Abstract
The coregulated PHO5 and PHO8 genes in Saccharomyces cerevisiae provide typical examples for the role of chromatin in promoter regulation. It has been a long-standing question why the cofactors Snf2 and Gcn5 are essential for full induction of PHO8 but dispensable for opening of the PHO5 promoter. We show that this discrepancy may result from different stabilities of the two promoter chromatin structures. To test this hypothesis, we used our recently established yeast extract in vitro chromatin assembly system, which generates the characteristic PHO5 promoter chromatin. Here we show that this system also assembles the native PHO8 promoter nucleosome pattern. Remarkably, the positioning information for both native patterns is specific to the yeast extract. Salt gradient dialysis or Drosophila embryo extract does not support proper nucleosome positioning unless supplemented with yeast extract. By competitive assemblies in the yeast extract system we show that the PHO8 promoter has greater nucleosome positioning power and that the properly positioned nucleosomes are more stable than those at the PHO5 promoter. Thus we provide evidence for the correlation of inherently more stable chromatin with stricter cofactor requirements.
Eukaryotic DNA is packaged into chromatin with the nucleosome as the basic building block (25, 30). This chromatin structure is a means of packaging large amounts of DNA into the nucleus but is also involved in the regulation of gene expression (15, 19, 34, 44). In this regard, the precise position of nucleosomes can be critical, and a very recent study of Saccharomyces cerevisiae shows that about 70% of all nucleosomes are clearly positioned, most prominently in gene regulatory regions (53). Especially in these regions, positioned nucleosomes can take on a positive or negative role in gene transcription by modulating the accessibility of DNA sequences for DNA binding factors (24, 29, 38, 39, 45, 51).
The PHO5 and PHO8 promoters in yeast constitute two well-studied examples for promoter regions with positioned nucleosomes (3, 4, 47). Their positioned nucleosomes contribute to the repressed state and are the in vivo substrate for remodeling processes that generate extensive nuclease-hypersensitive sites upon induction (2, 3, 51). A series of in vivo studies by our own and other laboratories has increased our understanding of the mechanism of remodeling leading to the activated state (41, 47). In particular, activation of the PHO5 and PHO8 genes upon phosphate starvation leads to the loss of histone DNA contacts, i.e., to histone eviction (1, 8, 40; P. Korber et al., submitted for publication), at their promoter regions. Remodeling at the PHO5 promoter is more extensive than at the PHO8 promoter, resulting in a hypersensitive site of 600 base pairs affecting four nucleosomes (3). In this case it was shown that histones leave by a mechanism in trans (9, 23) involving the histone chaperone Asf1, which is thought to provide the histone acceptor (1; Korber et al., submitted). As Asf1 also plays a role in the induction pathway of PHO8 (1; Korber et al., submitted), it is likely that histone eviction occurs here via a trans mechanism as well.
Even though both promoters are coregulated by the same transactivator, Pho4, and share the above features, chromatin remodeling at each promoter has cofactor requirements of differing stringencies. At the PHO8 promoter, no remodeling is detectable in the absence of a functional SWI/SNF complex, and in a strain without Gcn5 histone acetyltransferase activity there is only locally restricted remodeling that does not support induction of PHO8 activity (18). In contrast, at the PHO5 promoter the chromatin structure is still completely remodeled without Gcn5 and/or SWI/SNF activity, although with a kinetic delay (5, 12, 35, 40; T. Luckenbach et al., submitted for publication). It remains an open question why the PHO8 promoter is strictly dependent on one specific pathway of chromatin remodeling as defined by the cofactors SWI/SNF and Gcn5 whereas the PHO5 promoter can be opened via redundant pathways.
We showed previously that exchanging the DNA sequence, which is assembled into nucleosome −2 at the PHO5 promoter, for a strongly nucleosome-positioning satellite DNA fragment largely abolished chromatin opening (46). It was concluded that the inherent stability of a positioned promoter nucleosome directly affects the inducibility of the promoter. In light of this and other studies, we have speculated for some time that the differences in cofactor requirements for chromatin opening at the PHO5 and PHO8 promoters reflect differences in the stability of the chromatin substrate for the two remodeling processes (33). The inherent stability of a nucleosome may be especially relevant for a remodeling mechanism leading to histone eviction in trans as it requires the complete disruption of histone-DNA contacts. However, nucleosome stability and therefore this hypothesis are not easily testable with classical in vivo techniques.
As an alternative approach, we recently established an in vitro chromatin assembly system based on yeast whole-cell extracts supplemented with purified histones. This system is capable of generating extensive regular nucleosomal arrays with physiological spacing and proper nucleosomal positioning at the yeast PHO5 promoter (22). Here we show that this system also properly positions the nucleosomes at the PHO8 promoter. This in vitro approach now allowed us to directly compare the stabilities of the positioned nucleosomes at both promoters.
In contrast to the PHO5 promoter, properly positioned nucleosomes at the PHO8 promoter were assembled under more-diverse conditions and under conditions of limiting histones, even without the generation of extensive nucleosomal arrays. In addition, thermally induced loss of nucleosome positioning at the PHO8 promoter occurred more slowly. We conclude that the PHO8 promoter has greater positioning power and that the properly positioned nucleosomes are more stable than at the PHO5 promoter. This supports our concept that the stringent remodeling pathway required at the PHO8 promoter is dictated by a more stable chromatin structure.
Moreover, we show that the determinants responsible for nucleosome positioning (the positioning information) are unlikely to be solely intrinsic to the DNA sequence but probably rely on additional factors provided by the yeast whole-cell extract. Neither an in vitro chromatin assembly reaction using Drosophila melanogaster embryo extract nor one using salt gradient dialysis could generate the proper nucleosome positioning. Nonetheless, chromatin preassembled with either system was induced to rapidly adopt the proper nucleosome positioning by the addition of yeast extract.
MATERIALS AND METHODS
Extract preparation.
Yeast extracts were prepared as described previously (22). Briefly, cells were grown to an optical density at 600 nm of 2 to 4, harvested, and washed with extraction buffer [0.2 M Tris-HCl, pH 7.5, 10 mM MgSO4, 20% glycerol, 1 mM EDTA, 390 mM (NH4)2SO4, 1 mM dithiothreitol (DTT), and Complete protease inhibitor without EDTA; Roche Applied Science]. The pellets were shock frozen and cells lysed by grinding in liquid nitrogen. After slow thawing and clearing by centrifugation, proteins were precipitated with 337 mg/ml (NH4)2SO4, resuspended in dialysis buffer (20 mM HEPES-KOH, pH 7.5, 20% glycerol, 50 mM NaCl, 1 mM EGTA, 5 mM DTT, and Complete protease inhibitor without EDTA) and dialyzed three times for 30 min against the same buffer.
Drosophila extract was prepared as described previously (6, 10). Briefly, dechorionated Drosophila preblastoderm embryos (0 to 90 min) were homogenized in low-salt buffer (10 mM HEPES, pH 7.6, 10 mM KCl, 1.5 mM MgCl2, 0.5 mM EGTA, 10% glycerol, 1 mM DTT, 0.2 mM phenylmethylsulfonyl fluoride) and cleared by centrifugation.
In vitro chromatin assembly.
The DNA templates for all chromatin assembly reactions were circular, supercoiled, 10-kb plasmids that are derivatives of plasmid pCB/wt (LEU2) (13), where the TRP1 marker in pCB/wt is replaced by the LEU2 marker, and either the PHO5 open reading frame (ORF) plus the 1,311-bp upstream region or the PHO8 ORF plus the 1,661-bp upstream region was inserted analogously to the PHO5 insertion in pCB/wt. Mutation of the phosphate-controlled upstream activation sites (UASp) was as described previously (33, 51). Both plasmids were combined at an equimolar ratio in all in vitro assembly reactions.
Drosophila embryo extract assembly.
Chromatin assembly with Drosophila embryo extracts was performed according to published procedures (6, 49). A standard chromatin assembly reaction mixture contained 0.9 μg DNA and 40 to 80 μl Drosophila extract in a total of 150 μl assembly buffer (80 mM KCl, 10 mM Tris-HCl, pH 7.6, 1.5 mM MgCl2, 0.5 mM EGTA, 10% glycerol, 1 mM DTT) supplemented with a regenerative energy system of 3 mM ATP-MgCl2, 30 mM creatine phosphate (Sigma), and 5 ng/μl creatine kinase (Roche Applied Science) and was incubated for 6 h at 26°C.
Yeast extract assembly.
Chromatin assembly with yeast extracts was performed as described previously (22). In brief, 1.8 μg DNA, 300 μg of extract protein, and 6 μg of Drosophila histone octamers were incubated in 150 μl assembly buffer [20 mM HEPES, pH 7.5, 80 mM KCl, 25 mM (NH4)2SO4, 1.5 mM MgCl2, 0.5 mM EGTA, 12% glycerol, 2.5 mM DTT], supplemented with a regenerative energy system of 3 mM ATP-MgCl2, 30 mM creatine phosphate (Sigma), and 5 ng/μl creatine kinase (Roche Applied Science), for up to 6 h at 30°C.
Salt gradient dialysis assembly.
Salt gradient dialysis was performed as described previously (26). A typical assembly reaction mixture contained 4 μg DNA, 4 μg bovine serum albumin, and 3.6 to 4.4 μg Drosophila histone octamers in 50 μl high-salt buffer (10 mM Tris-HCl, pH 7.6, 2 M NaCl, 1 mM EDTA, 1 mM β-mercaptoethanol, 0.05% Nonidet P-40) and was dialyzed for 12 to 16 h while diluting the concentration of NaCl to 0.05 M.
Adding yeast extract to preassembled chromatin.
Yeast extract (100 to 900 μg protein for Drosophila embryo extract chromatin or 3 to 500 μg protein for salt gradient dialysis chromatin) was added to 2 μg DNA preassembled into chromatin along with a fresh complement of the regenerative energy system and further incubated at 30°C for up to 6 h.
Chromatin analysis.
Nucleus preparation using strain CY338 (pho4:URA3 derivative of CY337 [42]) and chromatin analysis by micrococcal nuclease (MNase) ladders and DNase I digestion with indirect end labeling were carried out as described previously (3, 17). The ApaI-BamHI fragment upstream of the PHO5 gene was used as a probe for DNase I mapping of the PHO5 promoter and the XhoI-PvuII fragment at the beginning of the PHO8 open reading frame for mapping of the PHO8 promoter. The probes hybridizing within the PHO5 or PHO8 promoter region correspond to the BstEII-DraI fragment of the PHO5 promoter or to a PCR fragment of the PHO8 promoter using the primers TGGAACTACTTGCGAATATG and ACGCCTTCTTCTAGTAGGAA, respectively. In all DNase I mapping experiments chromatin samples were digested with a range of DNase I concentrations. However, due to space limitations only one lane or a few representative lanes are shown in the figures.
RESULTS
The yeast extract assembly system generates the native nucleosome positioning at the PHO8 promoter in vitro.
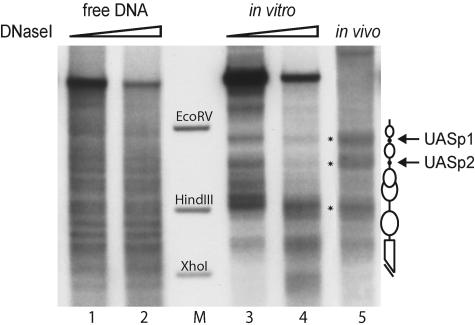
We recently established an in vitro assembly system using yeast whole-cell extracts and purified Drosophila histones that is able to generate proper nucleosome positions at the PHO5 promoter (22). In order to compare the promoter structures of the PHO5 and PHO8 promoters in vitro, we tested this chromatin assembly system also with a DNA template that contains the PHO8 locus. A direct comparison of this in vitro-assembled chromatin with the chromatin structure at the PHO8 promoter in vivo revealed a virtually identical nucleosome pattern that was clearly different from free DNA (Fig. 1). Characteristic for the PHO8 promoter chromatin pattern are two hypersensitive sites at the positions of the UASp elements flanking a nucleosome and a third hypersensitive site located close to a HindIII site (Fig. 1, schematic and lane M). Differences in the DNase I pattern further upstream of the promoter (in the upper part of the lanes) reflect differences in the template DNA, i.e., vector versus chromosomal sequences.
FIG. 1.
In vitro chromatin assembly with yeast extract generates the native chromatin structure at the PHO8 promoter. Limited DNase I digestion and secondary cleavage with BglII for indirect end labeling were performed with free DNA (lanes 1 and 2), chromatin assembled with yeast extract in vitro for 6 h (lanes 3 and 4), and wild-type yeast nuclei (lane 5). The marker bands correspond to the EcoRV-BglII, HindIII-BglII, and XhoI-BglII fragments of the PHO8 promoter (lane M). Schematics of the chromatin structure at the PHO8 promoter are on the right side of the gel. Ovals denote nucleosomes, black dots UASp elements, and the broken bar the open reading frame. Asterisks in the gel refer to the most distinguishing bands of the PHO8 promoter chromatin pattern. All samples were digested with a range of DNase I concentrations (ramps on top of the lanes). However, due to space limitations only representative lanes are shown.
The kinetics of nucleosome positioning at the PHO5 and PHO8 promoters are different in a de novo assembly reaction in vitro.
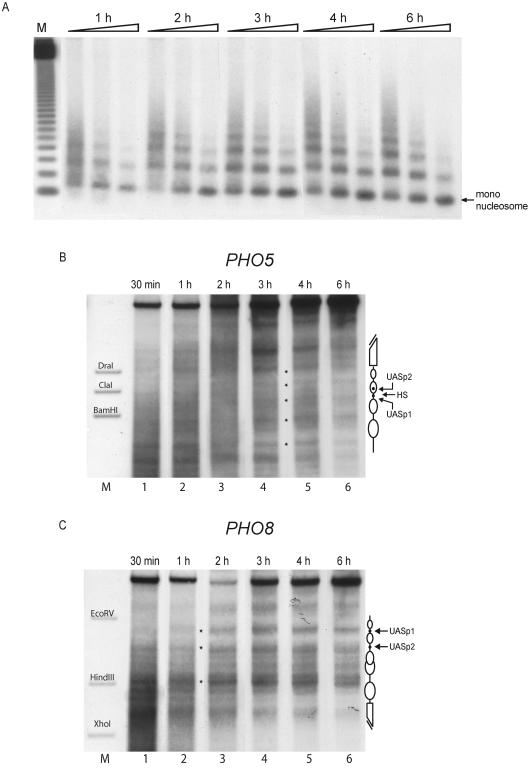
The generation of extensive nucleosomal arrays as well as proper nucleosome positioning at the PHO5 promoter in vitro is a slow process taking up to 6 hours (22). We compared the kinetics of nucleosome assembly and positioning at both the PHO5 and the PHO8 promoters by combining both templates at equimolar ratio in the same assembly reaction. The extent of chromatin assembly was monitored by determining MNase digestion without secondary cleavage (Fig. 2A) and nucleosome positioning by DNase I digestion with indirect end labeling (Fig. 2B and C).
FIG. 2.
In a de novo in vitro assembly reaction nucleosomes become positioned more rapidly at the PHO8 promoter than at the PHO5 promoter. Assembly kinetics of a yeast extract in vitro assembly reaction with plasmids containing the PHO5 and PHO8 loci in the same reaction were monitored at the indicated time points by MNase digestion with specific probing for the PHO8 promoter region (A) and DNase I mapping probing for the PHO5 (B) or the PHO8 (C) promoter. Equivalent results as in panel A were also obtained by using a PHO5 promoter probe (not shown). Ramps on top of the lanes represent increasing MNase digestion times. Lane M (A) shows a 123-bp ladder (Gibco). The markers in panels B and C correspond to the ApaI-BamHI, ApaI-ClaI, and ApaI-DraI fragments of the PHO5 promoter and the EcoRV-BglII, HindIII-BglII, and XhoI-BglII fragments of the PHO8 promoter, respectively. Schematics of the chromatin structure at the PHO5 and PHO8 promoters are on the right side of the gels. Ovals denote nucleosomes, black dots UASp elements, and broken bars the open reading frame. HS in panel B denotes the linker region between nucleosomes −2 and −3 at the PHO5 promoter. Due to space limitations only representative lanes are shown.
The generation of extensive nucleosomal arrays as detected by a probe for the PHO8 promoter took 3 to 4 hours (Fig. 2A). Similar results were obtained using a probe for the PHO5 promoter (data not shown); these results are in good agreement with our previously published data (22). Again in keeping with our published data, the proper nucleosome positioning over the PHO5 promoter was not discernible prior to 3 hours and very clearly established only after 6 hours of assembly (Fig. 2B). Surprisingly, the generation of the native chromatin structure at the PHO8 promoter was much more rapid. The nucleosomes became properly positioned already after 30 min to 1 hour (Fig. 2C).
The presence of the UASp elements does not influence the nucleosome-positioning kinetics in vitro.
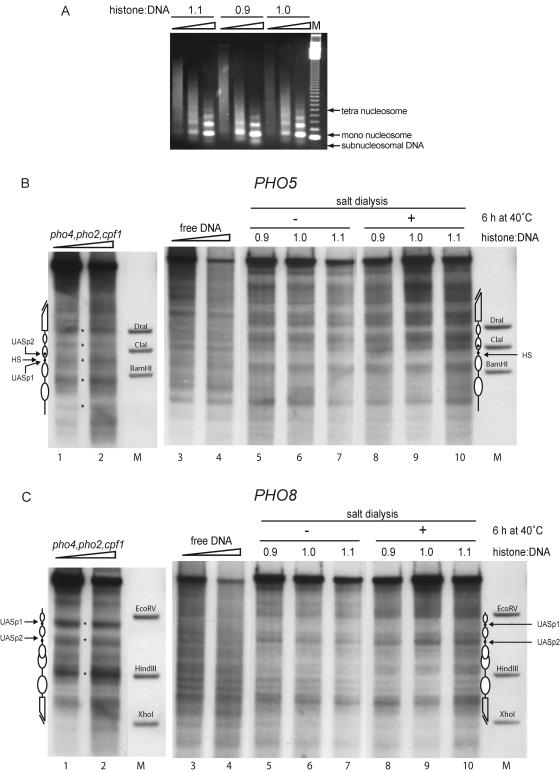
At the PHO5 promoter one of two UASp is intranucleosomal, whereas at the PHO8 promoter both UASp are located in linker regions. The UASp elements are the binding sites for Pho4 and contain an E box, a rather abundant DNA recognition sequence in eukaryotes (43). The possibility was considered that a fortuitous E box binding protein from the yeast whole-cell extract could bind to the intranucleosomal site in the PHO5 promoter, compete with the assembly of a nucleosome there, and thus negatively influence the assembly kinetics. Conversely, such a factor could help to position the nucleosomes at the PHO8 promoter due to binding in the linker regions. This offered a potentially straightforward but possibly artifactual explanation for the differences observed in the in vitro nucleosome positioning kinetics at the PHO5 and PHO8 promoters. We controlled for this by using templates with mutations of the UASp in either promoter. Deletion of the E boxes did not alter the assembly kinetics (data not shown). Moreover, using a yeast extract from a strain triply deleted for all the proteins known to bind to the UASp elements or other regions at both promoters, i.e., Cpf1, Pho2, and Pho4, also had no effect at either promoter on nucleosome positioning (Fig. 3 B and C; lanes 1 and 2).
FIG. 3.
Chromatin assembly by salt gradient dialysis does not generate the proper chromatin structure at the PHO5 or the PHO8 promoter. (A) Chromatin assembled by salt gradient dialysis at histone-to-DNA mass ratios of 0.9, 1.0, and 1.1 (as indicated) was subjected to MNase digestion (ramps on top of lanes represent increasing MNase digestion times) and visualized by ethidium bromide staining. Lane M shows a 123-bp ladder (Gibco). (B and C) The same chromatin preparations as in panel A were subjected to DNase I mapping and probed for the PHO5 (B) or the PHO8 (C) promoter before and after a 6-h incubation at 40°C (as indicated). DNase I mapping of free DNA (lanes 3 and 4) as well as chromatin assembled with yeast extract made from a pho4 pho2 cpf1 triple-mutant strain (lanes 1 and 2) is shown for comparison. Ramps on top of lanes 1 to 4 in panels B and C denote increasing DNase I concentrations. Marker bands (lanes M), schematics, and asterisks are as defined for Fig. 2. Due to space limitations only representative lanes are shown.
The DNA sequence information alone is not sufficient to position the nucleosomes at the yeast PHO5 and PHO8 promoters in salt gradient dialysis chromatin assembly.
The differences in the assembly kinetics were the first indication for a stronger nucleosome positioning power of the PHO8 promoter compared to the PHO5 promoter. We wondered if this positioning power was due to strong positioning information in the PHO8 promoter DNA sequence that may already be apparent in salt gradient dialysis chromatin assemblies. This in vitro chromatin assembly protocol has the advantage of working with purified components, i.e., only DNA and histones, and has been widely used in order to study the DNA sequence dependence of nucleosome formation (reference 52 and references therein). It was possible that the proper nucleosome positioning at the PHO8 promoter would be generated also in such an uncatalyzed system whereas positioning at the PHO5 promoter would be less defined through the DNA sequence alone and would have to rely on factors from the yeast extract.
We assembled chromatin in vitro by salt gradient dialysis and tested several ratios of histones to DNA. Nucleosomal arrays as assayed by MNase digestion were generated at ratios of histones to DNA ranging from 0.9 to 1.1 (Fig. 3A). A band of subnucleosomal migration position, which is an indicator of incomplete chromatin assembly, was less prominent at the higher ratios of histones to DNA, arguing for a more complete assembly under these conditions.
With the same chromatin preparations as in Fig. 3A we performed DNase I mapping and probed for both the PHO5 and the PHO8 promoter regions subsequently on the same blot membrane (Fig. 3B and C). The resulting nucleosomal patterns of the chromatin generated by salt gradient dialysis were distinct patterns and unlike the patterns of free DNA but were different from the proper yeast patterns. In particular, the PHO5 promoter region containing the hypersensitive site (corresponding to the linker between nucleosomes −2 and −3 in the native pattern) was prominently protected in chromatin assembled by salt dialysis, and the region of nucleosome −3 contained a strong band. Furthermore, a band at the position of the ClaI marker band is visible both in the patterns of free DNA and in the salt dialysis chromatin, but this region is protected by nucleosome −2 in the yeast pattern (Fig. 3B; compare lanes 1 and 2 with lanes 3 to 10). Notably, however, in chromatin assembled by salt dialysis a region similar to nucleosome −1 of the native pattern was also protected in the chromatin generated by salt dialysis suggesting that a strong nucleosome-positioning DNA sequence may be involved in determining this position. At the PHO8 locus the hypersensitive site at UASp1 in the yeast pattern was strongly protected in salt dialysis chromatin, while the hypersensitive sites at the position of UASp2, at the position of the HindIII marker band, and at the beginning of the open reading frame were present in both patterns (Fig. 3C; compare lanes 1 and 2 with 5 to 10).
It is known that the assembly of chromatin by salt dialysis need not result in nucleosomes occupying the energetically most favorable positions right away. Many protocols apply an additional heat shifting step at 37 to 55°C in order to allow the nucleosomes to adopt their preferred positions (14, 31, 32, 37). We heat shifted the salt dialysis assembly chromatin by incubation for up to 6 hours at 40°C. This did not lead to a significant change in the pattern at both promoters (Fig. 3B and C, compare lanes 5 to 7 with 8 to 10).
The nucleosome-positioning information is specific to the yeast extract.
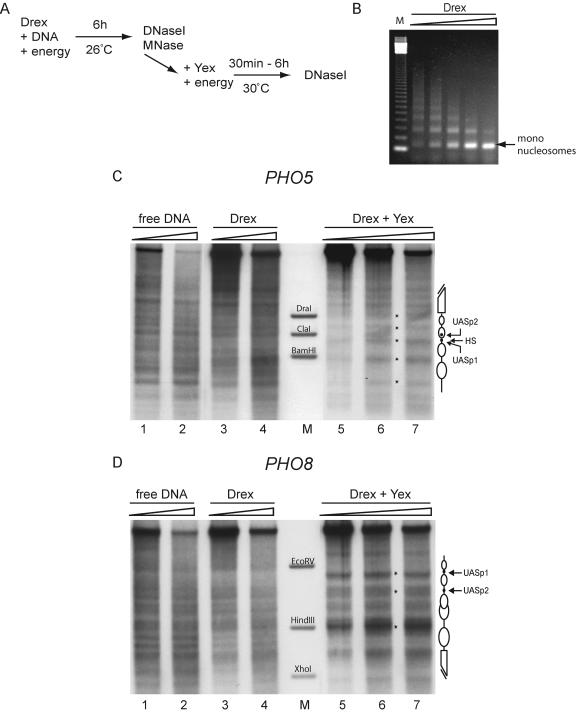
We concluded that the DNA sequence, as read only by histone interactions under conditions of salt gradient dialysis with or without thermal shifting, is not sufficient to correctly position the nucleosomes at either promoter. We wondered whether the presence of remodeling machines would make the crucial difference. A yeast whole-cell extract contains ATP-dependent remodeling activities and was sufficient to generate the proper nucleosome positioning (Fig. 1) (22). We therefore tested whether the histone-DNA interactions, as read by another set of remodeling machines, would work in the same way. We used the well-established cell-free chromatin reconstitution system based on Drosophila embryo extracts that are rich in chromatin-remodeling activities (6, 7, 20) (Fig. 4). As published previously (6, 7, 20), this system assembled arrays of regularly spaced nucleosomes on both plasmids (Fig. 4B and data not shown). However, the characteristic chromatin structure at both promoters was not generated in this system. Instead, the resulting pattern was very similar to the pattern of free DNA (Fig. 4C and D). We controlled also in this system for interference of a fortuitous E box binding protein from the Drosophila extract. Mutating the UASp at either promoter did not alleviate the inability of the Drosophila extract to generate the characteristic yeast pattern (data not shown). In all three chromatin assembly systems employed we use histones from Drosophila embryos, and therefore differences in the generated nucleosomal patterns cannot be due to the source of histones.
FIG. 4.
A Drosophila embryo extract assembly system cannot position the nucleosomes at the yeast PHO5 and PHO8 promoters. (A) Scheme of the assembly reaction. Chromatin was generated by incubating Drosophila embryo extract (Drex) with DNA and an energy regenerating system for 6 h at 26°C. The chromatin was analyzed by MNase digestion and DNase I mapping (DNase I) either directly or after addition of yeast extract (Yex) and a fresh energy mixture and incubation for up to six more hours at 30°C. (B) The chromatin generated with the Drosophila embryo extract system was subjected to MNase digestion and visualized by ethidium bromide staining. The ramp on top of the lanes represents increasing amounts of digestion time, and lane M shows a 123-bp ladder (Gibco). (C and D) Free DNA (lanes 1 and 2) or chromatin prior to (lanes 3 and 4) or after (lanes 5 to 7) addition of yeast extract and further incubation for 3 h was analyzed by DNase I mapping with probing for the PHO5 (C) or the PHO8 (D) promoter. Schematics, asterisks, and marker lanes (M) are as defined for Fig. 2. Ramps on top of the lanes denote increasing DNase I concentrations.
The addition of yeast extract to chromatin preassembled by Drosophila extract can shift the nucleosomes to the proper positions.
In order to see if the lack of proper positioning in Drosophila extract-assembled chromatin could be compensated by the yeast extract, we preassembled the PHO5 and PHO8 plasmids into chromatin using the Drosophila extract system and then added yeast extract and incubated up to six more hours. Strikingly, this indeed generated the proper patterns at both promoters (Fig. 4C and D, lanes 5 to 7). Nucleosome repositioning at both promoters by addition of the yeast extract was completed after just 30 min, and the chromatin retained the same pattern for up to six more hours of incubation (data not shown).
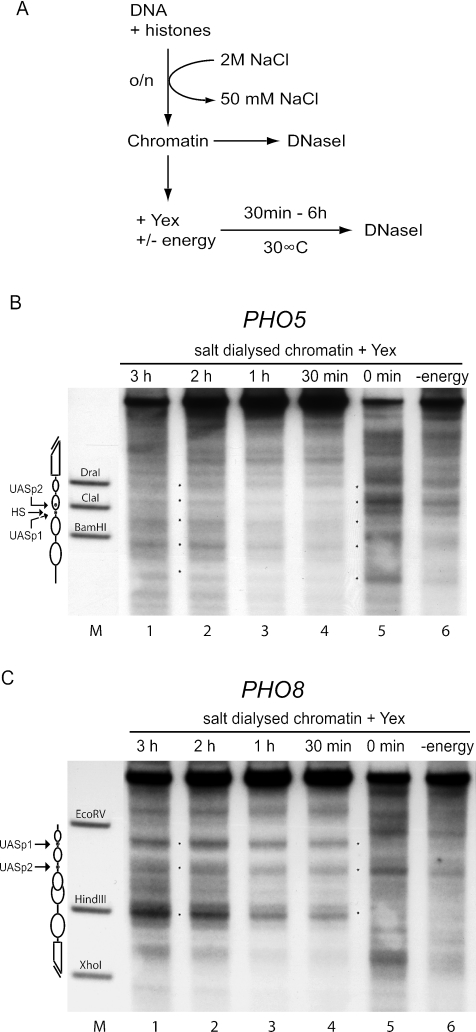
The addition of yeast extract to chromatin preassembled by salt gradient dialysis also repositions nucleosomes to the native chromatin patterns.
As chromatin preassembled with Drosophila extract could be properly repositioned by the addition of yeast extract, we also wanted to test chromatin preassembled by salt gradient dialysis in the same type of experiment. Indeed, we saw a clear shift of the chromatin pattern to properly positioned nucleosomes at both promoters (Fig. 5B and C, lanes 1 to 4). This shift in positioning was energy dependent as no change was seen in the absence of ATP (Fig. 5B and C, lanes 6). Again, the kinetics of the pattern switch were very rapid, and the switch was complete within 30 min at both promoters while no major changes in the pattern occurred after incubation for up to 6 hours (data not shown).
FIG. 5.
Nucleosomes preassembled by salt gradient dialysis become rapidly and properly repositioned after the addition of yeast extract. (A) Reaction scheme. Chromatin was generated by mixing DNA and histone octamers at a mass ratio of 1.1 and overnight (o/n) salt gradient dialysis from 2 M to 50 mM NaCl. The resulting chromatin was analyzed by DNase I mapping (DNase I) either right away or after an additional incubation with yeast extract (Yex) with or without an energy-regenerating system (energy) for up to 6 h at 30°C. (B and C) Chromatin after addition of yeast extract for the indicated times in the presence (lanes 1 to 5) or for 3 h in the absence (lane 6) of energy was analyzed by DNase I mapping with probing for the PHO5 (B) or the PHO8 (C) promoter. Marker lanes (M), schematics, and asterisks are as defined for Fig. 2. Due to space limitations only representative lanes are shown.
Proper nucleosome positioning at the PHO5 promoter is dependent on higher degrees of chromatin assembly than at the PHO8 promoter.
The kinetics of de novo chromatin assembly with yeast extracts showed that proper nucleosome positioning at the PHO5 promoter always correlated with the establishment of extensive nucleosomal arrays (Fig. 2) (22). In contrast, the proper PHO8 promoter pattern was generated already at time points when nucleosomal arrays as detected by monitoring MNase digestion were less extensive (Fig. 2A and C). The same was true when we tried to shift the pattern of chromatin preassembled with Drosophila embryo extract. Such an assembly can lead to more or less extensive nucleosomal arrays depending on the Drosophila extract and the buffer conditions (data not shown). The repositioning of the pattern at the PHO8 promoter to the proper chromatin structure by the addition of yeast extract was largely independent of the extent of nucleosomal arrays as preassembled by the Drosophila extract. However, the repositioning at the PHO5 promoter worked properly only if extensive nucleosomal ladders were preassembled by the Drosophila extract (data not shown).
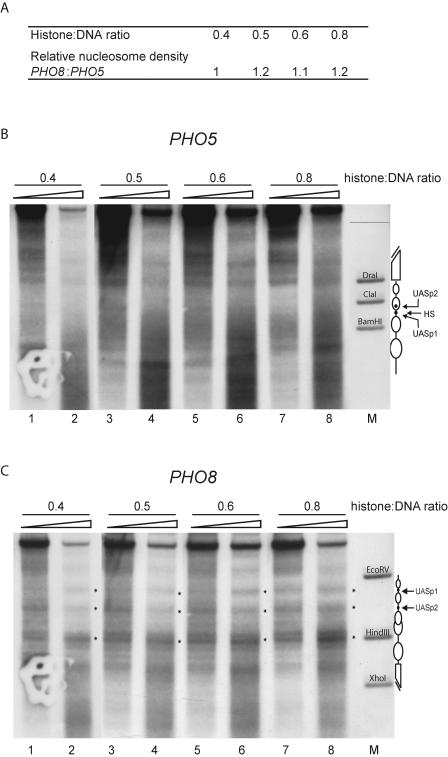
These two findings pointed to a requirement for extensive nucleosomal arrays in order to generate the proper PHO5 promoter pattern in vitro. We wanted to test this interpretation directly and generated chromatin templates with different degrees of assembly states by controlling the histone-to-DNA mass ratio in salt gradient dialysis. Both promoter regions were present in the same assembly reaction at equimolar ratio. We knew already that the degree of chromatin assembly by salt gradient dialysis at a histone-to-DNA mass ratio of 1.1 was sufficient to allow proper repositioning of nucleosomes at both the PHO5 and the PHO8 promoters (Fig. 5). In order to determine if the PHO5 pattern could not be generated under conditions of underassembled chromatin in which the PHO8 pattern was not compromised, we generated chromatin with lower histone-to-DNA ratios (0.4 to 0.8).
First we tested the different chromatin preparations by limited MNase digestion. The extent of the resulting MNase ladders increased with the histone-to-DNA mass ratio, confirming that we had generated chromatin preparations with different degrees of assembly (data not shown). Then we checked if the local states of chromatin assembly were similar at the PHO5 and PHO8 promoters by specific probing for these regions (Fig. 6A and data not shown). We quantified for each chromatin preparation and for each specific promoter probe the DNA remaining after MNase digestion and the total DNA without MNase digestion. The ratio of these two signals is a measure of how much of the DNA was protected from MNase and therefore nucleosomal under the given conditions. It was very similar for the PHO5 and PHO8 promoters in all tested chromatin preparations (Fig. 6A), implying that the histone-to-DNA mass ratio correlated with the same assembly state at both promoters and ruling out a more extensive local assembly of nucleosomes at the PHO8 promoter.
FIG. 6.
The PHO8 promoter has higher nucleosome-positioning power than the PHO5 promoter. (A) Chromatin was generated by salt gradient dialysis with increasing histone octamer-to-DNA mass ratios. The ratio of DNA protected from limited MNase digestion versus total DNA, reflecting the nucleosome density after chromatin assembly, was determined by specific probing for both the PHO5 and the PHO8 promoters (data not shown). The quotient of this ratio for the PHO8 promoter to that for the PHO5 promoter is given for the histone octamer-to-DNA mass ratios used in the respective salt gradient dialysis assembly reaction. (B and C) The same chromatin preparations as in panel A were mixed with yeast extract as in Fig. 5, incubated for 90 min at 30°C, and then subjected to DNase I mapping and probed for the PHO5 (B) or the PHO8 (C) promoter. Schematics, asterisks, and marker lanes are as defined for Fig. 2. Ramps on top of the lanes denote increasing DNase I concentrations.
Next we added yeast extract to each chromatin preparation and assayed the resulting nucleosome positions at both promoters by DNase I mapping. In all these underassembled chromatin preparations, nucleosomes at the PHO5 promoter did not become properly positioned (Fig. 6B, lanes 1 to 8). However, at the PHO8 promoter the correct positioning of the nucleosomes was already discernible even at low histone-to-DNA mass ratios (Fig. 6C, lanes 1 to 4) and became more distinctive with increasing mass ratios (Fig. 6C, lanes 5 to 8). Thus, the proper nucleosome positioning at the PHO5 promoter required higher degrees of chromatin assembly than the proper pattern shift at the PHO8 promoter.
Nucleosomes at the PHO8 promoter are more resistant to thermally induced loss of positioning than nucleosomes at the PHO5 promoter.
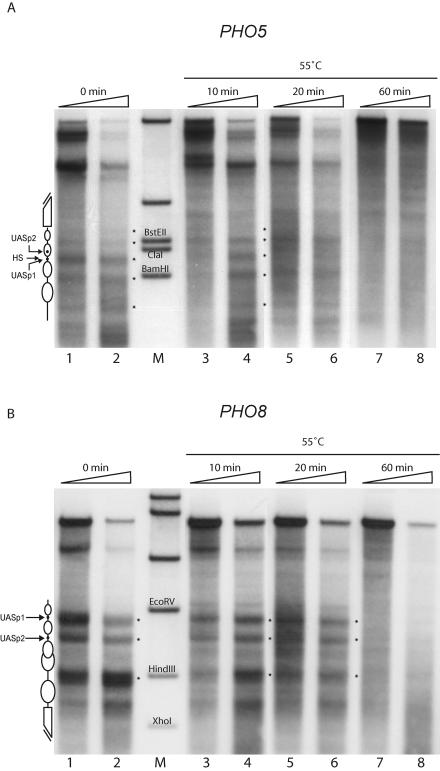
We wanted to test if the observed differences in positioning power at the PHO5 and PHO8 promoters could also be observed with native chromatin as assembled by yeast cells in vivo. It is not feasible to compare the assembly and positioning processes at both loci in vivo as in our in vitro system. However, we prepared yeast nuclei and used them as an in vitro substrate for the reverse process of nucleosome assembly, i.e., the loss of positioned nucleosome structure. It has previously been reported that nucleosomes exhibit temperature-dependent mobility on DNA (14, 31, 32, 37, 50). We prepared yeast nuclei and incubated them at 55°C. At three time points during this incubation we assayed changes in chromatin structure by DNase I mapping and specific probing for both the PHO8 and PHO5 promoter regions (Fig. 7). At the PHO5 promoter, the nucleosome pattern was largely lost after 20 min (Fig. 7A, lanes 5 and 6) whereas the nucleosomes at the PHO8 promoter were still properly positioned at this time point (Fig. 7B, lanes 5 and 6). After 1 hour the pattern was lost at both promoters (lanes 7 and 8). This showed that the nucleosomes at the PHO8 promoter have a higher kinetic stability towards thermally induced loss of nucleosome positioning.
FIG. 7.
Using in vivo-assembled chromatin, nucleosomes at the PHO8 promoter are found to be more resistant to temperature-induced loss of nucleosome positioning than those at the PHO5 promoter. (A and B) Yeast nuclei were incubated at 55°C for the indicated times, subjected to DNase I mapping, and probed for the PHO5 (A) or the PHO8 (B) promoter. Schematics and asterisks are as defined for Fig. 2. Ramps on top of the lanes denote increasing DNase I concentrations. The bands in the marker lanes (M) are generated by restriction digestion of genomic DNA and correspond to the restriction sites indicated on the gel. Additional bands outside the region of interest stem from the digestion of the PHO5 locus with BglII, used for secondary digestion of PHO8 (A), and from the digestion of the PHO8 locus with ApaI, used for secondary digestion of PHO5 (B).
DISCUSSION
The PHO8 promoter has greater nucleosome positioning power, and properly positioned PHO8 promoter nucleosomes are more stable, than their PHO5 counterparts.
Ever since the differential cofactor requirements of the coregulated PHO5 and PHO8 promoters were recognized (5, 16, 18), there has been speculation about the underlying cause for this difference. One attractive possibility is that differences inherent in the chromatin structures render one promoter more amenable to chromatin remodeling and chromatin opening than the other. Comparing both the PHO5 and the PHO8 promoter sequences in the same in vitro assembly reactions, we now provide evidence that the PHO8 promoter has more nucleosome-positioning power to generate the proper nucleosomal structure than the PHO5 promoter. In addition, at the PHO8 promoter nucleosomes adopt more-stable positions compared to the positioned nucleosomes at the PHO5 promoter.
This conclusion is twofold. First, we argue that the proper nucleosome positions at the PHO8 promoter have a higher stability relative to alternative positions in the same region than the proper positions at the PHO5 promoter have relative to alternative positions in that region. This relative stability of nucleosome positions is equivalent to the “nucleosome-positioning power” of the corresponding DNA regions (28). Importantly, this relative stability (intramolecular) should not be confused with the stability of positioned nucleosomes at the PHO8 promoter relative to the stability of positioned nucleosomes at the PHO5 promoter (intermolecular). The intermolecular stability comparison is made on an absolute scale, and we therefore refer to it as “absolute stability.” Absolute stability and positioning power need not correlate with each other. A DNA sequence of sufficient length may have a very high overall affinity for nucleosomes, i.e., confer high absolute stability, but several of the possible nucleosome positions may have very similar stabilities, i.e., there would be low positioning power. Nonetheless, it is reported for short in vitro-selected DNA fragments that the overall affinity of a DNA sequence for nucleosomes goes together with the ability to position nucleosomes either rotationally or translationally (28). In the same sense we argue secondly that the higher positioning power of the PHO8 promoter also goes together with higher absolute stability compared to the PHO5 promoter.
This twofold argument is based on the following. For one, nucleosomes at the PHO8 promoter become properly positioned even under conditions of limiting histone octamers whereas the proper positioning at the PHO5 promoter was not generated (Fig. 6B and C). Under such conditions of low nucleosome density there is competition of all possible nucleosome positions with each other for nucleosome assembly and only those positions will be occupied that are significantly more stable than others. Therefore, the proper positions at the PHO8 promoter are more stable than all alternative positions in this region. Secondly, under the same conditions of limiting histone octamers we also compared the overall assembly states of the PHO5 and PHO8 promoter regions in the same assembly reaction tube. This assays the overall, or average, affinity for nucleosomes of both regions regardless of the particular nucleosome positions. Both average affinities were very similar (Fig. 6A). This provides a common reference point for both promoter regions: the average affinities at each region are similar for the PHO8 and PHO5 promoters. As the proper positions at the PHO8 promoter are more stable than the average under the limiting conditions whereas the proper positions at the PHO5 promoter are not, we can therefore conclude that the proper positions at the PHO8 promoter are more stable than the proper positions at the PHO5 promoter.
These conclusions are valid only if the assembly conditions lead to an equilibrium state. We published previously that yeast extract assembly generates equilibrium positions, as prolonged incubation under conditions of sustained nucleosome mobility did not alter the final pattern (22). The same independence of the generated chromatin patterns from prolonged incubation times was confirmed for all assembly reactions presented here.
The difference in nucleosome stability correlates with differential cofactor requirements.
Complete chromatin remodeling at the PHO8 promoter proceeds through a dedicated pathway, stringently involving the cofactors Snf2 and Gcn5 (18). The chromatin transition upon induction of PHO5 also seems to involve these as it is delayed in their absence (5, 12, 35, 40; Luckenbach et al., submitted). However, after prolonged induction, chromatin remodeling still goes to completion, arguing for redundant pathways that can support promoter opening even without Snf2 and Gcn5. The same is true in the absence of Ino80, Asf1, Swr1, Isw1, Isw2, Chd1, Rad54, Mot1, and other cofactors (9, 16, 21; Korber et al., submitted; Luckenbach et al., submitted; our unpublished data). Further, while chromatin opening affects four nucleosomes at the PHO5 promoter, only one nucleosome is fully affected at the PHO8 promoter, resulting in much weaker promoter strength (3, 4).
These differences between promoter opening at PHO5 and PHO8 are unlikely to be due to differences in the recruitment of cofactors as both promoters are regulated by the same transactivator, Pho4. They could be a direct consequence of the strength of the UASp elements at each promoter. However, exchanging the UASp elements at the PHO8 promoter for those of the PHO5 promoter did not alter the extent of chromatin opening at the PHO8 promoter (33).
We rather think that properties of the substrate for the chromatin remodeling reaction, i.e., the positioned nucleosome structure, make one promoter chromatin more amenable to remodeling than the other. Especially for a mechanism leading to histone eviction in trans, the absolute stability of nucleosomes would be an important feature as a remodeler needs to completely disassemble the nucleosome. As discussed earlier (23), we cannot rule out at this point that a remodeling mechanism leading to histone eviction in trans may not involve an initial phase of nucleosome sliding. For such a phase the intramolecular relative stability of positioned nucleosomes compared to alternative positions along the same DNA molecule would be relevant. In both cases, not only the thermodynamic stabilities of proper and alternative positions but also the kinetic energy barrier for dislocating nucleosomes from their starting positions, i.e., their kinetic stability, have to be considered.
We show here that the absolute stability, the intramolecular relative stability, and the kinetic stability of the properly positioned nucleosomes at the PHO5 promoter are lower compared to those at the PHO8 promoter. Such an inherent instability supports our earlier suggestion that the PHO5 promoter chromatin resembles a “loaded spring” (22). This “loaded spring” will open up if provided with the right trigger, i.e., Pho4 and factors recruited by Pho4, and this process seems to be energetically so favorable that it is supported by a redundant set of cofactors.
The nature of the nucleosome positioning information at the PHO5 and PHO8 promoters.
Even though positioned nucleosomes have been described in vivo for a long time and have been shown to play important roles in gene regulation (24, 29, 45, 46, 51, 53), the molecular nature of the positioning information remains largely unresolved. The DNA sequence certainly plays an important role, but there is no algorithm available to reliably predict nucleosome positions from the DNA sequence alone (52). Most studies on the role of DNA sequence in nucleosome positioning are done in vitro by using salt gradient dialysis, which may reflect the equilibrium positioning of the H3/H4 tetramers in the range of 0.75 to 1 M salt (for a review of this argument see reference 52). This may be why these preferred positions in vitro need not coincide with the positions observed in vivo (11, 36, 48). Indeed it has been shown that >95% of a eukaryotic genome did not sufficiently constrain nucleosome positioning in salt gradient dialysis reconstitution (27). In the case of the PHO5 and PHO8 promoters, we also show here that salt gradient dialysis does not reflect the same preferences for nucleosome positioning seen under physiological conditions. Nonetheless, salt gradient dialysis may properly assemble a subregion of the PHO8 promoter that was previously characterized as especially repressive (between UASp2 and the HindIII site [33]), as well as a somewhat shifted nucleosome −1 at the PHO5 promoter. As both these regions are not completely remodeled in vivo (3, 4), this may point to strong nucleosome positioning sequences as involved in such chromatin structures.
Our yeast extract in vitro assembly system provides for the first time a strong tool with which to identify the nucleosome positioning information. The yeast extract contains something that is able to induce the proper positioning in assembly reactions that otherwise do not contain or support the right positioning information. This shifting of positions in preassembled chromatin is much faster than de novo assembly starting from free DNA. Therefore, it seems that the positioning of nucleosomes is uncoupled from their loading onto the DNA and that the loading of nucleosomes is the slow step in de novo assembly (22). We also show that the nucleosome positioning is energy dependent. This energy dependence may indicate that a remodeling complex is part of the positioning information or that ATP-dependent remodeling is necessary to overcome the kinetic barrier of nucleosome repositioning.
At this point we can only speculate about the molecular nature of the nucleosome-positioning information beyond the DNA sequence information. It may be a specific or unspecific DNA binding protein, a chromatin remodeling complex, or a combination of several types of factors. It is remarkable that this information appears to be species specific as it is not contained in a Drosophila embryo extract. As we have many copies of the PHO5 and PHO8 promoters in our assembly reactions, we think it unlikely that the yeast extracts used can contain enough promoter-specific factors to induce proper positioning in all these copies. Therefore we speculate that the nucleosome-positioning information should be an abundant factor acting genome wide, e.g., HMG proteins or remodeling factors, rather than sequence-specific DNA binding proteins.
At this point we cannot rigorously distinguish whether the thermodynamic equilibrium that correlates with proper nucleosome positioning is sufficiently determined by the histone-DNA contacts under certain buffer conditions and whether the yeast extract just enables the nucleosomes to adopt this equilibrium state or whether the yeast extract truly affects the equilibrium. The observation that the Drosophila embryo extract system cannot generate the native positioning even though the histones, DNA sequence, and buffer are the same as in the yeast system and even though it is rich in remodeling activities (6, 7, 20) strongly argues for the latter case. Nonetheless, it is possible that only a certain set of remodelers or other activities can catalyze the shift to the proper positions without necessarily affecting the equilibrium. In any case, remodeling complexes seem to be involved, and it is an attractive hypothesis that species-specific sets of remodeling machines may set up nucleosome positioning genome wide. Experiments are under way to reveal the molecular nature of the nucleosome positioning information at the PHO5 and PHO8 promoters and across the whole genome.
Acknowledgments
We are grateful to Andrea Schmid and Dorothea Blaschke for expert technical assistance, to Peter Becker for critically reading the manuscript, to Gregor Gilfillan for advice on English style, and to all members of the Hörz group for helpful discussions.
This work was supported by the Deutsche Forschungsgemeinschaft (Transregio 5) and the 6th Framework Programme of the European Union (Epigenome Network of Excellence). C.B.H. was supported by Familien Hede Nielsens Fond and Harboefonden.
REFERENCES
- 1.Adkins, M. W., S. R. Howar, and J. K. Tyler. 2004. Chromatin disassembly mediated by the histone chaperone Asf1 is essential for transcriptional activation of the yeast PHO5 and PHO8 genes. Mol. Cell 14:657-666. [DOI] [PubMed] [Google Scholar]
- 2.Almer, A. and W. Hörz. 1986. Nuclease hypersensitive regions with adjacent positioned nucleosomes mark the gene boundaries of the PHO5/PHO3 locus in yeast. EMBO J. 5:2681-2687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Almer, A., H. Rudolph, A. Hinnen, and W. Hörz. 1986. Removal of positioned nucleosomes from the yeast PHO5 promoter upon PHO5 induction releases additional upstream activating DNA elements. EMBO J. 5:2689-2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barbaric, S., K. D. Fascher, and W. Hörz. 1992. Activation of the weakly regulated PHO8 promoter in S. cerevisiae: chromatin transition and binding sites for the positive regulator protein Pho4. Nucleic Acids Res. 20:1031-1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barbaric, S., J. Walker, A. Schmid, J. Q. Svejstrup, and W. Hörz. 2001. Increasing the rate of chromatin remodeling and gene activation—a novel role for the histone acetyltransferase Gcn5. EMBO J. 20:4944-4951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Becker, P. B., T. Tsukiyama, and C. Wu. 1994. Chromatin assembly extracts from Drosophila embryos. Methods Cell Biol. 44:207-223. [DOI] [PubMed] [Google Scholar]
- 7.Becker, P. B., and C. Wu. 1992. Cell-free system for assembly of transcriptionally repressed chromatin from Drosophila embryos. Mol. Cell. Biol. 12:2241-2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boeger, H., J. Griesenbeck, J. S. Strattan, and R. D. Kornberg. 2003. Nucleosomes unfold completely at a transcriptionally active promoter. Mol. Cell 11:1587-1598. [DOI] [PubMed] [Google Scholar]
- 9.Boeger, H., J. Griesenbeck, J. S. Strattan, and R. D. Kornberg. 2004. Removal of promoter nucleosomes by disassembly rather than sliding in vivo. Mol. Cell 14:667-673. [DOI] [PubMed] [Google Scholar]
- 10.Bonte, E., and P. B. Becker. 1999. Preparation of chromatin assembly extracts from preblastoderm Drosophila embryos. Methods Mol. Biol. 119:187-194. [DOI] [PubMed] [Google Scholar]
- 11.Buttinelli, M., E. Dimauro, and R. Negri. 1993. Multiple nucleosome positioning with unique rotational setting for the Saccharomyces cerevisiae 5s rRNA gene in vitro and in vivo. Proc. Natl. Acad. Sci. USA 90:9315-9319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dhasarathy, A., and M. P. Kladde. 2005. Promoter occupancy is a major determinant of chromatin remodeling enzyme requirements. Mol. Cell. Biol. 25:2698-2707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fascher, K. D., J. Schmitz, and W. Hörz. 1993. Structural and functional requirements for the chromatin transition at the PHO5 promoter in Saccharomyces cerevisiae upon PHO5 activation. J. Mol. Biol. 231:658-667. [DOI] [PubMed] [Google Scholar]
- 14.Flaus, A., and T. J. Richmond. 1998. Positioning and stability of nucleosomes on MMTV 3′LTR sequences. J. Mol. Biol. 275:427-441. [DOI] [PubMed] [Google Scholar]
- 15.Fyodorov, D. V., and J. T. Kadonaga. 2001. The many faces of chromatin remodeling: SWItching beyond transcription. Cell 106:523-525. [DOI] [PubMed] [Google Scholar]
- 16.Gaudreau, L., A. Schmid, D. Blaschke, M. Ptashne, and W. Hörz. 1997. RNA polymerase II holoenzyme recruitment is sufficient to remodel chromatin at the yeast PHO5 promoter. Cell 89:55-62. [DOI] [PubMed] [Google Scholar]
- 17.Gregory, P. D., and W. Hörz. 1999. Mapping chromatin structure in yeast. Methods Enzymol. 304:365-376. [DOI] [PubMed] [Google Scholar]
- 18.Gregory, P. D., A. Schmid, M. Zavari, M. Münsterkötter, and W. Hörz. 1999. Chromatin remodelling at the PHO8 promoter requires SWI-SNF and SAGA at a step subsequent to activator binding. EMBO J. 18:6407-6414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grunstein, M. 1997. Histone acetylation in chromatin structure and transcription. Nature 389:349-352. [DOI] [PubMed] [Google Scholar]
- 20.Kamakaka, R. T., M. Bulger, and J. T. Kadonaga. 1993. Potentiation of RNA polymerase-II transcription by gal4-VP16 during but not after DNA replication and chromatin assembly. Genes. Dev. 7:1779-1795. [DOI] [PubMed] [Google Scholar]
- 21.Kent, N. A., N. Karabetsou, P. K. Politis, and J. Mellor. 2001. In vivo chromatin remodeling by yeast ISWI homologs Isw1p and Isw2p. Genes Dev. 15:619-626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Korber, P., and W. Hörz. 2004. In vitro assembly of the characteristic chromatin organization at the yeast PHO5 promoter by a replication independent extract system. J. Biol. Chem. 279:35113-35120. [DOI] [PubMed] [Google Scholar]
- 23.Korber, P., T. Luckenbach, D. Blaschke, and W. Horz. 2004. Evidence for histone eviction in trans upon induction of the yeast PHO5 promoter. Mol. Cell. Biol. 24:10965-10974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kornberg, R. D., and Y. Lorch. 1995. Interplay between chromatin structure and transcription. Curr. Opin. Cell Biol. 7:371-375. [DOI] [PubMed] [Google Scholar]
- 25.Kornberg, R. D., and Y. Lorch. 1999. Twenty-five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell 98:285-294. [DOI] [PubMed] [Google Scholar]
- 26.Längst, G., E. J. Bonte, D. F. Corona, and P. B. Becker. 1999. Nucleosome movement by CHRAC and ISWI without disruption or trans-displacement of the histone octamer. Cell 97:843-852. [DOI] [PubMed] [Google Scholar]
- 27.Lowary, P. T., and J. Widom. 1997. Nucleosome packaging and nucleosome positioning of genomic DNA. Proc. Natl. Acad. Sci. USA 94:1183-1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lowary, P. T., and J. Widom. 1998. New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning. J. Mol. Biol. 276:19-42. [DOI] [PubMed] [Google Scholar]
- 29.Lu, Q., L. L. Wallrath, and S. C. R. Elgin. 1995. The role of a positioned nucleosome at the Drosophila melanogaster hsp26 promoter. EMBO J. 14:4738-4746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Luger, K., A. W. Mader, R. K. Richmond, D. F. Sargent, and T. J. Richmond. 1997. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature 389:251-260. [DOI] [PubMed] [Google Scholar]
- 31.Luger, K., T. J. Rechsteiner, and T. J. Richmond. 1999. Preparation of nucleosome core particle from recombinant histones. Methods Enzymol. 304:3-19. [DOI] [PubMed] [Google Scholar]
- 32.Meersseman, G., S. Pennings, and E. M. Bradbury. 1992. Mobile nucleosomes—a general behavior. EMBO J. 11:2951-2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Münsterkötter, M., S. Barbaric, and W. Hörz. 2000. Transcriptional regulation of the yeast PHO8 promoter in comparison to the coregulated PHO5 promoter. J. Biol. Chem. 275:22678-22685. [DOI] [PubMed] [Google Scholar]
- 34.Narlikar, G. J., H. Y. Fan, and R. E. Kingston. 2002. Cooperation between complexes that regulate chromatin structure and transcription. Cell 108:475-487. [DOI] [PubMed] [Google Scholar]
- 35.Neef, D. W., and M. P. Kladde. 2003. Polyphosphate loss promotes SNF/SWI- and Gcn5-dependent mitotic induction of PHO5. Mol. Cell. Biol. 23:3788-3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Negri, R., M. Buttinelli, G. Panetta, V. De Arcangelis, E. Di Mauro, and A. Travers. 2001. Sequence dependence of translational positioning of core nucleosomes. J. Mol. Biol. 307:987-999. [DOI] [PubMed] [Google Scholar]
- 37.Pennings, S., G. Meersseman, and E. M. Bradbury. 1991. Mobility of positioned nucleosomes on 5S rDNA. J. Mol. Biol. 220:101-110. [DOI] [PubMed] [Google Scholar]
- 38.Perlmann, T., and O. Wrange. 1988. Specific glucocorticoid receptor binding to DNA reconstituted in a nucleosome. EMBO J. 7:3073-3079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Piña, B., U. Brüggemeier, and M. Beato. 1990. Nucleosome positioning modulates accessibility of regulatory proteins to the mouse mammary tumor virus promoter. Cell 60:719-731. [DOI] [PubMed] [Google Scholar]
- 40.Reinke, H., and W. Hörz. 2003. Histones are first hyperacetylated and then lose contact with the activated PHO5 promoter. Mol. Cell 11:1599-1607. [DOI] [PubMed] [Google Scholar]
- 41.Reinke, H., and W. Hörz. 2004. Anatomy of a hypersensitive site. Biochim. Biophys. Acta 1677:24-29. [DOI] [PubMed] [Google Scholar]
- 42.Richmond, E., and C. L. Peterson. 1996. Functional analysis of the DNA-stimulated ATPase domain of yeast SWI2/SNF2. Nucleic Acids Res. 24:3685-3692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Robinson, K. A., and J. M. Lopes. 2000. Survey and summary: Saccharomyces cerevisiae basic helix-loop-helix proteins regulate diverse biological processes. Nucleic Acids Res. 28:1499-1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Roeder, R. G. 2005. Transcriptional regulation and the role of diverse coactivators in animal cells. FEBS Lett. 579:909-915. [DOI] [PubMed] [Google Scholar]
- 45.Schild, C., F. X. Claret, W. Wahli, and A. P. Wolffe. 1993. A nucleosome-dependent static loop potentiates estrogen-regulated transcription from the Xenopus vitellogenin B1 promoter in vitro. EMBO J. 12:423-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Straka, C., and W. Hörz. 1991. A functional role for nucleosomes in the repression of a yeast promoter. EMBO J. 10:361-368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Svaren, J., and W. Hörz. 1997. Transcription factors vs nucleosomes: regulation of the PHO5 promoter in yeast. Trends Biochem. Sci. 22:93-97. [DOI] [PubMed] [Google Scholar]
- 48.Tanaka, S., M. Zatchej, and F. Thoma. 1992. Artificial nucleosome positioning sequences tested in yeast minichromosomes: a strong rotational setting is not sufficient to position nucleosomes in vivo. EMBO J. 11:1187-1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tsukiyama, T., P. B. Becker, and C. Wu. 1994. ATP-dependent nucleosome disruption at a heat-shock promoter mediated by binding of GAGA transcription factor. Nature 367:525-532. [DOI] [PubMed] [Google Scholar]
- 50.Ura, K., H. Kurumizaka, S. Dimitrov, G. Almouzni, and A. P. Wolffe. 1997. Histone acetylation: influence on transcription, nucleosome mobility and positioning, and linker histone-dependent transcriptional repression. EMBO J. 16:2096-2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Venter, U., J. Svaren, J. Schmitz, A. Schmid, and W. Hörz. 1994. A nucleosome precludes binding of the transcription factor Pho4 in vivo to a critical target site in the PHO5 promoter. EMBO J. 13:4848-4855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Widom, J. 2001. Role of DNA sequence in nucleosome stability and dynamics. Q. Rev. Biophys. 34:269-324. [DOI] [PubMed] [Google Scholar]
- 53.Yuan, G. C., Y. J. Liu, M. F. Dion, M. D. Slack, L. F. Wu, S. J. Altschuler, and O. J. Rando. 2005. Genome-scale identification of nucleosome positions in S. cerevisiae. Science 309:626-630. [DOI] [PubMed] [Google Scholar]