myc, and an anti-α-tubulin control. (B) Phosphorylation and activation of NDR2 by endogenous MST3. HA-NDR2 was expressed in HEK293F cells and transfected with pcDNA3.1 myc-C1-MOB1A and/or pTER-shMST3. Two days after transfection, the cells were starved for 24 h and subsequently stimulated for 10 min with TPA (100 ng/ml) prior to harvesting. HA-NDR2 was immunoprecipitated with anti-HA 12CA5 monoclonal antibody and assayed for kinase activity using the NDR peptide substrate. Bars represent the means ± standard deviations of duplicate immunoprecipitates and are representative of two independent experiments. All cell lysates were subjected to SDS-PAGE and immunoblotted with anti-P-T442, anti-HA, anti-MST3, anti-myc, and the anti-α-tubulin control.
To investigate the effects of MST3 on NDR kinase activity, we used the inducible membrane targeting method as described above. We transfected HEK293F cells with HA-NDR2, myc-C1-MOB1A, and pTER-shMST3 and determined the activation of NDR and hydrophobic motif phosphorylation (Fig. 5B). After incubation with TPA for 10 min, the activation of NDR was significantly diminished (∼50%) in samples cotransfected with pTER-shMST3. The activation was only partially inhibited, because we were unable to achieve full knockdown of MST3. In agreement with our results obtained with endogenous NDR, Thr442 phosphorylation of HA-NDR2 showed a clear dependence on MST3, indicating that membrane recruitment of the NDR/MOB complex is crucial for NDR activation and phosphorylation by MST3.
Activation and phosphorylation of MOB1-binding-deficient NDR2.
To obtain further insights into the role of MOB1 in the activation process of NDR, we examined its activation and phosphorylation following OA treatment using the MOB1-binding-deficient mutant NDR2Y32A (3). Despite the fact that NDR2Y32A was phosphorylated on Thr442 to the same extent as wild-type NDR2, activation of the kinase was only 2- to 3-fold, compared with 30-fold for the wild-type control (Fig. 6). This result shows that MOB1 binding is required for full activation and correlates well with the results obtained in vitro, arguing that MST3 functions as an upstream kinase and that MOB1 releases autoinhibition.
FIG. 6.
Activation and phosphorylation of MOB1-binding-deficient NDR2. HEK293F cells were transfected with HA-tagged NDR2 and NDR2Y32A and variants and stimulated for 1 h with 1 μM OA or solvent prior to harvesting. The NDR kinase variants were then immunoprecipitated (100 μg of cell-free protein) with anti-HA 12CA5 monoclonal antibody and assayed for kinase activity using the NDR peptide substrate. Bars represent the means ± standard deviations of duplicate immunoprecipitates and are representative of two independent experiments. Additionally, all lysates were subjected to SDS-PAGE and immunoblotted using anti-P-T442, anti-P-S282, and anti-HA 12CA5 antibodies.
Colocalization of MST3 and NDR.
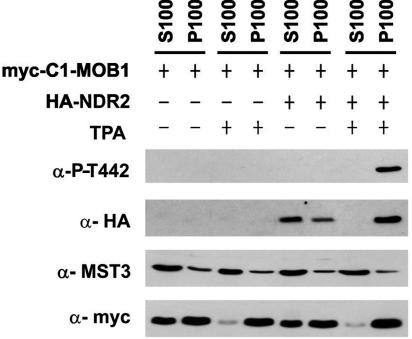
We examined the intracellular localization of GFP-MST3, HA-MST, HA-MST3KR, and myc-NDR2 in COS-7 cells. MST3 has been shown previously to localize predominantly to cytoplasmic structures (19) but translocates to the nucleus upon caspase cleavage. We confirmed the localization of full-length MST3 under our experimental conditions with two different epitope tags (GFP and HA) (Fig. 7A). To evaluate whether NDR2 and MST3 localize to similar cytoplasmic structures, we coexpressed MST3 and NDR2 in COS-7 cells. Examination by confocal microscopy demonstrated a largely overlapping localization of MST3 and NDR2 to cytoplasmic and membrane structures (Fig. 7B), indicating that these proteins indeed are able to interact in vivo. Colocalization itself is not reflected by an increase in NDR phosphorylation, whereas NDR localization to the membrane results in increased Thr442/4 phosphorylation, suggesting subcellular localization is an important component of NDR regulation (17). We further investigated biochemically whether the proteins form a stable complex. As expected, employing the myc-C1-MOB1A construct, TPA stimulation led to recruitment of HA-NDR2 to the membrane fraction, thereby activating and phosphorylating NDR2 at the hydrophobic motif (see also the report of Hergovich et al. [17]). However, the distribution of endogenous MST3 was unchanged under these conditions (Fig. 8). The major population of MST3 was cytoplasmic, but a significant portion was associated with the membrane. To ensure that the proteins are indeed membrane associated and not cellular aggregates, we also performed a control experiment where we pretreated samples with fractionation buffer containing 1% Triton (see Fig. S2 in the supplemental material). Further, MST3 was apparently not detected in HA-NDR2 immunoprecipitates under various conditions (e.g., TPA stimulation of myc-C1-MOB1A-cotransfected cells) (data not shown). This suggests that MST3 and NDR2 did not form a stable complex under our conditions and that membrane recruitment of the NDR/MOB1 complex is crucial for NDR phosphorylation by MST3 in vivo.
FIG. 7.
Colocalization of NDR2 and MST3 in COS-7 cells. (A) Localization of MST3. COS-7 cells expressing GFP-MST3 (upper panels), HA-MST3wt (middle panel), or HA-MST3KR (lower panels) were processed for immunofluorescence using either no antibody (upper right) or anti-HA Y11 (lower right). Anti-HA antibody was visualized using anti-rabbit antibody-FITC (green) and DNA stained with 1 μM TO-PRO-3 iodide (Molecular Probes Inc.) (blue; left panels). (B) Colocalization of NDR2 and MST3. COS-7 cells expressing HA-MST3 and myc-NDR2 were processed for immunofluorescence using anti-HA Y11 and anti-myc 9E10. Anti-HA Y11 antibody was visualized using anti-rabbit antibody-FITC (green), and anti-myc antibody was visualized using anti-mouse antibody-Texas Red (red). Representative pictures of HA-MST3 and myc-NDR2 localization are shown.
FIG. 8.
Fractionation of HA-NDR and MST3 after membrane recruitment of myc-C1-MOB1A. HEK293F cells were transfected with the HA-NDR2 and myc-C1-MOB1A constructs indicated and subjected to S100/P100 fractionation (S, cytoplasm; P, membrane), before immunoblotting with anti-P-T444, anti-HA 12CA5, anti-MST3, and anti-myc antibodies.
NDR promotes cleavage of MST3.
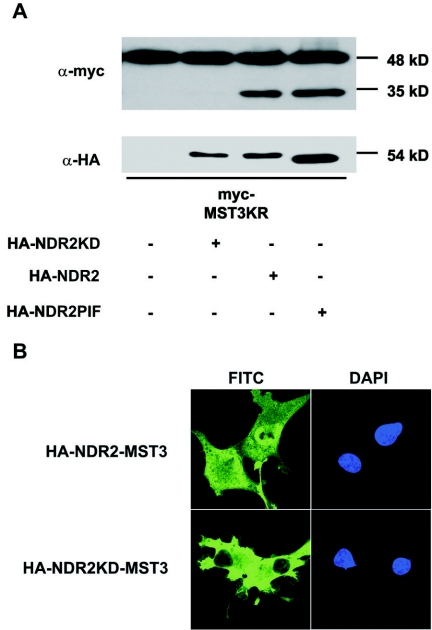
It has been reported that members of the MST family are frequently cleaved following caspase activation (8, 19, 21, 37). Based on our initial observations (data not shown) that most of the wild-type MST3 protein was efficiently cleaved but the kinase-dead MST3KR was only cleaved to a minor degree in samples harvested 36 h after transfection in COS-7 cells, we asked whether MST3 cleavage depends on the activity of full-length MST3 and whether this regulation involves NDR (Fig. 9A). We cotransfected COS-7 cells with kinase-dead MST3KR alone or together with HA-tagged NDR2, NDR2KD, and NDR2-PIF. Cotransfection of wild-type NDR2 and NDR2-PIF resulted in significantly higher cleavage of MST3KR than in cells transfected only with MST3KR, whereas cotransfection of MST3KR with kinase-dead NDR2 had no detectable effect on the cleavage of full-length MST3KR (Fig. 9A). This suggests that cleavage of MST3 can be induced by NDR activity. This is also clearly reflected by the localization of NDR-MST3 fusion proteins. MST3 was shown recently to contain both a nuclear localization signal (amino acids 278 to 294) and a nuclear export signal located in the C-terminal region (amino acids 335 to 386) downstream of the kinase domain. The nuclear export signal is cleaved (residue 314) after caspase activation, which leads to nuclear translocation of the kinase (24). In COS-7 cells, NDR2-MST3 fusion proteins showed both nuclear and cytoplasmic localization, whereas an NDR2KD-MST3 fusion was localized almost exclusively in the cytoplasm (Fig. 9B). Our results indicate that NDR can function in a regulatory feedback loop controlling MST3 cleavage.
FIG. 9.
NDR promotes cleavage of MST3 in COS-7 cells. (A) Induction of MST3 cleavage by overexpression of catalytically active NDR2. Lysates of COS-7 cells transfected as indicated with myc-MST3KR, HA-NDR2, HA-NDR2KD, and HA-NDR2-PIF were harvested after 36 h and subjected to Western blotting using anti-HA and anti-myc antibodies. (B) Localization of NDR2-MST3 fusion proteins. COS-7 cells expressing HA-NDR-MST3 (upper panel) or HA-NDR2KD-MST3 (lower panel) were processed for immunofluorescence 24 h after transfection using anti-HA Y11 (right panels). Anti-HA antibody was visualized using anti-rabbit antibody-FITC (green); DNA stained with DAPI (blue; left panels). Representative pictures of HA-NDR2-MST3 and HA-NDR2KD-MST3 localization are shown.
DISCUSSION
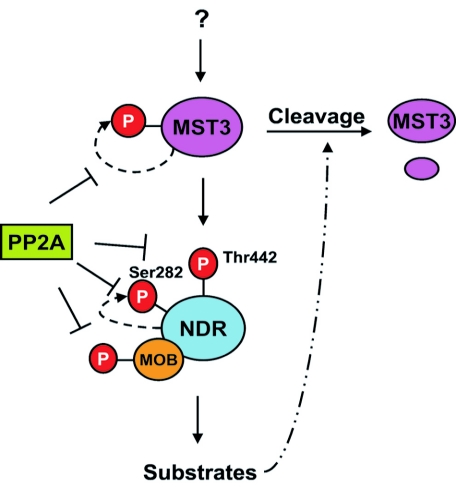
In this report we provide evidence that MST3 functions as a hydrophobic motif kinase of NDR protein kinase, and our current model for NDR regulation is summarized in Fig. 10. This is the first demonstration that a Ste20-like kinase specifically phosphorylates the hydrophobic motif (Thr442), and not the activation segment phosphorylation site, of NDR. We have established that NDR2 is phosphorylated by activated MST3, but not MST3KR (see below), specifically on the hydrophobic motif, leading to an approximately 10-fold activation of the kinase. Based on the catalytic domain conservation of mammalian Ste20-like kinases and yeast Ste20-like kinases, it is likely that the mechanism of NDR family kinase hydrophobic motif phosphorylation by Ste20-like kinases is conserved and is the basis for the Cdc15p-Dbf2p, Kic1p-Cbk1p, and HIPPO-WARTS interactions reported in yeast and flies (28, 33, 43, 51). We propose that NDR2 hydrophobic motif phosphorylation functions in a manner analogous to the reported disorder-to-order conformational transitions described for PKB (52). A similar disorder-to-order transition mechanism is probable for the other NDR-related kinases. It will be interesting to investigate whether other members of the Ste20 group of protein kinases function as hydrophobic motif kinases for other AGC kinases.
FIG. 10.
Model of NDR protein kinase activation by multisite phosphorylation. MOB as well as activated MST is required for full phosphorylation and activation of NDR. MOB binding stimulates auto- and transphosphorylation of NDR by releasing autoinhibition, whereas MST3 phosphorylation at the hydrophobic motif results in an active conformation of the kinase. The kinase complex is negatively regulated by PP2A or PP2A-related phosphatase. Activated NDR kinase in turn promotes cleavage of MST3.
Previous results revealed that MOB proteins play a critical role in the activation of several NDR kinase homologues. Mob1p was shown to be required for the activation and phosphorylation of Dbf2p by Cdc15p (28), and Mob2p is required for Cbk1p function (6, 50). Recent studies showed that human MOB proteins interact with NDR1 and -2 (3, 11). MOB1A binding induces the release of autoinhibition and thus stimulates autophosphorylation at the activation segment phosphorylation site of NDR kinase (3). Here we provide the first evidence for a functional interaction between mammalian Ste20-like kinase, MOB1A protein, and NDR2 kinase. In vitro, all three proteins MST3, MOB1A, and NDR2, are required for full kinase activity. MOB1A binding is essential for the release of NDR autoinhibition, thus stimulating autophosphorylation and transphosphorylation activity. As expected, the NDR2-AIS mutant showed full activation in the presence of activated MST3, independent of MOB1A binding, supporting the proposed mechanism. Additionally, we provide evidence that the functional interaction between MST3 and NDR2 occurs in vivo. Cotransfection of MST3 and NDR2 in COS-7 cells resulted in an increase in Thr442 phosphorylation, whereas cotransfection of MST3KR and NDR2 reduced phosphorylation of the hydrophobic motif. Nevertheless, these in vivo changes were lower than those observed after activation of the kinase with OA.
Targeting of NDR to the membrane provides evidence for the importance of subcellular localization in the activation mechanism. Membrane targeting of NDR results in increased hydrophobic motif phosphorylation as well as activation of the kinase (17), which is dependent on the MST3 activity in HEK293F cells. However, our results suggest that the interaction of NDR2 and MST3 is likely to be transient. Recruitment of NDR to the membrane via myc-C1-MOB1 resulted in phosphorylation of the kinase. MST3 was not corecruited, but a significant portion of MST3 was already associated with the membrane. This suggests that the interaction of MST3 with NDR2 is not stable, similar to the interaction reported between MST1/2 and the NDR relative LATS1/Warts (5, 21). However, it has also been reported for other kinase substrate interactions that the affinity for their substrate is weak (4) and below the detection limit of immunoprecipitation (35). We propose that membrane recruitment of NDR is an essential part of the NDR activation mechanism and that the phosphorylation of NDR by MST3 occurs at the membrane. However, future experiments will have to address the question of what signals are responsible for membrane recruitment of the NDR/MOB complex in vivo. It is also possible that NDR is targeted to the membrane to phosphorylate proteins specifically localized to this region of the cell.
MST3 purified from OA-treated cells showed significantly more activity in the cell-free assays compared with the enzyme isolated from untreated cells. This suggests that MST3 activity is regulated by phosphorylation that can be initiated by treating cells with the PP2A inhibitor OA. The mechanism of MST3 regulation remains to be fully elucidated, but the available results provide an explanation for the dramatic effects of OA on NDR activity. Previously, we found that MOB1A binding to NDR is facilitated apparently by treatment of cells with OA (3). We conclude that a PP2A-like phosphatase negatively regulates both kinases and MOB1A function; the key issue in the future will be to understand the regulation of the phosphatase in this signaling network.
Significantly, we provide the first evidence that cleavage of MST3 can, at least partially, be induced by NDR in a regulatory feedback loop. NDR2 overexpression can induce cleavage of MST3 and MST3KR, whereas the kinase-dead NDR2K119A had no effect on cleavage. Interestingly, caspases 3, 7, 8, and 9 are processed in cancer cells infected with an adenovirus expressing the human LATS2 kinase, a close relative of NDR. Further, it was shown that the Drosophila Warts kinase together with the Ste20-like kinase Hippo regulate the levels of DIAP1 (Drosophila inhibitor of apoptosis) (16, 20, 34, 45, 51). Mammalian IAP proteins are known inhibitors of caspase activation (26, 41) but also play a role in signal transduction and cell cycle regulation (1, 39).
Genetic studies with yeast, worms, and flies have shown that many of the components of the NDR signaling pathway are highly conserved, such as the scaffold protein Pag1p (mor2p, sax-2, FURRY), the Ste20-like kinase Kic1p (gck-1), the adapter protein Hym1p (dMO25), the activator protein Mob2p (dMOB proteins), and the transcription factor Ace2p (Krueppel-like transcription factor) (43). Based on the conserved protein components, the NDR signaling pathway is more closely related to the signaling pathway established for Cbk1p than to Dbf2p (which appears to be more closely related to LATS). Our work on the human NDR suggests that conservation is also maintained with the mammalian counterparts of some of these proteins. However, despite the high conservation of the proteins, the processes they regulate are likely to differ due to species variation between yeast (e.g., regulation of chitinase expression) and higher organisms. Further work is required to fully elucidate the functions of these different proteins. However, the involvement of the yeast NDR kinases in the MEN (mitotic exit network) and RAM (regulation of Ace2p transcription and morphogenesis) network (43) raises the possibility of a cell-intrinsic regulation of NDR activity, which might explain why all attempts so far (growth hormones, oxidative stress, UV, ceramide, fluoride, or serum) to identify a ligand/hormone or endogenous stimuli have failed (2).
Significant insights into the physiological roles of NDR kinases have been obtained using yeast, worm, and fly genetic screens. Early studies from yeast found that NDR family kinases play a significant role in the regulation of actin and tubulin cytoskeletal organization during polarized growth and cytokinesis and are central components of the mitotic exit network (43). Worm NDR regulates cell spreading and neurite outgrowth of chemosensory neurons (53). The fly NDR is reported to be important for the integrity of cellular outgrowths, such as wing hairs and bristles (14). Recently, it was shown that NDR kinase activity plays an important role in the regulation of dendritic branching and tiling in worms and flies (12, 13). These results predict that mammalian NDR also plays a significant role in the control of cell division and morphogenesis. Identification of NDR-specific substrates will help to define the physiological roles of this conserved signaling pathway.
Supplementary Material
Acknowledgments
We thank Chiun-Jye Yuan for providing the HA-MST3 and HA-MST3KR constructs and René Bernards for the pTER construct.
M.R.S. is supported by the Krebsliga beider Basel, R.T. is supported by the Krebsliga Schweiz (KLS 01342-02-2003), and A.H. is supported by the Roche Research Foundation. The Friedrich Miescher Institute is part of the Novartis Research Foundation.
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Altieri, D. C. 2003. Survivin, versatile modulation of cell division and apoptosis in cancer. Oncogene 22:8581-8589. [DOI] [PubMed] [Google Scholar]
- 2.Bichsel, S. J. 2000. Regulation of NDR, a nuclear serine/threonine kinase. Diploma thesis. University of Basel, Basel, Switzerland.
- 3.Bichsel, S. J., R. Tamaskovic, M. R. Stegert, and B. A. Hemmings. 2004. Mechanism of activation of NDR (nuclear Dbf2-related) protein kinase by the hMOB1 protein. J. Biol. Chem. 279:35228-35235. [DOI] [PubMed] [Google Scholar]
- 4.Borders, C. L., M. J. Snider, R. Wolfenden, and P. L. Edmiston. 2002. Determination of the affinity of each component of a quaternary transition state analogue complex of creatine kinase. Biochemistry 41:6995-7000. [DOI] [PubMed] [Google Scholar]
- 5.Chan, E. H., M. Nousiainen, R. B. Chalamalasetty, A. Schafer, E. A. Nigg, and H. H. Sillje. 2005. The Ste20-like kinase Mst2 activates the human large tumor suppressor kinase Lats1. Oncogene 24:2076-2086. [DOI] [PubMed] [Google Scholar]
- 6.Colman-Lerner, A., T. E. Chin, and R. Brent. 2001. Yeast Cbk1 and Mob2 activate daughter-specific genetic programs to induce asymmetric cell fates. Cell 107:739-750. [DOI] [PubMed] [Google Scholar]
- 7.Dan, I., S. E. Ong, N. M. Watanabe, B. Blagoev, M. M. Nielsen, E. Kajikawa, T. Z. Kristiansen, M. Mann, and A. Pandey. 2002. Cloning of MASK, a novel member of the mammalian germinal center kinase III subfamily, with apoptosis-inducing properties. J. Biol. Chem. 277:5929-5939. [DOI] [PubMed] [Google Scholar]
- 8.Deng, Y., A. Pang, and J. H. Wang. 2003. Regulation of mammalian STE20-like kinase 2 (MST2) by protein phosphorylation/dephosphorylation and proteolysis. J. Biol. Chem. 278:11760-11767. [DOI] [PubMed] [Google Scholar]
- 9.De Souza, P. M., and M. A. Lindsay. 2004. Mammalian Sterile 20-like kinase 1 and the regulation of apoptosis. Biochem. Soc. Trans. 32:485-488. [DOI] [PubMed] [Google Scholar]
- 10.De Souza, P. M., H. Kankaanranta, A. Michael, P. J. Barnes, M. A. Giembycz, and M. A. Lindsay. 2002. Caspase-catalyzed cleavage and activation of Mst1 correlates with eosinophil but not neutrophil apoptosis. Blood 99:3432-3438. [DOI] [PubMed] [Google Scholar]
- 11.Devroe, E., H. Erdjument-Bromage, P. Tempst, and P. A. Silver. 2004. Human Mob proteins regulate the NDR1 and NDR2 serine-threonine kinases. J. Biol. Chem. 279:24444-24451. [DOI] [PubMed] [Google Scholar]
- 12.Emoto, K., Y. He, B. Ye, W. B. Grueber, P. N. Adler, L. Y. Jan, and Y. N. Jan. 2004. Control of dendritic branching and tiling by the Tricornered-kinase/Furry signaling pathway in Drosophila sensory neurons. Cell 119:245-256. [DOI] [PubMed] [Google Scholar]
- 13.Gallegos, M. E., and C. I. Bargmann. 2004. Mechanosensory neurite termination and tiling depend on SAX-2 and the SAX-1 kinase. Neuron 44:239-249. [DOI] [PubMed] [Google Scholar]
- 14.Geng, W., B. He, M. Wang, and P. N. Adler. 2000. The tricornered gene, which is required for the integrity of epidermal cell extensions, encodes the Drosophila nuclear DBF2-related kinase. Genetics 156:1817-1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Graves, J. D., K. E. Draves, Y. Gotoh, E. G. Krebs, and E. A. Clark. 2001. Both phosphorylation and caspase-mediated cleavage contribute to regulation of the Ste20-like protein kinase Mst1 during CD95/Fas-induced apoptosis. J. Biol. Chem. 276:14909-14915. [DOI] [PubMed] [Google Scholar]
- 16.Harvey, K. F., C. M. Pfleger, and I. K. Hariharan. 2003. The Drosophila Mst ortholog, Hippo, restricts growth and cell proliferation and promotes apoptosis. Cell 114:457-467. [DOI] [PubMed] [Google Scholar]
- 17.Hergovich, A., S. J. Bichsel, and B. A. Hemmings. 2005. Human NDR kinases are rapidly activated by MOB proteins through recruitment to the plasma membrane. Mol. Cell. Biol. 25:8259-8272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hou, M. C., D. A. Guertin, and D. Mc Collum. 2004. Initiation of cytokinesis is controlled through multiple modes of regulation of the Sid2p-Mob1p kinase complex. Mol. Cell. Biol. 24:3262-3276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang, C. Y., Y. M. Wu, C. Y. Hsu, W. S. Lee, M. D. Lai, T. J. Lu, C. L. Huang, T. H. Leu, H. M. Shih, H. I. Fang, D. R. Robinson, H. J. Kung, and C. J. Yuan. 2002. Caspase activation of mammalian sterile 20-like kinase 3 (Mst3). Nuclear translocation and induction of apoptosis. J. Biol. Chem. 277:34367-34374. [DOI] [PubMed] [Google Scholar]
- 20.Jia, J., W. Zhang, B. Wang, R. Trinko, and J. Jiang. 2003. The Drosophila Ste20 family kinase dMST functions as a tumor suppressor by restricting cell proliferation and promoting apoptosis. Genes Dev. 17:2514-2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lai, Z. C., X. Wei, T. Shimizu, E. Ramos, M. Rohrbaugh, N. Nikolaides, L. L. Ho, and Y. Li. 2005. Control of cell proliferation and apoptosis by Mob as tumor suppressor, Mats. Cell 120:675-685. [DOI] [PubMed] [Google Scholar]
- 22.Lee, K. K., T. Ohyama, N. Yajima, S. Tsubuki, and S. Yonehara. 2001. MST, a physiological caspase substrate, highly sensitizes apoptosis both upstream and downstream of caspase activation. J. Biol. Chem. 276:19276-19285. [DOI] [PubMed] [Google Scholar]
- 23.Lee, K. K., and S. Yonehara. 2002. Phosphorylation and dimerization regulate nucleocytoplasmic shuttling of mammalian STE20-like kinase (MST). J. Biol. Chem. 277:12351-12358. [DOI] [PubMed] [Google Scholar]
- 24.Lee, W. S., C. Y. Hsu, P. L. Wang, P. L., C. Y. Huang, C. H. Chang, and C. J. Yuan. 2004. Identification and characterization of the nuclear import and export signals of the mammalian Ste20-like protein kinase 3. FEBS Lett. 572:41-45. [DOI] [PubMed] [Google Scholar]
- 25.Lin, J. L., H. C. Chen, H. I. Fang, D. Robinson, H. J. Kung, and H. M. Shih. 2001. MST4, a new Ste20-related kinase that mediates cell growth and transformation via modulating ERK pathway. Oncogene 20:6559-6569. [DOI] [PubMed] [Google Scholar]
- 26.Liston, P., W. G. Fong, and R. G. Korneluk. 2003. The inhibitors of apoptosis: there is more to life than Bcl2. Oncogene 22:8568-8580. [DOI] [PubMed] [Google Scholar]
- 27.Lowe, E. D., M. E. Noble, V. T. Skamnaki, N. G. Oikonomakos, D. J. Owen, and L. N. Johnson. 1997. The crystal structure of a phosphorylase kinase peptide substrate complex: kinase substrate recognition. EMBO J. 16:6646-6658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mah, A. S., J. Jang, and R. J. Deshaies. 2001. Protein kinase Cdc15 activates the Dbf2-Mob1 kinase complex. Proc. Natl. Acad. Sci. USA 98:7325-7330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Manning, G., D. B. Whyte, R. Martinez, T. Hunter, and S. Sudarsanam. 2002. The protein kinase complement of the human genome. Science 298:1912-1934. [DOI] [PubMed] [Google Scholar]
- 30.Millward, T. A., P. Cron, and B. A. Hemmings. 1995. Molecular cloning and characterization of a conserved nuclear serine (threonine) protein kinase. Proc. Natl. Acad. Sci. USA 92:5022-5026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Millward, T. A., C. W. Heizmann, B. W. Schafer, and B. A. Hemmings. 1998. Calcium regulation of Ndr protein kinase mediated by S100 calcium-binding proteins. EMBO J. 17:5913-5922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Millward, T. A., D. Hess, and B. A. Hemmings. 1999. Ndr protein kinase is regulated by phosphorylation on two conserved sequence motifs. J. Biol. Chem. 274:33847-33850. [DOI] [PubMed] [Google Scholar]
- 33.Nelson, B., C. Kurischko, J. Horecka, M. Mody, P. Nair, L. Pratt, A. Zougman, L. D. McBroom, T. R. Hughes, C. Boone, and F. C. Luca. 2003. RAM: a conserved signaling network that regulates Ace2p transcriptional activity and polarized morphogenesis. Mol. Biol. Cell 14:3782-3803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pantalacci, S., N. Tapon, and P. Leopold. 2003. The Salvador partner Hippo promotes apoptosis and cell-cycle exit in Drosophila. Nat. Cell Biol. 5:921-927. [DOI] [PubMed] [Google Scholar]
- 35.Piehler, J. 2005. New methodologies for measuring protein interactions in vivo and in vitro. Curr. Opin. Struct. Biol. 15:4-14. [DOI] [PubMed] [Google Scholar]
- 36.Pombo, C. M., T. Tsujita, J. Kyriakis, J. V. Bonventre, and T. Force. 1997. Activation of the Ste-20-like oxidant stress response kinase-1 during the initial stages of chemical anoxia-induced necrotic cell death. J. Biol. Chem. 272:29372-29379. [DOI] [PubMed] [Google Scholar]
- 37.Praskova, M., A. Khoklatchev, S. Ortiz-Vega, and J. Avruch. 2004. Regulation of the MST1 kinase by autophosphorylation, by the growth inhibitory proteins, RASSF1 and NORE1, and by Ras. Biochem. J. 381:453-462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Preisinger, C., B. Short, V. De Corte, E. Bruyneel, A. Haas, R. Kopajtich, J. Gettemans, and F. A. Barr. 2004. YSK1 is activated by the Golgi matrix protein GM130 and plays a role in cell migration through its substrate 14-3-3ζ. J. Cell Biol. 164:1009-1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Richter, B. W., and C. S. Duckett. 2000. The IAP proteins: caspase inhibitors and beyond. Sci. STKE 2000(44):PE1. [DOI] [PubMed] [Google Scholar]
- 40.Robinson, M. J., S. A. Stippec, E. Goldsmith, M. A. White, and M. H. Cobb. 1998. A constitutively active and nuclear form of the MAP kinase ERK2 is sufficient for neurite outgrowth and cell transformation. Curr. Biol. 8:1141-1150. [DOI] [PubMed] [Google Scholar]
- 41.Salvesen, G. S., and J. M. Abrams. 2004. Caspase activation—stepping on the gas or releasing the brakes? Lessons from humans and flies. Oncogene 23:2774-2784. [DOI] [PubMed] [Google Scholar]
- 42.Stegert, M. R., R. Tamaskovic, S. J. Bichsel, A. Hergovich, and B. A. Hemmings. 2004. Regulation of NDR2 protein kinase by multi-site phosphorylation and the S100B calcium-binding protein. J. Biol. Chem. 279:23806-23812. [DOI] [PubMed] [Google Scholar]
- 43.Tamaskovic, R., S. J. Bichsel, and B. A. Hemmings. 2003. NDR family of AGC kinases—essential regulators of the cell cycle and morphogenesis. FEBS Lett. 546:73-80. [DOI] [PubMed] [Google Scholar]
- 44.Tamaskovic, R., S. J. Bichsel, H. Rogniaux, M. R. Stegert, and B. A. Hemmings. 2003. Mechanism of Ca2+-mediated regulation of NDR protein kinase through autophosphorylation and phosphorylation by an upstream kinase. J. Biol. Chem. 278:6710-6718. [DOI] [PubMed] [Google Scholar]
- 45.Udan, R. S., M. Kango-Singh, R. Nolo, C. Tao, and G. Halder. 2003. Hippo promotes proliferation arrest and apoptosis in the Salvador/Warts pathway. Nat. Cell Biol. 5:914-920. [DOI] [PubMed] [Google Scholar]
- 46.Ura, S., N. Masuyama, J. D. Graves, and Y. Gotoh. 2001. Caspase cleavage of MST1 promotes nuclear translocation and chromatin condensation. Proc. Natl. Acad. Sci. USA 98:10148-10153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ura, S., N. Masuyama, J. D. Graves, and Y. Gotoh. 2001. MST1-JNK promotes apoptosis via caspase-dependent and independent pathways. Genes Cells 6:519-530. [DOI] [PubMed] [Google Scholar]
- 48.van de Wetering, M., I. Oving, V. Muncan, M. T. Pon Fong, H. Brantjes, D. van Leenen, F. C. Holstege, T. R. Brummelkamp, R. Agami, and H. Clevers. 2003. Specific inhibition of gene expression using a stably integrated, inducible small-interfering RNA vector. EMBO Rep. 4:609-615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Verde, F., D. J. Wiley, and P. Nurse. 1998. Fission yeast orb6, a ser/thr protein kinase related to mammalian rho kinase and myotonic dystrophy kinase, is required for maintenance of cell polarity and coordinates cell morphogenesis with the cell cycle. Proc. Natl. Acad. Sci. USA 95:7526-7531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Weiss, E. L., C. Kurischko, C. Zhang, K. Shokat, D. G. Drubin, and F. C. Luca. 2002. The Saccharomyces cerevisiae Mob2p-Cbk1p kinase complex promotes polarized growth and acts with the mitotic exit network to facilitate daughter cell-specific localization of Ace2p transcription factor. J. Cell Biol. 158:885-900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wu, S., J. Huang, J. Dong, and D. Pan. 2003. Hippo encodes a Ste-20 family protein kinase that restricts cell proliferation and promotes apoptosis in conjunction with Salvador and Warts. Cell 114:445-456. [DOI] [PubMed] [Google Scholar]
- 52.Yang, J., P. Cron, V. M. Good, V. Thompson, B. A., Hemmings, and D. Barford. 2002. Crystal structure of an activated Akt/protein kinase B ternary complex with GSK3-peptide and AMP-PNP. Nat. Struct. Biol. 9:940-944. [DOI] [PubMed] [Google Scholar]
- 53.Zallen, J. A., E. L. Peckol, D. M. Tobin, and C. I. Bargmann. 2000. Neuronal cell shape and neurite initiation are regulated by the Ndr kinase SAX-1, a member of the Orb6/COT-1/Warts serine/threonine kinase family. Mol. Biol. Cell 11:3177-3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.