Abstract
Fermentation of capers (the fruits of Capparis sp.) was studied by molecular and culture-independent methods. A lactic acid fermentation occurred following immersion of caper berries in water, resulting in fast acidification and development of the organoleptic properties typical of this fermented food. A collection of 133 isolates obtained at different times of fermentation was reduced to 75 after randomly amplified polymorphic DNA (RAPD)-PCR analysis. Isolates were identified by PCR or 16S rRNA gene sequencing as Lactobacillus plantarum (37 isolates), Lactobacillus paraplantarum (1 isolate), Lactobacillus pentosus (5 isolates), Lactobacillus brevis (9 isolates), Lactobacillus fermentum (6 isolates), Pediococcus pentosaceus (14 isolates), Pediococcus acidilactici (1 isolate), and Enterococcus faecium (2 isolates). Cluster analysis of RAPD-PCR patterns revealed a high degree of diversity among lactobacilli (with four major groups and five subgroups), while pediococci clustered in two closely related groups. A culture-independent analysis of fermentation samples by temporal temperature gradient electrophoresis (TTGE) also indicated that L. plantarum is the predominant species in this fermentation, in agreement with culture-dependent results. The distribution of L. brevis and L. fermentum in samples was also determined by TTGE, but identification of Pediococcus at the species level was not possible. TTGE also allowed a more precise estimation of the distribution of E. faecium, and the detection of Enterococcus casseliflavus (which was not revealed by the culture-dependent analysis). Results from this study indicate that complementary data from molecular and culture-dependent analysis provide a more accurate determination of the microbial community dynamics during caper fermentation.
Caper berries are the fruits of Capparis species (mainly Capparis spinosa L.), a Mediterranean shrub cultivated for its buds and fruits. The caper berry resembles a large grape with white stripes, or actually more like a teeny tiny watermelon. Fermented capers are often served as an appetizer with meat, olives, cheese, and nuts, or as a complement to salads, pasta, and other foods. The main producers of fermented caper berries are Mediterranean countries, especially Greece, Italy, Turkey, Morocco, and Spain, and the final products are exported mainly to central European countries, the United States, and the United Kingdom as a delicatessen product. Fermentation of caper fruits is often done by traditional artisanal ways (20). The fruits are collected during the months of June and/or July and immersed in tap water, where the fermentation takes place for approximately 5 to 7 days in a temperature range of 23 to 43°C. After this, fermented capers are placed in brine and distributed for consumption. Traditional fermented capers are highly appreciated for their unique organoleptic properties. Production of fermented caper fruits, like other natural vegetable fermentation, is a spontaneous lactic acid fermentation based on an empirical process which relies upon microorganisms present in the raw material and processing environment (33). Despite the economic and social importance of caper fermentations in Mediterranean regions, only limited information is available concerning the microbiota involved. Our present knowledge of microbial diversity in fermented capers is based mainly on microbiological counts of lactic acid bacteria, total bacteria, coliforms, and yeasts (1, 25-28).
Conventional culture-dependent methods of strain characterization by physiological and biochemical tests and molecular techniques, such as ribotyping, amplified fragment polymorphism, and randomly amplified polymorphic DNA, can give significant insight on specific isolates and microbial populations occurring in caper berry manufacture. However, cultivation can over- or underestimate the microbial diversity, as media may not be sufficiently selective for monitoring population dynamics. Novel molecular approaches, using molecular methods such as PCR amplification of the 16S rRNA gene in combination with denaturing gradient gel electrophoresis (DGGE) or temperature gradient gel electrophoresis is commonly performed in microbial ecology (2-5, 15, 16). These methods have provided a new insight into microbial diversity and allow a more rapid, high-resolution description of microbial communities than that provided by the traditional approach of isolation of microorganisms. The use of 16S rRNA gene and PCR-DGGE or PCR-temperature gradient gel electrophoresis has often been combined with sequencing and subsequent identification of the species present in a sample.
In this study, the dynamics of the microbial community responsible for the fermentation of caper berries was investigated for the first time by using a polyphasic approach combining microbial enumerations with culture media, randomly amplified polymorphic DNA (RAPD) and temporal temperature gradient gel electrophoresis (TTGE) fingerprinting of total community DNA and sequencing of partial 16S rRNA genes.
MATERIALS AND METHODS
Caper sampling.
The samples analyzed in this study were obtained from traditional caper berry fermentations carried out by a local industry (Aceitunera Jiennense S.A., Jaén, Spain). Caper berries (1.5- to 2.5-cm diameter) were placed in 150-liter vats filled with tap water and allowed to ferment at ambient temperature for 6 days. Then concentrated NaCl was added to the vats to achieve a final salt concentration of 10% (wt/vol). Sampling was performed from two vats under aseptic conditions at 24-h intervals (one sample per vat per time point) during the 6 days of fermentation and after 24 h of storage in brine (day 7).
Enumeration of microorganisms.
Serial dilutions of caper samples were used for microbial enumerations with the following media: plate count agar (Scharlab, Barcelona, Spain) for estimation of total aerobic mesophilic bacteria; MRS agar (Scharlab) for lactic acid bacteria (LAB); McConkey agar (Scharlab) for enterobacteria; potato dextrose agar (Merck) containing (per liter) 14 mg of tartaric acid (Sigma), 50 mg of chloramphenicol (Sigma), and 50 mg of rose bengal (Sigma) for yeasts and molds. All enumerations were done by plate counting. Portions (0.1 ml) of appropriate dilutions were spread plated in triplicate. Counts were obtained after 48 h and 5 days of incubation at 30°C. Results were calculated as the means of three determinations.
For LAB isolation, the plates were incubated under anaerobic conditions at 32°C. A total of 133 colonies were randomly picked from MRS agar plates. The isolates were purified, checked for catalase activity and gram reaction, and microscopically examined prior to storage in liquid culture using 20% glycerol at −80°C.
Isolates with coccoid morphology were tested for the capacity to grow in brain heart infusion (BHI) broth (Scharlab) at temperatures of 10°C and 45°C. The capacity to grow at 37°C in BHI broth adjusted to pH 9.6 with NaOH or in BHI supplemented with NaCl to 6.5% final concentration as well as growth and esculin hydrolysis on bile esculin agar (Scharlab) were also tested.
Preparation of DNA from pure culture.
Total DNA was extracted from overnight cultures by the method of De los Reyes-Gavilan et al. (13). Two-milliliter samples were centrifuged at 7,000 × g for 10 min. The cell pellets were resuspended in 200 μl of a solution of lysozyme (20 mg/ml) in TES buffer (50 mM Tris-HCl, pH 8.0, 1 mM EDTA, 8.56% [wt/vol] sucrose) containing 10 μl of a mutanolysin solution (1 U/μl) and were incubated for 1 h at 37°C. Fifty microliters of 20% sodium dodecyl sulfate was then added to each preparation to lyse the cells. Total DNA was then purified by repeated extraction with phenol-chloroform-isoamyl alcohol (25:24:1), followed by final extraction with chloroform-isoamyl alcohol (24:1). The DNA was precipitated with isopropanol, washed with 70% ethanol, and vacuum dried. The resulting pellets were resuspended in 200-μl portions of a solution containing 50 μg/ml RNase and incubated for 15 min at 37°C. DNA concentration and quality were assessed by measuring optical density at 260 and 280 nm with a Beckman DU670 spectrophotometer.
DNA isolation from fermented capers.
Samples of the fermentation broth (1.5 ml each) were centrifuged for 5 min at 12,000 rpm in a microcentrifuge. The cell pellets were kept overnight at −80°C. Cells were resuspended in 400 μl TESL buffer (25 mM Tris-HCl, pH 8.0, 10 mM EDTA, 20% sucrose, and 20 mg/ml lysozyme), and 20 μl of 1-U/μl mutanolysin (Sigma) was added to each sample. Samples were vortexed for 1 min and incubated for 1 h at 37°C. Fifty microliters of a proteinase K solution (10 mg/ml) was added to each tube, and the tubes were incubated for 50 min at 50°C and then for 10 min at 65°C. To lyse the cell membranes and separate DNA from cell debris and protein, 100 μl of 20% sodium dodecyl sulfate, 700 μl of phenol, pH 8.0 (Sigma), and 100 μl of chloroform-isoamyl alcohol were then added to each sample. The samples were mixed by inversion followed by microcentrifugation for 10 min at 12,000 × g, 4°C. The aqueous phase was transferred to a new tube, and another extraction was performed with phenol and chloroform-isoamyl alcohol followed by one extraction with chloroform. The resulting aqueous phase was precipitated by the addition of isopropanol. Samples were held at −20°C for at least 1 h and centrifuged again. DNA pellets were washed twice with 70% ethanol, dried briefly under a vacuum, dissolved in 50 μl water, and stored at −80°C. The quality of the DNA recovered was routinely checked on agarose gels, and DNA was quantified spectrophotometrically as previously described.
RAPD-PCR amplification.
RAPD-PCR analysis was performed as described previously (6) using the primers M13 (5′-GAG GGT GGC GGT TCT-3′) and B10 (5′-GGT GAC GCA G-3′) in separate reactions. DNA was amplified with primer M13 for 35 cycles (94°C for 1 min; 40°C for 20 s, ramp to 72°C at 0.6°C/s for 2 min; 72°C for 2 min) and with primer B10 for 30 cycles (94°C for 1 min, 34°C for 1 min, ramp to 72°C at 0.6°C/s for 2 min; 72°C for 2 min). The gels were stained in ethidium bromide and photographed on a UV transilluminator. Photopositives were scanned into a computer and subsequently analyzed using the Bionumerics software, version 2.5 (Applied Maths, Kortrijk, Belgium). RAPD-PCR patterns were grouped by means of cluster analysis with the Pearson product moment correlation coefficient and the unweighted pair group method using arithmetic averages (UPGMA). The RAPD patterns obtained with both primers were analyzed together using Bionumerics software to obtain a single dendrogram.
Identification of the isolates.
Molecular identification was carried out by PCR. The isolates were identified by using species-specific primers for Enterococcus faecalis (E1/E2), Enterococcus faecium (F1/F2) (14), Lactobacillus plantarum (PLANT1/LOWLAC), and Lactobacillus fermentum (FERM1/LOWLAC) (10). Isolates that tested positive in PCR amplification for L. plantarum were then tested by multiplex PCR for identification of L. plantarum, Lactobacillus paraplantarum, and Lactobacillus pentosus with recA gene-based primers paraF (5′-GTC ACA GGC ATT ACG AAA AC-3′), pentF (5′-CAG TGG CGC GGT TGA TAT C-3′), planF (5′-CCG TTT ATG CGG AAC ACC TA-3′), and pREV (5′-TCG GGA TTA CCA AAC ATC AC-3′) as described by Torriani et al. (32). The following strains were used as references: Enterococcus faecalis CECT 481T, E. faecium CECT 410T, L. plantarum CECT 220, L. paraplantarum CECT 5783, L. pentosus CECT 4023, and L. fermentum CECT 4007 (CECT strains are from the Colección Española de Cultivos Tipo [http://www.cect.org], Burjassot, Valencia, Spain). The PCR products were analyzed by electrophoresis on an agarose gel in Tris-borate-EDTA buffer at 100 V, and stained with ethidium bromide solution (0.5 μg/ml).
For 16S rRNA gene sequencing, genomic DNAs of strains were used to amplify the almost complete 16S rRNA gene. Amplification of the 16S rRNA gene sequence by PCR was performed using the following primers: 5′-AGAGTTTGATCMTGGCTC-3′ (forward), corresponding to conserved Escherichia coli 16S rRNA positions 8 to 25 (8), and 5′-CNCGTCCTTCATCGCCT-3′ (reverse), corresponding to conserved E. coli 23S rRNA positions 44 to 60 (9). PCR was carried out according to the method of Cibik et al. (11). The product of amplification was purified by using a Quantum Prep PCR Kleen Spin column (Bio-Rad, Madrid, Spain) and then sequenced by using the primers Sp3 (5′-TACGCATTTCACCKCTACA-3′, position 684 reverse), Sp4 (5′-CTCGTTGCGGGACTTAAC-3′, position 1089 reverse), and Sp5 (5′-GNTACCTTGTTACGACTT-3′, position 1492 reverse) according to the method of Weisburg et al. (34) in a CEQ 2000 XL DNA analysis system (Beckman Coulter, California). The partial sequences of the 16S rRNA gene were determined by using CEQ 2000 dye terminator cycle sequencing with a quick start kit (Beckman Coulter, California) according to the manufacturer's instructions. The sequence data were analyzed with a CEQ DNA analysis system (version 4.0). The overlapping sequences obtained with SP3; SP4 and SP5 were merged using theLaser gene program, version 5.05 (DNASTAR, Inc., Madison, WI). A search for homology of the DNA sequence was done using the BLAST algorithm available at the National Center for Biotechnology Information.
PCR amplification of the microbial community 16S rRNA gene.
TTGE samples were prepared by performing two successive PCR amplifications (nested PCR) by using the primer pairs described elsewhere (24). First, a 700-bp fragment of the 16S rRNA gene including the V3 region was amplified. The reaction mixture (100 μl) contained reaction buffer (10 mM Tris-HCl, 1.5 mM MgCl2, 50 mM KCl [final concentrations]), each deoxynucleoside triphosphate at a concentration of 200 μM, 60 pmol of primer W01 (5′-AGA GTT TGA TC[A/C] TGG CTC-3′), 60 pmol of primer W012 (5′-TAC GCA TTT CAC C[G/T]C TAC A-3′), ∼50 ng of total DNA, and 2.5 U of Taq DNA polymerase (Amersham Biosciences). The amplification program was 96°C for 2 min; 30 cycles of 96°C for 1 min, 50°C for 30 s, and 72°C for 1 min; and finally, 72°C for 2 min. Second, the 700-bp fragment was used to amplify the V3 region with the following primers (24): HDA1-GC (5′-CGC CCG GGG CGC GCC CCG GGC GGG GCG GGG GCA CGG GGG GAC TCC TAC GGG AGG CAG CAG T-3′, the GC clamp sequence is in bold) and HDA2 (5′-GTA TTA CCG CGG CTG CTG GCA-3′). The reaction mixture (100 μl) consisted of a reaction buffer (10 mM Tris-HCl, 1.5 mM MgCl2, 50 mM KCl [final concentrations]), each deoxynucleoside triphosphate at a concentration of 200 μM, 60 pmol of each primer, 1 μl of the amplified 700-bp fragment, and 2.5 U of Taq DNA polymerase. The amplification program was 94°C for 2 min; 30 cycles of 94°C for 1 min, 58°C for 30 s, and 72°C for 1 min; and finally, 72°C for 7 min. The sizes and quantities of PCR products were determined by 2% agarose gel electrophoresis.
TTGE analysis.
PCR products obtained from the V3 region amplification as described above were subjected to TTGE analyses. TTGE was performed by using the Dcode universal mutation detection system (Bio-Rad). Polyacrylamide (8%) gels were prepared and run with 1× TAE buffer (40 mM Tris base, 20 mM glacial acetic acid, 1 mM EDTA, pH 8.3) diluted from 50× TAE buffer. Gels were prepared with 8% (wt/vol) acrylamide stock solutions (acrylamide-bisacrylamide, 37.5:1) and a final urea concentration of 7 M. The final electrophoresis conditions were 70 V for 16 h with an initial temperature of 63°C and a final temperature of 70°C (the temperature was increased 0.4°C per h). Samples (20 μl) of PCR products were deposited in wells. A magnetic stirrer was used to mix the buffers and improve the temperature gradient homogeneity. After runs, gels were stained for 30 min with ethidium bromide (0.5 μg/ml of 1× TAE buffer), rinsed for 20 min in 1× TAE buffer, and photographed on a UV transillumination table.
DNA sequencing and analysis of sequence data.
TTGE fragments were excised with a sterile scalpel. The DNA of each fragment was eluted in 20 μl of sterile water overnight at 4°C. One microliter of the eluted DNA of each TTGE band was reamplified using primers HDA1-GC and HDA2. The success of this procedure was checked by electrophoresing 3-μl portions of the PCR products in TTGE gel as described above with caper-amplified DNA as control.
To determine the closet known relatives of the partial 16S rRNA gene sequences obtained, searches of public data libraries (RDP and GenBank) were performed by using the BLAST and RDP programs.
RESULTS
Changes of microbial counts during caper fermentation.
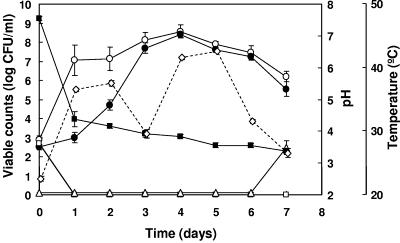
Microbial counts during fermentation of capers were detected by plating serial dilutions of samples: total mesophilic bacteria, LAB, Enterobacteriaceae, and yeasts (Fig. 1). Viable counts of total mesophilic bacteria as well as LAB were low at time zero (ca. 103 CFU/ml), but total mesophilic counts increased markedly during the first day of fermentation. This was followed by a slower increase to yield the highest counts at day 4. LAB counts increased very slowly during the first day of fermentation, followed by rapid increases at days 2 to 4. From day 4 on, LAB counts were almost identical to counts of total viable cells, indicating that they constitute the predominant group in the fermentation. Both total viable cells and LAB counts decreased gradually during the remaining period of fermentation. Enterobacteriaceae were also detected at time zero of fermentation but disappeared from day 1 on. This may be due to the large pH drop from 7.50 to 4.37 occurring during the first day of fermentation and the continued acidification of the fermentation broth down to pH 3.55 at the end of fermentation. Yeasts were detected only at the end of fermentation, when salt was added to the fermented capers to reach a 10% brine.
FIG. 1.
Changes in microbial counts during the fermentation of capers. Total aerobic mesophilic bacteria (○), lactic acid bacteria (•), Enterobacteriaceae (□), and yeasts (▵) were determined. Changes in pH (▪) and temperature (⋄) are also presented. Average data from two vats are shown. Error bars represent standard deviations.
Identification and cluster analysis of bacterial isolates.
A collection of 133 bacterial isolates obtained from MRS agar plates (including 103 rod-shaped bacteria and 26 cocci) collected during the fermentation period was used as starting material for RAPD-PCR amplification. After comparison of RAPD profiles and elimination of repeated isolates coming from the same day of fermentation with identical RAPD profiles, a total of 58 bacilli and 17 cocci were selected for species identification and cluster analysis.
Identification and cluster analysis of bacilli.
By means of PCR amplification with species-specific probes having rRNA genes as targets, 6 isolates were identified as Lactobacillus fermentum (Table 1) and 43 isolates tested positive for Lactobacillus plantarum. The later were further identified by multiplex PCR amplification of recA gene-based primers as L. plantarum (37 isolates), L. paraplantarum (1 isolate), and L. pentosus (5 isolates) (Table 1). The rest of the isolates could not be amplified by PCR and were identified as Lactobacillus brevis by 16S rRNA gene sequencing.
TABLE 1.
Identification of bacteria isolated through the fermentation period during caper manufacture
| Species | Identification method | Isolate(s) |
|---|---|---|
| Lactobacillus plantarum | Multiplex PCR amplification | Lb1g, Lb2g, Lb3, Lb4g, Lb5g, Lb5p, Lb12g, Lb14p, Lb17, Lb23, Lb25, Lb26, Lb27, Lb29, Lb30, Lb31g, Lb32g, Lb35, Lb38p, Lb42, Lb46, L3g, L3p, L4, L6, L10, L11g, L11p, L12g, L12p, L14g, P4, P7, P11g, P16, P17g, P21 |
| Lactobacillus paraplantarum | Multiplex PCR amplification | Lb20g |
| Lactobacillus pentosus | Multiplex PCR amplification | Lb11g, Lb19p, Lb47, P19g, P19p |
| Lactobacillus fermentum | PCR amplification | Lb34, Lb44, Lb45, P15g, P15p, P18g |
| Lactobacillus brevis | 16S rRNA gene sequencing | Lb13, Lb14g, Lb15, Lb21, L9, L13, L15, L16, L17 |
| Pediococcus pentosaceus | 16S rRNA gene sequencing | S4, S5, S7, S8, S9, S10, S12, S13, S14, P1, P2, P3, P6, P9 |
| Pediococcus acidilactici | 16S rRNA gene sequencing | S15 |
| Enterococcus faecium | PCR amplification | S1, S11 |
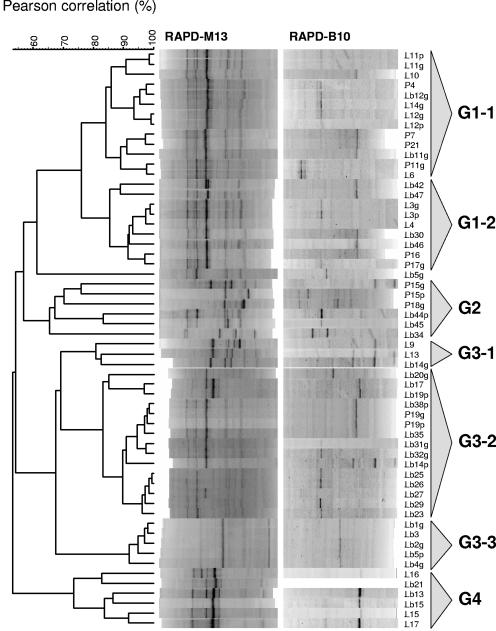
Cluster analysis of RAPD-PCR patterns revealed a high degree of diversity among lactobacilli, which could be separated into four different groups (G1 to G4) and one single isolate (Lb5g) of L. plantarum (Fig. 2).
FIG. 2.
Dendrogram obtained after RAPD-PCR analysis of rod-shaped lactic acid bacteria isolated from caper fermentation. Merged RAPD-PCR patterns obtained after amplification with primers M13 and B10 were grouped by means of the Pearson product moment correlation coefficient and UPGMA cluster analysis with Bionumerics software.
Group G1 includes 20 isolates of L. plantarum and 2 isolates of L. pentosus, distributed in subgroups G1-1 and G1-2. Subgroup G1-1 includes 13 genetically close isolates (L11p, L11g, L10, P4, Lb12g, L14g, L12g, L12p, P7, P21, Lb11g, P11g, and L6) with a similarity coefficient of 84.17%. Similarly, the 9 isolates includes in subgroup G1-2 (Lb42, Lb47, L3g, L3p, L4, Lb30, Lb46, P16, and P17g) are also genetically close and show a similarity coefficient of 85.45%.
Group G2 includes all six isolates of L. fermentum (P15g, P15p, P18g, Lb44p, Lb45, and Lb34). These are genetically distant and show a similarity coefficient of 65.51%.
Group G3 includes 23 isolates that cluster in three subgroups (G3-1 to G3-3). Subgroup G3-1 is composed of three isolates of L. brevis (L9, L13, and Lb14g) with an 80% similarity coefficient. Subgroup G3-2 includes 15 isolates genetically close that show a similarity coefficient of 83.13%: 11 isolates of L. plantarum (Lb17, Lb38p, Lb35, Lb31g, Lb32g, Lb14p, Lb25, Lb26, Lb27, Lb29, and Lb23), 3 isolates of L. pentosus (Lb19p, P19g, and P19p), and the single isolate L. paraplantarum Lb20g. Subgroup G3-3 is the most homogeneous and includes 5 isolates of L. plantarum (Lb1g, Lb3, Lb2g, Lb5p, and Lb4g) that are genetically very close and show a similarity coefficient of 91.89%.
Group G4 is more heterogeneous and includes six isolates of L. brevis (L16, Lb21, Lb13, Lb15, L15, and L17) with a similarity coefficient of 73%.
Identification and cluster analysis of cocci.
Preliminary physiological tests indicated that all isolates were able to grow at 45°C, but only two isolates (S1 and S11) were able to grow at 10°C as well as in media containing 6.5% NaCl or buffered at pH 9.6. They were also able to hydrolyze esculin. Both isolates were identified as Enterococcus faecium by PCR amplification with specific probes (Table 1). The remaining isolates were unable to grow at 10°C or at pH 9.6. Fourteen of them (S4, S5, S7, S8, S9, S10, S12, S13, S14, P1, P2, P3, P6, and P9) were able to hydrolyze esculin and to grow in media containing 6.5% NaCl. They were identified as Pediococcus pentosaceus by 16S rRNA gene sequencing (Table 1). Finally, isolate S15 was unable to hydrolyze esculin and showed a weak growth capacity in the presence of 6.5% NaCl. This isolate was identified as Pediococcus acidilactici by 16S rRNA gene sequencing.
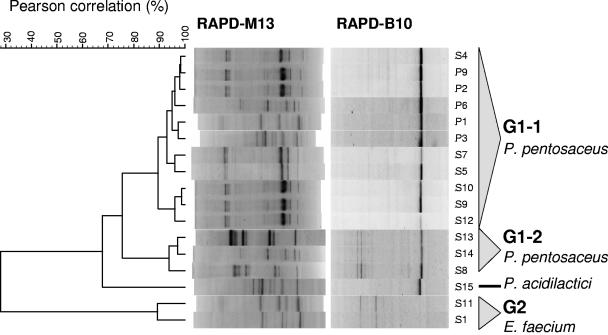
Cluster analysis of RAPD-PCR patterns obtained for cocci revealed two main groups, G1 and G2 (Fig. 3). Group G1 includes all isolates of P. pentosaceus distributed in two subgroups plus the single isolate of P. acidilactici (S15). Isolates included in subgroup G1-1 (S4, P9, P2, P6, P1, P3, S7, S5, S10, S9, and S12) are genetically close and show a similarity coefficient of 89.55%, while isolates in subgroup G1-2 (S13, S14, and S8) are genetically very close and show a higher similarity coefficient of 93.77%. Group G2 includes E. faecium isolates (S1 and S11) with a similarity coefficient of 89.24%.
FIG. 3.
Dendrogram obtained after RAPD-PCR analysis of lactic acid bacteria with coccoid morphology isolated from caper fermentation. Merged RAPD-PCR patterns obtained after amplification with primers M13 and B10 were grouped by means of the Pearson product moment correlation coefficient and UPGMA cluster analysis with Bionumerics software.
Microbiological profile of the fermentation.
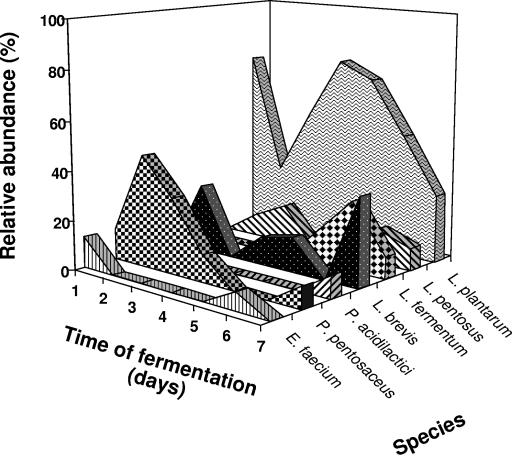
Based on the identity of the most frequently isolated bacteria, a microbiological profile of the fermentation was constructed (Fig. 4). Results clearly indicated that L. plantarum is the predominant species during the fermentation. Other lactobacilli like L. paraplantarum, L. pentosus, and L. brevis had a much lower relative abundance. Furthermore, L. fermentum was only detected at the end of the fermentation (days 6 and 7). The pediococci, represented by P. pentosaceus, were also abundant at the beginning of the fermentation (days 1 to 3) but were clearly displaced by L. plantarum after day 4. Both P. pentosaceus and P. acidilactici were also detected at the end of the fermentation. Enterococci were represented by E. faecium, which was detected at the beginning and also at the end of the fermentation (day 6), although the relative abundance was very low.
FIG. 4.
Relative abundance of the different lactic acid bacteria identified in caper fermentation.
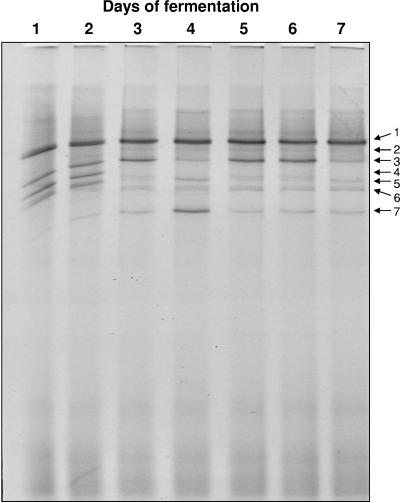
Estimation of the microbial diversity of caper fermentation by TTGE.
As work complementary to the results from bacterial isolation and identification, the microbial diversity of caper fermentation was studied by a culture-independent approach based on total DNA extraction followed by nested PCR amplification of bacterial 16S rRNA genes and separation by TTGE. The electrophoretic profiles of samples taken at different times during the fermentation are shown in Fig. 5. The most intense DNA band appearing through the whole fermentation period corresponds to L. plantarum, in agreement with results from culture-dependent analysis. Other lactobacilli were also identified, like L. brevis (which is also present during the whole fermentation period, although it showed a much lower intensity) and L. fermentum. Pediococci were also detected along the fermentation, and the corresponding band reached its highest intensity at day 4. A band corresponding to Enterococcus sp. was also detected during the first two days of fermentation. Two additional bands of lower intensities, corresponding to Enterococcus casseliflavus and E. faecium were also detected. In particular, the band corresponding to E. faecium clearly persisted from days 3 to 7 of fermentation.
FIG. 5.
Temporal temperature gradient gel electrophoresis of PCR-amplified 16S DNA from samples taken at different times during caper fermentation (lanes 1 to 7). DNAs from the different bands were extracted, sequenced, and compared with a database for identification: 1, Lactobacillus plantarum; 2, Enterococcus casseliflavus; 3, L. fermentum; 4, Enterococcus sp.; 5, L. brevis; 6, E. faecium; 7, Pediococcus sp.
DISCUSSION
Fermented caper berries are highly appreciated as a condiment in Mediterranean cuisine. Nevertheless, the microbiological aspects of this traditional fermentation are still largely unknown (1, 26-28). In the present study, microbial communities of naturally fermented caper berries were revealed by using a combination of classical and molecular techniques. The results obtained following conventional methods indicate that lactic acid bacteria dominate this fermentation. Enterobacteriaceae were also detected at time zero of fermentation but disappeared from day 1 on. This may be due to the large pH decrease from 7.5 to 4.37 occurring during the first day of fermentation and the continued acidification of the fermentation broth down to pH 3.55 at the end of fermentation. The low pH of fermented capers is therefore a key factor for safety and adequate preservation of the final product.
A collection of bacterial isolates obtained during the fermentation period was used as starting material for this study. Biochemical characterization, although useful for identification purposes, has a poor discriminating capability, and molecular techniques must be used with typing purposes. Of the various typing methods available, those based on PCR product analysis, such as RAPD technique, are reported to be simple and rapid to perform and to provide good levels of discrimination; they are therefore considered the most suitable when a large number of strains must be analyzed. After comparison of RAPD profiles and elimination of repeated isolates coming from the same day of fermentation with identical RAPD profiles, a total of 58 bacilli and 17 cocci were selected for species identification and cluster analysis. Following molecular identification methods using species-specific PCR primers and 16S rRNA gene sequencing, the 75 isolates were identified as L. plantarum, L. paraplantarum, L. pentosus, L. brevis, L. fermentum, P. pentosaceus, P. acidilactici, and E. faecium. About 49% of isolates were L. plantarum. This species showed greatest genetic diversity by RAPD-PCR analysis. This greater genotypic differentiation could be the result of the higher number of isolates belonging to these species used for comparison, which would increase the likelihood of finding more distantly related taxonomic units.
Results from this study indicate that caper fruit fermentation shares many characteristics with other vegetable fermentations, where L. plantarum is usually the predominant species as well, both because of its higher acid tolerance and its ability to degrade sugars which are present in vegetables (12, 17, 19, 23, 30). However, while Leuconostoc species are also present in other vegetable fermentations, they were not detected in caper fermentations. A similar profile has been described in Almagro eggplant fermentation, where Lactobacillus species were also predominant (31). The absence of Leuconostoc could be attributed to the rapid decrease of pH, since this bacterium is not able to grow below pH 4.5 (23). It should be noticed as well that this process takes place during a very warm time in the year, and the exposure of the containers to the solar radiation allows the fermentation to take place at temperatures over 40°C. On the contrary, growth of Leuconostoc appears be favored in fermentations that take place at low temperatures, between 8 and 18°C (29), and therefore, the high temperatures in caper fermentations could very well inhibit their growth.
The application of molecular methods in the field of microbiology allows a better understanding of the ecology of food fermentations. Since the results are simple to interpret and mistakes in the identification of species due to the problems related to biochemical tests are avoided, dominant strains responsible for the main transformations during fermentations can be easily detected and identified. The use of culture-independent methods is a recent trend in the study of the microbial ecology of foods (18). PCR-DGGE fingerprinting has been used to study microbial communities in different fermented foods including pozol, cassava, fermented sausages, and dairy products (15). TTGE, which has been used to characterize other types of microbial populations (7, 24), is a technique that works on the same basic principle than DGGE but without the requirement for a chemical denaturing gradient, thus producing more reproducible data (35). In the present work, TTGE was used to monitor the population dynamics during natural fermentation of capers. DNA was amplified by nested PCR, and gels were visually inspected to identify the bands representing the populations involved in the fermentation. To circumvent the biases inherent in subjective interpretation, the presence of the bands was confirmed by direct sequencing. When the results obtained from both traditional plating and TTGE were analyzed, it became evident that the fermentation was characterized by strong LAB activity. The profiles obtained by TTGE allowed us to differentiate the main groups of lactic acid bacteria (L. plantarum, L. brevis, L. fermentum, Pediococcus sp., Enterococcus sp., E. faecium, and E. casseliflavus). These results also show that the predominant species in this fermentation process is L. plantarum, which was represented by a highly intense band throughout the fermentation.
Comparison of results from molecular and culture-dependent studies revealed similar species composition for caper fermentation, and all species of Lactobacillus were correctly identified in both cases. However, Pediococcus could only be identified at species level by culture-dependent methods, while TTGE provided a more accurate picture of the distribution of this bacterium through the fermentation. E. faecium was also identified in both cases, but once more, its distribution during fermentation could only be determined by TTGE. Another enterococcal species (E. casseliflavus) could only be detected by TTGE analysis. These results suggest that TTGE analysis can offer better results for detection of species that are present at lower concentrations. However, it should also be considered that amplification results may largely be dependent on the extraction efficacy for different bacterial species and food matrices. Results from other works also revealed significant differences in the microbial compositions of fermented foods depending on the use of culture-dependent or culture-independent methods (5, 16, 21, 22), suggesting that polyphasic studies should be used to better understand the microbial ecology of foods.
The results presented in this work revealed the microbial complexity of caper berry fermentation. This would be expected for a natural fermentation, which relies solely on the microbiota of the fruit surface, manufacturing plant, and other natural sources. Once the predominant species have been identified, bacterial isolates with selected properties could specifically be used for development of starter cultures to avoid problems of stuck fermentations and to achieve a more homogeneous flavor of the fermented product.
Acknowledgments
This work was supported by the Research Plan of the Junta de Andalucía (research group AGR230) and the Research Programme of the University of Jaén. R.P.P. received a fellowship from the Spanish Ministry of Education.
We also acknowledge the Technical Services of this University for support with DNA sequencing.
REFERENCES
- 1.Alvarruiz, A., M. Rodrigo, J. Miquel, V. Girer, A. Feria, and R. Vila. 1990. Influence of brining and packing conditions on product quality of capers. J. Food Sci. 55:196-198. [Google Scholar]
- 2.Ampe, F., and E. Miambi. 2000. Cluster analysis, richness and biodiversity indexes derived from denaturing gradient gel electrophoresis fingerprints of bacterial communities demonstrate that traditional maize fermentations are driven by the transformation process. Int. J. Food Microbiol. 60:91-97. [DOI] [PubMed] [Google Scholar]
- 3.Ampe, F., N. Ben Omar, C. Moizan, C. Wacher, and J.-P. Guyot. 1999. Polyphasic study of the spatial distribution of microorganisms in Mexican pozol, a fermented maize dough, demonstrates the need for cultivation-independent methods to investigate traditional fermentations. Appl. Environ. Microbiol. 65:5464-5473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ampe, F., A. Sirvent, and N. Zakhia. 2001. Dynamics of the microbial community responsible for traditional sour cassava starch fermentation studied by denaturing gradient gel electrophoresis and quantitative rRNA hybridization. Int. J. Food Microbiol. 65:45-54. [DOI] [PubMed] [Google Scholar]
- 5.Ben Omar, N., and F. Ampe. 2000. Microbial community dynamics during production of the Mexican fermented maize dough pozol. Appl. Environ. Microbiol. 66:3664-3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ben Omar, N., A. Castro, R. Lucas, H. Abriouel, N. M. K. Yousif, C. M. A. P. Franz, W. H. Holzapfel, R. Pérez-Pulido, M. Martínez-Cañamero, and A. Gálvez. 2004. Functional and safety aspects of enterococci isolated from different Spanish foods. Syst. Appl. Microbiol. 27:118-130. [DOI] [PubMed] [Google Scholar]
- 7.Bosshard, P., Y. Santini, D. Grüter, R. Stettler, and R. Bachofen. 2000. Bacterial diversity and community composition in the chemocline of the meromictic alpine Lake Cadagno as revealed by 16S rDNA analysis. FEMS Microbiol. Ecol. 31:173-182. [DOI] [PubMed] [Google Scholar]
- 8.Brosius, J., M. L. Palmer, P. J. Kennedy, and H. F. Noller. 1978. Complete nucleotide sequence of a 16S ribosomal RNA gene from Escherichia coli. Proc. Natl. Acad. Sci. USA 75:4801-4805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brosius, J., T. J. Dull, and H. F. Soller. 1980. Complete nucleotide sequence of a 23S ribosomal RNA gene from Escherichia coli. Proc. Natl. Acad. Sci. USA 77:201-204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chagnaud, P., K. Machinis, L. A. Coutte, A. Marecat, and A. Mercenier. 2001. Rapid PCR-based procedure to identify lactic acid bacteria: application to six common Lactobacillus species. J. Microbiol. Methods 44:139-148. [DOI] [PubMed] [Google Scholar]
- 11.Cibik, R., E. Lepage, and P. Tailliez. 2000. Molecular diversity of Leuconostoc mesenteroides and Leuconostoc citreum isolated from traditional French cheeses as revealed by RAPD fingerprinting, 16S rDNA sequencing and 16S rDNA fragment amplification. Syst. Appl. Microbiol. 23:267-278. [DOI] [PubMed] [Google Scholar]
- 12.Daeschel, M. A., R. E. Andersson, and H. P. Fleming. 1987. Microbial ecology of fermenting plant materials. FEMS Microbiol. Rev. 46:357-367. [Google Scholar]
- 13.De los Reyes-Gavilan, C. G., G. K. Y. Limsowtin, P. Tailliez, L. Séchaud, and J. P. Accolas. 1992. A Lactobacillus helveticus-specific DNA probe detects restriction fragment length polymorphisms in this species. Appl. Environ. Microbiol. 58:3429-3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dutka-Malen, S., S. Evers, and P. Courvalin. 1995. Detection of glycopeptide resistance genotypes and identification to the species level of clinically relevant enterococci by PCR. J. Clin. Microbiol. 33:1628-1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ercolini, D. 2004. PCR-DGGE fingerprinting: novel strategies for detection of microbes in food. J. Microbiol. Methods 56:297-314. [DOI] [PubMed] [Google Scholar]
- 16.Ercolini, D., G. Moschetti, G. Blaiotta, and S. Coppola. 2001. The potential of a polyphasic PCR-DGGE approach in evaluating microbial diversity of natural whey cultures for water-buffalo mozzarella cheese production: bias of culture-dependent and culture-independent analyses. Syst. Appl. Microbiol. 24:610-617. [DOI] [PubMed] [Google Scholar]
- 17.Garrido Fernández, A., P. García García, and M. Brenes Balbuena. 1995. Olive fermentations, p. 593-627. In H.-J. Rehm and G. Reed (ed.), Enzymes, biomass, food and feed. VCH, New York, N.Y.
- 18.Giraffa, G. 2004. Studying the dynamics of microbial populations during food fermentation. FEMS Microbiol. Rev. 28:251-260. [DOI] [PubMed] [Google Scholar]
- 19.Leal-Sánchez, M. V., J. L. Ruiz-Barba, A. H. Sánchez, L. Rejano, R. Jiménez-Díaz, and A. Garrido. 2003. Fermentation profile and optimization of green olive fermentation using Lactobacillus plantarum LPCO10 as a starter culture. Food Microbiol. 20:421-430. [Google Scholar]
- 20.Luna, F., and M. Perez. 1985. La tapanera o alcaparra. Cultivo y aprovechamiento. Ministerio de Agricultura, Pesca y Alimentación, Madrid, Spain.
- 21.Meroth, C. B., J. Walter, C. Hertel, M. J. Brandt, and W. P. Hammes. 2003. Monitoring the bacterial population dynamics in sourdough fermentation processes by using PCR-denaturing gradient gel electrophoresis. Appl. Environ. Microbiol. 69:475-482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miambi, E., J. P. Guyot, and F. Ampe. 2003. Identification, isolation and quantification of representative bacteria from fermented cassava dough using an integrated approach of culture-dependent and culture-independent methods. Int. J. Food Microbiol. 82:111-120. [DOI] [PubMed] [Google Scholar]
- 23.Montaño, A., A. de Castro, and L. Rejano. 1992. Transformaciones bioquímicas durante la fermentación de productos vegetales. Grasas Aceites 43:352-360. [Google Scholar]
- 24.Ogier, J.-C., O. Son, A. Gruss, P. Tailliez, and A. Delacroix-Buchet. 2002. Identification of the bacterial microflora in dairy products by temporal temperature gradient gel electrophoresis. Appl. Environ. Microbiol. 68:3691-3701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Özcan, M. 1999. Pickling and storage of caperberries (Capparis spp.). Z. Lebensm. Unters. Forsch. 208:379-382. [Google Scholar]
- 26.Özcan, M. 2001. Pickling caper flower buds. J. Food Qual. 24:261-269. [Google Scholar]
- 27.Özcan, M., and A. Akgül. 1999. Pickling process of capers (Capparis spp.) flower buds. Grasas Aceites 50:94-99. [Google Scholar]
- 28.Ózcan, M., and A. Akgül. 1999. Storage quality in different brines of pickled capers (Capparis spp.) flower buds. Grasas Aceites 50:269-274. [Google Scholar]
- 29.Pederson, C. S., and M. N. Albury. 1954. The influence of salt and temperature on the microflora of sauerkraut fermentation? Food Technol. 8:1-5. [Google Scholar]
- 30.Ruiz-Barba, J. L., D. P. Cathcart, P. J. Warner, and R. Jímenez-Díaz. 1994. Use of Lactobacillus plantarum LPCO10, a bacteriocin producer, as a starter culture of Spanish-style green olive fermentations. Appl. Environ. Microbiol. 60:2059-2064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sánchez, I., L. Palop, and C. Ballesteros. 2000. Biochemical characterization of lactic acid bacteria isolated from spontaneous fermentation of ‘Almagro’ eggplants. Int. J. Food Microbiol. 59:9-17. [DOI] [PubMed] [Google Scholar]
- 32.Torriani, S., G. E. Felis, and F. Dellaglio. 2001. Differentiation of Lactobacillus plantarum, L. pentosus, and L. paraplantarum by recA gene sequence analysis and multiplex PCR assay with recA gene-derived primers. Appl. Environ. Microbiol. 67:3450-3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Valdés, B., S. Talavera, and E. Fernández-Galiano. 1987. Flora vascular de Andalucía Occidental. Ketras editores S.A., Barcelona, Spain.
- 34.Weisburg, W. G., S. M. Barns, D. A. Pelletier, and D. J. Lane. 1991. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173:697-703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yoshino, K., K. Nishigaki, and Y. Husimi. 1991. Temperature sweep gel electrophoresis: a simple method to detect point mutations. Nucleic Acids Res. 19:3153. [DOI] [PMC free article] [PubMed] [Google Scholar]