Abstract
Due to the difficulty of multiple deletions using the Cre/loxP system, a simple, markerless multiple-deletion method based on a Cre/mutant lox system combining a right-element (RE) mutant lox site with a left-element (LE) mutant lox site was employed for large-scale genome rearrangements in Corynebacterium glutamicum. Eight distinct genomic regions that had been identified previously by comparative analysis of C. glutamicum R and C. glutamicum 13032 genomes were targeted for deletion. By homologous recombination, LE and RE mutant lox sites were integrated at each end of a target region. Highly efficient and accurate deletions between the two chromosomal mutant lox sites in the presence of Cre recombinase were realized. A deletion mutant lacking 190 kb of chromosomal regions, encoding a total of 188 open reading frames (ORFs), was obtained. These deletions represent the largest genomic excisions in C. glutamicum reported to date. Despite the loss of numerous predicted ORFs, the mutant exhibited normal growth under standard laboratory conditions. The Cre/loxP system using a pair of mutant lox sites provides a new, efficient genome rearrangement technique for C. glutamicum. It should facilitate the understanding of genome functions of microorganisms.
The whole-genome sequences of more than 200 organisms have been deciphered since the first genome sequence was determined in 1995. These genome sequences have become an important resource for a more comprehensive understanding of cellular life. The availability of whole-genome sequences allows us to decipher the roles of thousands of genes. Corynebacterium glutamicum is a well-known industrial strain widely used for the production of amino acids, nucleic acids, and organic acids (18, 23). It has had two strains sequenced: R (3,314,179 bp [our unpublished data]) and ATCC 13032 (3,309,401 bp [14] or 3,282,708 bp [17]). Based on whole-genome sequences, strain reconstruction studies for improved industrial applications have been initiated (28). In the field of bacterial genomics and metabolic engineering in the postgenome era, the concept of minimum genome factories (MGFs) has been proposed (16). These can be defined as recombinant strains whose metabolism has been streamlined to the optimal minimal subset in order to maximize product formation for targeted applications. C. glutamicum is one of the most widely used bacteria for bioindustry, and the improvement of its genome is important for enhanced production of biochemicals.
To implement the concept of MGFs by the rearrangement of bacterial genomes, molecular biology tools that make multiple excisions and insertions possible are a prerequisite. For Escherichia coli, the development of genomic engineering techniques utilizing bacteriophage recombinases, homologous recombination, or transposable elements has been reported recently (6, 10, 12, 20, 39). By use of such techniques, the construction of a deletion mutant whose genome size was reduced by 1.38 Mb was reported (12). However, despite its industrial usefulness, similarly useful techniques have yet to be developed for C. glutamicum. In our laboratory, comprehensive studies on C. glutamicum, including isolation and characterization of new transposable elements and development of Cre/loxP-mediated genome deletion systems, are being undertaken (7, 16, 32, 33, 35, 38). The utilization of such techniques could greatly contribute to creation of a C. glutamicum-based MGF.
The Cre/loxP recombination system is a simple two-component system currently recognized as a powerful DNA recombination tool (21). Cre recombinase catalyzes reciprocal site-specific recombination between two loxP sites. It does not require any host cofactors or accessory proteins (9). When two loxP sites are in the same orientation on a linear DNA molecule, Cre-mediated intramolecular recombination resolves with the excision of the loxP-flanking region. Though we have previously accomplished successful genetic manipulation of C. glutamicum with this system, one major disadvantage remains. One loxP site is left on the genome even after recombination occurs, and it may interfere with subsequent rounds of recombination (32).
Recently, several methods to control the recombination of Cre/loxP have been reported (1, 2). The loxP site is composed of an asymmetric 8-bp spacer flanked by 13-bp inverted repeats (13). Cre protein binds to the 13-bp repeat, mediating the recombination within the 8-bp spacer (22). In these studies, nucleotide changes were introduced into the left 13-bp element (LE mutant lox site) or the right 13-bp element (RE mutant lox site). Recombination between the LE and RE mutant lox sites produces the wild-type loxP site and a mutant lox site containing both the LE and the RE (LE+RE mutant lox site) that is poorly recognized by Cre. By using this LE-and-RE mutant lox site(s), successful recombination in plant or mouse genomes was maintained (1, 2). This LE-and-RE mutant system has many potential uses, because after the initial recombination, subsequent rounds of recombination hardly ever occur.
To solve the problem of interference by the remaining loxP site, we used LE-and-RE mutant lox sites and successfully developed a new large-segment deletion method for the C. glutamicum genome that makes multiple reactions possible. Eleven genomic regions that were not essential for cell survival were identified, and successive deletions of eight of these regions were carried out in the same strain. A total of 190 kb of genomic regions, encoding 188 predicted open reading frames, was deleted. We confirmed that the lost genomic regions were not essential for cell survival under normal laboratory conditions.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The bacterial strains used in this study are listed in Table 1. E. coli was grown aerobically at 37°C in Luria-Bertani (LB) medium (31). C. glutamicum R was cultivated at 33°C in complex medium or minimal medium, each containing 4% glucose (15). Antibiotics were used at the following concentrations: for E. coli, 50 μg of kanamycin (Km) (Wako Pure Chemical, Osaka, Japan)/ml, 50 μg of chloramphenicol (Cm) (Wako)/ml, and 200 μg of spectinomycin (Sp) (Sigma-Aldrich, MO)/ml; for C. glutamicum, 50 μg of Km/ml, 5 μg of Cm/ml, and 200 μg of Sp/ml.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s) | Source or reference |
|---|---|---|
| Strains | ||
| E. coli | ||
| JM109 | recA1 endA1 gyrA96 thi hsdR17 supE44 relA1 Δ(lac-proAB)/F′ [traD36 proAB+lacIqlacZΔM15] | TAKARA |
| SCS110 | dam dcm endA1 supE44 hsdR17 thi leu rpsL1 lacY galK galT ara tonA thr Δ(lac-proAB)/F′ [traD36 proAB+lacIqlacZΔM15] | TOYOBO |
| C. glutamicum | ||
| R | Wild-type strain | Lab collection |
| RMD1 | R strain with SSI8 deleted | This work |
| RMD(190) | R strain with multiple deletions of SSI1 to 3 and SSI6 to 10 | This work |
| 13032 | Wild-type strain | ATCC |
| C. efficiens YS-314 | Wild-type strain | NBRC |
| Plasmids | ||
| pMC1871 | Tcr; lacZ/MCS; pBR322 ori; cloning vector | Accession no. L08936 |
| ploxSp | Spr; MCS (with loxP site); pMB1/M13 ori | 33 |
| ploxKm | Kmr; MCS (with loxP); pMB1/M13 ori | 33 |
| pRElox66Sp | Spr; MCS (with lox66 site); pMB1/M13 ori | This work |
| pRElox66Spblue | Spr; MCS (with lox66 site) carrying lacP::lacZ; pMB1/M13 ori | This work |
| pLElox71Km | Kmr; MCS (with lox71); pMB1/M13 ori | This work |
| pCRA405 | Cmr; MCS (with XhoI); pMB1/M13 ori; pBL1/coryneform bacterial ori; shuttle vector | 32 |
| pCRA406 | Cmr; MCS (with XhoI); Cre; pMB1/M13 ori; pBL1/coryneform bacterial ori | 32 |
| pCRA418 | Kmr; MCS (with lox71 site); GR1; pMB1/M13 ori | This work |
| pCRA419 | Spr; MCS (with lox66 site) carrying lacP::lacZ; GR2; pMB1/M13 ori | This work |
DNA manipulations.
E. coli plasmid DNA was isolated using a Qiaprep spin kit (QIAGEN, Hilden, Germany) according to the manufacturer's instructions. Restriction enzymes and T4 DNA ligase (TAKARA Bio Inc., Shiga, Japan) were used as recommended by the manufacturer. E. coli was transformed by the CaCl2 method (31). Transformation and integration of C. glutamicum were performed by electroporation as previously described (37). Purified DNA extracted from E. coli strain SCS110 with 1 μg of an integrative plasmid or 50 ng of the replicative plasmid was introduced into C. glutamicum cells using Gene Pulsar (Bio-Rad, Richmond, CA). After electroporation, cells were incubated at 33°C for 2 h in 1 ml complex medium and then transferred to complex medium plates containing appropriate reagents. β-Galactosidase activity was detected on complex medium plates containing 200 μg/ml 5-bromo-4-chloro-3-indolyl-β-galactopyranoside (X-Gal) (Nacalai Tesque, Kyoto, Japan). DNA concentrations were measured at 260 nm using a Beckman DU640 spectrophotometer (Beckman Coulter, CA).
Plasmids.
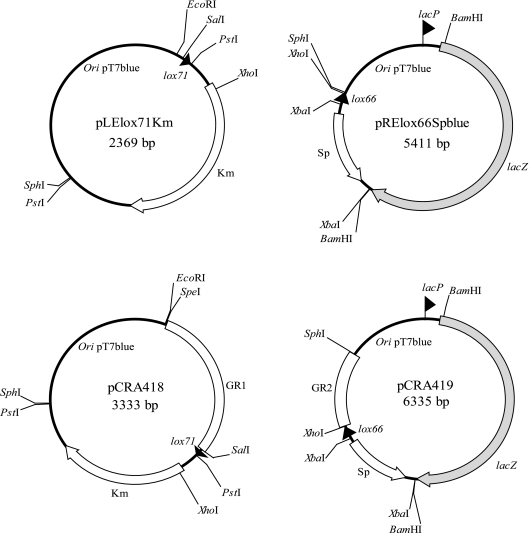
The plasmids and PCR primers used in this work are listed in Tables 1 and 2, respectively. The 34-bp LE and RE mutant lox sites were introduced into the genome of C. glutamicum via homologous recombination. For this purpose, two types of integrative plasmids, pCRA418 and pCRA419, were constructed (Fig. 1). They are suicide vectors to introduce LE and RE mutant lox sites into the chromosome. First, ploxSp and ploxKm were linearized by PCR using primer pairs loxSpF-RElox66R and LElox71F-loxKmR, respectively, and phosphorylated. Then they were circularized and used to transform E. coli. The plasmids extracted from E. coli were designated pRElox66Sp and pLElox71Km (Fig. 1). A BamHI fragment containing a lacZ gene derived from pMC1871 was ligated to the BamHI site of pRElox66Sp downstream of a lac promoter, resulting in pRElox66Spblue (Fig. 1). Two kinds of 1-kb DNA fragments, GR1(SSI8) and GR2(SSI8), were amplified by PCR using C. glutamicum R genomic DNA and primer pairs GR1-F(SSI8)-GR1-R(SSI8) and GR2-F(SSI8)-GR2-R(SSI8), respectively. GR1(SSI8) was digested with EcoRI and SalI and then ligated to the corresponding sites of pLElox71Km. GR2(SSI8) was digested with XhoI and SphI and then ligated to the corresponding sites of pRElox66Spblue. The resulting plasmids were designated pCRA418(SSI8) and pCRA419(SSI8), respectively. Both plasmids were integrated into C. glutamicum R by electroporation (32). Cells that integrated both mutant lox sites into their genomes were selected with corresponding antibiotics (Km, 50 μg/ml; Sp, 200 μg/ml) and confirmed by direct cell PCR. For the deletion reaction, the resultant cells were transformed by the Cre expression plasmid pCRA406 and the transformants were selected with chloramphenicol (5 μg/ml). For subsequent deletions, short DNA segments of approximately 1 kb carrying the 5′- and 3′-flanking regions of target regions were amplified by PCR and were displaced with GR1(SSI8) and GR2(SSI8) of pCRE418(SSI8) and pCRE419(SSI8) with SpeI and SalI or XhoI and SphI, respectively.
TABLE 2.
Oligonucleotide DNA primers used in this study
| Primer | Sequencea | Position in chromosome |
|---|---|---|
| GR1-F(SSI1) | 5′-atactagTAGCTCAGTTGGTAGAGCAC | 29090 |
| GR1-R(SSI1) | 5′-atgtcgacGGAAAACCGAGACTCCTATC | 29860 |
| GR2-F(SSI1) | 5′-atctcgaGCCACGCATTCAAGCAATCA | 43815 |
| GR2-R(SSI1) | 5′-atgcatgcACCTTGCTGTCGAATGTACG | 44556 |
| GR1-F(SSI2) | 5′-atactAGTCTAACCTGAGTATGGCAAGA | 69374 |
| GR1-R(SSI2) | 5′-atgtcgacTGTCTAGTACTGCCGATCGA | 70347 |
| GR2-F(SSI2) | 5′-atctcgagATCAAGAGCAGCCAGTGCAT | 81562 |
| GR2-R(SSI2) | 5′-atgcaTGCCATTGAAGAACTCGGCCAGT | 82583 |
| GR1-F(SSI3) | 5′-atactagtAGTCCACGACGCAATTATCG | 131135 |
| GR1-R(SSI3) | 5′-atgtcgacAGTTTCTAACGGAACGGCAC | 132117 |
| GR2-F(SSI3) | 5′-atctcgagCGACCCGAATATAGGTCTCA | 187812 |
| GR2-R(SSI3) | 5′-gcATGCCACCACTTCTACCTC | 188719 |
| GR1-F(SSI6) | 5′-atactagtAAGATTGCGCAGACTGAGCA | 700769 |
| GR1-R(SSI6) | 5′-atgtcgacTTCAAGACAGCACTCCAAGCT | 701786 |
| GR2-F(SSI6) | 5′-atctcgagCGTGCGTGGAATCAAACACT | 715258 |
| GR2-R(SSI6) | 5′-atgcatgcACGTAGTAGTTCTTGTCGCG | 716297 |
| GR1-F(SSI7) | 5′-atactagtTAGCTGCCAGGAACACTAGA | 792026 |
| GR1-R(SSI7) | 5′-atgtcgaCACCCAGCATCATGGCTTTCT | 792907 |
| GR2-F(SSI7) | 5′-atctcgaGATCAGGCATTGGCTAGTCCT | 808304 |
| GR2-R(SSI7) | 5′-atgcatgcCTACCTGCACCAATTCCAAC | 809341 |
| GR1-F(SSI8) | 5′-atgaattcactagTCTTCATCCTCAGCGGAATCA | 2067324 |
| GR1-R(SSI8) | 5′-atgtcgacTTTCGACAACTATGGGCACG | 2068290 |
| GR2-F(SSI8) | 5′-atctcgaGATGATGAGATTGCCGACGCA | 2111028 |
| GR2-R(SSI8) | 5′-atgcatgcAGTCGAGATTCTTGAAGGG | 2111942 |
| GR1-F(SSI9) | 5′-atactagtGCAATCGTCTGGTTCTCTGT | 2523132 |
| GR1-R(SSI9) | 5′-atgtCGACCTTGACCTCTACTTCTGCTG | 2524141 |
| GR2-F(SSI9) | 5′-GTTCTCAGCTCTGTCAACAG | 2540567 |
| GR2-R(SSI9) | 5′-tagcaTGCCATCAGCTGTGGTAGGAAGA | 2541696 |
| GR1-F(SSI10) | 5′-atactagTAACGGCAGTACCTATGTGGT | 2551837 |
| GR1-R(SSI10) | 5′-atgtcgacTGCGCAATGACACCACGATA | 2552788 |
| GR2-F(SSI10) | 5′-atctcgaGCTCACCCAGTGACAACATCA | 2570010 |
| GR2-R(SSI10) | 5′-atgcatgcCCAGCTGTAGATCTCTACCA | 2571014 |
| LElox71F | 5′-taccgttcgtatagcatacattatacgaagttat | |
| loxKmR | 5′-gtcgactctagaggatcccc | |
| loxSpF | 5′-ctcgagacctgcaggcatgcaa | |
| RElox66R | 5′-taccgttcgtataatgtatgctatacgaagttat |
Capital letters stand for nucleotides representing chromosomal sequences.
FIG. 1.
Construction of integrative plasmids for multiple deletion. pCRA418 and pCRA419 contain the genomic sequences named GR1 and GR2, respectively. Using them, the 34-bp LE and RE mutant lox sites were introduced into the genome of C. glutamicum via homologous recombination.
Sequencing.
Sequencing was performed using an ABI PRISM 3100 Genetic Analyzer with a BigDye Terminator v3.1 cycle sequencing kit (both from Applied Biosystems, Foster City, CA) according to the manufacturer's instructions. DNA sequence data were analyzed using the Genetyx (Tokyo, Japan) WIN program.
PFGE.
Intact Corynebacterium genomic DNA was prepared as follows. A 10-ml volume of overnight culture at an optical density at 610 nm (OD610) of 7 to 8 was centrifuged at 1,000 × g for 10 min at 4°C. Cells were collected and washed twice with STE buffer (pH 8.5; 10 mM NaCl, 1 mM EDTA, 20 mM Tris-HCl). Cells resuspended in 1 ml of STE buffer were digested with 10 mg of lysozyme (Sigma) and incubated for 3 h at 37°C. A 0.1-ml volume of suspension was mixed with 0.9 ml of 1% melting agarose in 1× TAE (40 mM Tris-acetate, 1 mM EDTA) buffer. Aliquots of the molten mixture were pipetted and solidified in a mold. The solid sample blocks were incubated for 15 h in 2 ml of lysis buffer (1 mg/ml proteinase K, 500 mM EDTA [pH 8.5], 10 mM Tris-HCl [pH 7.5], 1% N-laurylsarcosine), transferred to 2 ml of 50 mM EDTA (pH 7.5) solution, and incubated for 12 h at 4°C. Sample blocks were transferred to a new 50 mM EDTA solution and incubated for 12 h at 4°C. After incubation, samples were transferred to 1× TAE buffer and preserved. Pulsed-field gel electrophoresis (PFGE) was performed by using the CHEF-DR II system (Bio-Rad) with 0.8% SeaKem GTG agarose (Cambrex, NJ) in 1× TAE buffer. The running time was 74 h, the temperature 14°C, the angle 106°, the pulse time 35 min, and the voltage 2 V/cm. After electrophoresis, gels were stained with ethidium bromide and DNA was detected.
RESULTS
Scheme for deletion of the C. glutamicum R genome using the Cre/mutant lox system.
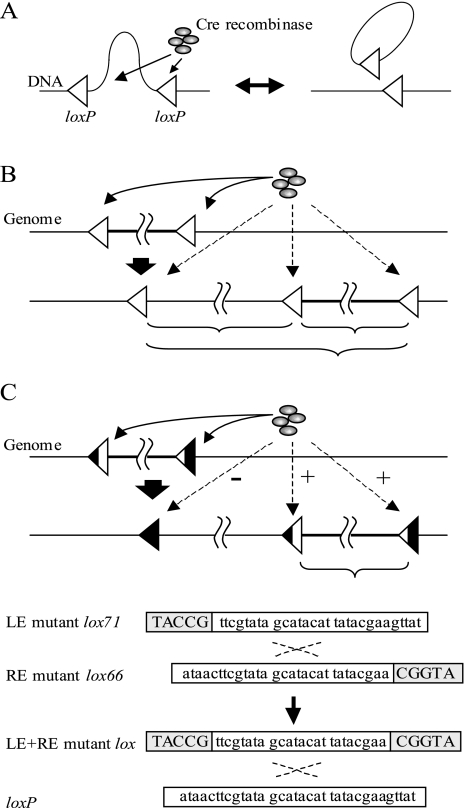
Normally, Cre catalyzes reciprocal site-specific recombination between two loxP sites (Fig. 2A). After recombination occurs, one loxP site is left on the genome and may interfere with subsequent rounds of reaction (Fig. 2B). The Cre/mutant lox system can avoid this problem. The loxP site is known to be composed of an asymmetric 8-bp spacer flanked by 13-bp inverted repeats (Fig. 2C). The spacer is a recombination region, and the inverted repeats are Cre recombinase binding sites (9). When recombination occurs, the inverted repeats are exchanged between two loxP sites (9). This mechanism is also utilized for the Cre/mutant lox system to avoid interference with subsequent rounds of reaction. One loxP is mutated in the left inverted repeat (LE mutant lox71), and the other loxP is mutated in the right inverted repeat (RE mutant lox66). Due to the exchange of inverted repeats, recombination between the LE mutant lox71 site and the RE mutant lox66 site produces a wild-type loxP site and a LE+RE mutant lox site (1). This LE+RE mutant site is poorly recognized by Cre recombinase because of the double mutation on Cre recombinase binding sites. By using this property, successive deletions of eight 10- to 56-kb genomic regions were achieved in each strain.
FIG. 2.
Diagrams of Cre/loxP and Cre/mutant lox systems. (A) Recombination between loxP sites. (B) Interference of loxP sites. After a deletion reaction using the Cre/loxP system, one loxP site remains on the genome. It interferes with subsequent rounds of Cre/loxP recombination. (C) Recombination between a LE mutant site and a RE mutant site produces a wild-type loxP site and a double-mutant (LE+RE) site. The black regions of the triangles represent sites at which nucleotide sequence changes occurred. Since the LE+RE mutant lox exhibits reduced binding affinity for Cre recombinase, it scarcely interferes with the other lox sequences. Nucleotide sequences of loxP and mutant lox sites are given under the diagram of the Cre/mutant lox system. The gray boxes also indicate sites at which nucleotide sequences changed.
In previous work, we identified 11 strain-specific islands (SSIs) of C. glutamicum R that are not essential for cell survival. They were individually deleted using the Cre/loxP system, and no significantly different phenotype was found for the deletion mutants (34). In this study, we chose SSI1 to -3 and SSI6 to -10, carrying a total of 188 genes, as targets to develop a multiple-deletion system based on the Cre/mutant lox system (34). The functions of most of the genes in these regions are annotated as unknown or hypothetical.
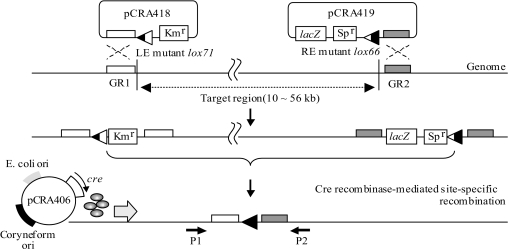
Three plasmids were used to excise each SSI from the genome. pCRA418 and pCRA419 are suicide plasmids that introduce the LE and RE mutant lox sites into the chromosome via homologous recombination. pCRA406 is a replicative Cre recombinase expression plasmid for C. glutamicum. The general scheme for deletion is depicted in Fig. 3.
FIG. 3.
Schematic representation of Cre/mutant lox-mediated deletion of a large segment of the C. glutamicum R genome. Kmr and Spr indicate kanamycin and spectinomycin resistance genes, respectively; lacZ encodes β-galactosidase. GR1 and GR2 are short segments of the C. glutamicum R genome. These segments were amplified by PCR and integrated into plasmids for homologous recombination. Horizontal black arrows (P1 and P2) under the chromosome represent PCR primers.
Deletion of SSI8 using the Cre/mutant lox system.
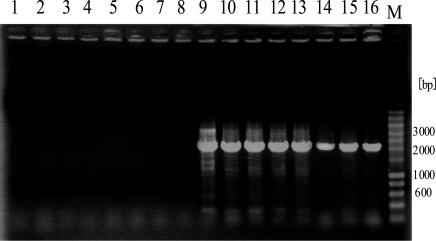
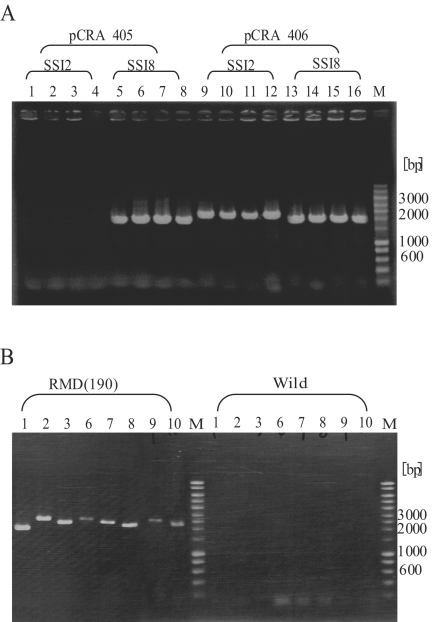
First, both pCRA418(SSI8) and pCRA419(SSI8) were integrated into 5′ and 3′ flanking regions of a target SSI8. Then cells with corresponding antibiotic resistance were selected, and chromosomal modification was verified by direct cell PCR. The resultant mutants were transformed with pCRA406 and selected based on their chloramphenicol resistance. Cre recombinase is constitutively expressed under the control of the lac promoter on pCRA406 (5). Successfully deleted strains lost lacZ and both Sp and Km resistance genes along with each SSI. By transformation with pCRA406, 300 to 500 yellow colonies were obtained on complex medium plates containing chloramphenicol and X-Gal, indicating the loss of the lacZ gene. Forty-eight of these colonies were transferred to a new plate and monitored for kanamycin and spectinomycin resistance. All of them were kanamycin and spectinomycin sensitive (data not shown). These results suggested that the target region was successfully deleted. As a control, pCRA405, which has exactly the same sequences as pCRA406 but does not carry the cre gene, was also used for transformation. All transformants were blue, indicating the presence of the lacZ gene. Cells that lost β-galactosidase activity and antibiotic resistance were selected, and the deletion of SSI8 was confirmed by PCR using P1(SSI8) and P2(SSI8). Since the distance between P1(SSI8) and P2(SSI8) was approximately 52 kb, it was difficult to amplify this region by PCR. However, the region was easily amplified after the deletion reaction, because the distance was reduced to only 2 kb. The ∼2-kb DNA fragments were successfully amplified, and no DNA fragments were observed from the Cre− strain (Fig. 4). Finally, the PCR products with P1(SSI8) and P2(SSI8) were sequenced, and GR1(SSI8) was directly connected to GR2(SSI8) via a LE+RE mutant lox site. These results indicated the excision of SSI8 by the Cre/mutant lox system. The deletion strain was designated RMD1.
FIG. 4.
Direct cell PCR results for Cre− and Cre+ strains. Eight colonies each of pCRA405 and pCRA406 transformants were chosen. Since the lengths of GR1 and GR2 are approximately 1.1 and 0.9 kb, respectively, the distance between GR1 and GR2 of the deletion strain of SSI8 is calculated to be about 2 kb but that of the pCRA405 strain is about 52 kb. Lanes 1 to 8, Cre− strains; lanes 9 to 16, Cre+ strains; lane M, marker.
Construction of a multiple-deletion strain.
Since excision of SSI8 by the Cre/mutant lox system was successful, multiple excisions of other SSIs were performed similarly. SSI2 was chosen as the second deletion target. In order to remove the plasmid expressing Cre recombinase, RMD1 cells were cultivated for 24 h in complex medium without antibiotics and plated. Approximately 2 to 5% of cells showed chloramphenicol sensitivity due to the loss of pCRA406. One of them was used in SSI2 deletion. pCRA418(SSI2) and pCRA419(SSI2) were constructed by replacement of GR1(SSI8) and GR2(SSI8) with GR1(SSI2) and GR2(SSI2). Both pCRA418(SSI2) and pCRA419(SSI2) were integrated into 5′ and 3′ flanking regions of a target SSI2, and the resultant mutants were transformed with pCRA406. As a result, 200 to 300 yellow colonies appeared on a complex medium plate containing chloramphenicol and X-Gal, and the 48 colonies transferred to a new plate completely lost kanamycin and spectinomycin resistance (data not shown). To confirm the SSI2 deletion, PCR was carried out. By using P1(SSI2) and P2(SSI2), a DNA fragment of approximately 2.1 kb was successfully amplified from the transformant of pCRA406 but not from the transformant of pCRA405 (Fig. 5A). When P1(SSI8) and P2(SSI8) were used, amplification of a fragment of approximately 2.0 kb was observed in all samples (Fig. 5A). PCR products amplified with P1(SSI2) and P2(SSI2) were isolated and sequenced. Sequence data indicated that GR1(SSI2) was directly connected to GR2(SSI2) via a mutant LE+RE lox site (data not shown).
FIG. 5.
Direct cell PCR results for the double-deletion strain and the multiple-deletion strain. (A) LE and RE mutant lox sites were integrated into RMD1, and pCRA405 or pCRA406 was transformed. P1 and P2 primers for each SSI were used for direct cell PCR. The fact that transformants were obtained for pCRA406 only indicates successful amplification of 2-kb DNA fragments derived from SSI2 and SSI8 deletion. Lanes 1 to 8, Cre− strains; lanes 9 to 16, Cre+ strains; lane M, marker. (B) By using P1 and P2 primers for each SSI, corresponding PCR products of eight SSIs were observed with the multiple-deletion strain only. Lane numbers correspond to SSI numbers; lane M, marker.
Since two genomic regions were deleted within the same cells by using the Cre/mutant lox system, subsequent deletion of SSI1, -3, -6, -7, -9, and -10 was performed. These SSIs were deleted with almost equivalent efficiency. The constructed multiple-deletion mutant was designated RMD(190). The lengths of the SSIs are as follows (DDBJ accession numbers are in parentheses): SSI1 (AB185495), 15.6 kb; SSI2 (AB193029), 11.1 kb; SSI3 (AB193030), 56.2 kb; SSI6 (AB193033), 16.2 kb; SSI7 (AB193034), 16.5 kb; SSI8 (AB193035), 45.1 kb; SSI9 (AB193036), 16.3 kb; and SSI10 (AB193037), 14.6 kb. The total size of the deletion was about 190 kb. DNA fragments of 1.9 to 2.5 kb were amplified by using strain RMD(190) as the PCR template with P1 and P2 primers corresponding to each SSI (Fig. 5B). The eight new chromosomal joints, formed by SSI deletions, were verified by sequencing of PCR products (data not shown). Even though a predicted total of 188 genes were lost from its genome, strain RMD(190) formed colonies on plates and showed no significant difference from the wild type (data not shown). Microscopic observation also showed no significant differences from the wild type strain. The genes predicted in the regions are listed in Table 3.
TABLE 3.
Genes in deleted regions
| NCBI COGa | Annotation by KEEG | No. of genes |
|---|---|---|
| Information storage | Transcriptional regulator | 9 |
| Putative transposase | 8 | |
| Putative DNA invertase | 3 | |
| Hypothetical protein | 3 | |
| Putative regulatory protein | 2 | |
| Putative antirepressor | 1 | |
| Putative single-strand binding protein | 1 | |
| Cellular processes | Hypothetical protein | 16 |
| Cation-transporting ATPase | 5 | |
| Putative transferase | 2 | |
| Two-component system response regulator | 2 | |
| Putative two-component sensor kinase | 2 | |
| ClpP protease family protein | 1 | |
| Benzoate dioxygenase large subunit | 1 | |
| Putative monooxygenase | 1 | |
| Metabolism | Hypothetical protein | 10 |
| Putative dehydrogenase | 7 | |
| Putative transport protein | 6 | |
| Putative reductase | 6 | |
| Enoyl-CoA hydratase | 2 | |
| Putative phenylacetic acid degradation protein PaaB | 2 | |
| Polysaccharide deacetylase | 1 | |
| Aminopeptidase N | 1 | |
| AMP nucleosidase | 1 | |
| Probable oligoribonuclease | 1 | |
| Putative phenylacetate-CoA ligase | 1 | |
| Putative beta-ketoadipyl CoA thiolase | 1 | |
| Poorly characterized | Hypothetical protein | 12 |
| Putative reductase | 4 | |
| Putative phenylacetic acid degradation protein PaaD | 1 | |
| Putative phenylacetic acid degradation protein PaaA | 1 | |
| Putative phenylacetic acid degradation protein PaaC | 1 | |
| Not in COG | Hypothetical protein | 67 |
| Putative transposase | 3 | |
| Putative cadmium resistance protein | 2 | |
| Putative DNA-binding protein | 1 | |
| Total | 188 |
COG, clusters of orthologous groups of proteins.
Comparison of genome sizes.
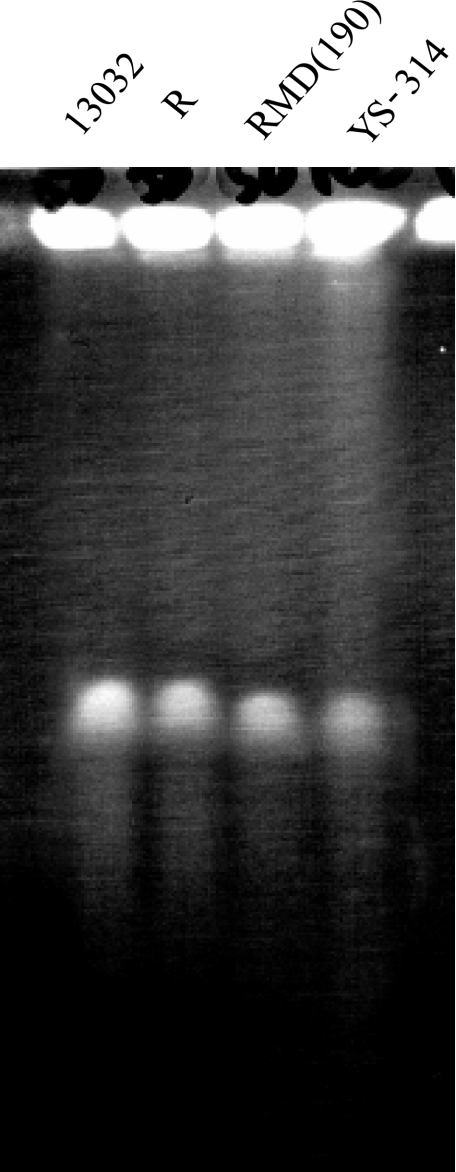
Since the size of the C. glutamicum R genome is approximately 3.31 Mb and strain RMD(190) lacks a total of 190 kb, the size of the RMD(190) genome is estimated to be approximately 3.12 Mb. Corynebacterium efficiens YS-314 is the closest relative of C. glutamicum, and its genome size is approximately 3.15 Mb. In the PFGE analysis, the band position of RMD(190) genomic DNA was almost the same as that of YS-314, which confirmed the reduction of genome size (Fig. 6).
FIG. 6.
PFGE of strains. Genomic DNAs of C. glutamicum ATCC 13032, R, and RMD(190) and C. efficiens YS-314 were run on a 0.8% agarose gel. The genome sizes of ATCC 13032, R, and YS-314 are 3.31, 3.31, and 3.15 Mb, respectively. The calculated genome size of RMD(190) was predicted as 3.12 Mb. DNAs were visualized by ethidium bromide staining and UV irradiation after electrophoresis.
Growth characteristics of RMD(190).
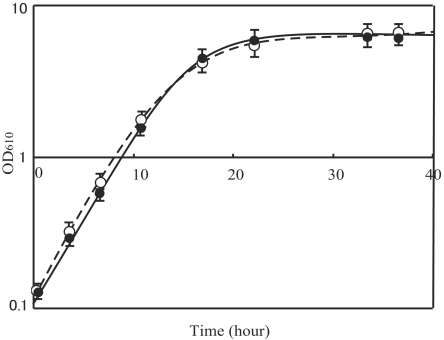
The wild type and RMD(190) were cultivated in minimal medium in which glucose was the sole carbon source. Cells were initially cultured in 1.0 ml minimal medium, centrifuged, and washed twice with 5 ml minimal medium. They were then diluted in approximately 1.5 ml of minimal medium, and 1 ml of each solution was inoculated in 100 ml minimal medium. Despite the loss of numerous genes, no significant differences in the growth rate or the final OD610 were observed between RMD(190) and the wild type (Fig. 7). However, no competition experiment between the 190-kb-deletion strain and the wild type was performed.
FIG. 7.
Growth curves of wild-type C. glutamicum and RMD(190). Cultures were grown in triplicate in minimal medium. Each 100 ml of medium inoculated with overnight cultures was grown at 33°C with shaking. Open and closed circles, wild type and RMD(190), respectively.
DISCUSSION
The Cre/loxP system is a powerful tool for in vivo genome rearrangement in prokaryotic and eukaryotic cells (2, 3, 29, 32, 36, 39). However, it has the disadvantage that a loxP sequence is left on the genome after reaction and may interfere with subsequent rounds of recombination. To overcome this, a new, simple large-segment-deletion method using LE and RE mutant lox, which is markerless and makes multiple deletions possible for Corynebacterium glutamicum, was developed. Since this method is based on the Cre/loxP system, the recombination efficiency is very high and does not require any host cofactors or accessory proteins.
LE and RE mutant lox sites were initially devised to integrate foreign DNA into a plant or mouse genome using Cre recombinase (1, 2). Many types of mutant lox can be designed. Albert et al. report a series of experiments which identified three sets of mutant lox sites that favor the forward over the reverse direction of recombination (1). We chose and applied the combination of mutant lox71 and lox66 for the multiple-deletion method due to the efficiency of targeted recombination between the mutant lox sites. Both mutant lox71 and lox66 retain the function for DNA recombination, but the recognition by Cre recombinase is slightly reduced. When loxP sites with mutations in the LE and RE are used for recombination, Cre recombinase recombines the DNA between a site with a mutation in the LE and a site with a mutation in the RE, producing one wild-type loxP site and one lox site with mutations in both the LE and the RE (LE+RE mutant lox site). The LE+RE mutant lox site showed dramatically reduced binding affinity for Cre recombinase. When mutant lox71 and lox66 were used as the LE and RE mutant lox sites, recombination between the wild-type site and the LE+RE mutant lox site was three- to sevenfold less than that of the lox71 and lox66 sites (1). Recombination between the lox71 or lox66 and the LE+RE mutant lox site would be much less than that of the lox71 and lox66 sites. Indeed in this experiment, LE+RE mutant lox sites on the genome did not interfere with lox71 and lox66 for subsequent rounds of deletion, and precise excision of the target region only was observed. We successfully used the Cre/mutant lox system for the recombination of the C. glutamicum genome. Since the Cre/loxP system is used to recombine DNA in many types of cells, this system could be applied to DNA recombination in various organisms.
As another point, the selection of target regions is important for the multiple-deletion experiments. Essential or significant genes are normally scattered in prokaryotic genomes. Large-segment deletion is accompanied by the loss of numerous genes. If the mutant lost any essential genes by deletion, it would be impossible to isolate it. To avoid deleting these essential genes, we used strain-specific regions as deletion targets. These regions were identified by comparative genomics. Thousands of these regions (islands) were identified in the common backbone of the C. glutamicum R genome (32). We have already identified 11 SSIs larger than 10 kb and designated them SSI1 to SSI11 (34). The regions were probably shaped on the genome by integration and deletion through evolutionary processes. The existence of similar regions in E. coli is also known (20). Genes found in these regions are often annotated as encoding antibiotic resistance factors, bacteriocins, or proteins with specific metabolic functions (11, 24, 27, 30). These regions are probably deleted with little effect on cell survival. We have already reported the successful deletion of 11 SSIs individually, and all deletion mutants survived with no significant phenotypic change (34). However, the effect of multiple deletions of SSIs was unclear. In this study, multiple deletion of SSI1 to -3 and SSI6 to -10 was performed, and no significant phenotypic change was observed under laboratory conditions. It is an interesting basis for analyzing genomic and gene functions.
Genomes probably contain many genes that are not essential for cell survival (4, 25). A popular approach to the identification of nonessential genes is to construct gene disruption mutants. For Bacillus subtilis, construction of comprehensive gene knockouts on a genomic scale is complete (19). For E. coli and Saccharomyces cerevisiae, libraries of single-gene deletions using a PCR-based mutagenesis approach are nearly complete (8, 26). However, although this approach is useful in identifying the roles of various important genes and potentially facilitates the understanding of gene function, it requires numerous deletion experiments. Furthermore, some difficulties in the study of the effects of multiple gene disruptions remain.
The utilization of multiple-large-segment-deletion methods should help solve these problems. These methods facilitate the investigation of unknown gene functions and contribute to the elucidation of new gene functions that cannot be understood by individual gene disruption. C. glutamicum is one of the most widely used bacteria for bioindustry. The utilization of such large-segment-deletion methods should greatly contribute to the investigation of gene and genome functions and the creation of improved cells for bioindustry.
Acknowledgments
We thank Roy H. Doi (University of California, Davis) and C. Omumasaba for critical reading of the manuscript. We are also grateful to M. Wada for technical support.
This study was carried out as part of The Project for Development of a Technological Infrastructure for Industrial Bioprocesses of New Industrial Science and Technology Frontiers by the Ministry of Economy, Trade and Industry (METI) and funded by the New Energy and Industrial Technology Development Organization (NEDO).
REFERENCES
- 1.Albert, H., E. C. Dale, E. Lee, and D. W. Ow. 1995. Site-specific integration of DNA into wild-type and mutant lox sites placed in the plant genome. Plant J. 7:649-659. [DOI] [PubMed] [Google Scholar]
- 2.Araki, K., M. Araki, and K. Yamamura. 1997. Targeted integration of DNA using mutant lox sites in embryonic stem cells. Nucleic Acids Res. 25:868-872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Campo, N., M. L. Daveran-Mingot, K. Leenhouts, P. Ritzenthaler, and P. Le Bourgeois. 2002. Cre-loxP recombination system for large genome rearrangements in Lactococcus lactis. Appl. Environ. Microbiol. 68:2359-2367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Casjens, S. 2003. Prophages and bacterial genomics: what have we learned so far? Mol. Microbiol. 49:277-300. [DOI] [PubMed] [Google Scholar]
- 5.Eikmanns, B. J., E. Kleinertz, W. Liebl, and H. Sahm. 1991. A family of Corynebacterium glutamicum/Escherichia coli shuttle vectors for cloning, controlled gene expression, and promoter probing. Gene 102:93-98. [DOI] [PubMed] [Google Scholar]
- 6.Fukiya, S., H. Mizoguchi, and H. Mori. 2004. An improved method for deleting large regions of Escherichia coli K-12 chromosome using a combination of Cre/loxP and lambda Red. FEMS Microbiol. Lett. 234:325-331. [DOI] [PubMed] [Google Scholar]
- 7.Garbe, T. R., N. Suzuki, M. Inui, and H. Yukawa. 2004. Inhibitor-associated transposition events in Corynebacterium glutamicum. Mol. Genet. Genomics 271:729-741. [DOI] [PubMed] [Google Scholar]
- 8.Giaever, G., A. M. Chu, L. Ni, C. Connelly, L. Riles, S. Veronneau, S. Dow, A. Lucau-Danila, K. Anderson, B. Andre, A. P. Arkin, A. Astromoff, M. El-Bakkoury, R. Bangham, R. Benito, S. Brachat, S. Campanaro, M. Curtiss, K. Davis, A. Deutschbauer, K. D. Entian, P. Flaherty, F. Foury, D. J. Garfinkel, M. Gerstein, D. Gotte, U. Guldener, J. H. Hegemann, S. Hempel, Z. Herman, D. F. Jaramillo, D. E. Kelly, S. L. Kelly, P. Kotter, D. LaBonte, D. C. Lamb, N. Lan, H. Liang, H. Liao, L. Liu, C. Luo, M. Lussier, R. Mao, P. Menard, S. L. Ooi, J. L. Revuelta, C. J. Roberts, M. Rose, P. Ross-Macdonald, B. Scherens, G. Schimmack, B. Shafer, D. D. Shoemaker, S. Sookhai-Mahadeo, R. K. Storms, J. N. Strathern, G. Valle, M. Voet, G. Volckaert, C. Y. Wang, T. R. Ward, J. Wilhelmy, E. A. Winzeler, Y. Yang, G. Yen, E. Youngman, K. Yu, H. Bussey, J. D. Boeke, M. Snyder, P. Philippsen, R. W. Davis, and M. Johnston. 2002. Functional profiling of the Saccharomyces cerevisiae genome. Nature 418:387-391. [DOI] [PubMed] [Google Scholar]
- 9.Gopaul, D. N., F. Guo, and G. D. Van Duyne. 1998. Structure of the Holliday junction intermediate in Cre-loxP site-specific recombination. EMBO J. 17:4175-4187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goryshin, I. Y., T. A. Naumann, J. Apodaca, and W. S. Reznikoff. 2003. Chromosomal deletion formation system based on Tn5 double transposition: use for making minimal genomes and essential gene analysis. Genome Res. 13:644-653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hacker, J., G. Blum-Oehler, I. Muhldorfer, and H. Tschape. 1997. Pathogenicity islands of virulent bacteria: structure, function and impact on microbial evolution. Mol. Microbiol. 23:1089-1097. [DOI] [PubMed] [Google Scholar]
- 12.Hashimoto, M., T. Ichimura, H. Mizoguchi, K. Tanaka, K. Fujimitsu, K. Keyamura, T. Ote, T. Yamakawa, Y. Yamazaki, H. Mori, T. Katayama, and J. Kato. 2005. Cell size and nucleoid organization of engineered Escherichia coli cells with a reduced genome. Mol. Microbiol. 55:137-149. [DOI] [PubMed] [Google Scholar]
- 13.Hoess, R. H., M. Ziese, and N. Sternberg. 1982. P1 site-specific recombination: nucleotide sequence of the recombining sites. Proc. Natl. Acad. Sci. USA 79:3398-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ikeda, M., and S. Nakagawa. 2003. The Corynebacterium glutamicum genome: features and impacts on biotechnological processes. Appl. Microbiol. Biotechnol. 62:99-109. [DOI] [PubMed] [Google Scholar]
- 15.Inui, M., S. Murakami, S. Okino, H. Kawaguchi, A. A. Vertès, and H. Yukawa. 2004. Metabolic analysis of Corynebacterium glutamicum during lactate and succinate productions under oxygen deprivation conditions. J. Mol. Microbiol. Biotechnol. 7:182-196. [DOI] [PubMed] [Google Scholar]
- 16.Inui, M., Y. Tsuge, N. Suzuki, A. A. Vertès, and H. Yukawa. 2005. Isolation and characterization of a native composite transposon, Tn14751, carrying 17.4 kilobases of Corynebacterium glutamicum chromosomal DNA. Appl. Environ. Microbiol. 71:407-416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kalinowski, J., B. Bathe, D. Bartels, N. Bischoff, M. Bott, A. Burkovski, N. Dusch, L. Eggeling, B. J. Eikmanns, L. Gaigalat, A. Goesmann, M. Hartmann, K. Huthmacher, R. Krämer, B. Linke, A. C. McHardy, F. Meyer, B. Möckel, W. Pfefferle, A. Pühler, D. A. Rey, C. Rückert, O. Rupp, H. Sahm, V. F. Wendisch, I. Wiegräbe, and A. Tauch. 2003. The complete Corynebacterium glutamicum ATCC 13032 genome sequence and its impact on the production of l-aspartate-derived amino acids and vitamins. J. Biotechnol. 104:5-25. [DOI] [PubMed] [Google Scholar]
- 18.Kinoshita, S. 1985. Glutamic acid bacteria, p. 115-146. In A. L. Demain and N. A. Solomon (ed.), Biology of industrial microorganisms. Cummings, London, United Kingdom.
- 19.Kobayashi, K., S. D. Ehrlich, A. Albertini, G. Amati, K. K. Andersen, M. Arnaud, K. Asai, S. Ashikaga, S. Aymerich, P. Bessieres, F. Boland, S. C. Brignell, S. Bron, K. Bunai, J. Chapuis, L. C. Christiansen, A. Danchin, M. Debarbouille, E. Dervyn, E. Deuerling, K. Devine, S. K. Devine, O. Dreesen, J. Errington, S. Fillinger, S. J. Foster, Y. Fujita, A. Galizzi, R. Gardan, C. Eschevins, T. Fukushima, K. Haga, C. R. Harwood, M. Hecker, D. Hosoya, M. F. Hullo, H. Kakeshita, D. Karamata, Y. Kasahara, F. Kawamura, K. Koga, P. Koski, R. Kuwana, D. Imamura, M. Ishimaru, S. Ishikawa, I. Ishio, D. Le Coq, A. Masson, C. Mauel, R. Meima, R. P. Mellado, A. Moir, S. Moriya, E. Nagakawa, H. Nanamiya, S. Nakai, P. Nygaard, M. Ogura, T. Ohanan, M. O'Reilly, M. O'Rourke, Z. Pragai, H. M. Pooley, G. Rapoport, J. P. Rawlins, L. A. Rivas, C. Rivolta, A. Sadaie, Y. Sadaie, M. Sarvas, T. Sato, H. H. Saxild, E. Scanlan, W. Schumann, J. F. Seegers, J. Sekiguchi, A. Sekowska, S. J. Seror, M. Simon, P. Stragier, R. Studer, H. Takamatsu, T. Tanaka, M. Takeuchi, H. B. Thomaides, V. Vagner, J. M. van Dijl, K. Watabe, A. Wipat, H. Yamamoto, M. Yamamoto, Y. Yamamoto, K. Yamane, K. Yata, K. Yoshida, H. Yoshikawa, U. Zuber, and N. Ogasawara. 2003. Essential Bacillus subtilis genes. Proc. Natl. Acad. Sci. USA 100:4678-4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kolisnychenko, V., G. Plunkett III, C. D. Herring, T. Feher, J. Posfai, F. R. Blattner, and G. Posfai. 2002. Engineering a reduced Escherichia coli genome. Genome Res. 12:640-647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuhn, R., and R. M. Torres. 2002. Cre/loxP recombination system and gene targeting. Methods Mol. Biol. 180:175-204. [DOI] [PubMed] [Google Scholar]
- 22.Lee, L., and P. D. Sadowski. 2001. Directional resolution of synthetic Holliday structures by the Cre recombinase. J. Biol. Chem. 276:31092-31098. [DOI] [PubMed] [Google Scholar]
- 23.Malumbres, M., L. M. Mateos, and J. F. Martin. 1995. Microorganisms for amino acid production: Escherichia coli and corynebacteria, p. 423-469. In Y. H. Hui and G. G. Kachatourians (ed.), Food biotechnology microorganisms, vol. 2. VCH Publishers, New York, N.Y. [Google Scholar]
- 24.McClelland, M., L. Florea, K. Sanderson, S. W. Clifton, J. Parkhill, C. Churcher, G. Dougan, R. K. Wilson, and W. Miller. 2000. Comparison of the Escherichia coli K-12 genome with sampled genomes of a Klebsiella pneumoniae and three Salmonella enterica serovars, Typhimurium, Typhi and Paratyphi. Nucleic Acids Res. 28:4974-4986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mira, A., H. Ochman, and N. A. Moran. 2001. Deletional bias and the evolution of bacterial genomes. Trends Genet. 17:589-596. [DOI] [PubMed] [Google Scholar]
- 26.Mori, H., K. Isono, T. Horiuchi, and T. Miki. 2000. Functional genomics of Escherichia coli in Japan. Res. Microbiol. 151:121-128. [DOI] [PubMed] [Google Scholar]
- 27.Ochman, H., and I. B. Jones. 2000. Evolutionary dynamics of full genome content in Escherichia coli. EMBO J. 19:6637-6643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ohnishi, J., M. Hayashi, S. Mitsuhashi, and M. Ikeda. 2003. Efficient 40°C fermentation of l-lysine by a new Corynebacterium glutamicum mutant developed by genome breeding. Appl. Microbiol. Biotechnol. 62:69-75. [DOI] [PubMed] [Google Scholar]
- 29.Qin, M., C. Bayley, T. Stockton, and D. W. Ow. 1994. Cre recombinase-mediated site-specific recombination between plant chromosomes. Proc. Natl. Acad. Sci. USA 91:1706-1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Riley, M., and M. H. Serres. 2000. Interim report on genomics of Escherichia coli. Annu. Rev. Microbiol. 54:341-411. [DOI] [PubMed] [Google Scholar]
- 31.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 32.Suzuki, N., Y. Tsuge, M. Inui, and H. Yukawa. 2005. Cre/loxP-mediated deletion system for large genome rearrangements in Corynebacterium glutamicum. Appl. Microbiol. Biotechnol. 67:225-233. [DOI] [PubMed] [Google Scholar]
- 33.Suzuki, N., H. Nonaka, Y. Tsuge, S. Okayama, M. Inui, and H. Yukawa. Multiple large segment deletion method for Corynebacterium glutamicum. Appl. Microbiol. Biotechnol., in press. [DOI] [PubMed]
- 34.Suzuki, N., S. Okayama, H. Nonaka, Y. Tsuge, M. Inui, and H. Yukawa. 2005. Large-scale engineering of the Corynebacterium glutamicum genome. Appl. Environ. Microbiol. 71:3369-3372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tsuge, Y., K. Ninomiya, N. Suzuki, M. Inui, and H. Yukawa. 2005. A new insertion sequence, IS14999, from Corynebacterium glutamicum. Microbiology 151:501-508. [DOI] [PubMed] [Google Scholar]
- 36.Van Deursen, J., M. Fornerod, B. Van Rees, and G. Grosveld. 1995. Cre-mediated site-specific translocation between nonhomologous mouse chromosomes. Proc. Natl. Acad. Sci. USA 92:7376-7380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vertès, A. A., M. Inui, M. Kobayashi, Y. Kurusu, and H. Yukawa. 1993. Presence of mrr- and mcr-like restriction systems in coryneform bacteria. Res. Microbiol. 144:181-185. [DOI] [PubMed] [Google Scholar]
- 38.Vertès, A. A., M. Inui, M. Kobayashi, Y. Kurusu, and H. Yukawa. 1994. Isolation and characterization of IS31831, a transposable element from Corynebacterium glutamicum. Mol. Microbiol. 11:739-746. [DOI] [PubMed] [Google Scholar]
- 39.Yu, B. J., B. H. Sung, M. D. Koob, C. H. Lee, J. H. Lee, W. S. Lee, M. S. Kim, and S. C. Kim. 2002. Minimization of the Escherichia coli genome using a Tn5-targeted Cre/loxP excision system. Nat. Biotechnol. 20:1018-1023. [DOI] [PubMed] [Google Scholar]