Abstract
Bacteriophage K1F specifically infects Escherichia coli strains that produce the K1 polysaccharide capsule. Like several other K1 capsule-specific phages, K1F encodes an endo-neuraminidase (endosialidase) that is part of the tail structure which allows the phage to recognize and degrade the polysaccharide capsule. The complete nucleotide sequence of the K1F genome reveals that it is closely related to bacteriophage T7 in both genome organization and sequence similarity. The most striking difference between the two phages is that K1F encodes the endosialidase in the analogous position to the T7 tail fiber gene. This is in contrast with bacteriophage K1-5, another K1-specific phage, which encodes a very similar endosialidase which is part of a tail gene “module” at the end of the phage genome. It appears that diverse phages have acquired endosialidase genes by horizontal gene transfer and that these genes or gene products have adapted to different genome and virion architectures.
Many strains of Escherichia coli produce the K1 antigen, which is a thick polysaccharide capsule composed mainly of α-2,8-linked polysialic acid (46). This capsule contributes to pathogenicity by allowing the bacteria to evade the immune system and cross barriers (i.e., the blood-brain barrier). The K1 capsule may also act as a defense against certain phages, such as T7, which recognizes structures beneath the capsule, by physically blocking adsorption (43). Some phages, on the other hand, require the K1 capsule for infection (19). Such phages typically possess virion-bound endo-neuraminidase (endosialidase) that degrades the capsule by cleaving the α-2,8 bond, allowing the phage access to the bacterial surface. Phage-encoded endosialidase genes that have been cloned and sequenced include K1E (28), K1-5 (41), 63D (30), and K1F (37), all of which share significant sequence similarity to one another.
Analysis of several phage genome sequences has led to the classification of the T7 supergroup, the members of which have common morphological, biological, and genomic characteristics (20; for a review on the T7 group, see reference 32). T7 supergroup phages in which the genomes have been sequenced include coliphages T7 (13), T3 (34), and K1-5 (42); yersiniophages φA1122 (16) and φYe03-12 (35); salmonellaphage SP6 (12, 42); cyanophage P60 (4); vibriophage VpV262 (20); Pseudomonas phages gh-1 (25) and KMV (26); and roseophage SI01 (40). Also, the genome sequence of Pseudomonas putida KT2440 revealed a prophage with considerable sequence similarity to members of the T7 supergroup, although it is not known if this phage is still functional (52). It has become apparent that within the T7 supergroup there are subgroups of phages that are more similar to each other than to other T7 supergroup members. One of these subgroups contains those that are very similar to T7 itself and includes T3, φA1122, φYe03-12, and the slightly more distant gh-1. Another subgroup includes salmonellaphage SP6 and K1-5, which can infect either K1 or K5 E. coli strains (42). The SP6 subgroup has diverged significantly in sequence similarity from the T7 subgroup and has somewhat different genome organization, but nevertheless the members of this group probably share similarities in replication to T7. Unlike the T7 subgroup, SP6 and K1-5 have a tail gene module at the end of the linear genome that encodes capsule-degrading tail proteins. In K1-5, one of these genes is the endosialidase that is responsible for degrading the K1 capsule. Members of the T7 subgroup, on the other hand, encode tail fibers that simply recognize host lipopolysaccharide and do not have any known capsule-degrading activity.
K1F was first isolated from sewage in 1984 (51) and was used to identify polysialic acid in neuronal membranes as well as for detecting the K1 antigen in E. coli (53). K1F encodes a phage-bound endosialidase that was shown to be part of the tail structure of the virion (37). The endosialidase is autocatalytically truncated, in which 152 amino acids are cleaved from the C terminus to generate active enzyme (33). The enzyme has also been shown to form trimers, which seem to be common among phage tail proteins, including the T7 tail fiber (48). Other than its host range, little else is known about the biology of K1F. In this work, we determined the complete nucleotide sequence of the K1F genome and found that it is another member of the T7 supergroup. Unlike K1-5, K1F is more closely related to T7, T3, φA1122, φYeO3-12, and gh-l, and we therefore assign it as a member of the T7 subgroup and not the SP6 subgroup. Also unlike K1-5, the K1F endosialidase is not encoded in a module at the end of the genome but is located in the analogous position to the T7 tail fiber. The K1F endosialidase has an N-terminal head-binding domain with similarity to the T7 tail fiber. It appears that K1F evolved from a close T7 ancestor in which the C-terminal portion of the tail fiber was replaced by an endosialidase domain, allowing it to replicate on K1 strains of E. coli.
The K1F genome.
The K1F genome sequence was determined by a combination of shotgun sequencing and primer walking (Fig. 1). Open reading frames (ORFs) were determined using a combination of visual inspection and translated BLAST searches (1) and by analysis of the predicted translated sequence using GCG-lite (11). The genome consists of a single double-stranded DNA molecule of 39,704 bp with a GC content of 50%. We have annotated 42 open reading frames (see Table S1 in the supplemental material), which are all transcribed on one strand of the DNA molecule. The K1F genome is flanked by terminal repeats of 179 bp. It is a tightly packed genome with 88% of the sequence predicted to encode proteins. We use the same gene-numbering system, starting from left to right on the genetic map, as that of T7 since the two genomes are similar in both organization and sequence. ORFs that are not present, or have no sequence similarity, to a previously characterized T7-like phage are simply named by the amino acid length (i.e., orf156). Like T7, the genome can be divided into three regions: early, middle, and late.
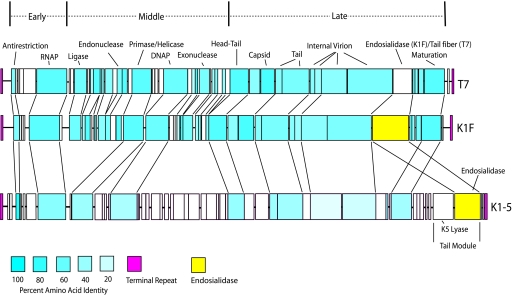
FIG. 1.
ORF-by-ORF comparison of the genomes of T7, K1F, and K1-5. The blue shading indicates the percent amino acid identity of the open reading frames. Open reading frames throughout the K1F genome show considerably more sequence similarity to T7 than to K1-5. The exception is the endosialidase gene (shown in yellow), which is highly similar to that of K1-. K1-5 and SP6 are members of two different phage subgroups that have both acquired an endosialidase gene allowing them to replicate on K1 strains.
The first gene predicted to be transcribed on the K1F genome is 0.3, which is likely to be involved in antirestriction (49). Other than the endosialidase (see below), this the only open reading frame carried by K1F that is more similar to phages K1-5 and SP6 than to T7 and could reflect differences in the host strains that they infect. Characteristic of the T7-like phages, K1F encodes an RNA polymerase (RNAP; 1.0) which is responsible for transcription of most of the phage genes but also is involved in other functions, such as translocating phage DNA into the cell (17), as well as DNA replication, maturation, and packaging (55). A DNA ligase gene analogous to that of T7, 1.3, is present and represents the end of the early genes. K1F carries fewer genes in the early region and is missing equivalents to T7 genes 0.4, 0.5, 0.7, and 1.2.
The middle region encodes mainly proteins involved in DNA metabolism and includes a host RNA polymerase inhibitor (2.0), single-stranded DNA binding protein (2.5), endonuclease (3.0), lysozyme (3.5), primase/helicase (4.0), DNA polymerase (DNAP; 5.0), and exonuclease (6.0). All are closely related to one of the close-knit T7 family members, including T7, T3, and YeO3-12. This region carries several smaller putative T7-like genes, including 1.6, 1.7, 1.8, 5.5, 5.7, and 6.7; the function of these ORFs is unknown. ORFs 50, 156, 71, 50, 57, and 122 have no significant sequence similarity to anything in the databases.
Several bacterium and bacteriophage genomes have been found to contain group I introns, including phages phiI and W31, which are possibly members of the T7 supergroup (2). Both have a group I intron inserted 156 bp from the end of the DNAP gene. K1F also contains a putative group I intron within its DNAP gene, also 156 bp from the end (positions 14937 to 15534 on the genome). Like phi and W31, the intron encodes a 131-amino-acid homing endonuclease very similar (77% identity) to I-TsII (for a review, see reference 18). T3, T7, and φYeO3-12 also contain ORFs with similarity to homing endonucleases (34); however, these do not appear to be part of introns and are located in intergenic regions.
The late region of the T7 family encodes the virion structural proteins as well as many of the proteins involved in maturation and cell lysis. The organization of the K1F structural genes is nearly identical to that of T7. The first ORF is the head-tail connector (8.0), followed by those coding for the scaffolding protein (9.0), capsid (10.0), tail tube (11.0 and 12.0), internal virion proteins (13.0 to 16.0), and the tail protein (17.0; see below). Following the genes coding for the major structural proteins are those involved in lysis and maturation, which include genes 17.5, 18, 18.5, and 19.0.
Perhaps the most striking difference between K1F and T7 is the tail protein, gp17. The gene coding for this product in T7, T3, φA1122, and φYeO3-12 encodes the tail fiber protein which is involved in recognition and adsorption to the host LPS. The K1F counterpart, however, has only a small region of amino acid similarity to the T7 tail fiber at the N-terminal head-binding portion (37). The central catalytic region is highly similar to those of other phage endosialidases (57% amino acid identity to 63D, 64% to CUS-3, and 80% to K1-5/K1E) which are involved in both recognition and depolymerization of the K1 polysaccharide capsule.
The initial report of the cloning of the endosialidase gene showed a length of 2,763 bp (37). However a sequencing error was discovered, and it was found that the actual gene length is 3,195 bp and that the C terminus of the protein product is postranslationally cleaved (33). Our results confirm a gene length of 3,195 bp.
Like most of the members of the closely related T7 family, K1F is flanked by terminal repeats suggesting similar replication strategies. Immediately following the left terminal repeat is an equivalent to the transcriptional termination site known as the CJ terminator (bases 179 to 186), where RNAP, complexed with lysozyme, pauses at the concatemer junction during maturation and packaging (29, 54, 55).
Most gene transcription among T7 supergroup phages is driven by phage-encoded RNAPs which typically recognize very specific promoter sequences. K1F encodes an RNAP similar to that of T7 (64% amino acid identity), so we predicted that K1F might have a similar promoter consensus sequence. By mainly visual analysis, we were able to identify 10 putative promoters, most of which have T7 homologues (see Table S2 in the supplemental material). The consensus sequence from −8 to +2 is identical to that of T7. However bases −9 through −12, which are important for promoter specificity, differ. In T7 any change from the consensus C at −9 results in an inactive promoter (21). Since K1F has a T at this position, it is unlikely that the T7 RNA polymerase will initiate transcription from a K1F promoter and it is likely that the two RNAPs have different promoter specificities.
This exlusivity of promoter specificity seems to be the trend among the T7-like phages (24, 42, 45), and it seems that there must be some selective pressure that resulted in this feature.
K1F appears to have fewer phage promoters throughout the middle region of its genome and is missing equivalents to T7 promoters φ1.1a, φ1.1b, φ3.8, φ4c, φ4.3, and φ4.7. (see Fig. S2 in the supplemental material). The late region of K1F appears to have all of the T7 analogues. We identified a φOR promoter in K1F but not a φOL promoter. φOR is involved in packaging (6, 7) and possibly replication (39). A phage-specific promoter was found immediately upstream of the RNA polymerase gene in K1F, suggesting that there is some autoregulation of this gene; phage gh-1 is the only other T7-like phage to have a phage-specific promoter in this region.
The T7 genome contains three strong σ70 promoters responsible for early gene transcription, including phage RNA polymerase. We have annotated one such promoter in K1F (positions 924 to 951).
The end of transcription of the early genes in T7 is marked by a rho-independent terminator (TE) immediately following the ligase gene (23). K1F has a sequence in the equivalent position (7128 to 7161) characteristic of such terminators that could serve this function. Efficient termination of host RNAP in T7 requires gene 0.7, which encodes a kinase that phosphorylates E. coli RNAP (38). Since K1F lacks this gene, we have to assume that this terminator alone is enough to end transcription. K1F also has a potential analogue to the T7 Tφ terminator (positions 22314 to 22352) which terminates transcription of phage RNAP.
The primary origin of replication in T7 is downstream of the φ1.1a and φ1.1b promoters, which are used by T7 RNAP to initiate replication of the leading strand (14, 15). K1F has a relatively large noncoding region in the analogous position on the genome but does not have analogues to promoters φ1.1a and φ1.1b. However K1F promoter φ1.3 is actually in a position such that it could serve both to initiate replication as well as to initiate transcription of downstream gene(s). In addition, it is known that T7 does not require the primary origin for growth, and replication can also be initiated from secondary origins associated with promoters φ6.5, φ13, and φOR (39). These are all present in K1F, and any one of these could potentially serve as the primary origin.
Conclusions.
Double-stranded DNA phages are known to have mosaic genome structures (3, 22, 36) that are composed of functional modules which can be genes, clusters of genes, or domains within a gene that are acquired through horizontal transfer. Such mosaicism is evident in all of the members of the T7 supergroup. This makes classification of these phages particularly difficult based on classical criteria (27). However, based on overall genome organization and sequence similarity, it is clear that K1F is more closely related to T7, T3, φA1122, φYe03-12, and gh-1 within the larger T7 supergroup. K1-5, on the other hand, has a somewhat different genome organization and has more sequence similarity to SP6, and they together make up their own subgroup within the T7 supergroup. There are just two genes in K1F that show more sequence similarity to K1-5 than to any member of the T7 subgroup: one is gene 0.3, and the other, discussed below, is the gene coding for endosialidase.
Like all K1-specific phages studied at the molecular level, K1F encodes a tail-associated endosialidase, a key determinant in host specificity that allows the phages to penetrate the polysaccharide capsule. The K1F endosialidase shares sequence similarity to the N-terminal portion of the T7 tail fiber protein, the region that is responsible for attachment to the virion, and it appears that K1F arose from a T7-like ancestor in which an endosialidase gene became fused to a portion of the tail fiber. Another phage-encoded endosialidase, that of phage 63D, which possesses a longer tail structure and is probably not a member of the T7 supergroup (30), has an N-terminal domain with sequence similarity to the minor structural protein of salmonellaphage MB78 (9), which perhaps could play the role connecting the endosialidase to the head. Genome sequencing of E. coli K1 strain RS218 has uncovered yet another probable K1-specific prophage, CUS-3, which is also not a member of the T7 supergroup but is more similar to the lysogenic phages HK620 and P22 (10). It is not known if this phage is still able to go through a lytic cycle, but CUS-3 does encode an endosialidase which shares sequence similarity to the central portion of the endosialidases described above. The N-terminal 120-amino-acid sequence of the CUS-3 endosialidase is nearly identical (90% amino acid identity) to the head-binding domain of the HK620 tailspike (8) and the well-studied p22 tailspike (47) as well as the head-binding domains of Sf6 (5), ST64T (31), and APSE-1 (50). Again, the fusion of an endosialidase gene to a portion of a tail gene appears to have occurred in an ancestor of CUS-3. The K1-5 endosialidase differs from the above three in that it has no head-binding domain at all and is possibly linked to the head by separate polypeptide, a feature that may allow the phage to fit two different tail proteins to its head (42). It appears that phage-encoded endosialidases have been adapted to a variety of phage structural architectures, particularly with regard to the mechanisn in which they are linked to the virion.
The genomes of several members of the T7 supergroup members have now been sequenced, and a clearer picture of how these viruses evolved is coming into view. However the pool of phages examined so far is somewhat skewed toward those that infect laboratory strains, which often have no capsule or a greatly reduced amount. There are many potential T7-like phages that infect a wide range of hosts that produce chemically different capsules (44). Further work needs to be done to determine how these phages have evolved to infect diverse bacteria.
Nucleotide sequence accession number.
The GenBank accession number for the K1F genome is DQ111067.
Supplementary Material
Acknowledgments
This research was supported by the Intramural Research Program of the National Institute of Mental Health, National Institutes of Health.
We thank Eric Vimr for supplying strains and Ian Molineux and Sankar Adhya for helpful discussions.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Bonocora, R. P., and D. A. Shub. 2004. A self-splicing group I intron in DNA polymerase genes of T7-like bacteriophages. J. Bacteriol. 186:8153-8155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Botstein, D. 1980. A theory of modular evolution for bacteriophages. Ann. N. Y. Acad. Sci. 354:484-490. [DOI] [PubMed] [Google Scholar]
- 4.Chen, F., and J. Lu. 2002. Genomic sequence and evolution of marine cyanophage P60: a new insight on lytic and lysogenic phages. Appl. Environ. Microbiol. 68:2589-2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chua, J. E. H., P. A. Manning, and R. Morona. 1999. The Shigella flexneri bacteriophage Sf6 tailspike protein (TSP)/endorhamnosidase is related to the bacteriophage P22 TSP and has a motif common to exo- and endoglycanases, and C-5 epimerases. Microbiology 145:1649-1659. [DOI] [PubMed] [Google Scholar]
- 6.Chung, Y.-B., and D. C. Hinkle. 1990. Bacteriophage T7 DNA packaging. I. Plasmids containing a T7 replication origin and the T7 concatemer junction are packaged into transducing particles during phage infection. J. Mol. Biol. 216:911-926. [DOI] [PubMed] [Google Scholar]
- 7.Chung, Y.-B., and D. C. Hinkle. 1990. Bacteriophage T7 DNA packaging. II. Analysis of the DNA sequences required for packaging using a plasmid transduction assay. J. Mol. Biol. 216:927-938. [DOI] [PubMed] [Google Scholar]
- 8.Clark, A. J., W. Inwood, T. Cloutier, and T. S. Dhillon. 2001. Nucleotide sequence of coliphage HK620 and the evolution of lambdoid phages. J. Mol. Biol. 311:657-679. [DOI] [PubMed] [Google Scholar]
- 9.Datta, P., P. Mallik, A. N. Ghosh, and M. Chakravorty. 2005. Temperature sensitive mutation in the 38 kDa minor structural protein gene of phage MB78 interferes with phage morphogenesis. Virus Genes 30:197-207. [DOI] [PubMed] [Google Scholar]
- 10.Deszo, E. L., S. M. Steenbergen, D. I. Freedberg, and E. R. Vimr. 2005. Escherichia coli K1 polysialic acid O-acetyltransferase gene, neuO, and the mechanism of capsule form variation involving a mobile contingency locus. Proc. Natl. Acad. Sci. USA 102:5564-5569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Devereux, J., P. Haeberli, and O. Smithies. 1984. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 12:387-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dobbins, A. T., M. George, Jr., D. A. Basham, M. E. Ford, J. M. Houtz, M. L. Pedulla, J. G. Lawrence, G. F. Hatfull, and R. W. Hendrix. 2004. Complete genomic sequence of the virulent Salmonella bacteriophage SP6. J. Bacteriol. 186:1933-1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dunn, J. J., and F. W. Studier. 1983. Complete nucleotide sequence of bacteriophage T7 DNA and the locations of T7 genetic elements. J. Mol. Biol. 166:477-535. [DOI] [PubMed] [Google Scholar]
- 14.Fuller, C. W., and C. C. Richardson. 1985. Initiation of DNA replication at the primary origin of bacteriophage T7 by purified proteins. Site and direction of initial DNA synthesis. J. Biol. Chem. 260:3185-3196. [PubMed] [Google Scholar]
- 15.Fuller, C. W., and C. C. Richardson. 1985. Initiation of DNA replication at the primary origin of bacteriophage T7 by purified proteins. Initiation of bidirectional synthesis. J. Biol. Chem. 260:3197-3206. [PubMed] [Google Scholar]
- 16.Garcia, E., J. M. Elliott, E. Ramanculov, P. S. G. Chain, M. C. Chu, and I. J. Molineux. 2003. The genome sequence of Yersinia pestis bacteriophage φA1122 reveals an intimate history with the coliphage T3 and T7 genomes. J. Bacteriol. 185:5248-5262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garcia, L. R., and I. J. Molineux. 1995. Rate of translocation of bacteriophage T7 DNA across the membranes of Escherichia coli. J. Bacteriol. 177:4066-4076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gimble, F. S. 2000. Invasion of a multitude of genetic niches by mobile endonuclease genes. FEMS Microbiol. Lett. 185:99-107. [DOI] [PubMed] [Google Scholar]
- 19.Gross, R. J., T. Cheasty, and B. Rowe. 1977. Isolation of bacteriophages specific for the K1 polysaccharide antigen of Escherichia coli. J. Clin. Microbiol. 6:548-550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hardies, S. C., A. M. Comeau, P. Serwer, and C. A. Suttle. 2003. The complete sequence of marine bacteriophage VpV262 infecting Vibrio parahaemolyticus indicates that an ancestral component of a T7 viral supergroup is widespread in the marine environment. Virology 310:359-371. [DOI] [PubMed] [Google Scholar]
- 21.Imburgio, D., M. Rong, K. Ma, and W. T. McAllister. 2000. Studies of promoter recognition and start site selection by T7 RNA polymerase using a comprehensive collection of promoter variants. Biochemistry 39:10419-10430. [DOI] [PubMed] [Google Scholar]
- 22.Juhala, R. J., M. E. Ford, R. L. Duda, A. Youlton, G. F. Hatfull, and R. W. Hendrix. 2000. Genomic sequences of bacteriophages HK97 and HK022: pervasive genetic mosaicism in the lambdoid bacteriophages. J. Mol. Biol. 299:27-51. [DOI] [PubMed] [Google Scholar]
- 23.Kieffer, M., N. Neff, and M. J. Chamberlin. 1977. Transcriptional termination at the end of the early region of bacteriophages T3 and T7 is not affected by polarity suppressors. J. Virol. 22:548-552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Klement, J. F., M. B. Moorefield, E. Jorgensen, J. E. Brown, S. Risman, and W. T. McAllister. 1990. Discrimination between bacteriophage T3 and T7 promoters by the T3 and T7 RNA polymerases depends primarily upon a three base-pair region located 10 to 12 base-pairs upstream from the start site. J. Mol. Biol. 215:21-29. [DOI] [PubMed] [Google Scholar]
- 25.Kovalayova, I. V., and A. M. Kropinski. 2003. The complete genomic sequence of lytic bacteriophage gh-1 infecting Pseudomanas putida—evidence for close relationship to the T7 group. Virology 311:305-315. [DOI] [PubMed] [Google Scholar]
- 26.Lavigne, R., M. V. Bourkaltseva, J. Robben, N. N. Sykilinda, L. P. Kurochkina, B. Grymonprez, B. Jonckx, V. N. Krylov, V. V. Mesyanzhinov, and G. Volckaert. 2003. The genome of bacteriophage φKMV: a T7-like lytic phage infecting Pseudomonas aeruginosa. Virology 312:49-59. [DOI] [PubMed] [Google Scholar]
- 27.Lawrence, J. G., G. F. Hatfull, and R. W. Hendrix. 2002. Imbroglios of viral taxonomy: genetic exchange and failings of phonetic approaches. J. Bacteriol. 184:4891-4905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Long, G. S., J. M. Bryant, P. W. Taylor, and J. P. Luzio. 1995. Complete nucleotide sequence of the gene encoding bacteriophage E endosialidase: implications for K1E endosialidase structure and function. Biochem. J. 309:543-550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lyakhov, D. L., B. He, X. Zhang, F. W. Studier, J. J. Dunn, and W. T. Mcallister. 1998. Pausing and termination of bacteriophage T7 RNA polymerase. J. Mol. Biol. 26:28-40. [DOI] [PubMed] [Google Scholar]
- 30.Machida, Y., K. Miyake, K. Hattori, S. Yamamoto, M. Kawase, and S. Iijima. 2000. Structure and function of a novel coliphage-associated sialidase. FEMS Microbiol. Lett. 182:333-337. [DOI] [PubMed] [Google Scholar]
- 31.Mmolawa, P. T., H. Schmieger, C. P. Tucker, and M. W. Heuzenroeder. 2003. Genomic structure of the Salmonella enterica serovar Typhimurium DT 64 bacteriophage ST64T: evidence for modular genetic architecture. J. Bacteriol. 185:3473-3475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Molineux, I. J. 2005. The T7 group. In The bacteriophages, 2nd ed., in press. Oxford University Press, Oxford, United Kingdom.
- 33.Muhlenhoff, M., K. Stummeyer, M. Grove, M. Sauerborn, and R. Gerardy-Schahn. 2003. Proteolytic processing and oligomerization of bacteriophage-derived endosialidases. J. Biol. Chem. 278:12634-12644. [DOI] [PubMed] [Google Scholar]
- 34.Pajunen, M. I., M. R. Elizondo, M. Skurnik, J. Kieleczawa, and I. J. Molineux. 2002. Complete nucleotide sequence and likely recombinatorial origin of bacteriophage T3. J. Mol. Biol. 319:1115-1132. [DOI] [PubMed] [Google Scholar]
- 35.Pajunen, M. I., S. J. Kiljunen, M. E.-L. Söderholm, and M. Skurnik. 2001. Complete genomic sequence of the lytic bacteriophage φYeO3-12 of Yersinia enterocolitica serotype O:3. J. Bacteriol. 183:1928-1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pedulla, M. L., M. E. Ford, J. M. Houtz, T. Karthikeyan, C. Wadsworth, J. A. Lewis, D. Jacobs-Sera, J. Falbo, J. Gross, N. R. Pannunzio, W. Brucker, V. Kumar, J. Kandasamy, L. Keenan, S. Bardarov, J. Kriakov, J. G. Lawrence, W. R. Jacobs, Jr., R. W. Hendrix, and G. F. Hatfull. 2003. Origins of highly mosaic mycobacteriophage genomes. Cell 113:171-182. [DOI] [PubMed] [Google Scholar]
- 37.Petter, J. G., and E. R. Vimr. 1993. Complete nucleotide sequence of the bacteriophage K1F tail gene encoding endo-N-acylneuraminidase (endo-N) and comparison to an endo-N homolog in bacteriophage PK1E. J. Bacteriol. 175:4354-4363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pfennig-Yeh, M. L., H. Ponta, M. Hirsch-Kauffman, H. J. Rahmsdorf, P. Herrlich, and M. Schweiger. 1978. Early T7 gene expression: rates of RNA synthesis and degradation, protein kinase dependent termination of transcription, and efficiency of translation. Mol. Gen. Genet. 166:127-140. [DOI] [PubMed] [Google Scholar]
- 39.Rabkin, S. D., and C. C. Richardson. 1990. In vivo analysis of the initiation of bacteriophage T7 DNA replication. Virology 174:585-592. [DOI] [PubMed] [Google Scholar]
- 40.Rohwer, F., A. Segall, G. Steward, V. Seguritan, M. Breitbart, F. Wolven, and F. Azam. 2000. The complete genomic sequence of the marine phage Roseophage SIO1 shares homology with nonmarine phages. Limnol. Oceanogr. 45:408-418. [Google Scholar]
- 41.Scholl, D., S. Rogers, S. Adhya, and C. R. Merril. 2001. Bacteriophage K1-5 encodes two different tail fiber proteins, allowing it to infect and replicate on both K1 and K5 strains of Escherichia coli. J. Virol. 75:2509-2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Scholl, D., J. Kieleczawa, P. Kemp, J. Rush, C. C. Richardson, C. Merril, S. Adhya, and I. J. Molineux. 2004. Genomic analysis of bacteriophages SP6 and K1-5, an estranged subgroup of the T7 supergroup. J. Mol. Biol. 335:1151-1171. [DOI] [PubMed] [Google Scholar]
- 43.Scholl, D., S. Adhya, and C. Merril. 2005. Escherichia coli's K1 capsule is a barrier to bacteriophage T7. Appl. Environ. Microbiol. 71:4872-4874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Scholl, D., and C. Merril. 2005. Phage: role in bacterial pathogenesis and biotechnology, p. 400-414. ASM Press, Washington, D.C.
- 45.Shin, I., J. Kim, C. R. Cantor, and C. Kang. 2000. Effects of saturation mutagenesis of the phage SP6 promoter on transcription activity, presented by activity logos. Proc. Natl. Acad. Sci. USA 97:3890-3895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Silver, R. P., and E. R. Vimr. 1990. The bacteria, vol. 11. Molecular basis of bacterial pathogenesis, p. 39-60. Academic Press, Inc., New York, N.Y. [Google Scholar]
- 47.Steinbacher, S., S. Miller, U. Baxa, N. Budisa, A. Weintraub, R. Seckler, and R. Huber. 1997. Phage P22 tailspike protein: crystal structure of the head-binding domain at 2.3 Å, fully refined structure of the endorhamnosidase at 1.56 Å, and the molecular basis of O-antigen recognition and cleavage. J. Mol. Biol. 267:865-880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Steven, A. C., B. L. Trus, J. V. Maizel, M. Unser, D. A. Parry, J. S. Wall, J. Hainfeld, and F. W. Studier. 1988. Molecular substructure of a viral receptor-recognition protein. The gp17 tail-fiber of bacteriophage T7. J. Mol. Biol. 200:351-365. [DOI] [PubMed] [Google Scholar]
- 49.Studier, F. W. 1975. Gene 0.3 of bacteriophage T7 acts to overcome the DNA restriction system of its host. J. Mol. Biol. 94:283-295. [DOI] [PubMed] [Google Scholar]
- 50.van der Wilk, F., A. M. Dullemans, M. Verbeck, and J. F. Heuvel. 1999. Isolation and characterization of APSE-1, a bacteriophage infecting the secondary endosymbiont of Acyrthosiphon pisum. Virology 262:104-113. [DOI] [PubMed] [Google Scholar]
- 51.Vimr, E. R., R. D. McCoy, H. F. Vollger, N. C. Wilkison, and F. A. Troy. 1984. Use of prokaryotic-derived probes to identify poly(sialic acid) in neonatal neuronal membranes. Proc. Natl. Acad. Sci. USA 81:1971-1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Weinel, C., K. E. Nelson, and B. Tummler. 2002. Global features of the Pseudomonas putida KT2440 genome sequence. Environ. Microbiol. 4:809-818. [DOI] [PubMed] [Google Scholar]
- 53.Whitfield, C., E. R. Vimr, J. W. Costerton, and F. A. Troy. 1984. Protein synthesis is required for in vivo activation of polysialic acid capsule synthesis in Escherichia coli K1. J. Bacteriol. 159:321-328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang, X., and F. W. Studier. 1997. Mechanism of inhibition of T7 RNA polymerase by T7 lysozyme. J. Mol. Biol. 269:964-981. [DOI] [PubMed] [Google Scholar]
- 55.Zhang, X., and F. W. Studier. 2004. Multiple roles of T7 RNA polymerase and T7 lysozyme during bacteriophage T7 infection. J. Mol. Biol. 340:707-730. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.