Abstract
The genome-probing microarray (GPM) was developed for quantitative, high-throughput monitoring of community dynamics in lactic acid bacteria (LAB) fermentation through the deposit of 149 microbial genomes as probes on a glass slide. Compared to oligonucleotide microarrays, the specificity of GPM was remarkably increased to a species-specific level. GPM possesses about 10- to 100-fold higher sensitivity (2.5 ng of genomic DNA) than the currently used 50-mer oligonucleotide microarrays. Since signal variation between the different genomes was very low compared to that of cDNA or oligonucleotide-based microarrays, the capacity of global quantification of microbial genomes could also be observed in GPM hybridization. In order to assess the applicability of GPMs, LAB community dynamics were monitored during the fermentation of kimchi, a traditional Korean food. In this work, approximately 100 diverse LAB species could be quantitatively analyzed as actively involved in kimchi fermentation.
Since human beings began eating fermented foods, via “the oldest biotechnology,” lactic acid bacteria (LAB) have been of major concern to microbiologists (19, 21, 48). LAB play crucial roles in the manufacturing of fermented milk products, vegetables, and meat as well as in the processing of other products such as wine. LAB have been studied extensively in order to gain a better understanding of their roles in these diverse processes, especially with an aim to manipulate the fermentation process. LAB are now among the best-characterized microorganisms with respect to their genetics, physiology, and applications (30). However, recent ecological investigations have revealed that the microbial communities of most foods are more diverse and complex than they were generally thought to be (19). Thus, development of more sensitive, quantitative, and culture-independent tools is needed in order to explore the microbial ecology of foods (8, 22). Although molecular investigations have been widely applied to LAB fermentations (1, 3, 11, 32, 38, 52), PCR-based molecular procedures are known to introduce certain biases such as primer mismatches to multitemplates (27, 36), Taq errors (2), biased template-to-product ratios (36), and microvariation artifacts (46). Thus, additional means to quantitatively analyze LAB involvement in fermentation are warranted.
DNA microarrays are emerging genomic technologies that are commonly applied to the exploration of genome-wide transcriptional profiles (56). Their application has recently been extended into the realms of environmental microbiology and microbial ecology as a substitute for existing molecular tools (24). Whereas opportunities to use this technology to address important questions in microbial ecology are abundant, several practical limitations slow its implementation (12). First, until very recently, most prior microarray researchers have utilized PCR amplification for detecting genes from the natural environment due to the low sensitivity of the developed DNA microarrays (33, 34, 40, 49, 53). The PCR-based microarrays were criticized because PCR amplification of the environmental DNA produces biased template-to-product ratios and as a result, fail to quantitatively reflect community composition (35). Second, although the 16S rRNA gene-based method (6, 7, 44, 54) is a valuable tool for determining phylogenetic relationships among different bacteria, it provides poor resolution at the species level (13, 28) and insufficient sequence information for determining the positive signal when it is used as a short oligonucleotide probe. Third, the heterogeneity of 16S rRNA between multiple copies in a strain can sometimes mask the appropriate probe of a specific species. Currently, DNA-DNA relatedness provides higher resolution than small-subunit rDNA sequencing (20) and is considered to be the cornerstone in determining the species boundary (55), rather than Stackebrandt and Goebel's 97% rule (different species share less than 97% of 16S rRNA identity) (47).
Reverse sample genome probing (RSGP) is a DNA macroarray method that characterizes community composition based on whole-genome DNA-DNA hybridization (23). Although RSGP has provided valuable insights into microbial population dynamics in situ from various environments, a miniaturized microarray was suggested to be far more efficient due to the limited capacity of RSGP in its current format (59). Zhang et al. (58) recently reported the construction of a microarray fabricated with an Escherichia coli reference collection for exploring genetic diversity within the species. This “library on a slide” was used to determine the presence or absence of pathogenicity-related genes in E. coli strains. Although Cho and Tiedje (9) did pioneering work on genome-based species identification via microarrays, there has not yet been any microarray spotted with bacterial genomes as probes and validated for ecological applications. The accurate and precise printing by robots of miniaturized genome-probing microarrays (GPMs) on nonporous substrates coupled with fluorescent detection could produce several key advances such as guaranteed high reproducibility when a number of environmental samples are analyzed (17) as well as on the ability to work with small amounts of retrievable biomass (60). In this study, we have used GPMs to monitor the population dynamics of lactic acid bacteria (LAB) during the fermentation of kimchi, a traditional food in Korea. As a probe, each GPM contained genomic DNA isolated from 149 different strains, including 138 type strains of LAB, with four replicates. The results of GPM hybridization with fluorescently labeled bulk community DNA suggested the practicality and applicability of this newly invented specific, sensitive, and quantitative tool for the estimation of cultivable bacteria from different environments. The underlying rationale of GPM hybridizations is discussed from a phylogenomic viewpoint.
MATERIALS AND METHODS
Bacterial strains, growth conditions, and kimchi samples.
For monitoring the dynamics of LAB in kimchi fermentation, 149 strains (137 type strains of LAB, 2 control type strains, and 10 isolated strains in the laboratory) were collected as probes (see Table S1 in the supplemental material). In the previous literature the presence of LAB in kimchi was searched for by using denaturing gradient gel electrophoresis (DGGE) analysis on a variety of kimchi types, and culture-dependent analysis was done using agar plates made from filtered kimchi soup (data not shown). Strains used in this study were obtained from the Korea Type Culture Collection and the German Collection of Microorganisms and Cell Cultures. Each strain was grown under the conditions suggested by the collections. Cells at the mid-exponential phase of their healthiest state were quickly harvested and frozen at −80°C for the extraction of DNA.
For testing purposes, five bags of kimchi (10 kg) were purchased from the distributor of the best-selling kimchi brand in Korea, Chongga (Doosan Corp.; http://www.chongga.com). We obtained kimchi just after it was made in the factory and stored it at 4°C. Fifty milliliters (five bags of 10 ml each) of kimchi soup were sampled periodically (about every 3 days). Cell numbers and the pH of each sample were directly determined, and the rest of sample was stored at −80°C for the extraction of bulk community DNA. Serial dilutions of kimchi soup samples in 0.85% NaCl were used for LAB enumerations with MRS-glucose (Difco) agar media (15).
Nucleic acid extraction and quantification.
The genomic DNAs of pure cultures used for arraying on glass slides and the bulk community DNAs from kimchi were isolated using the bead-beating method as described previously (57). All DNA samples were treated with RNase A (Sigma, St. Louis, MO) and were analyzed on agarose gels stained with ethidium bromide prior to microarray fabrication and hybridization. Extracted DNAs were further purified using an UltraClean Microbial DNA Isolation Kit (Mo Bio Laboratories, Solana Beach, CA) with the following modifications. The bead-beating step was excluded, and DNA solution was added to solution MD1 instead of MicroBead solution. After the manufacturer's protocol was followed, the community DNA concentrations were determined in triplicate using a spectrophotometer (Nanodrop Technologies, Rockland, DE).
Microarray construction, labeling, and hybridization.
The genomic DNAs were diluted to a final concentration of 400 ng μl−1 in 0.1× Tris-EDTA buffer. Five microliters of each probe genome was transferred to a 384-well microplate and mixed with 5 μl of 2× microarray spotting solution (ArrayIt; Telechem International, Inc., Sunnyvale, CA) for printing. At a spacing distance of 250 μm, the probes were arrayed onto 25- by 75-mm Superamine glass slides (Telechem) with one pin using a PixSys 5500 printer (Cartesian Technologies, Inc., Irvine, CA) at 55 to 58% relative humidity. Each probe set was printed in quadruplicate. The exact location of each genomic DNA in the glass slide is listed in Table S1 in the supplemental material. The slides were cross-linked by exposure to 120 mJ of UV irradiation (UV Stratalinker 1800; Stratagene, La Jolla, CA). Immediately following UV cross-linking, the DNA was denatured by immersion of the slides in deionized water at 95°C for 2 min. The microarrays were then rinsed briefly in 95% ethanol, air dried at room temperature, and stored dry in a clean slide box at room temperature.
In order to label the genomic DNA and bulk community DNA, the BioPrime DNA Labeling System was modified as follows: 15 μl of various concentrations of DNA was mixed with 20 μl of 2.5× Random Primers solution in the kit and was then denatured by boiling for 2 min and immediately chilled on ice. When the baseline sensitivity of the GPM was determined, serial dilutions of genomic DNA ranging from 0.1 to 2,000 ng were made and labeled. The denatured genomic DNA solution was then mixed with 15 μl of a labeling reaction solution containing 5 mM dATP, 5 mM dTTP, 5 mM dGTP, 2.5 mM dCTP (New England Biolabs, Beverly, MA), 2.5 mM Cy5 dUTP (Amersham Pharmacia Biotech, Piscataway, NJ), and 40 U of Klenow fragment (Invitrogen, Carlsbad, CA). The reaction mixture was incubated at 37°C for 3 h. The labeled target DNA was purified using a QIAQuick PCR purification column (QIAGEN, Valencia, CA), concentrated in a Speedvac for 1 h, and resuspended in 4.35 μl of deionized water for hybridization.
All microarray hybridizations were performed in triplicate (a total of 12 replicates per genomic DNA probe), unless otherwise noted, to facilitate statistical analyses. The hybridization solution contained 4.35 μl of labeled DNA, 8.75 μl of formamide (50%, vol/vol), 3× SSC (1× SSC is 150 mM NaCl and 15 mM trisodium citrate), 1.25 μg of unlabeled herring sperm DNA (Promega, Madison, WI), and 0.3% sodium dodecyl sulfate (SDS) in a total volume of 17.5 μl. A reduced volume (7.5 μl) of the hybridization mixture was deposited directly onto the slides and covered with a coverslip (10 by 15 mm; Sigma). Fifteen microliters of 3× SSC was dispensed into the hydration wells on either side of the hybridization chambers (Corning, Inc., Corning, N.Y.). The microarray slide was placed into a hybridization chamber, boiled for 5 min to denature the hybridization solution, and immediately plunged into the temperature-adjusted water bath for overnight hybridization. After hybridization, each microarray slide was taken out, and the coverslip was immediately removed in wash solution 1 (1× SSC and 0.2% SDS). Slides were washed using wash solution 1, wash solution 2 (0.1× SSC and 0.2% SDS), and wash solution 3 (0.1× SSC) for 5 min each at room temperature prior to drying. The slides were dried by centrifugation.
Microarray scanning and data analysis.
A GenePix 4000A microarray scanner set (Axon instruments, Union City, CA) was used for scanning GPMs at a resolution of 10 μm. Visual displays of hybridization results presented here are representative images which have been contrast adjusted using PowerPoint 2003 (Microsoft) or PhotoShop 7.0 (Adobe). For consistent scanning of all hybridized slides, the laser power and photomultiplier tube (PMT) gain were adjusted to 1,000 V. Scanned image displays were analyzed through quantitation of the pixel density (intensity) of each hybridization spot using GenePix version 6.0 software (Axon instruments). A grid of individual circles defining the location of each DNA spot on the array was superimposed onto the image to indicate each fluorescent spot that was to be quantified. Mean signal intensity was automatically determined for each spot. The local background signal was also automatically subtracted from the hybridization signal of each individual spot. Subsequently, for each probe, the signal-to-noise ratio (SNR) was calculated according to the following formula (25): SNR = (IP − IPB) × ISD−1, where IP is the mean pixel intensity of all replicate probe spots, and IPB is the mean background signal intensity; ISD is the standard deviation of background in which the “background” measurement refers to the local spot background intensity, and the “standard deviation of background” was calculated across all pixels as measured by the GenePix software. The SNRs from 12 replicates were then averaged to represent the SNR of a particular probe. Probes for which the SNR was equal to or greater than 2.0 were considered positive (33). Statistical analysis was performed using Excel 2003 (Microsoft) and Sigmaplot 8.0 (Jandel Scientific, San Rafael, CA). For global normalization, the SNR of each probe was normalized against the SNR of 10 ng of spiked E. coli genomic DNA on the same experimental slide according to the following formula: nSNR = SNR × [(IEcoli − IEcoliB) × IEcoliSD−1]−1, where nSNR is the normalized SNR of the specific probe, IEcoli is the mean pixel intensity of all E. coli probe spots, IEcoliB is the mean pixel intensity of the local background area around all E. coli probe spots, and IEcoliSD is the standard deviation of IEcoliB. Furthermore, in order to show more clearly whether a certain microorganism is a major component in the sample, relative SNRs were obtained by dividing the normalized SNR by the mean value of the normalized SNR in the same kimchi samples according to the following formula: rSNR = nSNR × (SSNR × Nprobe−1)−1, where rSNR is the relative SNR of the specific probe, SSNR is the sum of nSNRs in the sample, and Nprobe is the number of probes. A spreadsheet of Excel data was visualized by ArrayColor.exe (http://microarray.kaist.ac.kr). Using this software, we were able to produce more yellow squares from lower values of normalized/relative SNRs and more red squares from higher values.
DGGE and construction of a phylogenetic tree.
PCR-DGGE analysis of the 16S rRNA gene was conducted with the same bulk community DNA from the kimchi samples. Extraction of genomic DNA and PCR-DGGE with the bacteria-specific primer set (518r and 338f with a GC clamp) were carried out as described elsewhere (26). Major DGGE bands were excised with a razor blade and sequenced to gain information on the bacterial composition of kimchi. The 16S rDNA sequences for the phylogenetic tree (see Fig. 5) were obtained from the GenBank database, and multiple alignments were performed by the Clustal X program (50). The evolutionary distances were calculated using the Jukes and Cantor method. The phylogenetic tree was constructed by using a neighbor-joining method (41) in the MEGA 2 program (31).
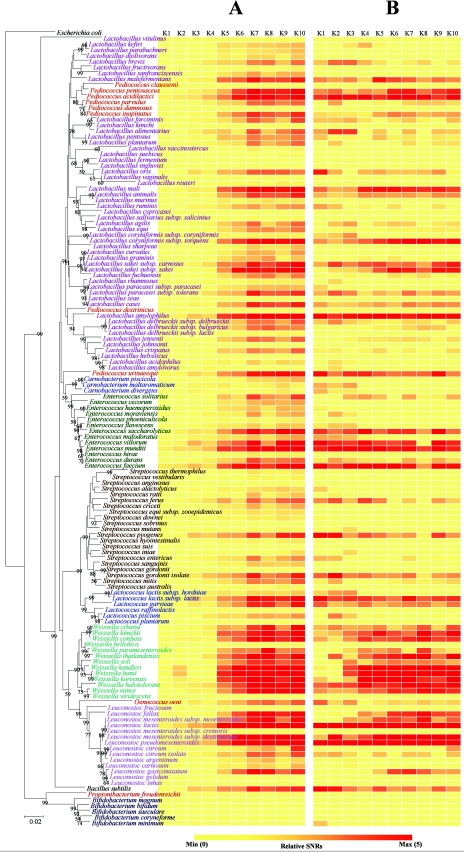
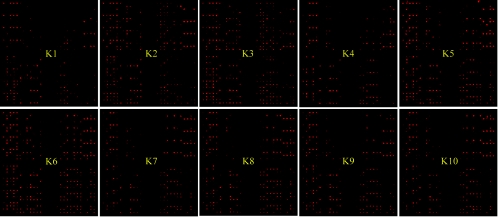
FIG.5.
Quantities of LAB in kimchi samples detected with GPMs at a PMT gain of 700 V. Microarray hybridization patterns with the labeled genomic DNAs from kimchi (samples K1 to K10) are shown in each column. Each row represents the hybridization signal observed for each LAB when 1 μg of genomic DNA from the kimchi (see column) was used for hybridization. The SNRs from 12 replicates were then averaged to represent the SNR for a particular probe. Normalized and relative SNRs were visualized by ArrayColor.exe (http://microarray.kaist.ac.kr), which produces more yellow squares from lower values of normalized/relative SNRs and more red squares from higher values. (A) Normalized SNR values of LAB in 1 μg of bulk community DNA extracted from each phase of kimchi fermentation. For global normalization, normalized SNRs were obtained by dividing the SNR value from each spot by the SNR of 10 ng of spiked E. coli genomic DNA on the same experimental slide. (B) Relative SNRs obtained by dividing the normalized SNR by the mean value of the normalized SNR in the same kimchi samples. A phylogenetic tree indicating the relationships of LAB was harmonized manually with the two SNR pictures. For NCBI numbers of LAB 16S rRNA used in the phylogenetic tree, see Table S1 in the supplemental material.
RESULTS
Specificity of GPM.
To determine the conditions under which species differentiation can be achieved, microarray hybridizations were conducted using various concentrations of genomic DNA from Enterococcus mundtii as the target and probes at 27, 37, 47, and 57°C in the presence of 50% formamide. Temperatures ranging from 27 to 57°C did not have significant effects on the species discrimination capability of the GPMs. At 27°C, slight cross-hybridizations ranging from 1 to 12% of the SNRs were observed between the target species and its phylogenetic neighbor species (Enterococcus hirae, 1.4 to 4.2%; Enterococcus faecium, 2.4 to 11.5%; Enterococcus durans, 3.1 to 6.5%). However, at 37°C, hybridization specificity was improved and was absolutely specific to the probe for the strain of the target species (i.e., E. mundtii) (Table 1). Nonspecific cross-hybridization was reduced to background levels at 37°C (less than 2% of signal intensity and SNR 3) (Table 1). Increasing the temperature above 37°C did not have a substantial impact on hybridization specificity. Rather, at high temperatures (47 and 57°C), significant decreases in hybridization signal intensities were observed.
TABLE 1.
Hybridization specificity of GPMs at 37°C with 50% formamide
| Genome probe | Hybridized templateb | 16S rRNA gene identity (%) | SNR percentagec |
|---|---|---|---|
| Weissella confusaa | Weissella confusa MR2* | 100 | 95 |
| Weissella koreensisa | 95 | 4 | |
| Oenococcus oenia | 90 | 0 | |
| Bacillus subtilisa | 85 | 0 | |
| Escherichia colia | Weissella confusaa | 78 | 0 |
| Enterococcus mundtiia | 78 | 0 | |
| Lactococcus lactisa | 76 | 0 | |
| Bacillus subtilisa | 78 | 0 | |
| Oenococcus oenia | Weissella kimchia | 92 | 0 |
| Weissella confusaa | 91 | 0 | |
| Leuconostoc mesenteroides subsp. dextranicuma | 85 | 0 | |
| Bacillus subtilisa | 81 | 0 | |
| Lactobacillus sakei subsp. sakeia | Lactobacillus sakei subsp. carnosusa | 100 | 63 |
| Lactobacillus fuchuensisa | 97 | 1 | |
| Lactococcus lactis subsp. lactisa | 88 | 1 | |
| Bacillus subtilisa | 87 | 0 | |
| Enterococcus mundtiia | Enterococcus mundtii KM4* | 100 | 102 |
| Enterococcus hiraea | 99 | 2 | |
| Carnobacterium pisciolaa | 93 | 0 | |
| Bacillus subtilisa | 90 | 0 | |
| Leuconostoc mesenteroides subsp. dextranicuma | Leuconostoc mesenteroides subsp. dextranicum K31* | 100 | 87 |
| Leuconostoc mesenteroides subsp. mesenteroidesa | 100 | 37 | |
| Leuconostoc citreuma | 97 | 1 | |
| Bifidobacterium bifiduma | Bifidobacterium coryneformea | 95 | 1 |
| Propionibacterium freudenreichiia | 84 | 0 | |
| Weissella confusaa | 76 | 0 | |
| Bacillus subtilisa | 79 | 0 | |
| Leuconostoc citreuma | Leuconostoc citreum NCTC3351 | 100 | 47 |
| Leuconostoc lactisa | 98 | 2 | |
| Oenococcus oenia | 90 | 0 | |
| Bacillus subtilisa | 83 | 0 |
Type strain.
Strains that were isolated from kimchi and identified as being of the same species by sequencing the 16S rRNA gene are indicated by an asterisk.
SNR percentage = (SNR of each reference/SNR of the target template) × 100.
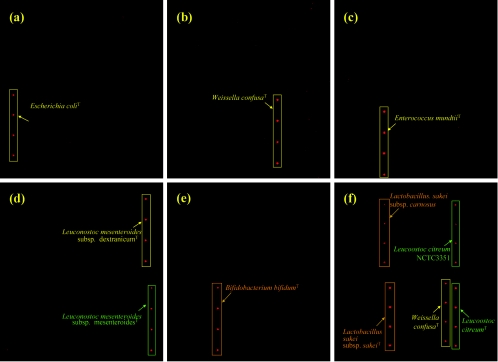
Under hybridization conditions of 37°C and 50% formamide, GPM hybridization specificity was further examined using seven representative species including Weissella confusa, Lactobacillus sakei subsp. sakei, Leuconostoc citreum, Leuconostoc mesenteroides subsp. dextranicum, Bifidobacterium bifidum, Oenococcus oeni, and E. coli as target templates. Strong signals were obtained only for the genomic DNA of species corresponding to the labeled target (Fig. 1 and Table 1). Little to no cross-hybridization (0 to 4%; nSNRs of all cross-hybridizations were under 3) was observed for nontarget species, thus indicating that species-specific hybridizations may be achieved with GPMs under the described conditions. In order to assess the subspecies level specificity of GPM-based hybridization, genomic DNA extracted from several strains of the same species sharing more than 99% homology of 16S rDNA sequences was also spotted on the slide. Strain differentiation could not be achieved either by using the described conditions or by elevating the hybridization temperature. Additionally, signal cross-hybridization was observed among different strains of the same species. Signal intensity of the probe of Leuconostoc citreum NCTC3351 to the template of Leuconostoc citreum type strain and the probe of Lactobacillus sakei subsp. carnosus type strain to the template of Lactobacillus sakei subsp. sakei type strain were 47 and 63%, respectively. The strains isolated from kimchi that were identified as the same species by 16S rRNA gene sequencing (Table 1, asterisks) showed a variety of very similar signal intensities (87 to 102%) for the type strains of the species.
FIG. 1.
Fluorescence images showing hybridization specificity of GPMs. Levels of specificity obtained with 500 ng of labeled target genomes from (a) E. coli type strain, (b) W. confusa type strain, (c) E. mundtii type strain, (d) Leuconostoc mesenteroides subsp. mesenteroides type strain, (e) B. bifidum type strain, and (f) Lactobacillus sakei subsp. sakei type strain, W. confusa type strain, and Leuconostoc citreum type strain. Target genomes are marked with squares and arrows. T, type strain.
Since real environments are composed of a variety of different microorganisms, evaluation of GPM specificity with mixed genomes is indispensable. To determine whether specificity of the GPM-based hybridization is influenced by the presence of other nontarget DNA, three different genomes (500 ng per species) including W. confusa, Lactobacillus sakei subsp. sakei, and Leuconostoc citreum were mixed, labeled, and hybridized to the GPMs (Fig. 1f). Specific hybridization signals were observed on corresponding probes showing SNRs of 86.3, 92.8, and 109.4, respectively. These results suggest that GPM-based hybridization could also be specific in the presence of other genomes.
Detection sensitivity of GPM-based hybridization.
Detection sensitivity of GPM-based hybridization was determined using genomic DNA extracted from pure cultures of W. confusa and E. mundtii. At 900 V PMT gain (laser power set at 100%), 1,430 ± 320 of the F635 median value (SNR of 3.45 ± 0.2) was observed using 5 ng of W. confusa and E. mundtii genomic DNA for the target genome. With 2.5 ng of the DNA, the target hybridization signal was substantially weaker but was still detectable (F635 median value, 658 ± 121; SNR, 1.92) (see Fig. S2 in the supplemental material). However, hybridization signals using 1 ng of genomic DNA were barely detectable above background levels. Therefore, the detection limit of GPMs with randomly labeled pure genomic DNA under the hybridization conditions was 2.5 ng.
The existence of alternative nontarget DNA might affect hybridization with target DNA, thereby decreasing the detection sensitivity (39). To evaluate the detection sensitivity in the presence of heterogeneous nontarget DNA, genomic DNA (2.5 to 1,000 ng) from E. mundtii was mixed with 1 μg of E. coli DNA and labeled with Cy5. The detection limit of GPMs in the presence of nontarget DNA was also 2.5 ng (approximately 2 SNR values), thus showing that the presence of another species's genome does not have a significant effect on the detection sensitivity of the GPMs.
Quantification of the GPM-based hybridization.
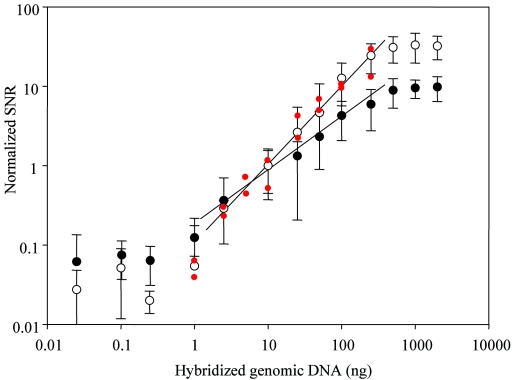
The capacity of the GPM-based hybridization to serve as a quantitative tool was explored by examining the relationship between the concentration of target genomic DNA and signal intensity on the corresponding spot. Genomic DNAs from W. confusa and E. mundtii were fluorescently labeled and hybridized with the microarrays. To increase the quantifying ability of the GPMs, the slides were scanned using different combinations of PMT gain. At 900 V PMT gain, the sensitivity became too high to measure more than 1 μg of genomic DNA and resulted in saturated signals. At 700 V PMT gain, less than 2.5 ng of genomic DNA was undetected; however, this amount was detectable at a PMT gain of 900 V. Strong linear relationships were observed for signal intensity and target genomic DNA concentrations ranging from 2.5 to 500 ng (r2 = 0.97), indicating that GPM hybridization could serve as a quantitative tool for the detection of bacterial species within a wide range of DNA concentrations. Larger SNR values (14.50 ± 9.11) from 10 ng of E. coli genomic DNA at 900 V PMT gain resulted in lower relative SNR values (ordinate) per hybridized genomic DNA (abscissa) than at a PMT gain of 700 V (Fig. 2). The quantitative capacity of the GPMs was also investigated with mixtures of DNA from 16 different bacteria at different concentrations (Fig. 2, legend). A remarkable linear relationship (r2 = 0.96) was observed between signal intensity and target genomic DNA concentrations within a range of 2.5 to 250 ng (Fig. 2, red spots). The variation of signal intensity from genome to genome was very low compared to gene-to-gene variation of cDNA signals and oligonucleotide-based microarrays (39).
FIG. 2.
Evaluation of the quantitative potential of GPM-based hybridization with genomic DNAs from W. confusa type strain and E. mundtii type strain. The log ratios of hybridization signals (normalized SNR) between the target genome and spiked control genomic DNA from E. coli type strain were calculated and plotted against the log of the concentrations of the genomic DNA. Slides were scanned using different PMT gain settings: 700 V (○) and 900 V (•). Quantitative capacity of GPMs was also investigated with mixtures of DNAs from 16 different, arbitrarily selected bacteria with different concentrations at a PMT gain of 700 V (red dots): 1 ng (each) of Enterococcus hemoperoxidus and Weissella kimchii, 2.5 ng (each) of Lactobacillus casei and O. oeni, 5 ng (each) of Streptococcus vestibularis and P. acidilactici, 10 ng (each) of Lactobacillus delbrueckii subsp. delbrueckii and Lactobacillus sakei subsp. sakei, 25 ng (each) of Leuconostoc mesenteroides subsp. mesenteroides and Propionibacterium freudenreichii subsp. freudenreichii, 100 ng (each) of Lactobacillus brevis and Bifidobacterium minimum, and 250 ng (each) of Lactobacillus plantarum and Streptococcus gordonii (the pairs are ordered such that the organism with the higher signal is listed first).
Enumeration of microorganisms and pH changes in kimchi fermentation.
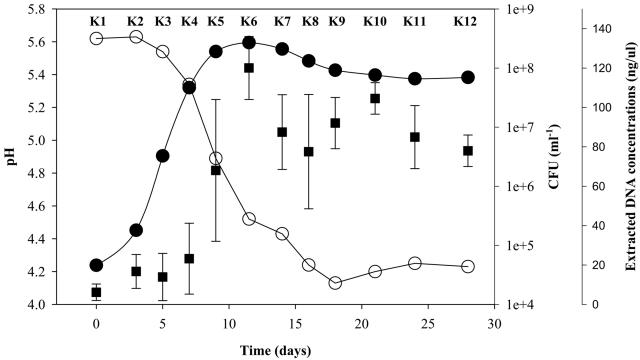
Prior to applying the specifically and quantitatively evaluated GPMs to kimchi, kimchi fermentation was preliminarily characterized by a rapid rise in the total number of LAB, which increased from an initial value of 4.6 × 104/ml to 2.7 × 108/ml within the first 12 days of ripening and which remained stable for the rest of the fermentation (Fig. 3). This large increase in LAB abundance correlated with the decrease in pH values in the early stages of the maturation (kimchi samples 1 to 4 [K1 to K4]); the pH reached 4.1 and was maintained after 18 days of fermentation. This was the lowest pH during the period of fermentation. To use GPMs to monitor the dynamics of microbial population during fermentation, bulk community DNAs were isolated from 5-ml samples that were taken periodically from kimchi soup (Fig. 3).
FIG. 3.
LAB growth (filled circles) and pH change (open circles) during kimchi fermentation. Each point of the total LAB counts and pH is the mean of three samplings. The concentration of bulk community DNA (filled squares) extracted from each sample was also plotted. Standard deviations are shown with error bars. Kimchi samples were named K1 to K12. Samples K1 to K10 were also used in the experiments shown Fig. 4 and 5.
Application of GPMs for profiling microbial communities in kimchi.
To monitor the dynamics of the microbial population during the fermentation of kimchi, 1 μg of the purified bulk community DNAs extracted from the soup samples was labeled and hybridized with the GPMs. No hybridization signal was observed for the E. coli probe in any kimchi sample. For global normalization, 10 ng of E. coli genomic DNA was spiked into 1 μg of bulk kimchi DNA prior to labeling. Overall, a variety of LAB displaying kaleidoscopic dynamics was detected in each kimchi sample (Fig. 4 and 5A). Under the experimental conditions employed, there was no signal saturation for any of these features. The number of positive signals in the late phase of fermented kimchi samples (K5 to K10) was apparently higher than the number in early phase samples (K1 to K4). The proportions of the probes that showed statistically significant positive signals were 18 to 29% (28 to 45 species) and 46 to 64% (71 to 99 species) in early and late stages of kimchi fermentation, respectively. A sudden change in the numbers of positive signals was especially observed at the transition between K4 and K5, and the increase of positives after K5 was relatively small. Accordingly, pH and total cell numbers also changed significantly in this transition. The average values of LAB nSNRs in early (K1 to K4) and late (K5 to K10) stages of kimchi fermentation were 2.16 to 3.39 and 9.24 to 18.52, respectively (Fig. 5). With the nSNRs, we were able to estimate the contents of each LAB species in 1 μg of bulk community DNA extracted from each phase of kimchi. As the fermentation of kimchi progressed, high degrees of nSNRs (nSNR of >10) were observed in most species belonging to the genus Weissella and Leuconostoc, with some belonging to the genera Lactobacillus, Enterococcus, Pediococcus, and Lactococcus. Seventeen species of Streptococcus were also present in small amounts (nSNR of <5), and no significant amounts of Bifidobacterium were found in kimchi. Most species in the genus Weissella were present in the late kimchi phase of fermentation with two exceptions, Weissella hellenica and Weissella viridescens. These results appear to be partly consistent with the previous culture-dependent observations that some species of Weissella, Leuconostoc, and Lactobacillus are abundant, frequently isolated microbial populations in kimchi. In addition, GPM revealed considerable differences in population quantities among close phylogenetic neighbors. For instance, Pediococcus acidilactici and Pediococcus pentosaceus were detected in samples with nSNRs higher than 10 in the later kimchi phase; however, Pediococcus claussenii was not. Abundances of E. faecium, E. durans, and E. hirae in the late phase of kimchi samples were also significantly different despite the fact that they are phylogenetically closely related microorganisms.
FIG. 4.
Representative fluorescence images showing GPM hybridization with kimchi samples (K1 to K10). The contrast of each image was automatically modulated with GenePix software to be more recognizable with the naked eye.
Comparison of GPM with PCR-dependent microbial community analysis.
Unlike the majority of other molecular ecological tools, GPM analysis is not dependent on PCR amplification. In order to estimate the community-profiling ability of GPM hybridization more clearly, we compared the results of GPM analysis to those of DGGE using the community DNAs of kimchi. Since DGGE only produces relative values, for direct comparison, SNR data of GPMs were divided by total values of each phase of kimchi fermentation to show the relative population composition (Fig. 5B). With the relative SNRs, LAB compositions between early and late phases of fermentation were compared. Many LAB species such as Lactobacillus brevis, Lactobacillus alimentarius, Lactobacillus oris, Lactobacillus maltaromicus, and Carnobacterium divergens were observed in the early phase of kimchi fermentation but disappeared in the late phase. However, relative abundances of P. acidilactici, Lactobacillus amylophilus, E. mundtii, E. faecium, Lactococcus lactis, Leuconostoc mesenteroides, and Leuconostoc pseudomesenteroides were maintained during the entire kimchi fermentation period. In relative analyses, most Weissella species were found to be abundant only in the late kimchi fermentation phase.
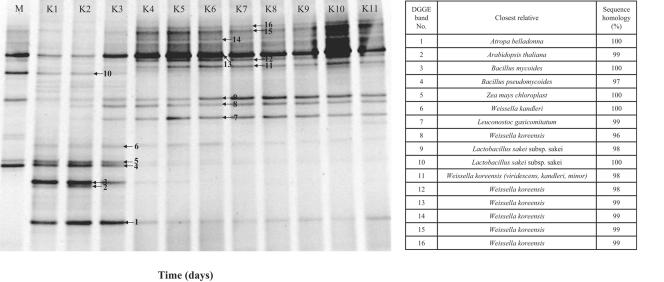
Bulk community DNAs used for GPM analysis were PCR amplified and analyzed with DGGE (Fig. 6). From DGGE analysis, we found that only 9 different microorganisms were detected, although by GPM analysis, we were able to find 99 microorganisms. The majority of bands (11/16) detected in DGGE analysis were LAB, although three bands were retrieved from eukaryotes, and two were from Bacillus. In all of the DGGE lanes, 5 to 10 prominent bands were observed with approximately 5 minor bands. Succession profiles of detected microorganisms were compared. After 5 days of fermentation at 4°C, several bands indicating the presence of LAB such as Weissella kandleri, Leuconostoc gasicomitatum, Lactobacillus sakei subsp. sakei, and Weissella koreensis were observed, and these increased in intensity as fermentation proceeded. In the early phase of kimchi fermentation (samples K1 to K4), only 18 to 29% of positive signals were detected, which is most likely due to the low occupancy of the lactic acid bacteria.
FIG. 6.
DGGE profiles of PCR-amplified 16S rDNA segments from periodically sampled kimchi (samples K1 to K10).
DISCUSSION
Kimchi, a kind of pickled (salted) vegetables, was first created as a traditional food in Korea around the seventh century (8). Without starter cultures, kimchi is made through lactic acid fermentation of Chinese cabbage at low temperatures to ensure proper ripening and preservation. It is processed with a variety of seasoning mixtures: red pepper powder, garlic, ginger, green onion, and radishes. Currently, more than 1.5 × 106 tons of kimchi is consumed each year in South Korea, a phenomenon which is rapidly growing and spreading to other countries in Asia. Despite the great impact of kimchi on Asian health, the microbiology of kimchi has only been explored by nonquantitative PCR-based pattern analyses (29). Since kimchi is representative of a typical open ecosystem, each batch of fermented kimchi has a different composition of bacteria depending on fermentation conditions and ingredients, which can be highly variable. In order to reveal correlations between kimchi fermentation and its bacterial composition, high-throughput analysis tools such as DNA microarrays are indispensable (21). Thus, GPM has been developed to meet this urgent need for a more sensitive, quantitative, and high-throughput analysis tool.
The theoretical background of GPMs established the criteria for estimating the total nucleotide sequence divergence of the microbial genome, which has been widely used to define species demarcations and relationships (42). Conventional DNA-DNA hybridization using microwell plates (10) or membrane filters (14) has several limitations which can be overcome by microarray hybridization using glass slides. (i) Results obtained in different laboratories or even in replicated experiments in the same laboratory appear discordant (43). (ii) Reciprocal experiments often yield nonisomorphic values. (iii) For practical applications, conventional DNA hybridization experiments have provided information regarding the degree of genome similarity of an isolated strain to one or a few reference strains rather than to a complete matrix of coefficients of genetic relatedness among all strains of the same group (see reference 45). Recently, Ramisse et al. (37) attempted to solve the limits of the DNA-DNA hybridization method by reverse sample genome probing (RSGP) using a nylon membrane. In their work, large numbers of clinical and environmental isolates could be quickly identified as belonging or not belonging to a particular species. However, GPMs using glass slides offer several important advantages over RSGP formats for characterizing microbial bioprocesses. The main advantage is that high-throughput and parallel analysis can be achieved with microarray hybridization. Theoretically, with GPMs at least 17,880 hybridizations (149 genomes × quadruplicate printing × 10 samples × triplicate experiments) could be executed while the dynamics of LAB in kimchi fermentation are monitored. This work might be too tedious and laborious to do with RSGP. Furthermore, printing genomic DNAs with the robot and precisely reading the signal intensity with the microscanner enable global quantification (Fig. 2).
Depending on the probe characteristics, various types of microbial diagnostic microarrays might be designed (4). Most of these microarrays are based on the detection of 16S rRNA genes. GPM may have higher specificity for species discrimination than the 16S rRNA gene-based cDNA chip or the oligomer chip for detection of specific microorganisms in natural environments. For the detection of the 16S rRNA gene, cross-hybridization to closely related nonspecific targets is nearly unavoidable due to the high similarity of 16S rRNA sequences. Thus, perfectly matched and mismatched oligonucleotide probes have been employed to obtain more specific signals using relative comparisons. However, this makes quantification and discrimination of real and false-positive signals quite difficult. In microarray experiments using long oligonucleotide probes under general hybridization conditions, nontarget genes showing more than 75 to 87% identities to probes are hybridized (59). In the case of GPM, all segments of the whole genome, which are much more divergent than the 16S rRNA gene, were employed for microarray hybridization. According to our phylogenomic studies, less than 1% of the open reading frames actually showed higher than 87% nucleotide sequence similarity between species that share 97% or greater similarity with regard to the 16S rRNA gene. (Comparison of total open reading frames in a genome to those of closely related bacterial strains in the same species or genus from Lactobacillus, Bacillus, Streptococcus, and Staphylococcus were executed for the analysis [data not shown].) This means that cross-hybridization of GPMs could not be considered significant over the species boundary. The result of specificity experiments also did not exhibit any reliable cross-hybridization (SNR of more than 3) among the species of the whole LAB library. Therefore, GPMs provide a higher level of resolution in differentiating species and a more reliable signal from the sum of the total genes of the whole genome than when a single gene is used.
In order to avoid PCR amplification, sensitivity is another critical parameter that impacts the effectiveness of the microarray-based approach for detecting genes in environmental samples. With GPMs, genomes involved in kimchi fermentation could be detected with 2.5 ng of genomic DNA in the presence of background DNA (0.25% of microbial composition). Denef et al. (16) achieved a detection sensitivity of 1% total community using tyramide signal amplification. Tiquia et al. (51), Loy et al. (33), and Bodrossy et al. (5) achieved a 5% detection limit, which is the same level obtained by Denef et al. (16) when tyramide signal amplification was avoided. The level of GPM detection sensitivity is thus a great advancement for the environmental microarray and should be sufficient for the detection of the dominant members of a microbial community. Taking the mean bacterial DNA content values (3.8 to 4.9 fg of DNA/cell [18]) into consideration, in principle, roughly 105 to 106 cells are needed to achieve reasonably strong hybridization. This value also indicates that GPM possesses about 10- to 100-fold higher sensitivity 50-mer DNA microarrays (39).
The quantitative capability of microarray-based hybridizations is another critical issue for environmental application. We observed a good linear relationship between hybridization signal intensity and target genome concentration in GPM hybridization (Fig. 2). Since the signal comes from the sum of several thousands of genes, signal variation from each gene might be diminished. We observed that signal variation from the same quantity of genomes is not significant (r2 = 0.96). When oligonucleotide or cDNA was used as a probe, gene-to-gene signal variation was kaleidoscopic up to one order, depending on probe and gene pairs (39). This means that, for the same copy number of genes, we could not detect the same level of signals; thus, quantification could be severely hindered when these methods are used. Furthermore, because PCR is not necessary for GPM analysis, we can precisely quantify microorganisms using GPMs in the microbial community without having to account for PCR bias. However, we should be cautious in interpreting the quantification of signals from GPM, since specificity for quantification of a microarray signal is still a contentious issue. Strains that are different from a type strain and belong to the same species showed reduced signal intensities in GPM (Fig. 1 and Table 1). When GPM is applied to environmental samples, the signal intensity of a spot would be the integral value of the signals from a number of strains showing different genome similarities to type strains. If strains possessing low genome similarity exist in the samples, the actual correlation for the quantification of cell numbers could be distorted. This difficulty can be reduced if the strains isolated from the target community are used as probes together with type strains, as in our experiment. This same limitation is an issue for other bacterial diagnostic microarrays using oligonucleotide or cDNA probes, since these probes hybridize to both perfectly matched genes as well as to genes with certain levels of mismatches. This means that the actual signal arises from a group of genes with a certain level of similarity (such as 87% in the case of the 50-mer oligonucleotide probe [4]).
Although there are several methods to characterize the contribution of LAB to human health and the dairy industry, no appropriate tool has been developed yet for the estimation of the comprehensive, quantitative dynamics of microbial populations during fermentation processes. In this work, diverse LAB communities (more than 100 species) could be observed to be actively involved in the fermentation of kimchi and its ripening during storage. Several Weissella species were the most dominant microflora in kimchi fermented at 4°C. This is a very distinctive observation considering that other LAB fermentation products such as artisanal cheeses (Lactobacillus) (38), malt whisky (Leuconostoc and Lactococcus) (52), Mexican maize dough (Streptococcus) (3), Italian sausages (Lactobacillus) (11), raw milk products (Lactococcus lactis) (32), and traditional sour cassava starch (Bifidobacterium, Lactococcus, Streptococcus, Enterococcus, and Lactobacillus) (1) have not been reported to be associated with the genus Weissella. GPM profiles of kimchi samples evolved significantly after 7 to 9 days of fermentation, showing that some Streptococcus and Lactobacillus species disappeared after the decrease in pH. No known molecular tools are available that can provide this kind of global picture of fermentation processes in a short time. Actually, DGGE experiments with the same samples used for GPM hybridization showed significant underestimation of the diversity of LAB (Fig. 6). These new results from GPM hybridization will greatly change our understanding of microbial ecology during LAB fermentation.
By exploiting GPMs to achieve more detailed pictures of the microbial community, improved fermentation processes could be developed to improve the quality of food products. GPMs could be also applied to elucidate the underlying mechanisms of various mixed culture bioreactors to achieve optimization and modification of these processes.
Supplementary Material
Acknowledgments
We thank Eugene Madsen for his contribution in improving the manuscript.
This work was supported by grant BDM0200524, grant NNM0100512, and the NRL research program (grant M10104000294-01J000012800) of the Korean Ministry of Science and Technology (MOST).
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Ampe, F., A. Sirvent, and N. Zakhia. 2001. Dynamics of the microbial community responsible for traditional sour cassava starch fermentation studied by denaturing gradient gel electrophoresis and quantitative rRNA hybridization. Int. J. Food Microbiol. 65:45-54. [DOI] [PubMed] [Google Scholar]
- 2.Becker, S., P. Boger, R. Oehlmann, and A. Ernst. 2000. PCR bias in ecological analysis: a case study for quantitative Taq nuclease assays in analyses of microbial communities. Appl. Environ. Microbiol. 66:4945-4953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.ben Omar, N., and F. Ampe. 2000. Microbial community dynamics during production of the Mexican fermented maize dough pozol. Appl. Environ. Microbiol. 66:3664-3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bodrossy, L., and A. Sessitsch. 2004. Oligonucleotide microarrays in microbial diagnostics. Curr. Opin. Microbiol. 7:245-254. [DOI] [PubMed] [Google Scholar]
- 5.Bodrossy, L., N. Stralis-Pavese, J. C. Murrell, S. Radajewski, A. Weilharter, and A. Sessitsch. 2003. Development and validation of a diagnostic microbial microarray for methanotrophs. Environ. Microbiol. 5:566-582. [DOI] [PubMed] [Google Scholar]
- 6.Call, D. R., M. K. Borucki, and F. J. Loge. 2003. Detection of bacterial pathogens in environmental samples using DNA microarrays. J. Microbiol. Methods 53:235-243. [DOI] [PubMed] [Google Scholar]
- 7.Chandler, D. P., G. J. Newton, J. A. Small, and D. S. Daly. 2003. Sequence versus structure for the direct detection of 16S rRNA on planar oligonucleotide microarrays. Appl. Environ. Microbiol. 69:2950-2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheigh, H. S., and K. Y. Park. 1994. Biochemical, microbiological, and nutritional aspects of kimchi (Korean fermented vegetable products). Crit. Rev. Food Sci. Nutr. 34:175-203. [DOI] [PubMed] [Google Scholar]
- 9.Cho, J. C., and J. M. Tiedje. 2001. Bacterial species determination from DNA-DNA hybridization by using genome fragments and DNA microarrays. Appl. Environ. Microbiol. 67:3677-3682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Christensen, H., O. Angen, R. Mutters, J. E. Olsen, and M. Bisgaard. 2000. DNA-DNA hybridization determined in micro-wells using covalent attachment of DNA. Int. J. Syst. Evol. Microbiol. 50:1095-1102. [DOI] [PubMed] [Google Scholar]
- 11.Cocolin, L., M. Manzano, C. Cantoni, and G. Comi. 2001. Denaturing gradient gel electrophoresis analysis of the 16S rRNA gene V1 region to monitor dynamic changes in the bacterial population during fermentation of Italian sausages. Appl. Environ. Microbiol. 67:5113-5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cook, K. L., and G. S. Sayler. 2003. Environmental application of array technology: promise, problems and practicalities. Curr. Opin. Biotechnol. 14:311-318. [DOI] [PubMed] [Google Scholar]
- 13.Dahllof, I. 2002. Molecular community analysis of microbial diversity. Curr. Opin. Biotechnol. 13:213-217. [DOI] [PubMed] [Google Scholar]
- 14.De Ley, J., and J. De Smedt. 1975. Improvements of the membrane filter method for DNA:rRNA hybridization. Antonie Leeuwenhoek 41:287-307. [DOI] [PubMed] [Google Scholar]
- 15.de Man, J., M. Rogosa, and M. E. Sharpe. 1960. A medium for the cultivation of lactobacilli. J. Appl. Bacteriol. 23:130-135. [Google Scholar]
- 16.Denef, V. J., J. Park, J. L. Rodrigues, T. V. Tsoi, S. A. Hashsham, and J. M. Tiedje. 2003. Validation of a more sensitive method for using spotted oligonucleotide DNA microarrays for functional genomics studies on bacterial communities. Environ. Microbiol. 5:933-943. [DOI] [PubMed] [Google Scholar]
- 17.Dorris, D. R., R. Ramakrishnan, D. Trakas, F. Dudzik, R. Belval, C. Zhao, A. Nguyen, M. Domanus, and A. Mazumder. 2002. A highly reproducible, linear, and automated sample preparation method for DNA microarrays. Genome Res. 12:976-984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ellenbroek, F. M., and T. E. Cappenberg. 1991. DNA synthesis and tritiated thymidine incorporation by heterotrophic freshwater bacteria in continuous culture. Appl. Environ. Microbiol. 57:1675-1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fleet, G. H. 1999. Microorganisms in food ecosystems. Int. J. Food Microbiol. 50:101-117. [DOI] [PubMed] [Google Scholar]
- 20.Forney, L. J., X. Zhou, and C. J. Brown. 2004. Molecular microbial ecology: land of the one-eyed king. Curr. Opin. Microbiol. 7:210-220. [DOI] [PubMed] [Google Scholar]
- 21.Giraffa, G. 2004. Studying the dynamics of microbial populations during food fermentation. FEMS Microbiol. Rev. 28:251-260. [DOI] [PubMed] [Google Scholar]
- 22.Giraffa, G., and E. Neviani. 2001. DNA-based, culture-independent strategies for evaluating microbial communities in food-associated ecosystems. Int. J. Food Microbiol. 67:19-34. [DOI] [PubMed] [Google Scholar]
- 23.Greene, E. A., and G. Voordouw. 2003. Analysis of environmental microbial communities by reverse sample genome probing. J. Microbiol. Methods 53:211-219. [DOI] [PubMed] [Google Scholar]
- 24.Guschin, D. Y., B. K. Mobarry, D. Proudnikov, D. A. Stahl, B. E. Rittmann, and A. D. Mirzabekov. 1997. Oligonucleotide microchips as genosensors for determinative and environmental studies in microbiology. Appl. Environ. Microbiol. 63:2397-2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Handran, S., S. Pickett, and D. Verdnik. 2002. Key considerations for accurate microarray scanning and image analysis, p. 83-98. In G. Kamberova (ed.), DNA image analysis: nuts and bolts. DNA Press, Salem, Mass.
- 26.Henckel, T., M. Friedrich, and R. Conrad. 1999. Molecular analyses of the methane-oxidizing microbial community in rice field soil by targeting the genes of the 16S rRNA, particulate methane monooxygenase, and methanol dehydrogenase. Appl. Environ. Microbiol. 65:1980-1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ishii, K., and M. Fukui. 2001. Optimization of annealing temperature to reduce bias caused by a primer mismatch in multitemplate PCR. Appl. Environ. Microbiol. 67:3753-3755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kakinuma, K., M. Fukushima, and R. Kawaguchi. 2003. Detection and identification of Escherichia coli, Shigella, and Salmonella by microarrays using the gyrB gene. Biotechnol. Bioeng. 83:721-728. [DOI] [PubMed] [Google Scholar]
- 29.Kim, T.-W., J.-Y. Lee, S.-H. Jung, Y.-M. Kim, J.-S. Jo, D.-K. Chung, H.-J. Lee, and H.-Y. Kim. 2002. Identification and distribution of predominant lactic acid bacteria in kimchi, a Korean traditional fermented food. J. Microbiol. Biotechnol. 12:635-642. [Google Scholar]
- 30.Konings, W. N., J. Kok, O. P. Kuipers, and B. Poolman. 2000. Lactic acid bacteria: the bugs of the new millennium. Curr. Opin. Microbiol. 3:276-282. [DOI] [PubMed] [Google Scholar]
- 31.Kumar, S., K. Tamura, I. B. Jakobsen, and M. Nei. 2001. MEGA2: molecular evolutionary genetics analysis software. Bioinformatics 17:1244-1245. [DOI] [PubMed] [Google Scholar]
- 32.Lafarge, V., J. C. Ogier, V. Girard, V. Maladen, J. Y. Leveau, A. Gruss, and A. Delacroix-Buchet. 2004. Raw cow milk bacterial population shifts attributable to refrigeration. Appl. Environ. Microbiol. 70:5644-5650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Loy, A., A. Lehner, N. Lee, J. Adamczyk, H. Meier, J. Ernst, K. H. Schleifer, and M. Wagner. 2002. Oligonucleotide microarray for 16S rRNA gene-based detection of all recognized lineages of sulfate-reducing prokaryotes in the environment. Appl. Environ. Microbiol. 68:5064-5081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Peplies, J., F. O. Glockner, and R. Amann. 2003. Optimization strategies for DNA microarray-based detection of bacteria with 16S rRNA-targeting oligonucleotide probes. Appl. Environ. Microbiol. 69:1397-1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Peplies, J., S. C. Lau, J. Pernthaler, R. Amann, and F. O. Glockner. 2004. Application and validation of DNA microarrays for the 16S rRNA-based analysis of marine bacterioplankton. Environ. Microbiol. 6:638-645. [DOI] [PubMed] [Google Scholar]
- 36.Polz, M. F., and C. M. Cavanaugh. 1998. Bias in template-to-product ratios in multitemplate PCR. Appl. Environ. Microbiol. 64:3724-3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ramisse, V., J. Balandreau, F. Thibault, D. Vidal, G. Vergnaud, and P. Normand. 2003. DNA-DNA hybridization study of Burkholderia species using genomic DNA macro-array analysis coupled to reverse genome probing. Int. J. Syst. Evol. Microbiol. 53:739-746. [DOI] [PubMed] [Google Scholar]
- 38.Randazzo, C. L., S. Torriani, A. D. Akkermans, W. M. de Vos, and E. E. Vaughan. 2002. Diversity, dynamics, and activity of bacterial communities during production of an artisanal Sicilian cheese as evaluated by 16S rRNA analysis. Appl. Environ. Microbiol. 68:1882-1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rhee, S. K., X. Liu, L. Wu, S. C. Chong, X. Wan, and J. Zhou. 2004. Detection of genes involved in biodegradation and biotransformation in microbial communities by using 50-mer oligonucleotide microarrays. Appl. Environ. Microbiol. 70:4303-4317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rudi, K., O. M. Skulberg, R. Skulberg, and K. S. Jakobsen. 2000. Application of sequence-specific labeled 16S rRNA gene oligonucleotide probes for genetic profiling of cyanobacterial abundance and diversity by array hybridization. Appl. Environ. Microbiol. 66:4004-4011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 42.Schleifer, K. H., and E. Stackebrandt. 1983. Molecular systematics of prokaryotes. Annu. Rev. Microbiol. 37:143-187. [DOI] [PubMed] [Google Scholar]
- 43.Selander, R. K., R. M. McKinney, T. S. Whittam, W. F. Bibb, D. J. Brenner, F. S. Nolte, and P. E. Pattison. 1985. Genetic structure of populations of Legionella pneumophila. J. Bacteriol. 163:1021-1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Small, J., D. R. Call, F. J. Brockman, T. M. Straub, and D. P. Chandler. 2001. Direct detection of 16S rRNA in soil extracts by using oligonucleotide microarrays. Appl. Environ. Microbiol. 67:4708-4716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sneath, P. H. A. 1983. Distortions of taxonomic structure from incomplete data on a restricted set of reference strains. J. Gen. Microbiol. 129:1045-1073. [Google Scholar]
- 46.Speksnijder, A. G., G. A. Kowalchuk, S. De Jong, E. Kline, J. R. Stephen, and H. J. Laanbroek. 2001. Microvariation artifacts introduced by PCR and cloning of closely related 16S rRNA gene sequences. Appl. Environ. Microbiol. 67:469-472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stackebrandt, E., and B. Goebel. 1994. Taxonomic note: a place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int. J. Syst. Bacteriol. 44:846-849. [Google Scholar]
- 48.Tannock, G. W. 2004. A special fondness for lactobacilli. Appl. Environ. Microbiol. 70:3189-3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Taroncher-Oldenburg, G., E. M. Griner, C. A. Francis, and B. B. Ward. 2003. Oligonucleotide microarray for the study of functional gene diversity in the nitrogen cycle in the environment. Appl. Environ. Microbiol. 69:1159-1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tiquia, S. M., L. Wu, S. C. Chong, S. Passovets, D. Xu, Y. Xu, and J. Zhou. 2004. Evaluation of 50-mer oligonucleotide arrays for detecting microbial populations in environmental samples. BioTechniques 36:664-670, 672, 674-675. [DOI] [PubMed] [Google Scholar]
- 52.van Beek, S., and F. G. Priest. 2002. Evolution of the lactic acid bacterial community during malt whisky fermentation: a polyphasic study. Appl. Environ. Microbiol. 68:297-305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang, D., L. Coscoy, M. Zylberberg, P. C. Avila, H. A. Boushey, D. Ganem, and J. L. DeRisi. 2002. Microarray-based detection and genotyping of viral pathogens. Proc. Natl. Acad. Sci. USA 99:15687-15692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang, D., L. Zhu, D. Jiang, X. Ma, Y. Zhou, and J. Cheng. 2004. Direct detection of 16S rRNA using oligonucleotide microarrays assisted by base stacking hybridization and tyramide signal amplification. J. Biochem. Biophys. Methods 59:109-120. [DOI] [PubMed] [Google Scholar]
- 55.Wayne, L., D. Brenner, R. Colwell, P. Grimont, O. Kandler, M. Krichevsky, L. Moore, W. Moore, R. Murray, E. Stackebrandt, M. Starr, and H. Truper. 1987. Report of the Ad Hoc Committee on Reconciliation of Approaches to Bacterial Systematics. Int. J. Syst. Bacteriol. 37:463-464. [Google Scholar]
- 56.Ye, R. W., T. Wang, L. Bedzyk, and K. M. Croker. 2001. Applications of DNA microarrays in microbial systems. J. Microbiol. Methods 47:257-272. [DOI] [PubMed] [Google Scholar]
- 57.Yeates, C., M. R. Gillings, A. D. Davison, N. Altavilla, and D. A. Veal. 1998. Methods for microbial DNA extraction from soil for PCR amplification. Biol. Proced. Online 1:40-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang, L., U. Srinivasan, C. F. Marrs, D. Ghosh, J. R. Gilsdorf, and B. Foxman. 2004. Library on a slide for bacterial comparative genomics. BMC Microbiol. 4:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhou, J. 2003. Microarrays for bacterial detection and microbial community analysis. Curr. Opin. Microbiol. 6:288-294. [DOI] [PubMed] [Google Scholar]
- 60.Zhou, J., and D. K. Thompson. 2002. Challenges in applying microarrays to environmental studies. Curr. Opin. Biotechnol. 13:204-207. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.