Abstract
1-Deoxy-d-xylulose 5-phosphate reductoisomerase (IspC) catalyzes the first committed step in the mevalonate-independent isopentenyl diphosphate biosynthetic pathway and is a potential drug target in some pathogenic bacteria. The antibiotic fosmidomycin has been shown to inhibit IspC in a number of organisms and is active against most gram-negative bacteria but not gram positives, including Mycobacterium tuberculosis, even though the mevalonate-independent pathway is the sole isopentenyl diphosphate biosynthetic pathway in this organism. Therefore, the enzymatic properties of recombinant IspC from M. tuberculosis were characterized. Rv2870c from M. tuberculosis converts 1-deoxy-d-xylulose 5-phosphate to 2-C-methyl-d-erythritol 4-phosphate in the presence of NADPH. The enzymatic activity is dependent on the presence of Mg2+ ions and exhibits optimal activity between pH 7.5 and 7.9; the Km for 1-deoxyxylulose 5-phosphate was calculated to be 47.1 μM, and the Km for NADPH was 29.7 μM. The specificity constant of Rv2780c in the forward direction is 1.5 × 106 M−1 min−1, and the reaction is inhibited by fosmidomycin, with a 50% inhibitory concentration of 310 nM. In addition, Rv2870c complements an inactivated chromosomal copy of IspC in Salmonella enterica, and the complemented strain is sensitive to fosmidomycin. Thus, M. tuberculosis resistance to fosmidomycin is not due to intrinsic properties of Rv2870c, and the enzyme appears to be a valid drug target in this pathogen.
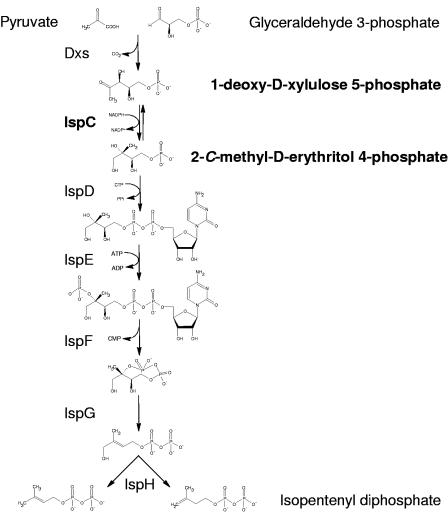
Fosmidomycin is a phosphonate antibiotic that is reported to be active against most gram-negative bacteria (22, 25, 37) but not gram-positive cocci or anaerobic species (25). In 1985 the antibiotic underwent phase I and II clinical trials for treating urinary tract infections (14, 15). However, the mechanism of action was unknown until 1989, when fosmidomycin was shown to inhibit bacterial isoprenoid biosynthesis (31). Nine years passed before it was demonstrated that fosmidomycin specifically inhibits IspC (16, 38), which catalyzes the first committed step in the mevalonate-independent isopentenyl diphosphate (IPP) biosynthetic pathway, also known as the 2-C-methyl-d-erythritol 4-phosphate (MEP) pathway or the 1-deoxy-d-xylulose 5-phosphate (DXP or DOXP) pathway. In this biosynthetic scheme (Fig. 1), pyruvate is condensed with glyceraldehyde 3-phosphate to form DXP, which is then converted to MEP by IspC and finally to IPP (1) via multiple steps which are not well characterized in Mycobacterium spp. The MEP pathway is unrelated to the mevalonate-dependent IPP biosynthetic pathway, which is found primarily in eukaryotes and archea (6), thus making it of great interest for drug development.
FIG. 1.
The MEP pathway. The reaction catalyzed by 1-deoxy-d-xylulose 5-phosphate reductoisomerase (IspC) is indicated in bold.
Interestingly, the MEP pathway is not confined to gram-negative organisms, as it appears to be utilized by a majority of bacteria, including most proteobacteria, some low-G+C gram positives, most high-G+C gram positives, and photosynthetic eukaryotes (6). The MEP pathway has been biochemically demonstrated in Mycobacterium phlei (27) and Mycobacterium tuberculosis (4), which are high-G+C gram positives, and there is no indication that the mevalonate-dependent IPP biosynthetic pathway exists in mycobacteria (5, 6), although some gram-positive organisms do appear to have both IPP biosynthetic pathways (6, 32).
IPP is a precursor of all known isoprenoid compounds, which are essential compounds in all domains of life (6). Mycobacteria, in which isoprenoid compounds play crucial roles in cell wall biosynthesis and energy metabolism, are no exception. For example, prenyl phosphates (Pol-P) are required for the biosynthesis of many of cell wall components, including decaprenyl-phosphorylarabinose and decaprenyl-phosphorylmannose, which have been shown to be precursors of the arabinan portions of mycolylarabinogalactan-petidoglycan, arabinogalactan, arabinomannan, and lipoarabinomannan (36). A polyprenyl diphosphate “carrier lipid” has also been implicated in the synthesis of the “linker unit” of mycobacterial arabinogalactan (20), and decaprenyl phosphate-containing lipid I and lipid II are fundamental to peptidoglycan synthesis in mycobacteria (19). In addition, menaquinones, the only lipoquinones involved in the electron transport chain found in mycobacteria (23), contain a naphthoate ring that is prenylated during biosynthesis. Thus, the synthesis of isoprenoids is an important early step in mycobacterial cell wall construction and oxidative phosphorylation. However, in our hands fosmidomycin has no activity against Mycobacterium tuberculosis at concentrations up to 200 μM.
There are several possible explanations for this phenomenon: (i) IspC in M. tuberculosis may be refractory to inhibition by fosmidomycin, as seemingly minor differences in primary structure may significantly alter the properties of an enzyme. For example, MurA in M. tuberculosis was found to have an aspartate at position 117 instead of the cysteine found in Escherichia coli (8), a difference that renders the enzyme insensitive to fosfomycin, a broad-spectrum phosphonate antibiotic structurally related to fosmidomycin, which is also inactive against M. tuberculosis (8). (ii) The antibiotic may not cross the permeability barrier presented by the mycobacterial cell wall. (iii) The antibiotic may be extruded from the cell or otherwise metabolized.
ispC genes have previously been cloned and expressed, and the enzymes have been at least partially characterized from α-proteobacteria (Zymomonas mobilis [10]), γ-proteobacteria (Escherichia coli [13, 17] and Pseudomonas aeruginosa [3]), cyanobacteria (Synechocystis sp. [26] and Synechococcus leopoliensis [21]) and plants (Mentha piperita [18] and Arabidopsis thaliana [30]). However, Streptomyces coelicolor (7) is the only gram-positive bacterium from which IspC has been cloned and partially characterized, and no information regarding the fosmidomycin sensitivity of IspC from these bacteria has been reported. Therefore, the enzymatic properties and fosmidomycin sensitivity of recombinant DXP reductoisomerase from M. tuberculosis were characterized.
MATERIALS AND METHODS
Materials and instrumentation.
H37Rv genomic DNA was provided by NIH National Institute of Allergy and Infectious Diseases (NIAID) contract NO1-A1-75320, “Tuberculosis Research Material and Vaccine Testing.” All PCR product and plasmid purifications were performed using QIAGEN kits (Valencia, CA). All antibiotics were purchased from Sigma-Aldrich (St. Louis, MO). Bio-Rex 70 cation exchange resin was obtained from Bio-Rad (Hercules, CA). DXP, racemic 1-deoxy-d-xylulose (DX), MEP, and racemic 2-C-methyl-d-erythritol (ME) were from Echelon Laboratories (Salt Lake City, UT). Fosmidomycin was obtained from M. Patel (GlaxoSmithKline, Collegeville, PA). All other chemicals and reagents were purchased from Sigma-Aldrich unless otherwise noted. Enzyme assays (50- or 100-μl final volume) were performed in 96-well, flat-bottom, half-area plates (Costar, Bethesda, MD) using a SpectraMax Plus 384 plate reader (Molecular Devices, Sunnyvale, CA).
Cloning, expression, and purification of RV2870c.
Based on the nucleotide sequence of the open reading frame Rv2870c, the following primers were designed and synthesized (Macromolecular Resources, Colorado State University): F, 5′-AATGATCATATGACCAACTCGACCGAC-3′, and R, 5′-AACAAGCTTTCAGGACCTTTCTAACG-3′. NdeI and HindIII restriction sites (underlined) were engineered in the N-terminal and C-terminal primers, respectively. PCR amplification of Rv2870c from DNA was performed using a Perkin-Elmer GeneAmp 2400 PCR system and rTth polymerase (PE Biosystems, Foster City, CA). The PCR product was digested with the indicated enzymes and cloned into the multiple cloning site of pET28a(+) (EMD Biosciences, Inc., San Diego, CA). The product, designated pAXRh, was used to transform E. coli DH5α subcloning cells (Life Technologies, Rockville, MD) for plasmid propagation. The plasmid was subsequently purified, analyzed by restriction endonuclease digestion, and sequenced (Macromolecular Resources, Colorado State University). The E. coli expression host BL21(DE3)pLysS (Novagen, Madison, WI) was transformed with pAXRh and grown to mid-log phase (optical density at 600 nm [OD600], ∼0.6) in Luria-Bertani (LB) broth supplemented with kanamycin (50 μg/ml) and chloramphenicol (34 μg/ml); 1 mM (final concentration) isopropyl-β-d-thiogalactopyranoside (IPTG) was added, and incubation was continued for 4 h at 30°C. Cells were then harvested and frozen at −20°C.
Frozen cells were thawed on ice in buffer A (50 mM morpholinepropanesulfonic acid [MOPS; pH 7.9], 200 mM NaCl, 10 mM MgCl2, 5 mM β-mercaptoethanol, and 10% glycerol) at 2 ml/g of cells and sonicated using a Sanyo Soniprep 150 (Integrated Services, TCP Inc., Palisades Park, NJ). Cell debris was pelleted by centrifugation at 20,000 × g for 20 min and discarded. The supernatant, containing the recombinant Rv2870c, was loaded on a Talon Co2+ immobilized metal affinity resin (Clontech, Palo Alto, CA), which was preequilibrated with buffer B (50 mM MOPS [pH 7.9], 200 mM NaCl, 1 mM MgCl2, 1 mM β-mercaptoethanol, and 10% glycerol) and rocked at 4°C for 20 min. The slurry was then packed in a column and washed with five bed volumes of buffer B containing 5 mM imidazole. The recombinant enzyme was eluted stepwise with 1-ml aliquots of buffer B containing 25, 50, 100, 150, and 200 mM imidazole. Fractions were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and those containing the most recombinant protein were combined and passed over a PD10 G-25 gel filtration column (Amersham, Piscataway, NJ) preequilibrated with buffer B minus NaCl to remove imidazole and NaCl. Recombinant Rv2870c was eluted with buffer B minus NaCl, divided into aliquots, and stored at −80°C. Protein concentrations were determined using a BCA protein assay kit (Pierce, Rockford, IL).
Enzyme assays.
A continuous assay format was used to monitor DXP reductoisomerase activity by following the oxidation of NADPH or reduction of NADP+ in the forward or reverse reaction, respectively (Fig. 1). In the forward assay, DXP and NADPH were incubated at 30°C in 100 mM MOPS, pH 7.9, 4 mM MgCl2, 0.01% 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate (CHAPS) buffer for 5 min prior to initiation of the reaction with Rv2870c. Reduction of DXP to MEP results in oxidation of NADPH and a decrease in signal at A340. To assay Rv2870c in the reverse direction, MEP and NADP+ were incubated in the same buffer and the reaction was initiated with Rv2870c. In this case, oxidation of MEP to DXP results in the concomitant reduction of NADP+, causing an increase in A340 signal. Negative control reaction mixtures for either assay contained substrates and buffer but no enzyme. Various concentrations of enzyme and substrates were used, the details of which are included below in the figure legends and text. All assays were done in either duplicate or triplicate, and the initial rates were determined by measuring the change in A340 at 30-s intervals throughout the course of the reaction.
In some cases endogenous divalent cations were removed from the recombinant protein by cation exchange using Bio-Rex 70 resin. Optimal concentrations for divalent cations were then determined using the in vitro assay conditions described above with the indicated concentration of divalent cation. Optimal pH was also determined; in this case three separate buffers covering a broad pH range were applied, and appropriate counter-ions were used to each buffer in pH increments of 0.5 unit from pH 5.0 to 6.5 (50 mM MES), pH 6.5 to 8.0 (50 mM MOPS), and pH 8.0 to 9.0 (50 mM 3-{[tris-(hydroxymethyl)-methyl]-amino}-propanesulfonic acid).
Complementation of disrupted chromosomal ispC in Salmonella enterica serovar Typhimurium with Rv2870c.
The chromosomal copy of Salmonella enterica serovar Typhimurium dxs, which encodes the enzyme catalyzing the first reaction in the MEP pathway, had previously been disrupted with a synthetic mevalonate pathway operon to generate strain RMC26 (35), which is blocked in the de novo biosynthesis of IPP and dimethylallyl diphosphate (DMAPP) via the MEP pathway but is able to synthesize both molecules from mevalonate. Therefore, the strain is an auxotroph, which can be satisfied by addition of either DX or mevalonate (MVA) to the growth medium. The ispC gene in RMC26 was then disrupted by insertion of a chloramphenicol acyltransferase (CAT) cassette to generate CT12 (RMC26 ispC::CAT) as previously described (35). This strain is also an auxotroph, which can be satisfied by addition of either ME or MVA but not DX to the growth medium. CT12 was electroporated with pAXRh, generating a strain designated CT25, and both strains were tested for auxotrophy and sensitivity to fosmidomycin by plating on LB agar containing 40 μg/ml kanamycin and 134 μM ME, 2 mM DX, 1 mM IPTG, or 100 μM fosmidomycin singly or in combination.
Other procedures:.
Restriction digests, ligations, and electroporations were done as described by Sambrook et al. (29) unless otherwise noted. BLAST searches were done on the National Center for Biotechnology Information website and the Mycobacterium tuberculosis Structural Genomics Consortium website using standard protein-protein BLAST (blastp). Alignments were done using multiple sequence alignments with hierarchical clustering (2) using the Multalin interface at the Institut National de la Recherche Agronomique (Toulouse, France) website.
RESULTS
Characteristics of M. tuberculosis IspC.
BLAST searches indicated that a single gene, Rv2870c, from M. tuberculosis encodes a protein with significant homology to known DXP reductoisomerases and has been annotated as such. Therefore, Rv2870c was cloned from M. tuberculosis genomic DNA and expressed in an E. coli host, and the resulting protein was purified to near homogeneity by immobilized metal affinity chromatography (Fig. 2). Rv2870c is predicted to be cytosolic, with a molecular mass of 42.8 kDa and an isoelectric point of 5.59 and contains a zinc-binding motif (Mycobacterium tuberculosis Structural Genomics Consortium website). The expressed protein appears to be somewhat larger than 45 kDa by SDS-PAGE analysis; this could, in part, be due to the presence of the polyhistidine tag and was observed consistently.
FIG. 2.
SDS-PAGE analysis of Rv2870c. Immobilized metal affinity chromatography-purified, recombinant Rv2870c was loaded in lane 1, and molecular mass markers were loaded in lane 2.
Reaction conditions.
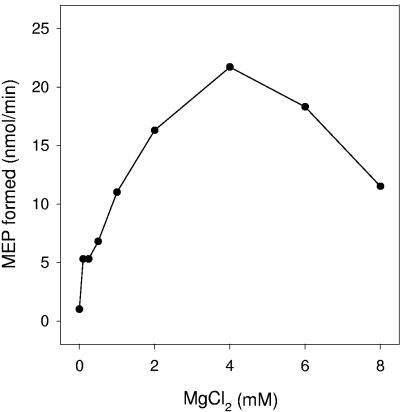
The reaction was absolutely dependent on the addition of Mg2+ ions, was completely abolished by the addition of 5 mM EDTA, and was optimal at 4 mM MgCl2 (Fig. 3). Mn2+, Ca2+, Co2+, Zn2+, and Fe2+ ions were unable to support activity at any concentration tested. The enzyme is active over a relatively narrow range of pH, with the optimal product formation near pH 7.9 (Fig. 4). In addition, the reaction was dependent on the addition of DXP and NADPH in the forward direction and MEP and NADP+ in the reverse direction.
FIG. 3.
Effect of Mg2+ concentration on Rv2870c activity. Endogenous divalent cations were removed from the recombinant protein by cation exchange using Bio-Rex 70 resin, and a continuous assay format was used to monitor 1-deoxy-d-xylulose 5-phosphate reductoisomerase activity by following the oxidation of NADPH. Reaction mixtures contained 300 μM DXP and 300 μM NADPH in 100 mM MOPS, pH 7.9, 0.01% CHAPS, and the indicated concentration of MgCl2. Reaction mixtures were incubated at 30°C for 5 min.
FIG. 4.
Effect of pH on Rv2870c activity. Reaction mixtures contained 300 μM DXP, 300 μM NADPH, 4 mM MgCl2, and 0.01% CHAPS. The buffer and pH used for each reaction mixture are indicated. Reaction mixtures were incubated at 30°C for 5 min.
Characterization of Rv2870c in the forward and reverse reactions.
The assay in the forward direction was linear with Rv2870c concentrations up to 200 nM in reaction mixtures containing 100 μM DXP and 200 μM NADPH. In subsequent experiments, Rv2870c was typically used at a concentration of 66 nM.
The effects of concentrations of DXP or NADPH on reaction rate were determined by varying the amount of one substrate while keeping the concentration of the second substrate fixed (Fig. 5). The KmDXP is calculated to be 47.1 ± 5.9 μM, and substrate inhibition was observed with a Ki of approximately 1.1 mM. The KmNADPH is 29.7 ± 2.7 μM, and there was no apparent substrate inhibition. These results provided guidelines for determination of the effects of fosmidomycin on the activity of M. tuberculosis IspC.
FIG. 5.
Effects of NADPH and DXP concentrations on Rv2870c activity. Reaction mixtures contained 100 mM MOPS, pH 7.9, 4 mM MgCl2, 0.01% CHAPS, and 66 nM Rv2870c. In some cases DXP was held at 100 μM and the concentration of NAPH was varied (A); in other cases NADPH was held at 100 μM and the concentration of DXP was varied (B). Reaction mixtures were incubated at 30°C for 5 min.
Linearity with respect to protein concentration (up to 250 nM) was also determined in the reverse assay; a concentration of 250 nM was chosen for subsequent experiments except where specifically noted. Generally, Rv2870c was not as active in the reverse compared to the forward direction. The initial rate observed in the forward reaction (860 pmol/min) was close to 10-fold higher than that seen in the reverse assay (86.8 pmol/min) at 200 nM Rv2870c. The effects of concentrations of MEP and NADP+ on reaction rates in the reverse direction were also determined (Fig. 6). The KmMEP is calculated to be 174.0 ± 28.1 μM, and the KmNADP+ is 560.5 ± 78.0 μM. No substrate inhibition was observed in either case.
FIG. 6.
Effects of NADP+ and MEP concentrations on Rv2870c activity in the reverse direction. Reaction mixtures contained 100 mM MOPS, pH 7.9, 4 mM MgCl2, 0.01% CHAPS, and 250 nM Rv2870c. In some cases MEP was held at 150 μM and the concentration of NADP+ was varied (A); in other cases NADP+ was held at 200 μM and the concentration of MEP was varied (B). Reaction mixtures were incubated at 30°C for 5 min.
Calculated kinetic parameters for Rv2870c in both the forward and reverse assays are shown in Table 1. The kcat values obtained in the forward reaction were significantly higher than those for the reverse, suggesting that the former is more efficient. This is also reflected in the specificity constants (kcat/Km), which were ∼106 and ∼104 M−1 min−1 for the forward and reverse assays, respectively.
TABLE 1.
Kinetic parameters for M. tuberculosis Rv2870c
| Reaction and substratea | Calculated parameter
|
|||
|---|---|---|---|---|
| Km (μM) | Vmax (pmol/min) | kcat (min−1) | kcat/Km (M−1 min−1) | |
| Forward reaction | ||||
| DXP | 47.1 ± 5.9 | 463.8 ± 22.7 | 70.3 | 1.5 × 106 |
| NADPH | 29.7 ± 2.7 | 311.9 ± 4.9 | 47.3 | 1.6 × 106 |
| Reverse reaction | ||||
| MEP | 174.0 ± 28.1 | 313.5 ± 15.6 | 12.5 | 7.2 × 104 |
| NADP+ | 560.5 ± 78.0 | 646.6 ± 46.4 | 25.9 | 4.62 × 104 |
Substrate concentrations for Km determinations for Rv2870c were as follows: for DXP, 100 μM NADPH and 40 to 700 μM DXP; for NADPH, 100 μM DXP and 40 to 700 μM NADPH; for MEP, 63 to 1000 μM MEP and 200 μM NADP+; for NADP+, 150 μM MEP and 63 to 1,000 μM NADP+. The concentration of Rv2870c in each reaction mixture was 66 nM and 250 nM for the forward and reverse reaction, respectively.
Inhibition of Rv2870c by fosmidomycin.
Recombinant Rv2870c is sensitive to fosmidomycin in the forward assay. The 50% inhibitory concentration (IC50) was calculated to be 0.310±0.045 μM from the data shown in Fig. 7. Fosmidomycin also inhibited the reverse reaction, with a calculated IC50 of 2.7± 0.3 μM (data not shown).
FIG. 7.
Effect of fosmidomycin concentration on Rv2870c activity. Reaction mixtures contained 100 mM MOPS, pH 7.9, 4 mM MgCl2, 0.01% CHAPS, 50 μM DXP, 50 μM NADPH, and 66 nM Rv2870c. Fosmidomycin was added at the indicated concentrations, and reaction mixtures were incubated at 30°C for 5 min.
Complementation of an ispC disruption in Salmonella enterica serovar Typhimurium.
The chromosomal copy of Salmonella enterica serovar Typhimurium dxs, which encodes the enzyme catalyzing the first reaction in the MEP pathway, was disrupted with a synthetic mevalonate pathway operon, generating strain RMC26 (see Materials and Methods), which is blocked in the de novo biosynthesis of IPP and DMAPP via the MEP pathway but is able to synthesize both molecules from either MVA or DX. The ispC gene, encoding the second enzyme in the MEP pathway, was subsequently disrupted to generate CT12 (RMC26 ispC::CAT). Thus, strain CT12 was predicted to be an auxotroph which can be satisfied by addition of either ME or MVA but not DX. CT12 was transformed with pAXRh, generating CT25, which was predicted to have the same phenotype as RMC26.
The engineered strain CT12 is auxotrophic for ME but was not viable in the presence of DX, as predicted (Table 2). However, when CT12 was transformed with pAXRh, to generate CT25, the phenotype was dramatically changed. Unlike CT12, CT25 is viable in the presence of DX with or without the addition of IPTG to induce Rv2870c expression. Thus, pAXRh was able to complement the ispC disruption. Viability in the absence of IPTG is likely due to low-level endogenous activation of the promoter on the plasmid. When fosmidomycin was added to the plates at 100 μM, CT25 did not grow in the presence or absence of IPTG.
TABLE 2.
Complementation of a Salmonella enterica serovar Typhimurium methylerythritol auxotroph with Rv2870ca
| Strain | Growth with addition to medium
|
|||||
|---|---|---|---|---|---|---|
| None | ME | DX | DX + IPTG | DX + fosmidomycin | DX + IPTG + fosmidomycin | |
| CT12 | − | + | − | − | − | − |
| CT25 | − | + | + | + | − | − |
The chromosomal copy of Salmonella enterica serovar Typhimurium dxs, which encodes the enzyme catalyzing the first reaction in the MEP pathway (Fig. 1), was disrupted with a synthetic mevalonate pathway operon, generating strain RMC26 (35); subsequently, the native ispC gene in RMC26 was disrupted by insertion of a chloramphenicol acyltransferase (CAT) cassette to generate CT12 (RMC26 ispC::CAT) and CT12 was transformed with a plasmid (pAXRh) bearing Rv2870c to generate CT25 (see Materials and Methods). Plates contained LB agar with kanamycin (40 μg/ml) and various additions as indicated. DX, ME, IPTG, and fosmidomycin were added at final concentrations of 2 mM, 134 μM, 1 mM, and 100 μM, respectively. Bacterial growth is indicated by +; no observable growth is indicated by −.
DISCUSSION
Due to the interest in the MEP pathway, both as a target for antimicrobials and as an herbicide, a significant number of experiments have shown that IspC from several organisms is inhibited by the antibiotic fosmidomycin, including that from A. thaliana (24), barley (3, 24), Plasmodium falciparum (11), Z. mobilis (10), and E. coli (13, 17). Since M. tuberculosis is highly resistant to fosmidomycin and it has previously been reported that a single amino acid change in MurA confers resistance to the phosphonate antibiotic fosfomycin in this pathogen (8), experiments were undertaken to determine whether this is also the case for the structurally related fosmidomycin.
Recombinant Rv2870c efficiently catalyzed the conversion of DXP to MEP in the presence of NADPH and the reverse reaction in the presence of NADP+. The enzymatic activity was dependent on the presence of Mg2+ ions and was abolished by the addition of EDTA under the standard reaction conditions. Interestingly, no other divalent cation tested, including Mn2+, Ca2+, Co2+, Zn2+, or Fe2+, was able to support the activity. This is unlike initial studies, where E. coli IspC was reported to prefer Mn2+ (33) or Co2+ (17), and subsequent studies found that Mg2+, Mn2+, and Co2+ were essentially equally effective as cofactors for recombinant IspC from E. coli, with Mg2+ being the probable normal cofactor in vivo (13). The enzyme from M. tuberculosis also exhibits a narrow range of optimal activity between pH 7.5 and 7.9. Enzymatic activity dropped dramatically between pH 7.9 and pH 8.0 in two buffer systems; the reason for this is not clear.
The KmDXP calculated from the data presented here is consistent with values reported for the enzyme from E. coli (99 μM [17] or 175 μM [13]), Z. mobilis (300 μM [10]), and S. coelicolor (190 μM [7]). The KmNADPH reported here is also similar to the values previously published for IspC from E. coli (0.5 to 1.0 μM [13] and 7.4 to 18 μM [17]), and the specificity constant of Rv2780c in the forward direction is similar to the value of 2.4 × 106 M−1 min−1 reported for recombinant IspC from E. coli (17). Koppisch et al. (13) appear to be the only other group to have determined Km values for substrates in the reverse reaction, and their value for KmMEP (390 μM) is reasonably close to the KmMEP calculated from the data presented here, although the value they calculated for KmNADP+ (30 μM) was approximately 18-fold lower than the value calculated from the Rv2870c data. Thus, it appears that recombinant Rv2870c and recombinant IspC from E. coli have similar kinetic constants, with the greatest difference being divalent cation requirements.
Having established the enzymatic characteristics of Rv2870c, it was possible to evaluate the protein for sensitivity to fosmidomycin. The experiments presented here show that Rv2870c is inhibited by the antibiotic at concentrations that are comparable to other DXP reductoisomerases, including those from A. thaliana (IC50, 280 nM [24]), barley (IC50, 700 nM [24]), P. aeruginosa (IC50, 150 nM [3]), and Z. mobilis (Ki, 600 nM [10]). IspC isolated from the apicomplexan P. falciparum appears to be significantly more sensitive to fosmidomycin, with a reported IC50 of 28 nM (11). Ki values for recombinant IspC from E. coli have been reported between 215 nM and 21 nM, depending on whether calculations are based on initial or final reaction velocity (13). Thus, M. tuberculosis is resistant to concentrations of fosmidomycin that are at least 3 orders of magnitude greater than the IC50 determined in vitro for Rv2870c, and resistance cannot be attributed to intrinsic properties of the enzyme itself.
There is circumstantial evidence that mycobacterial resistance to fosmidomycin may derive from limited uptake of the antibiotic. In E. coli, fosmidomycin is transported into the bacterium via the glycerol 3-phosphate transporter GlpT (28); glpT mutants are resistant to both fosmidomycin and fosfomycin (which are both phosphonate-containing antibiotics). In addition, when 80 fosmidomycin-resistant strains of P. aeruginosa were isolated, all were found to be deficient in GlpT activity (12). BLAST searches of the M. tuberculosis genome do not reveal any genes encoding proteins with significant similarity in primary structure to GlpT, supporting the hypothesis that mycobacterial resistance to fosmidomycin and, to some extent, fosfomycin, could be due to inefficient uptake. It is possible that efflux pumps and/or enzymes capable of drug modification also contribute to the observed resistance, as both categories of enzymes have been reported in M. tuberculosis (2, 34). An efflux pump (Fsr), capable of conferring fosmidomycin resistance when amplified, has been reported in E. coli (9). However, fsr was discovered as the result of efforts to find a gene related to fosmidomycin action other than glpT (9). So far, attempts to express E. coli glpT in Mycobacterium smegmatis generating a fosmidomycin-sensitive mycobacterium have been unsuccessful, as the protein was not expressed at detectable levels in the heterologous host (data not shown).
In order to confirm that Rv2870c is not intrinsically resistant to fosmidomycin, Rv2870c was expressed in a heterologous gram-negative host, known to be sensitive to the antibiotic, which had been engineered such that the chromosomal copy of ispC was inactivated, making the bacterial MEP pathway dependent on either exogenously supplied ME or a plasmid bearing a gene encoding an MEP synthase. Since ME phosphorylation is independent of the MEP pathway, its use in the growth medium renders the bacterium insensitive to fosmidomycin (35). Transformation with a plasmid bearing Rv2870c was able to alleviate the auxotrophy, and when the resulting strain was grown under conditions requiring the expression of the enzyme, the new strain was sensitive to fosmidomycin, confirming that Rv2870c encodes a MEP synthase and that the mycobacterial enzyme does not confer resistance to the antibiotic. Thus, the data are consistent with IspC being an attractive target for the development of novel drugs against tuberculosis.
Acknowledgments
This work was supported by a grant (AI49151) from NIAID, NIH, and a program project (AI46393) from the National Cooperative Drug Discovery Group, Opportunistic Infections in AIDS (NCDDG-OI), NIAID, NIH.
REFERENCES
- 1.Adam, P., S. Hecht, W. G. Eisenreich, J. Kaiser, T. Grawert, D. Arigoni, A. Bacher, and F. Rohdich. 2002. Biosynthesis of terpenes: studies on 1-hydroxy-2-methyl-2-(E)-butenyl 4-diphosphate reductase. Proc. Natl. Acad. Sci. USA 99:12108-12113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ainsa, J. A., E. Perez, V. Pelicic, F. X. Berthet, B. Gicquel, and C. Martin. 1997. Aminoglycoside 2′-N-acetyltransferase genes are universally present in mycobacteria: characterization of the aac(2′)-lc gene from Mycobacterium tuberculosis and the aac(2′)-ld gene from Mycobacterium smegmatis. Mol. Microbiol. 24:431-441. [DOI] [PubMed] [Google Scholar]
- 3.Altincicek, B., M. Hintz, S. Sanderbrand, J. Wiesner, E. Beck, and H. Jomaa. 2000. Tools for discovery of inhibitors of the 1-deoxy-D-xylulose 5-phosphate (DXP) synthase and DXP reductoisomerase: an approach with enzymes from the pathogenic bacterium Pseudomonas aeruginosa. FEMS Microbiol. Lett. 190:329-333. [DOI] [PubMed] [Google Scholar]
- 4.Bailey, A. M., S. Mahapatra, P. J. Brennan, and D. C. Crick. 2000. Purification and characterization of 1-deoxyxylulose 5-phosphate synthase from Mycobacterium tuberculosis. Glycobiology 10:46. [DOI] [PubMed] [Google Scholar]
- 5.Bailey, A. M., S. Mahapatra, P. J. Brennan, and D. C. Crick. 2002. Identification, cloning, purification, and enzymatic characterization of Mycobacterium tuberculosis 1-deoxy-D-xylulose 5-phosphate synthase. Glycobiology 12:813-820. [DOI] [PubMed] [Google Scholar]
- 6.Boucher, Y., and W. F. Doolittle. 2000. The role of lateral gene transfer in the evolution of isoprenoid biosynthesis pathways. Mol. Microbiol. 37:703-716. [DOI] [PubMed] [Google Scholar]
- 7.Cane, D. E., C. Chow, A. Lillo, and I. Kang. 2001. Molecular cloning, expression and characterization of the first three genes in the mevalonate-independent isoprenoid pathway in Streptomyces coelicolor. Bioorg. Med. Chem. 9:1467-1477. [DOI] [PubMed] [Google Scholar]
- 8.De Smet, K. A., K. E. Kempsell, A. Gallagher, K. Duncan, and D. B. Young. 1999. Alteration of a single amino acid residue reverses fosfomycin resistance of recombinant MurA from Mycobacterium tuberculosis. Microbiology 145:3177-3184. [DOI] [PubMed] [Google Scholar]
- 9.Fujisaki, S., S. Ohnuma, T. Horiuchi, I. Takahashi, S. Tsukui, Y. Nishimura, T. Nishino, M. Kitabatake, and H. Inokuchi. 1996. Cloning of a gene from Escherichia coli that confers resistance to fosmidomycin as a consequence of amplification. Gene 175:83-87. [DOI] [PubMed] [Google Scholar]
- 10.Grolle, S., S. Bringer-Meyer, and H. Sahm. 2000. Isolation of the dxr gene of Zymomonas mobilis and characterization of the 1-deoxy-D-xylulose 5-phosphate reductoisomerase. FEMS Microbiol. Lett. 191:131-137. [DOI] [PubMed] [Google Scholar]
- 11.Jomaa, H., J. Wiesner, S. Sanderbrand, B. Altincicek, C. Weidemeyer, M. Hintz, I. Turbachova, M. Eberl, J. Zeidler, H. K. Lichtenthaler, D. Soldati, and E. Beck. 1999. Inhibitors of the nonmevalonate pathway of isoprenoid biosynthesis as antimalarial drugs. Science 285:1573-1576. [DOI] [PubMed] [Google Scholar]
- 12.Kojo, H., Y. Shigi, and M. Nishida. 1980. Fr-31564, a new phosphonic acid antibiotic—bacterial resistance and membrane permeability. J. Antibiot. (Tokyo) 33:44-48. [DOI] [PubMed] [Google Scholar]
- 13.Koppisch, A. T., D. T. Fox, B. S. Blagg, and C. D. Poulter. 2002. E. coli MEP synthase: steady-state kinetic analysis and substrate binding. Biochemistry 41:236-243. [DOI] [PubMed] [Google Scholar]
- 14.Kuemmerle, H. P., T. Murakawa, H. Sakamoto, N. Sato, T. Konishi, and F. De Santis. 1985. Fosmidomycin, a new phosphonic acid antibiotic. Part II. 1. Human pharmacokinetics. 2. Preliminary early phase IIa clinical studies. Int. J. Clin. Pharmacol. Ther. Toxicol. 23:521-528. [PubMed] [Google Scholar]
- 15.Kuemmerle, H. P., T. Murakawa, K. Soneoka, and T. Konishi. 1985. Fosmidomycin: a new phosphonic acid antibiotic. Part I: phase I tolerance studies. Int. J. Clin. Pharmacol. Ther. Toxicol. 23:515-520. [PubMed] [Google Scholar]
- 16.Kuzuyama, T., T. Shimizu, S. Takahashi, and H. Seto. 1998. Fosmidomycin, a specific inhibitor of 1-deoxy-D-xylulose 5-phosphate reductoisomerase in the nonmevalonate pathway for terpenoid biosynthesis. Tetrahedron Lett. 39:7913-7916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuzuyama, T., S. Takahashi, M. Takagi, and H. Seto. 2000. Characterization of 1-deoxy-D-xylulose 5-phosphate reductoisomerase, an enzyme involved in isopentenyl diphosphate biosynthesis, and identification of its catalytic amino acid residues. J. Biol. Chem. 275:19928-19932. [DOI] [PubMed] [Google Scholar]
- 18.Lange, B. M., and R. Croteau. 1999. Isoprenoid biosynthesis via a mevalonate-independent pathway in plants: cloning and heterologous expression of 1-deoxy-D-xylulose-5-phosphate reductoisomerase from peppermint. Arch. Biochem. Biophys. 365:170-174. [DOI] [PubMed] [Google Scholar]
- 19.Mahapatra, S., T. Yagi, J. T. Belisle, B. J. Espinosa, P. J. Hill, M. R. McNeil, P. J. Brennan, and D. C. Crick. 2005. Mycobacterial lipid II is composed of a complex mixture of modified muramyl and peptide moieties linked to decaprenyl phosphate. J. Bacteriol. 187:2747-2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mikusova, K., M. Mikus, G. S. Besra, I. Hancock, and P. J. Brennan. 1996. Biosynthesis of the linkage region of the mycobacterial cell wall. J. Biol. Chem. 271:7820-7828. [DOI] [PubMed] [Google Scholar]
- 21.Miller, B., T. Heuser, and W. Zimmer. 1999. A Synechococcus leopoliensis SAUG 1402-1 operon harboring the 1-deoxyxylulose 5-phosphate synthase gene and two additional open reading frames is functionally involved in the dimethylallyl diphosphate synthesis. FEBS Lett. 460:485-490. [DOI] [PubMed] [Google Scholar]
- 22.Mine, Y., T. Kamimura, S. Nonoyama, M. Nishida, S. Goto, and S. Kuwahara. 1980. In vitro and in vivo antibacterial activities of FR-31564, a new phosphonic acid antibiotic. J. Antibiot. (Tokyo) 33:36-43. [DOI] [PubMed] [Google Scholar]
- 23.Minnikin, D. E. 1982. Lipids: complex lipids, their chemistry, biosynthesis and roles, p. 95-184. In C. Ratledge and J. Stanford (ed.), The biology of mycobacteria. Academic Press, London, England.
- 24.Mueller, C., J. Schwender, J. Zeidler, and H. K. Lichtenthaler. 2000. Properties and inhibition of the first two enzymes of the non-mevalonate pathway of isoprenoid biosynthesis. Biochem. Soc. Trans. 28:792-793. [PubMed] [Google Scholar]
- 25.Neu, H. C., and T. Kamimura. 1981. In vitro and in vivo antibacterial activity of FR-31564, a phosphonic acid antimicrobial agent. Antimicrob. Agents Chemother. 19:1013-1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Proteau, P. J., Y. H. Woo, R. T. Williamson, and C. Phaosiri. 1999. Stereochemistry of the reduction step mediated by recombinant 1-deoxy-D-xylulose 5-phosphate isomeroreductase. Org. Lett. 1:921-923. [DOI] [PubMed] [Google Scholar]
- 27.Putra, S. R., A. Disch, J. M. Bravo, and M. Rohmer. 1998. Distribution of mevalonate and glyceraldehyde 3-phosphate/pyruvate routes for isoprenoid biosynthesis in some gram-negative bacteria and mycobacteria. FEMS Microbiol. Lett. 164:169-175. [DOI] [PubMed] [Google Scholar]
- 28.Sakamoto, Y., S. Furukawa, H. Ogihara, and M. Yamasaki. 2003. Fosmidomycin resistance in adenylate cyclase deficient (cya) mutants of Escherichia coli. Biosci. Biotechnol. Biochem. 67:2030-2033. [DOI] [PubMed] [Google Scholar]
- 29.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 30.Schwender, J., C. Muller, J. Zeidler, and H. K. Lichtenthaler. 1999. Cloning and heterologous expression of a cDNA encoding 1-deoxy-D-xylulose-5-phosphate reductoisomerase of Arabidopsis thaliana. FEBS Lett. 455:140-144. [DOI] [PubMed] [Google Scholar]
- 31.Shigi, Y. 1989. Inhibition of bacterial isoprenoid synthesis by fosmidomycin, a phosphonic acid-containing antibiotic. J. Antimicrob. Chemother. 24:131-145. [DOI] [PubMed] [Google Scholar]
- 32.Takagi, M., T. Kuzuyama, S. Takahashi, and H. Seto. 2000. A gene cluster for the mevalonate pathway from Streptomyces sp. strain CL190. J. Bacteriol. 182:4153-4157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Takahashi, S., T. Kuzuyama, H. Watanabe, and H. Seto. 1998. A 1-deoxy-D-xylulose 5-phosphate reductoisomerase catalyzing the formation of 2-C-methyl-D-erythritol 4-phosphate in an alternative nonmevalonate pathway for terpenoid biosynthesis. Proc. Natl. Acad. Sci. USA 95:9879-9884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Takiff, H. E., M. Cimino, M. C. Musso, T. Weisbrod, R. Martinez, M. B. Delgado, L. Salazar, B. R. Bloom, and W. R. Jacobs. 1996. Efflux pump of the proton antiporter family confers low-level fluoroquinolone resistance in Mycobacterium smegmatis. Proc. Natl. Acad. Sci. USA 93:362-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Testa, C. A., R. A. Cornish, and C. D. Poulter. 2004. The sorbitol phosphotransferase system is responsible for transport of 2-C-methyl-d-erythritol into Salmonella enterica serovar Typhimurium. J. Bacteriol. 186:473-480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wolucka, B. A., M. R. McNeil, E. de Hoffmann, T. Chojnacki, and P. J. Brennan. 1994. Recognition of the lipid intermediate for arabinogalactan/arabinomannan biosynthesis and its relation to the mode of action of ethambutol on mycobacteria. J. Biol. Chem. 269:23328-23335. [PubMed] [Google Scholar]
- 37.Yokota, Y., T. Murakawa, and M. Nishida. 1981. In vitro synergism of Fr-31564, a new phosphonic acid antibiotic. J. Antibiot. (Tokyo) 34:876-883. [DOI] [PubMed] [Google Scholar]
- 38.Zeidler, J., J. Schwender, C. Muller, J. Wiesner, C. Weidemeyer, E. Beck, H. Jomaa, and H. K. Lichtenthaler. 1998. Inhibition of the non-mevalonate 1-deoxy-D-xylulose-5-phosphate pathway of plant isoprenoid biosynthesis by fosmidomycin. Zeitsch. Naturforsch. C 53:980-986. [Google Scholar]