Abstract
The switch from budding to filamentous growth is a key aspect of invasive growth and virulence for the fungal phytopathogen Ustilago maydis. The cyclic AMP (cAMP) signaling pathway regulates dimorphism in U. maydis, as demonstrated by the phenotypes of mutants with defects in protein kinase A (PKA). Specifically, a mutant lacking the regulatory subunit of PKA encoded by the ubc1 gene displays a multiple-budded phenotype and fails to incite disease symptoms, although proliferation does occur in the plant host. A mutant with a defect in a catalytic subunit of PKA, encoded by adr1, has a constitutively filamentous phenotype and is nonpathogenic. We employed serial analysis of gene expression to examine the transcriptomes of a wild-type strain and the ubc1 and adr1 mutants to further define the role of PKA in U. maydis. The mutants displayed changes in the transcript levels for genes encoding ribosomal proteins, genes regulated by the b mating-type proteins, and genes for metabolic functions. Importantly, the ubc1 mutant displayed elevated transcript levels for genes involved in phosphate acquisition and storage, thus revealing a connection between cAMP and phosphate metabolism. Further experimentation indicated a phosphate storage defect and elevated acid phosphatase activity for the ubc1 mutant. Elevated phosphate levels in culture media also enhanced the filamentous growth of wild-type cells in response to lipids, a finding consistent with PKA regulation of morphogenesis in U. maydis. Overall, these findings extend our understanding of cAMP signaling in U. maydis and reveal a link between phosphate metabolism and morphogenesis.
Development and virulence are regulated by cyclic AMP (cAMP)/protein kinase A (PKA) and mitogen-activated protein (MAP) kinase signaling pathways in several fungi, including the corn smut pathogen Ustilago maydis (13, 38, 46). Mating of haploid U. maydis cells, which are nonpathogenic and yeast-like, leads to the formation of infectious, dikaryotic hyphae. This process is regulated by two unlinked mating-type loci, a and b. Cell recognition and fusion of conjugation tubes are controlled by the a mating-type locus encoding a pheromone (mfa)-pheromone receptor (pra) system that acts through a conserved MAP kinase module (8). Several components of the MAP kinase module have been identified, including the MAP kinase kinase kinase Ubc4, the MAP kinase kinase Fuz7/Ubc5, the MAP kinase Ubc3/Kpp2, and the putative adaptor protein Ubc2 (1, 5, 46-48, 50). One target of the module is the pheromone response transcription factor Prf1, a high-mobility group domain protein that activates the transcription of the bW and bE genes encoded by the b mating-type locus (20, 26, 27, 34). The bE and bW homeodomain proteins dimerize to form a transcriptional regulator that controls the formation of the filamentous dikaryon and subsequent pathogenic development (7, 10).
The cAMP-dependent protein kinase signaling pathway also plays a major role in the control of morphogenesis in U. maydis. When cAMP levels are low, PKA is an inactive tetramer comprised of two regulatory and two catalytic subunits. When cAMP levels increase, cAMP binds to the regulatory subunits and induces a conformational change that causes subunit dissociation and activation of the catalytic subunits. Characterization of U. maydis mutants with defects in the genes for the catalytic and regulatory subunits of PKA revealed that mutants with high PKA activity have a budding phenotype but that those with low PKA activity display filamentous growth (15, 22). For example, mutants defective in the regulatory subunit of PKA (encoded by ubc1) display a multiple-budded phenotype, while those lacking adenylyl cyclase (encoded by uac1) or one of the catalytic subunits of PKA (encoded by adr1) are constitutively filamentous (6, 15, 22). The adr1 gene was identified initially in a screen for genes mediating resistance to dicarboximide and aromatic hydrocarbon fungicides (54). Although the PKA encoded by adr1 does not appear to be a direct target of these fungicides, a mutant with a defect in the regulatory subunit of PKA (ubc1) does show resistance as well as other phenotypes, such as osmotic sensitivity (59).
The mutants with defects in the components of the cAMP/PKA pathway also are unable to either proliferate in host tissue or induce tumor formation in planta. In fact, several critical events during infection, including the development of infection filaments within the host, the induction of tumors, hyphal fragmentation, and the formation of prespores, appear to require changes in the level of PKA activity, demonstrating that the cAMP pathway also plays an important role in virulence (6, 15, 22). Consistent with this idea, the novel protein Hgl1 was identified in a suppressor screen for mutations that restored budding growth to a filamentous adr1 mutant, and mutations in hgl1 block sporulation during infection (14).
The processes regulated by cAMP and PKA in Saccharomyces cerevisiae have been well characterized and include, in part, cell cycle progression, cell growth in response to carbon source, ribosome biogenesis, response to stress, accumulation of storage carbohydrates, pseudohyphal growth, and sporulation (71). The cAMP/PKA pathway has also been shown to regulate genes encoding the high-affinity iron uptake system (60). Furthermore, Giots et al. (21) recently identified phosphate as a nutrient signal that activates the cAMP/PKA pathway. In Schizosaccharomyces pombe, cAMP signaling controls gluconeogenesis, spore germination, and the influence of carbon and nitrogen starvation on mating (28). In filamentous saprophytic fungi as well as plant and animal pathogens, the cAMP/PKA pathway regulates morphogenesis (e.g., hyphal growth morphology, appressorium formation) and development (e.g., conidiation) among other processes (13, 38).
Considerable effort is still needed to dissect the processes activated by different levels of PKA activity, including the identification of new targets of PKA and other levels of cross-talk between the cAMP and MAP kinase pathways. In this paper, we describe a comparative analysis of RNA expression for a wild-type strain and mutants defective in the regulatory and catalytic subunits of PKA using the technique of serial analysis of gene expression (SAGE). SAGE involves the generation of short sequence tags (10 to 14 nucleotides) that represent individual transcripts and the use of large-scale sequencing to establish the frequency of occurrence of these tags as a measure of transcript levels (72). SAGE has been applied to define transcriptomes (73) and more extensively to explore transcription in normal and tumor cells (77), including the identification of targets of defined pathways (58). We chose SAGE for defining the U. maydis transcriptome because microarrays are not yet generally available and because we have successfully used SAGE to characterize the transcriptome of another basidiomycete fungal pathogen, Cryptococcus neoformans (40, 69, 70).
Our application of SAGE to U. maydis identified genes whose transcript levels vary with PKA activity. In addition to finding genes encoding metabolic functions and genes known to be regulated by mating type, we found that many genes encoding ribosomal proteins (RPs) have altered transcript levels in the mutant strains. We also discovered that some of the transcripts regulated by PKA encode phosphate acquisition and storage functions, and we confirmed the regulation of phosphate metabolism by demonstrating differential polyphosphate accumulation and acid phosphatase activity between mutant and wild-type cells. Given the role of the cAMP/PKA pathway in morphogenesis for U. maydis, we also examined possible connections between phosphate and the switch from budding to filamentous growth. In this case, we found that phosphate levels influence the filamentous growth of U. maydis that occurs in response to lipids (36), thus revealing further connections between morphogenetic signals and the cAMP pathway.
MATERIALS AND METHODS
Strains and growth conditions.
The following U. maydis strains were employed: the wild-type strain 521 (a1b1), the ubc1 mutant lacking the regulatory subunit of PKA (strain 111, a1b1 ubc1::hygBr), and the adr1 mutant lacking the catalytic subunit of PKA that is responsible for the majority of the activity (strain 002-10, a1b1 adr1::phlr) (15, 22). For SAGE library construction, 2-ml cultures of potato dextrose broth (PDB; Difco) were inoculated with single colonies and grown overnight at 30°C in a gyratory shaker (250 rpm). These cultures were inoculated into 250 ml of the same medium for subsequent incubation under the same conditions to obtain 107 cells per ml for budding strains or a similar optical density for filamentous strains. Cells were harvested by centrifugation at 4°C for budding strains or by vacuum filtration for filamentous strains and immediately flash frozen in a dry ice-ethanol bath. The strains were streaked on potato dextrose agar (PDA; Difco) plates containing 200 ng ml−1 rapamycin (Sigma-Aldrich) and incubated for 2 days at 30°C.
RNA isolation and analysis.
Frozen cell pellets were lyophilized overnight at −20°C until dry and resuspended in 15 ml of TRIZOL extraction buffer (GIBCO BRL). Total RNA was isolated according to the manufacturer's recommendations. Poly(A)+ RNA was isolated using the FastTrack 2.0 kit (Invitrogen). RNA blot preparation and hybridization were performed as described previously (63). Hybridization probes for genes represented by differently expressed tags were prepared by PCR amplification from genomic DNA using the primers listed in Table 1. These probes were labeled with an oligonucleotide labeling kit (Amersham Pharmacia Biotech, Inc.).
TABLE 1.
Primer pairs used to amplify probes for Northern analysis
| Preliminary gene designation | Tag | Primers | Amplicon size (bp) |
|---|---|---|---|
| Inorganic phosphate permease | TGTATCTTAC | 5′GACGACCCTGAGCTTATTGC3′ | 525 |
| 5′GACGAAGACGACGAAGAAGG3′ | |||
| Hmp1, cruciform DNA-binding protein | CCAATGAATA | 5′GCTGGTACCGTCAAGGAGAC3′ | 398 |
| 5′CAATGGTCGATGACCAAACA3′ | |||
| β-1,3-Glucan binding protein | CACTCGACCC | 5′GTGCTGGATGAGCAGTTTCA3′ | 537 |
| 5′GGTCGAGTGCATGTTGTTTG3′ | |||
| Hypothetical protein | GCAAGCACTG | 5′TGTCAGCGTCTCAGCACTTC3′ | 403 |
| 5′CTTGGTGACGCACTTCTTGA3′ | |||
| Hypothetical protein | TTTGATTCGT | 5′GGCAAGTCTGTTGCACTTGA3′ | 535 |
| 5′TACGCATGACGGTTTGTGAT3′ | |||
| Hypothetical protein | TGCGATCCCG | 5′GGTGGAGCAGAAGAGTGGAG3′ | 546 |
| 5′CCAAAGCCAAAAATTTCGAG3′ | |||
| Thiazole biosynthetic enzyme | AATCACGAAT | 5′TATATGGCGGACATGATGGA3′ | 438 |
| 5′AAGACCATGTGCCATTGTGA3′ | |||
| Acid phosphatase | ACGAACCTGA | 5′AACCGTACGCTTGTTCTGCT3′ | 343 |
| 5′AGAACAACGTGTTCGCAGTG3′ | |||
| Repellent protein | CGCTGCTTGC | 5′TGACCAACGAGAACAAGCTG3′ | 416 |
| 5′CGTCTGCTTAGGAGGAGTGG3′ | |||
| Sugar transporter | TTAGCCTTCT | 5′GGGCTGCTGTCTTCTTCATC3′ | 530 |
| 5′GATGAGATGGCCCTGTTTGT3′ |
SAGE analysis.
SAGE was performed as described by Velculescu et al. (72) using the protocol available at www.sagenet.org. Poly(A) RNA was converted to double-stranded cDNA using the Invitrogen synthesis kit and biotinylated oligo(dT18). Briefly, the cDNA was cleaved with NlaIII, the 3′-terminal cDNA fragments were bound to streptavidin beads (Dynal), and oligonucleotide linkers containing BsmFI restriction sites were ligated to the 5′ ends. The cDNA with linkers was released from the streptavidin beads by BsmFI digestion, and tags were ligated to one another, PCR amplified, concatemerized, and cloned into the SphI site of pZERO 1.0 (Invitrogen). Twenty-six PCR cycles were used to amplify ditags during library construction. Colonies were screened by PCR (M13F and M13R primers) to assess the average clone insert size and the percentage of nonrecombinants. Tag sequences were obtained by BigDye primer cycle sequencing and analysis on an ABI PRISM 3700 DNA analyzer. Sequence chromatograms were processed using Phred (16, 17), and vector sequence was detected using Cross_match (23). Fourteen-base-pair tags were extracted from the vector-clipped sequence, and an overall quality score for each tag was derived based on the cumulative Phred score. Duplicate ditags and linker sequences were removed as described previously (72). Only tags with a predicted accuracy of ≥99% were used in this study, and statistical differences between tag abundance in different libraries were determined using the methods of Audic and Claverie (4).
Tag identification.
Custom Perl scripts were used to make preliminary assignments of tags, and their abundance relative to that of putative transcripts was determined using release 2 (March 2004) of the genome sequence data for U. maydis (strain 521), assembled at the Broad Institute Center for Genome Research (http://www.broad.mit.edu/annotation/fungi/ustilago_maydis/index.html). Tag and gene identification also used the May 2005 version of the Munich Information Center for Protein Sequences (MIPS) Ustilago maydis database (MUMDB [http://mips.gsf.de/genre/proj/ustilago/]) and a database of ∼4,000 expressed sequence tags (ESTs) for this strain generously supplied by Barry Saville (University of Toronto) (52, 62). In some instances, more than one differentially expressed tag could be assigned to a candidate transcript, which may indicate alternative splicing events, differences in poly(A) site use, or partial NlaIII digestion during library preparation. In these instances, where the same trends in abundance were observed for such tags, they were assigned as a group to that transcript, and the cumulative abundance was reported along with the sequence of the tag located at the 3′-most NlaIII site. We restricted our studies to only those candidates for which an unambiguous tag assignment could be made.
For unannotated or uncharacterized transcripts, the coding regions and 1 kb of 5′ and 3′ flanking sequences were used for BLASTX searches, and the results were recorded for those genes that had significant similarity with other proteins in the nonredundant database at the National Center for Biotechnology Information (NCBI). Each BLASTX result was inspected individually, and expect values and tentative gene assignments were recorded for those tags that were found to correspond to the 3′-most NlaIII site within the putative open reading frame or within a 3′ untranslated region. Percent identity and percent similarity between the protein from the nonredundant database and the U. maydis sequences were also recorded when the expect value did not reflect the extent of similarity (mainly for small proteins).
Morphological response to phosphate concentration.
Strains 521, 111 (ubc1::hygBr a1b1), and 002-10 (adr1::phelor a1b1) were grown at 30°C in 5 ml of minimal medium (minus phosphate) supplemented with either 1% glucose, 1% Tween 40, or 1% glucose and 1% Tween 40, combined with either 7.35 or 250 mM KH2PO4. A concentration of 7.35 mM is the standard amount of KH2PO4 in U. maydis minimal medium. Strains were observed using differential-interference-contrast microscopy on a Zeiss Axioplan 2 microscope after 3 and 5 days. Three independent experiments were performed.
Polyphosphate staining.
Strains 521, 111, and 002-10 were grown overnight in 5 ml of PDB medium in a 30°C shaking incubator. Two milliliters of overnight culture was centrifuged at 14,000 rpm for 1 minute, and the medium was removed. The pellets were resuspended in 1 ml of 95% ethanol and incubated for 10 min (room temperature). The suspensions were recentrifuged at 14,000 rpm for 30 seconds, and the ethanol was aspirated. Cell pellets were resuspended in 0.05% toluidine blue O (Fisher Scientific) in 0.2 M acetic acid and incubated for 10 min at room temperature as described previously (49, 67). Cells were washed three to six times in sterile distilled water before observation using light microscopy. Four independent experiments were performed.
Acid phosphatase assay.
U. maydis strains 521, 111, and 002-10 were grown overnight in PDB at 30°C in a shaking incubator. Protein extractions were performed with Y-PER yeast protein extraction reagent (Pierce) as per the manufacturer's instructions. Protein concentrations were estimated using the Bio-Rad microassay (Bio-Rad Laboratories, Inc.) as per the manufacturer's instructions. Acid phosphatase assays were performed according to guidelines in Sigma procedure EC 3.1.3.2 (enzymatic assay of acid phosphatase) using 4-nitrophenol phosphate disodium salt hexahydrate (Sigma-Aldrich) as the substrate and 0.09 M citrate buffer solution (Sigma). A control was performed using 0.18 units/ml of prostatic acid phosphatase (Sigma-Aldrich). Protein extract (500 μl) from each strain was used in the assay. Proteins were extracted, and acid phosphatase assays were performed in triplicate (four independent experiments).
RESULTS
Overview of the SAGE libraries.
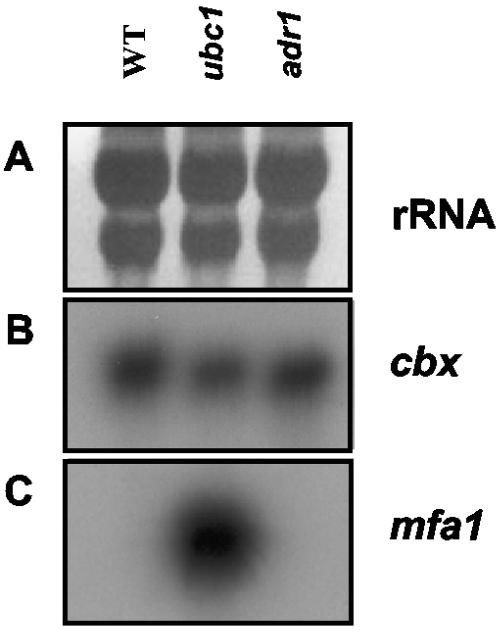
To examine RNA expression in the context of cAMP signaling in U. maydis, we used the wild-type strain 521 and mutants (in the same strain background) deficient in the regulatory (ubc1) or catalytic (adr1) subunits of PKA to generate SAGE libraries designated WT, UBC1, and ADR1. RNA quality was initially checked by RNA blot analysis with probes for the pheromone gene (mfa1), whose expression level is known to be conditioned by the cAMP pathway (27), and the constitutively expressed gene for succinate dehydrogenase (using the allele that confers resistance to carboxin [cbx]). The hybridization results (Fig. 1) revealed similar patterns of expression of cbx among the three strains, as well as the expected expression of mfa1 in the ubc1 mutant, and that the RNA was of sufficient quality for SAGE library construction. A summary of the collection of tags for each library is presented in Table 2. The data shown in this table reflect Phred scores that provide a 99% probability that each tag sequence is correct (see Materials and Methods) and indicate the expression profiles of the three strains compared in pairwise combinations. The percentages of tags differentially expressed between libraries ranged from 0.54% (UBC1 versus ADR1) to 0.92% (WT versus UBC1). It is interesting to note that the highest similarity (99.46%) was obtained when we compared the UBC1 and ADR1 expression profiles. This could indicate that gene expression is strongly affected when PKA activity is lost or not regulated and that in many cases, the alterations of the gene expression are similar when the PKA activity level is either higher or lower than normal. It should also be noted that a second catalytic subunit of PKA (Uka1) is present in U. maydis and that this enzyme could play a role under some conditions (15).
FIG. 1.
Evaluation of RNA for SAGE library preparation. The RNA quality for the wild-type strain 002 (lane WT) and the mutant strains 111 (ubc1::hygBr) and 002-10 (adr1::phelor) was tested prior to library construction. (A) rRNA (18S and 28S) bands as a loading control. (B) Hybridization with the constitutively expressed gene for succinate dehydrogenase (cbx). (C) Hybridization with the pheromone gene (mfa1).
TABLE 2.
Analysis of SAGE libraries
| Characteristic | WT | UBC1 | ADR1 |
|---|---|---|---|
| No. of sequences read | 2,304 | 1,920 | 1,536 |
| Total no. of tagsa | 50,033 | 46,990 | 43,629 |
| Tag families | |||
| No. (%) of singletons | 9,998 (65.7) | 11,184 (68.8) | 11,012 (70) |
| No. (%) with 2 to 9 tags | 4,548 (29.9) | 4,533 (27.9) | 4,402 (27.6) |
| No. (%) with 10 to 99 tags | 625 (4.1) | 512 (3.1) | 525 (3.3) |
| No. (%) with ≥100 tags | 40 (0.26) | 38 (0.23) | 25 (0.16) |
| Total no. of tags | 15,211 | 16,267 | 15,964 |
| Differentially expressed genes (P ≤ 0.01) | |||
| No. (%) in WT library | 231 (0.92) | 199 (0.8) | |
| No. (%) in UBC1 library | 140 (0.54) | ||
| No. (%) in ADR1 library |
Ninety-nine percent probability that each tag sequence is correct.
Identification of differentially expressed genes.
We were able to make preliminary tag assignments to predicted genes using EST data and the partially annotated genomic sequence for U. maydis available at the Broad Institute and MIPS. Of the 230 tags found to have differential abundance between the WT, UBC1, and ADR1 libraries at a threshold P value of less than 0.01, 52 (22.6%) were found to match two or more different locations in the genomic sequence of U. maydis (data not shown) and were not used for further analysis. A further 32 tags (14%) did not match any of the sequences in either the genomic or EST U. maydis database. Tags in this class may fail to match because of errors in the tag sequence (e.g., at the reverse transcription stage of library preparation), because of incomplete genomic or EST sequence data, or because the tags span an intron position and thus don't match the genomic sequence. The remaining 146 tags could be unambiguously correlated to candidate transcripts. BLASTX analysis of these transcripts, when we searched the nonredundant database at NCBI (May 2005), indicated that 16 tags (9%) did not result in a significant BLAST score (i.e., less than E-05); 34 (19%) had similarity to uncharacterized, hypothetical proteins; and 96 (54%) had similarity to genes with assigned functions.
The tags with differential abundance were organized into four categories based on their expression levels in the different libraries (Tables 3 to 6). Table 3 lists tags that were elevated in the ubc1 mutant (expected to have elevated PKA activity) and/or reduced in the adr1 mutant (expected to have low PKA activity). That is, these tags had higher frequencies in the UBC1 library or lower frequencies in the ADR1 library (UBC1 > WT [or ADR1], UBC1 > WT > ADR1, or UBC1 [or WT] > ADR1). Table 4 contains tags with reduced levels in the condition of high PKA activity and/or elevated levels in the condition of low PKA activity. These tags had higher frequencies in the ADR1 library or lower frequencies in the UBC1 library (ADR1 > WT [or UBC1], ADR1 > WT > UBC1, or ADR1 [or WT] > UBC1). Table 5 contains tags with lower levels in situations of loss of PKA activity or loss of regulation, i.e., tags with higher frequencies in the WT library (WT > UBC1 or [ADR1], WT > UBC1 > ADR1, or WT > ADR1 > UBC1). Finally, Table 6 contains tags with elevated levels in situations of loss of PKA activity or regulation, i.e., tags with higher frequencies in the UBC1 and ADR1 libraries (UBC1 or [ADR1] > WT, UBC1 > ADR1 > WT, or ADR1 > UBC1 > WT). The patterns of regulation of specific functional categories of genes identified by the tags are described in the following paragraphs.
TABLE 3.
Tags with elevated levels in the UBC1 library and/or reduced levels in the ADR1 library
| Tag sequence | Normalized frequencya
|
P valueb
|
MUMDB gene accession no.c | Preliminary gene product designation | ||||
|---|---|---|---|---|---|---|---|---|
| WT | UBC1 | ADR1 | WT vs UBC1 | WT vs ADR1 | UBC1 vs ADR1 | |||
| CGTAAAAAGC | 172 | 432 | 104 | 6.99E−30 | 2.50E−5 | 5.51E−50 | No hits | Hypothetical protein |
| TCCTTGATTT | 37 | 88 | 3 | 1.38E−6 | 3.70E−9 | 1.51E−23 | UM10186 | Conserved hypothetical protein |
| GCAAGCACTGd | 0 | 41 | 0 | 1.35E−14 | 7.37E−1 | 2.93E−13 | UM03392 | Hypothetical protein |
| TGTATCTTACd | 3 | 61 | 6 | 1.16E−16 | 4.06E−1 | 3.64E−13 | UM06490 | Inorganic phosphate permease |
| ACGAACCTGAd | 0 | 27 | 0 | 7.16E−10 | 7.37E−1 | 5.55E−9 | UM05803 | Acid phosphatase |
| CGTCAGACCGd | 215 | 239 | 133 | 2.50E−1 | 5.12E−6 | 2.18E−8 | UM00924 | Translation elongation factor 1a |
| CTTGGAAGAC | 5 | 29 | 2 | 1.04E−5 | 2.57E−1 | 2.07E−7 | UM05785 | Acyltransferase-like protein |
| ATGGAATCGA | 67 | 120 | 57 | 4.46E−5 | 3.53E−1 | 1.36E−6 | UM06404 | Thioredoxin peroxidase |
| CATCCTACAA | 55 | 116 | 61 | 6.42E−7 | 5.57E−1 | 2.41E−5 | UM05031 | 60S Ribosomal protein L10a |
| GTGAAAAGCA | 4 | 13 | 0 | 2.93E−2 | 4.65E−2 | 1.05E−4 | UM04248 | Conserved hypothetical protein |
| CACTCCCCTT | 55 | 65 | 31 | 3.33E−1 | 8.02E−3 | 4.04E−4 | UM00926 | Ribosomal protein L13A |
| ATACGTCTTT | 102 | 110 | 65 | 5.40E−1 | 3.18E−3 | 4.72E−4 | UM10196 | 60S Ribosomal protein L27-A |
| CCAAAGGCCG | 5 | 10 | 0 | 1.89E−1 | 2.48E−2 | 7.56E−4 | UM05782 | Capsule-associated protein |
| GCATCACTTA | 126 | 160 | 108 | 3.81E−2 | 2.17E−1 | 1.31E−3 | UM02714 | 40S ribosomal protein S4A/S4.1 |
| ACCTCACTGT | 14 | 36 | 14 | 8.16E−4 | 9.80E−1 | 1.40E−3 | UM05595 | Vacuolar transporter chaperone 1 (Vtc1) |
| GCTTTCGTAC | 2 | 12 | 1 | 2.82E−3 | 7.09E−1 | 1.57E−3 | UM00384 | Hypothetical protein |
| TCTACTGTCC | 11 | 8 | 0 | 4.96E−1 | 3.08E−4 | 2.81E−3 | UM01390 | Hypothetical protein |
| CTCGCATCTC | 3 | 28 | 10 | 1.45E−6 | 6.76E−2 | 3.17E−3 | UM00004 | Conserved hypothetical protein |
| CATCGTGTTT | 7 | 17 | 4 | 3.54E−2 | 3.89E−1 | 4.54E−3 | UM06405 | Conserved hypothetical protein |
| GACATCAGCG | 7 | 7 | 0 | 8.96E−1 | 7.08E−3 | 5.42E−3 | UM01681 | NADH-ubiquinone oxidoreductase |
| AACCAAAATG | 168 | 199 | 148 | 9.42E−2 | 2.41E−1 | 5.64E−3 | UM02440 | Ubiquitin fusion protein |
| CAATCTAAGC | 41 | 31 | 13 | 2.02E−1 | 7.67E−5 | 6.78E−3 | UM00862 | 60S ribosomal protein L14-B |
| CTTTTCTGTA | 470 | 607 | 519 | 1.01E−5 | 1.07E−1 | 7.49E−3 | UM03322 | 60S ribosomal protein L10 |
| GTCAATTATAd | 49 | 77 | 48 | 7.85E−3 | 9.38E−1 | 8.33E−3 | UM10146 | Guanine nucleotide binding protein |
| CTTTTGTAAC | 148 | 159 | 116 | 5.26E−1 | 4.13E−2 | 8.74E−3 | UM01635 | 40S ribosomal protein S27 |
| CAAGATAGCTd | 4 | 32 | 15 | 2.88E−7 | 1.14E−2 | 1.01E−2 | UM03411 | Endo-1,4 beta-d-xylanase |
| TCACATACTT | 83 | 68 | 43 | 1.98E−1 | 2.47E−4 | 1.70E−2 | No hitse | 40S ribosomal protein S29A |
| TATTATTTCA | 0 | 10 | 2 | 3.33E−4 | 2.01E−1 | 1.88E−2 | UM06383 | Hypothetical protein |
| TACATATGAT | 11 | 27 | 15 | 7.60E−3 | 4.58E−1 | 6.40E−2 | UM06230 | Vacuolar transporter chaperone 4 (Vtc4) |
Tag frequencies have been normalized to the size of the smallest library for comparison (ADR1, 43,629 tags).
Statistical significance of the differential tag frequencies between libraries.
MUMDB, MIPs Ustilago maydis database (http://mips.gsf.de/genre/proj/ustilago/).
SAGE tag differences were confirmed by Northern analysis.
Tag has an associated sequence in the U. maydis EST database.
TABLE 6.
Tags with higher levels in the UBC1 and ADR1 libraries
| Tag sequence | Normalized frequencya
|
P valueb
|
MUMDB gene accession no.c | Preliminary gene product designation | ||||
|---|---|---|---|---|---|---|---|---|
| WT | UBC1 | ADR1 | WT vs UBC1 | WT vs ADR1 | UBC1 vs ADR1 | |||
| CCAATGAATAd | 45 | 486 | 248 | 1.00E−105 | 3.62E−38 | 2.53E−19 | UM00496 | Hmp1, mismatch base pair and cruciform DNA recognition protein |
| ATCCTGTGAT | 129 | 346 | 272 | 8.54E−27 | 7.17E−14 | 2.36E−3 | UM00753 | Heat shock protein related to DDR48 |
| TTTCCCTAAT | 4 | 31 | 15 | 9.51E−7 | 1.14E−2 | 1.93E−2 | UM10587 | Conserved hypothetical protein |
| CTCGACACTC | 4 | 26 | 13 | 1.77E−5 | 3.13E−2 | 3.60E−2 | UM03169 | Ornithine aminotransferase |
| ACAACAGTAA | 12 | 40 | 29 | 3.72E−5 | 6.28E−3 | 1.85E−1 | UM01269 | Conserved hypothetical protein |
| TTTCTTGAGC | 0 | 11 | 8 | 1.61E−4 | 2.07E−3 | 4.84E−1 | UM10242 | Hypothetical protein |
| TACATTTATC | 26 | 56 | 42 | 5.22E−4 | 4.64E−2 | 1.61E−1 | UM10700 | 60S ribosomal protein L30-2 |
| TACTCGTATC | 85 | 130 | 125 | 1.05E−3 | 3.74E−3 | 7.53E−1 | UM03791 | Ums2, heat shock 70-kDa protein 2 |
| AAAGCTTGGC | 7 | 21 | 29 | 4.07E−3 | 9.21E−5 | 2.74E−1 | UM04268 | Saccharopine dehydrogenase |
| CAACGTCTGC | 0 | 6 | 6 | 6.05E−3 | 9.52E−3 | 9.01E−1 | UM02109 | Hypothetical protein |
| GATCTTGCTG | 0 | 6 | 6 | 6.05E−3 | 9.52E−3 | 9.01E−1 | UM05036 | Mixed-linked glucanase |
| TGCGATCCCGd | 7 | 19 | 109 | 9.97E−3 | 3.30E−27 | 3.12E−17 | UM03664 | Conserved hypothetical protein |
Tag frequencies have been normalized to the size of the smallest library for comparison (ADR1, 43,629 tags).
Statistical significance of the differential tag frequencies between libraries.
MUMDB, MIPs Ustilago maydis database (http://mips.gsf.de/genre/proj/ustilago/).
SAGE tag differences were confirmed by Northern analysis.
TABLE 4.
Tags with reduced levels in the UBC1 library and/or elevated levels in the ADR1 library
| Tag sequence | Normalized frequencya
|
P valueb
|
MUMDB gene accession no.c | Preliminary gene product designation | ||||
|---|---|---|---|---|---|---|---|---|
| WT | UBC1 | ADR1 | WT vs UBC1 | WT vs ADR1 | UBC1 vs ADR1 | |||
| CGCTGCTTGCd | 0 | 0 | 87 | 7.62E−1 | 1.27E−29 | 2.32E−28 | UM03924 | Repellent protein 1 precursor |
| CAATCTTACG | 3 | 0 | 78 | 1.41E−1 | 1.72E−22 | 1.67E−25 | UM05104 | Hypothetical protein |
| AATCACGAATd | 37 | 7 | 94 | 1.23E−6 | 2.12E−7 | 1.87E−21 | UM02278 | Thiazole biosynthetic enzyme |
| TTCTGTTCCG | 64 | 37 | 158 | 5.48E−3 | 2.42E−11 | 5.59E−20 | UM04922 | Aldo/keto reductase |
| TCTACAGCAGd | 119 | 19 | 112 | 2.66E−21 | 6.17E−1 | 1.74E−18 | No hitse | 6-Phosphogluconolactonase |
| TCTCGCTGCTd | 3 | 27 | 104 | 2.66E−6 | 1.36E−29 | 1.39E−12 | UM03665 | Aldehyde dehydrogenase family 7 member A1 |
| CGATATCTCT | 13 | 2 | 34 | 2.03E−3 | 1.38E−3 | 2.92E−9 | UM10073 | Lincomycin-condensing protein ImbA |
| AACAATCGAA | 0 | 0 | 25 | 7.62E−1 | 4.73E−9 | 1.12E−8 | UM05103b | Sulfate adenylyltransferase |
| CAAAGCAATT | 14 | 6 | 41 | 8.92E−2 | 1.19E−4 | 9.44E−8 | UM01885 | Alcohol dehydrogenase |
| TTAGCCTTCTd | 20 | 1 | 25 | 3.16E−6 | 4.43E−1 | 1.62E−7 | UM01656 | Probable sugar transporter |
| TCATCATTTT | 66 | 70 | 131 | 7.59E−1 | 1.44E−6 | 8.72E−6 | UM03085 | Manganese superoxide dismutase precursor |
| GCACCCATCT | 0 | 1 | 19 | 4.16E−1 | 4.63E−7 | 1.02E−5 | UM01052 | Hypothetical protein |
| ATTCTTCTTA | 21 | 12 | 42 | 1.10E−1 | 5.61E−3 | 2.26E−5 | UMd12-160 | Hypothetical protein |
| ACCAACAACT | 3 | 5 | 27 | 6.78E−1 | 3.40E−6 | 2.72E−5 | UM00816 | Thiamine biosynthesis protein |
| AAAGGCTCGC | 2 | 0 | 14 | 2.72E−1 | 9.13E−4 | 3.46E−5 | UM03728 | Conserved hypothetical protein |
| TTTGCTACGT | 15 | 4 | 24 | 6.77E−3 | 1.27E−1 | 4.61E−5 | UM04957f | Glucose oxidase |
| ATTGGTTCCG | 39 | 27 | 61 | 1.14E−1 | 2.40E−2 | 1.88E−4 | UM10339f | Fatty acyl coenzyme A synthase |
| GCATCTCAAT | 5 | 2 | 16 | 2.13E−1 | 1.47E−2 | 4.09E−4 | UM03103 | Putative ribose 5-phosphate isomerase |
| GCTTCTGGCG | 17 | 6 | 24 | 9.55E−3 | 2.90E−1 | 4.17E−4 | UM00138 | Translation elongation factor Tu |
| CTTTACGAAGd | 1 | 1 | 13 | 7.97E−1 | 3.84E−4 | 5.94E−4 | UM02703 | Fructose-bisphosphatase |
| AAATGTCGTC | 1 | 0 | 10 | 4.59E−1 | 3.08E−3 | 6.44E−4 | UM01775 | Conserved hypothetical protein |
| TGAGCCATAT | 27 | 10 | 31 | 3.73E−3 | 5.85E−1 | 8.27E−4 | UM02267 | Hypothetical protein |
| CTGCTCTCGT | 4 | 6 | 23 | 5.05E−1 | 1.35E−4 | 1.69E−3 | UM06138 | Probable PTR2 di- and tripeptide permease |
| CAATCGCCAC | 17 | 4 | 17 | 1.55E−3 | 9.51E−1 | 2.50E−3 | UM04702 | Holocytochrome c synthase |
| ACGCCGTGCT | 9 | 6 | 22 | 5.68E−1 | 1.24E−2 | 2.76E−3 | UM04972 | Related to NFU-1 protein (iron homeostasis) |
| GCTCCTTATC | 4 | 6 | 20 | 6.91E−1 | 7.60E−4 | 3.26E−3 | UM06052 | Aldose reductase |
| CGCGAGTTGT | 13 | 6 | 20 | 7.30E−2 | 2.13E−1 | 3.26E−3 | UM04207 | Hypothetical protein |
| GAGCAGATGAd | 55 | 25 | 49 | 4.21E−4 | 5.55E−1 | 4.42E−3 | UM04138 | Transaldolase TAL1 |
| AATTTTGATA | 49 | 33 | 60 | 7.54E−2 | 2.66E−1 | 4.92E−3 | UM04794 | Transcription initiation factor IIB |
| ATCTAATCTA | 9 | 7 | 22 | 7.50E−1 | 1.24E−2 | 5.81E−3 | UM01712 | Conserved hypothetical protein |
| TTTGGTCATT | 24 | 12 | 29 | 4.51E−2 | 4.33E−1 | 6.80E−3 | UM05969 | Short-chain alcohol dehydrogenase |
| ATTGGGGTCT | 20 | 10 | 26 | 6.30E−2 | 3.63E−1 | 7.18E−3 | UM05549 | Inositol 1-phosphate synthase |
| ATTTTCCATAd | 1 | 4 | 15 | 1.91E−1 | 9.38E−5 | 7.33E−3 | UM06332 | Egl1, endoglucanase 1 precursor |
| TGCGGTGACG | 16 | 3 | 13 | 1.41E−3 | 6.21E−1 | 8.24E−3 | UM03449 | Septin-3 |
| ATCCACGAAA | 0 | 1 | 9 | 4.16E−1 | 9.62E−4 | 8.28E−3 | UM01973 | Cyanate lyase |
| AATGTTTCTT | 5 | 11 | 27 | 1.29E−1 | 3.88E−5 | 8.56E−3 | UM05495 | Conserved hypothetical protein |
| TACCACTACG | 0 | 0 | 6 | 7.62E−1 | 9.52E−3 | 1.20E−2 | UM05550 | Exo-beta-1,3-glucanase |
| GGTGCTGCGG | 9 | 0 | 5 | 1.37E−3 | 3.28E−1 | 2.49E−2 | UM10589 | Cystathionine gamma-lyase |
| GCTCGCGCTC | 12 | 19 | 34 | 2.30E−1 | 7.52E−4 | 3.00E−2 | UM00595 | Aspartate aminotransferase |
| GATCCTGCTA | 31 | 9 | 21 | 2.29E−4 | 1.45E−1 | 3.01E−2 | UM04347 | Sexual differentiation process protein isp4 |
| CCCGTCAAGT | 1 | 3 | 10 | 3.37E−1 | 3.08E−3 | 4.15E−2 | UM05130 | Phosphoenolpyruvate carboxykinase |
| GAGTTTGTGT | 9 | 0 | 4 | 1.37E−3 | 1.95E−1 | 5.17E−2 | No hitse | Putative exosome 3′-5′ exonuclease complex |
| GCCGAGAAAC | 11 | 16 | 28 | 3.72E−1 | 5.47E−3 | 6.03E−2 | UM05733 | Conserved hypothetical protein |
| TGAACAAAAA | 17 | 28 | 42 | 1.04E−1 | 8.43E−4 | 8.48E−2 | UM00745 | Hypothetical protein |
| CGCAATGGAT | 0 | 2 | 6 | 2.26E−1 | 9.52E−3 | 1.46E−1 | UM02483 | Hypothetical protein |
Tag frequencies have been normalized to the size of the smallest library for comparison (ADR1, 43,629 tags).
Statistical significance of the differential tag frequencies between libraries.
MIPs Ustilago maydis database (http://mips.gsf.de/genre/proj/ustilago/).
SAGE tag differences were confirmed by Northern analysis.
Tag has an associated sequence in the U. maydis EST database.
Probable gene association considering the existence of a 3′ untranslated region longer than 500 bases.
TABLE 5.
Tags with reduced levels in the UBC1 and ADR1 libraries
| Tag sequence | Normalized frequenciesa
|
P valueb
|
MUMDB gene accession no.c | Preliminary gene product designation | ||||
|---|---|---|---|---|---|---|---|---|
| WT | UBC1 | ADR1 | WT vs UBC1 | WT vs ADR1 | UBC1 vs ADR1 | |||
| CACTCGACCCd | 58 | 0 | 0 | 1.07E−19 | 1.14E−18 | 7.59E−1 | UM02803 | β-1,3-Glucan binding protein |
| TTCGGCAAGG | 140 | 64 | 80 | 2.00E−8 | 3.34E−5 | 1.76E−1 | UM00919 | ADP, ATP carrier protein |
| TTGGTCATCT | 214 | 120 | 92 | 5.45E−8 | 4.47E−13 | 5.28E−2 | UM01318 | 40S ribosomal protein S12 |
| TTCTCTTTCG | 72 | 24 | 38 | 1.57E−7 | 7.44E−4 | 7.34E−2 | UM01505 | Histone H2B |
| ATGCAATGAT | 278 | 177 | 148 | 6.15E−7 | 6.40E−11 | 9.87E−2 | UM04632 | Ribosomal protein P2 |
| TTTGATTCGTd | 18 | 0 | 0 | 9.41E−7 | 2.04E−6 | 7.59E−1 | UM02804 | Conserved hypothetical protein |
| GCCGAAGAGG | 48 | 14 | 17 | 3.72E−6 | 6.81E−5 | 5.74E−1 | UM03931 | Hypothetical protein |
| GTAATCACTT | 335 | 233 | 285 | 6.65E−6 | 3.91E−2 | 1.99E−2 | UM02710 | Histone H4 |
| GAGGCGACCC | 25 | 5 | 14 | 4.87E−5 | 6.75E−2 | 2.75E−2 | UM03308 | Leucine aminopeptidase |
| TCGTAGGGCG | 57 | 23 | 24 | 7.28E−5 | 1.69E−4 | 9.03E−1 | UM05832 | Heat shock protein 10 (chaperonin CPN10) |
| GGCACCAAGG | 44 | 17 | 13 | 2.46E−4 | 2.29E−5 | 5.01E−1 | UM10051 | 60S ribosomal protein L31 |
| ACAGGATTTG | 115 | 69 | 76 | 3.08E−4 | 3.58E−3 | 5.36E−1 | UM01189 | Translation elongation factor eEF-1 |
| CCCCTCCTCA | 57 | 28 | 35 | 9.46E−4 | 2.02E−2 | 3.59E−1 | UM10286 | Nuclear transport factor 2 |
| TATCGCATTT | 40 | 17 | 15 | 1.05E−3 | 4.76E−4 | 7.67E−1 | UM04716 | Vacuolar H+ ATPase |
| AACAACTTTG | 14 | 2 | 3 | 1.16E−3 | 6.58E−3 | 6.18E−1 | UM04923 | Hypothetical protein |
| CACAACGGTG | 50 | 24 | 17 | 1.72E−3 | 3.21E−5 | 2.66E−1 | UM10621 | 60S ribosomal protein L32 |
| TTTCGGCCAT | 82 | 48 | 52 | 1.88E−3 | 7.76E−3 | 7.03E−1 | UM00686 | 60S ribosomal protein L22 |
| GAGGCTCAGC | 10 | 1 | 4 | 2.66E−3 | 9.14E−2 | 1.86E−1 | UM00856 | DNA-binding protein HEXBP |
| GCGAAGCGCT | 24 | 8 | 15 | 3.12E−3 | 1.29E−1 | 1.65E−1 | UM04882 | Conserved hypothetical protein |
| CAATCCACTG | 29 | 11 | 18 | 3.30E−3 | 1.10E−1 | 1.99E−1 | No hits | Prefoldin subunit 4 |
| TAAAATCGTC | 12 | 2 | 2 | 3.53E−3 | 5.60E−3 | 9.31E−1 | UM10267 | Hypothetical protein |
| TTCTTCGACA | 85 | 53 | 48 | 3.61E−3 | 8.33E−4 | 6.23E−1 | UM03237 | Ribosomal protein S20 |
| TGCACAAGTC | 21 | 6 | 6 | 3.72E−3 | 3.12E−3 | 9.01E−1 | UM10346 | rRNA processing-related protein |
| TGTGCTGTAA | 19 | 6 | 7 | 3.97E−3 | 1.53E−2 | 6.84E−1 | UM05503 | Vacuolar ATP synthase subunit H |
| AAGCTCCAGA | 28 | 11 | 9 | 4.79E−3 | 1.34E−3 | 6.44E−1 | UM05990 | 60S ribosomal protein L17 |
| GTACCCAAGG | 10 | 1 | 0 | 4.82E−3 | 1.08E−3 | 4.63E−1 | UM00851 | Nuclear segregation protein Bfr1 |
| TCTCTGTTTG | 10 | 1 | 4 | 4.82E−3 | 1.35E−1 | 1.86E−1 | No hitse | Mitochondrial 40S ribosomal protein mrp17 |
| GTATACAGCG | 10 | 1 | 5 | 4.82E−3 | 2.38E−1 | 1.02E−1 | UM06108 | Putative purine nucleoside phosphorylase |
| GCGGATTGCT | 10 | 1 | 3 | 4.82E−3 | 6.51E−2 | 3.30E−1 | UM03509 | Conserved hypothetical protein |
| CTGCCGAAGC | 41 | 20 | 14 | 5.83E−3 | 1.63E−4 | 2.75E−1 | UM03356 | Enolase (2-phosphoglycerate dehydratase) |
| AAACATTTCA | 59 | 34 | 46 | 6.74E−3 | 1.86E−1 | 1.86E−1 | UM03192 | Conserved hypothetical protein |
| GCATCGCTGC | 48 | 26 | 31 | 7.28E−3 | 5.09E−2 | 4.99E−1 | UM10296f | Cytochrome c oxidase |
| AGGTGGTTCT | 21 | 7 | 5 | 7.82E−3 | 1.23E−3 | 5.10E−1 | UM10035 | Conserved hypothetical protein |
| CGCCATAAAC | 40 | 20 | 17 | 7.87E−3 | 1.64E−3 | 5.79E−1 | UM03074 | Probable transcription factor BTF3a |
| TCATCCCCGG | 37 | 19 | 17 | 7.94E−3 | 4.36E−3 | 7.98E−1 | UM10701 | 60S ribosomal protein L37-A |
| GCACGCCGAC | 37 | 19 | 16 | 1.08E−2 | 3.49E−3 | 6.67E−1 | UM10607 | NADH-ubiquinone oxidoreductase |
| ATCTGCATCC | 63 | 39 | 31 | 1.33E−2 | 7.16E−4 | 3.37E−1 | UM10625 | 60S ribosomal protein L24 |
| ACGGTTTGTG | 8 | 1 | 0 | 1.55E−2 | 3.78E−3 | 4.63E−1 | UM10608 | Quinate permease |
| TGGCGTGGTG | 74 | 50 | 35 | 2.39E−2 | 1.09E−4 | 9.75E−2 | UM02577 | 6-Phosphogluconate dehydrogenase |
| TCCTTGGCCT | 116 | 86 | 75 | 2.86E−2 | 2.26E−3 | 3.67E−1 | UM02450 | Eukaryotic translation initiation factor 5A |
| AACCCCAAAA | 61 | 41 | 29 | 3.59E−2 | 4.96E−4 | 1.53E−1 | UM10197 | 40S ribosomal protein s23 |
| TCCGTTTTCC | 8 | 2 | 0 | 4.99E−2 | 3.78E−3 | 2.76E−1 | UM10053 | Carbonic anhydrase |
| GGTAACCAAG | 44 | 29 | 16 | 5.50E−2 | 1.50E−4 | 5.44E−2 | UM10361 | 60S ribosomal protein L38-1 |
| TGACGCCTAA | 10 | 4 | 1 | 6.58E−2 | 4.07E−3 | 2.55E−1 | UM01195 | Sulfide dehydrogenase |
Tag frequencies have been normalized to the size of the smallest library for comparison (ADR1, 43,629 tags).
Statistical significance of the differential tag frequencies between libraries.
MUMDB, MIPs Ustilago maydis database (http://mips.gsf.de/genre/proj/ustilago/).
SAGE tag differences were confirmed by Northern analysis.
Tag has an associated sequence in the U. maydis EST database.
Probable gene association considering the existence of a 3′ untranslated region longer than 500 bases.
Connections between ribosome biogenesis and PKA.
Our initial analysis of the SAGE data revealed a potential connection between cAMP signaling, ribosome biogenesis, and translation in U. maydis. Specifically, we identified tags matching genes for eight RPs and one translation elongation factor that were elevated in the UBC1 library (high PKA activity) and/or reduced in the ADR1 library (low PKA activity) (Table 3). We also found that tags for 12 other RP genes, one translation initiation factor, one translation elongation factor, and one rRNA processing-related protein have lower levels in the libraries from cells expected to show either high or low PKA activity (Table 5) and that the tag for one RP gene was somewhat elevated in the mutant libraries relative to that of the wild-type library (Table 6). Although the patterns of expression were not identical for all of the RP genes, the prominent pattern was that tags for RP genes generally had lower levels under the condition of low PKA activity (i.e., in the adr1 mutant) than in the libraries from the wild-type strain and/or the ubc1 mutant. The discovery of tags in the SAGE libraries for RP genes was expected because transcripts for these genes are generally abundant and therefore more likely to be detected by SAGE. For example, we have detected a similar set of genes in previous SAGE experiments with Cryptococcus neoformans (40, 70), and SAGE data for S. cerevisiae also identified transcripts for RPs as being abundant (73). The PKA regulation of tag levels for the RP genes suggests an important role for the cAMP pathway in cell growth in U. maydis.
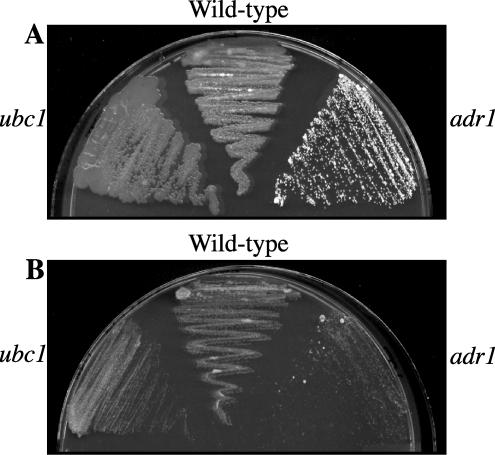
In S. cerevisiae, details of the connection between nutrient availability, PKA, and RP gene expression are emerging with the recent description of the control of transcription factors coregulated by the cAMP and TOR pathways. In particular, Schmelzle et al. (66) and Zurita-Martinez and Cardenas (78) found that mutations that hyperactivate the PKA pathway resulted in reduced sensitivity to the TOR inhibitor rapamycin and that both pathways signal together or in parallel to respond to nutrients and regulate RP genes. If a similar regulation by these pathways occurs in U. maydis, one would predict that mutants with altered PKA activity might also have a different response to rapamycin than the wild-type strain. To test this prediction, we examined the three strains used for SAGE library construction for growth on medium containing 200 ng/ml rapamycin. As shown in Fig. 2, the wild-type and ubc1 strains showed limited sensitivity to rapamycin. In contrast, the adr1 strain exhibited markedly reduced growth in the presence of the drug, suggesting that reduced PKA activity in the mutant conferred enhanced sensitivity. This result suggests that the TOR pathway may also influence RP gene expression in U. maydis and may be related to our general observation from the SAGE data that loss of PKA activity in the adr1 mutant results in reduced tag levels for genes encoding RPs.
FIG. 2.
Sensitivity of the adr1 mutant to rapamycin. Evaluation of the growth of wild-type U. maydis strain 521 and the mutants 111 (ubc1::hygBr) and 002-10 (adr1::phelor), after 2 days at 30°C on PDA plates (A) and PDA plates containing 200 ng/ml rapamycin (B). The growth of strain 002-10 is dramatically reduced on plates containing rapamycin.
Shared targets of cAMP signaling and mating.
The SAGE experiments also extended previous observations that the cAMP signaling pathway shares regulatory targets with the bW/bE homeodomain proteins that form a transcriptional regulator encoded by different alleles of the b mating-type locus during formation of the infectious dikaryon (10). Specifically, we found that tags that were elevated in the UBC1 library and/or reduced in the ADR1 library identified genes encoding an acyltransferase-like protein and a capsule-associated protein (Table 3). The expression of these genes (and three others) was previously found to be negatively regulated by the bW/bE complex in U. maydis (10). The idea of shared targets is reinforced by our additional observation that two tags elevated in the ADR1 library matched genes for endoglucanase 1 (egl1) and repellent protein 1 (rep1) (Table 4). The transcripts for these genes were previously shown to be up-regulated by the bW/bE complex (65, 75), and their filament-specific expression is consistent with the SAGE data. We also noted that a tag for the mfa1 gene was not detected in our SAGE analysis, even though the transcript level is clearly elevated in the ubc1 mutant (27) (Fig. 1). This is because inspection of the cDNA corresponding to this gene indicated the absence of an NlaIII restriction site necessary to generate a SAGE tag.
Differential expression of genes encoding metabolic functions in PKA mutants.
The SAGE tags revealed genes encoding several metabolic functions, including enzymes involved in carbohydrate metabolism (e.g., 6-phosphogluconolactonase, aldehyde dehydrogenase, alcohol dehydrogenase, ribose 5-phosphate isomerase, fructose-bisphosphatase, aldo-keto reductase, transaldolase, phosphoenolpyruvate carboxykinase, and others). Tags for these genes were generally elevated in the ADR1 library relative to those in the UBC1 and/or WT library (Table 4). The tags for two genes involved in amino acid metabolism (cystathionine gamma-lyase and aspartate aminotransferase) showed similar patterns of expression. Other tags that had reduced levels in the UBC1 library and/or were elevated in the ADR1 library identified genes encoding proteins with predicted transport functions, including two peptide transporters and a putative sugar transporter. One of these genes shows a high similarity to the Isp4 protein of Schizosaccharomyces pombe; the transcript for this gene was elevated during the sexual differentiation process (64), and the protein was later classified as an oligopeptide transporter (44). Finally, tags that were lower in the UBC1 library than in the ADR1 library identified genes predicted to encode fatty acyl coenzyme A synthase, inositol 1-phosphate synthase, and two enzymes related to vitamin metabolism (thiazole biosynthetic enzyme and thiamine biosynthesis protein).
Connections between PKA and genes implicated in morphogenesis.
Given the role of the cAMP pathway in the morphogenesis of U. maydis, we anticipated that the SAGE data would reveal tags for genes involved in influencing cell morphology (15, 22). In this category, we identified tags for genes encoding the following predicted proteins: a GTP binding protein, the repellent protein (Rep1), a septin, an exo-β-1,3 glucanase, a β-1,3-glucan binding protein, a predicted prefoldin protein, and a mixed-linked glucanase. These tags showed a variety of expression patterns. For example, some of these tags showed higher levels in the ADR1 library (Table 4), a result that may be consistent with the filamentous morphology of this mutant (e.g., repellent protein and exo-β-1,3 glucanase). Others were expressed only in the WT library (β-1,3-glucan binding protein) (Table 5) or in the mutant libraries (mixed-linked glucanase) (Table 6). The detection of the tag for the septin transcript (lower in UBC1) (Table 4) was interesting because septins are a conserved family of GTP-binding, filament-forming proteins with complex roles in cytokinesis and/or cell septation (43). A potential influence of PKA on septin expression may be relevant in U. maydis because a defect in the ubc1 gene causes an unusual multiple-bud phenotype due to a defect in the separation of mother and daughter cells. GTP-binding proteins are also important for morphogenesis in U. maydis, and we found that a tag for a putative G protein that is similar to Gβ subunits showed the opposite pattern of expression to the septin (higher in UBC1) (Table 3).
Elevated transcript levels of genes involved in phosphate acquisition in the ubc1 mutant.
A striking finding from the SAGE data was that a number of tags elevated in the UBC1 library (high PKA activity) and/or reduced in the ADR1 library (low PKA activity) identified genes predicted to encode proteins connected with phosphate metabolism. Six of these genes were orthologs of components of the phosphate (PHO) regulatory pathway that is involved in the acquisition of phosphate (Pi) in S. cerevisiae (57). In yeast, low extracellular Pi induces the expression of genes in the PHO pathway, including PHO84, which encodes a high-affinity Pi transporter localized to the plasma membrane, and PHO5, which encodes a repressible acid phosphatase localized to the periplasmic space (55). We found tags corresponding to similar PHO-related functions (phosphate transporter and acid phosphatase) to be higher in the UBC1 library, suggesting that a relationship exists between PKA signaling and a predicted PHO regulatory pathway in U. maydis (Table 3). Additional yeast genes regulated by the PHO system have been identified in microarray experiments, and these include the VTC (vacuolar transporter chaperone) gene family involved in poly(P) synthesis (53). Two of these gene products, Vtc1 and Vtc4, operate together as a complex and control vacuole acidification by the distribution of vacuolar ATPase. Inactivation of the VTC1 gene results in a reduced vacuole acidification as a consequence of reduced amounts of vacuolar ATPase on the vacuolar membranes (12). Our SAGE data identified tags (that were elevated in the UBC1 library) for orthologs of the VTC1 and VTC4 genes (Table 3). In contrast, we also found tags corresponding to a vacuolar H+ ATPase and a vacuolar ATP synthase that were elevated in the WT but not the UBC1 library (Table 5). An additional coregulated gene in yeast encodes a leucine aminopeptidase, and we identified a tag for a predicted ortholog in our SAGE data (Table 5).
The observation of a connection between cAMP signaling and phosphate metabolism prompted a closer inspection of other tags related to metabolic functions. In this context, we noted (mentioned earlier) that the two tags identifying genes encoding putative thiamine biosynthetic functions were apparently expressed in a pattern opposite to that of the tags for phosphate acquisition (Table 4). These tags were found at lower levels in the UBC1 library, suggesting that elevated PKA activity down-regulates the transcript levels for these genes. Thiamine is known to regulate phosphate metabolism and mating in Schizosaccharomyces pombe (18). For example, thiamine represses the expression of an acid phosphatase thought to play a role in dephosphorylating thiamine phosphates in growth substrates. Interestingly, both thiamine and the cAMP pathway influence pigmentation in Ustilago hordei (41). The observed altered expression of genes encoding functions for phosphate metabolism in cAMP signaling mutants and the results from U. hordei prompted a more detailed examination of the connections between these processes in U. maydis.
Phosphate concentration influences the morphological response to lipids.
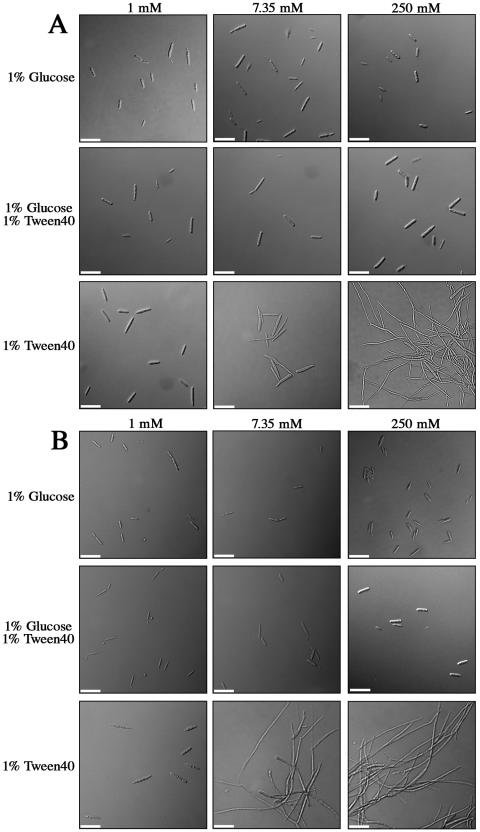
We initially examined the influence of phosphate on morphogenesis because of the known role of PKA in the regulation of dimorphism in U. maydis. Previously, we showed that the dimorphic transition from budding to filamentous growth can be triggered in a PKA-dependent manner by growth in lipids (36). During tests on the influence of phosphate, the wild-type strain 521 (a1b1) was grown in minimal medium (minus phosphate) supplemented with glucose, glucose and Tween 40, or Tween 40, with the addition of different levels of phosphate. Phosphate concentration did not affect the morphology of budding yeast cells after 3 or 5 days of growth in glucose or glucose-plus-Tween 40 medium (Fig. 3A and B). In contrast, an increased phosphate concentration directly correlated with increased filamentation when cells were grown in 1% Tween 40 for 3 days (Fig. 3A). Cells grown in 1 mM phosphate grew as budding yeast cells, whereas yeast cells grown in 7.35 mM phosphate were beginning to elongate at this time and cells grown in 250 mM phosphate were completely filamentous (Fig. 3A). Conversely, a reduced phosphate concentration resulted in a decrease in filamentation. Interestingly, the cells grown with Tween 40 and 7.35 mM phosphate resembled mating cells in the early stages of conjugation tube formation. After 5 days, cells grown in 1% Tween 40 and 1 mM phosphate maintained a budding morphology in contrast to the completely filamentous growth observed in both 7.35 and 250 mM phosphate (Fig. 3B). We previously described filament formation in Tween 40 after 5 days, which is consistent with these results (36). Overall, these observations indicated that phosphate levels influence the lipid-responsive dimorphic transition in U. maydis.
FIG.3.
Influence of phosphate on the morphology of U. maydis in Tween 40. Differential-interference-contrast micrographs of the wild-type U. maydis strain 002 (a1b1) grown in minimal medium (minus phosphate) supplemented with 1% glucose, 1% glucose plus 1% Tween 40, or 1% Tween 40. Cultures were also supplemented with 1, 7.35, or 250 mM KH2PO4. (A) Budding growth for the wild-type U. maydis strain 521 after 3 days in 1% glucose and 1% glucose plus 1% Tween 40 under all phosphate concentrations tested. In 1% Tween 40, increasing phosphate concentration results in increased filamentation. U. maydis grows as budding yeast cells at low phosphate concentrations (1 mM), begins to extend cell length in 7.35 mM phosphate, and grows as hyphal filaments in high phosphate concentration (250 mM). (B) Budding growth after 5 days in 1% glucose and 1% glucose plus 1% Tween 40 under all phosphate concentrations tested. At this time, however, cells growing in 1% Tween 40 supplemented with 1 mM phosphate remain as budding yeast cells, but the cells are completely filamentous when the medium contains 7.35 mM and 250 mM phosphate. Scale bars are 20 μm.
Polyphosphate accumulation is decreased in a ubc1 mutant.
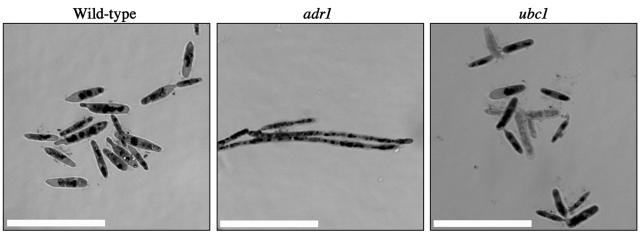
The expression of the PHO genes required for phosphate metabolism and the acquisition of phosphate in S. cerevisiae are induced under conditions of low cellular phosphate (53). This suggests that the ubc1 mutant, which shows increased expression of the PHO gene orthologs in U. maydis, has low levels of cellular phosphate. Excess phosphate is stored in the vacuole as linear polymers of orthophosphate, termed polyphosphate (67). Phosphate and polyphosphate can be visualized by staining with toluidine blue O dye and can be observed in the cytoplasm (phosphate) or as granules in the cell vacuole (polyphosphate) (49). We therefore stained the wild-type strain and the ubc1 and adr1 mutants with toluidine blue O to test whether altered regulation of the PHO genes in U. maydis resulted in increased or decreased polyphosphate accumulation. Consistent with the SAGE data, we found that approximately 25% of the cells of the ubc1 mutant lacked polyphosphate accumulations, in contrast to less than 2% of the wild-type or adr1 mutant cells (Fig. 4). The variability may be due to differences in the ages of the individual cells. We should note that the reduction in polyphosphate staining could be the result of cell wall permeability defects (i.e., uptake differences) in the ubc1 mutant rather than defects in storage. In general, however, the combination of the SAGE expression data and the staining assay suggests a connection between PKA and the maintenance of intracellular phosphate levels in U. maydis.
FIG. 4.
Reduced polyphosphate accumulation in the ubc1 mutant. Differential-interference-contrast micrographs of the wild-type strain 521 and the ubc1 and adr1 mutants grown overnight in PDB and stained for polyphosphate with toluidine blue O dye are shown. The ubc1 mutant showed a decrease in the amount of polyphosphate accumulation compared to that in the wild-type strain and the adr1 mutant. Specifically, 100 cells of each strain from separate experiments were scored as stained or unstained with the following results (percentages of stained cells): strain 521, 99% ± 0.0%; adr1 mutant, 98% ± 2.8%; and ubc1 mutant, 74% ± 6.4%. Scale bars = 20 μm.
Acid phosphatase activity is influenced by PKA.
The SAGE data revealed that transcripts for genes in the PHO regulatory pathway were elevated in the ubc1 mutant and/or reduced in the adr1 mutant, and this included a gene encoding an acid phosphatase with similarity to a gene from Aspergillus fumigatus (Table 3). To investigate whether PKA-regulated expression of the U. maydis gene correlated with an increase in acid phosphatase levels, the activity of the enzyme was measured in protein extracts from the wild-type U. maydis strain and the ubc1 and adr1 mutants (Materials and Methods). As predicted from the SAGE data, the specific activity for acid phosphatase in the ubc1 strain (0.0289 ± 0.0000) was elevated compared to that in the wild-type (0.0190 ± 0.0001) and adr1 strains (0.0174 ± 0.0002). Overall, these results support the implication by SAGE of PKA-dependent regulation of acid phosphatase activity. It should be noted that the assay measures total acid phosphatase activity, and analysis of the genome annotation indicates that there are other genes for putative acid phosphatases in U. maydis, besides the gene identified by SAGE. These other genes may encode enzymes that also contribute to the activity that we observed.
Confirmation of the SAGE results.
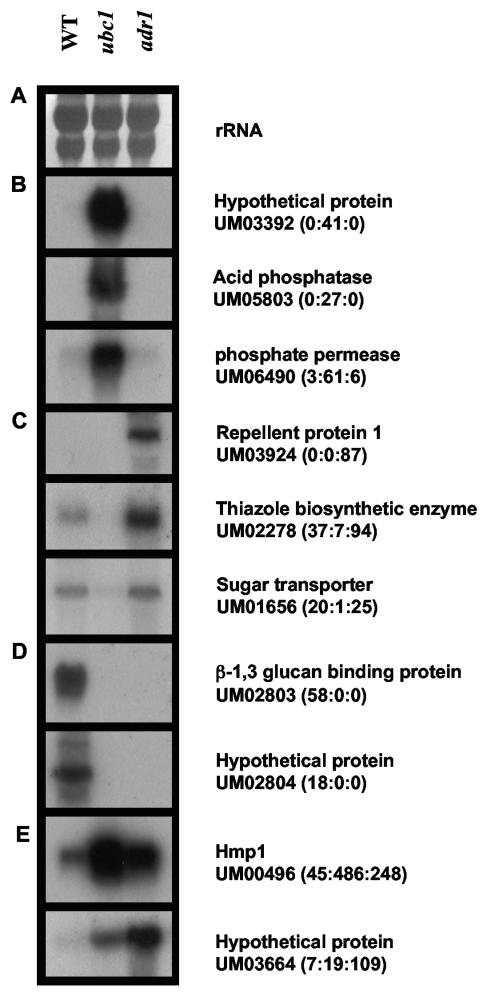
RNA blot analysis was used to independently confirm that the observed differences in tag levels reflected differences in transcript abundance. Ten pairs of primers (Table 1) were designed to amplify sequences containing tags that were differentially expressed (Tables 3 to 6). The amplified sequences were used as probes for RNA blot analysis, and a strong correspondence between SAGE tag frequencies and hybridization patterns was observed in all cases. Figure 5 shows examples of differentially expressed tags belonging to the four classes described in Tables 3 to 6. Compared to the rRNA loading controls in Fig. 5A, Fig. 5B shows three examples of genes with tags that were clearly elevated in the ubc1 mutant (acid phosphatase, inorganic phosphate permease, and a hypothetical protein). Figure 5C shows two examples of genes with elevated tags in the filamentous adr1 mutant (repellent protein 1 and thiazole biosynthetic enzyme) and an example of a gene with decreased tag abundance in the ubc1 mutant (predicted sugar transporter). Finally, Fig. 5D and E each show two examples of genes with tags that had reduced levels in the mutants (β-1,3-glucan binding protein and a hypothetical protein) or with tags that were elevated in the mutants (Hmp1 and a hypothetical protein), respectively. Overall, the RNA blot analysis confirms the differential RNA levels indicated by the SAGE results, and a similar confirmation was obtained for eight additional genes (data not shown).
FIG. 5.
RNA blot analysis of selected genes with differentially expressed tags among the WT, UBC1, and ADR1 libraries. (A) rRNA (18S and 28S) bands as a loading control. (B) Hybridization patterns of representative genes for which tags were elevated in the UBC1 library and/or down-regulated in the ADR1 library. (C) Hybridization of representative genes with tags with lower levels in the UBC1 library and/or higher levels in the ADR1 library. (D and E) Hybridization of representative genes for which tags were reduced and elevated, respectively, in the libraries from the mutants. The normalized frequencies of the tags for the genes in the three libraries are indicated after the MIPS U. maydis gene accession number in the order WT:UBC1:ADR1.
DISCUSSION
Here, we describe SAGE experiments designed to investigate the influence of mutations that interfere with the level or regulation of PKA activity on the transcriptome of U. maydis. The results revealed differences in the transcript levels for genes in several functional categories, including ribosome biogenesis, metabolism, phosphate acquisition, stress, mating, and morphogenesis. These results provide a broad view of the influence of PKA mutations on the transcriptome in U. maydis and reveal conserved features of cAMP signaling among fungi, in particular the involvement of PKA in the regulation of RP gene expression and phosphate metabolism.
Interestingly, a small subset of the genes identified by SAGE had previously been identified in studies employing suppressive subtractive hybridization (SSH) to compare transcript levels in cells of a wild-type strain and a uac1 strain defective in adenylyl cyclase (2, 19). The uac1 mutant displays a constitutively filamentous growth morphology that is similar to that of the adr1 mutant that we employed in this study, and both mutants are expected to have low PKA activity. However, the activity of the pathway may be affected to different extents in the two mutants, and therefore, the expression results are only partially comparable. This is because both the Uka1 and the Adr1 catalytic subunits are still present and potentially active in the uac1 mutant, while adenylyl cyclase and the Uka1 catalytic subunit are still potentially active in the adr1 mutant. In light of these considerations, some genes with elevated transcripts were found in both the adr1 and uac1 mutants (as identified by SSH and SAGE), including rep1 (repellant) and the gene for superoxide dismutase. Both methods also identified genes encoding phosphate uptake functions and a xylanase with elevated transcripts in budding cells. A striking difference in the data collected with the two methods was the predominance of RP genes revealed by SAGE. This difference may reflect the respective abilities of the techniques to detect differences in transcript abundance, perhaps due to the relatively abundant nature of RP transcripts or the subtle differences in transcript levels between strains. However, the difference may also suggest the more interesting possibility that the uac1 and adr1 mutations have disparate influences on RP gene expression because of the positions of their respective defects in the cAMP/PKA pathway.
Ribosomal-protein gene expression and PKA.
As expected from work with other organisms, particularly S. cerevisiae, the RP genes showed differences in transcript levels in the PKA mutants relative to those in the wild-type strain. The connections between nutrient acquisition, growth control, PKA, and RP gene expression are well established in yeast, and the SAGE data suggest that U. maydis has a similar regulatory network. Although the variations in tag levels were not dramatic, the general trend that we observed was that tags for 20 RP genes were found at reduced levels under the condition of low PKA activity (i.e., in the adr1 mutant) relative to those in the wild-type strain and/or the ubc1 mutant. RP genes are generally found to be differentially expressed by SAGE analysis when different growth or physiological conditions are compared (3, 56, 69).
The factors implicated in the regulation of RP synthesis in S. cerevisiae have been well characterized and include the interruption of secretory-pathway signals mediated by protein kinase C (39), nitrogen regulation via the TOR signaling pathway (11), carbon source utilization as regulated by the cAMP/PKA pathway (35, 51, 74), and phosphate sensing (21). Recent studies of the connection between PKA and RPs in yeast have provided a detailed view of the regulatory mechanisms that link nutrient sensing, the TOR and Ras/PKA pathways, cell growth, and cell cycle progression (32, 66, 78). The TOR pathway is known to control ribosome biogenesis in yeast, and TOR controls the subcellular location of the PKA subunit Tpk1 (45, 66). PKA appears to function downstream of TOR (66) or in a parallel pathway (78) to activate the transcription of RP genes. Martin et al. (45) have shown that this regulation of the expression of RP genes acts via the transcription factor FHL1 and the coregulators IFHI and CRF1. The SAGE data for U. maydis PKA mutants hint at interesting parallels for the roles of cAMP signaling in U. maydis and S. cerevisiae. Consistent with the yeast model, the tags for the RP genes that we detected were generally lower in the adr1 mutant defective in PKA activity than in the wild type, suggesting that U. maydis also has a connection between cAMP and the TOR pathway similar to that of S. cerevisiae in terms of nutrient sensing and the regulation of RP expression. We examined the rapamycin sensitivity of our mutants with defects in the cAMP pathway and found that the adr1 mutant was relatively more sensitive than the wild-type strain or the ubc1 mutant. This result suggests that the inhibition of TOR in a strain already lacking PKA may compromise growth by accentuating problems with the transcription of RP genes. This result is strikingly similar to the increased sensitivity to rapamycin that Zurita-Martinez and Cardenas (78) described for yeast mutants with defects in the catalytic subunit of PKA. Recent work with S. cerevisiae has also implicated the unusual prefoldin URI (unconventional prefoldin RPB5 interactor) in the TOR pathway (24), and we identified a tag for a prefoldin gene in the SAGE data (Table 5). Overall, these results suggest that the general role of PKA, and perhaps the TOR pathway, is conserved between ascomycete and basidiomycete fungi with regard to the regulation of RP gene expression.
Phosphate acquisition and PKA.
The SAGE data uncovered a relationship between cAMP signaling and phosphate sensing and acquisition in U. maydis. Specifically, a number of tags elevated under the condition of high PKA activity and/or reduced in the adr1 mutant with low PKA activity identified genes associated with phosphate metabolism. These included orthologs of S. cerevisiae components of the PHO regulatory pathway for the acquisition of phosphate (Pi) (57), such as transporters and an acid phosphatase as well as components of the VTC gene family involved in poly(P) synthesis (53). Two of these gene products, Vtc1 and Vtc4, operate together as a complex and control acidification of the vacuole (12). Our SAGE data identified tags for orthologs of the VTC1 and VTC4 genes that were elevated in the UBC1 library (Table 3). In contrast, we found tags corresponding to a vacuolar H+ ATPase and a vacuolar ATP synthase that were elevated in the WT but not the UBC1 library, suggesting that U. maydis may have similar components for controlling phosphate storage (Table 5). The SAGE data also indicated that the tag for a U. maydis ortholog of the yeast Pho84 phosphate sensor was elevated by ∼20-fold in the UBC1 library relative to that in the wild-type strain. The phosphate transporter Pho84 functions in manganese homeostasis in yeast and therefore influences the activity of manganese superoxide dismutase (30). We detected a tag for a putative manganese superoxide dismutase regulated by PKA in the SAGE data and noted that a connection between cAMP signaling and the response to stress has been well characterized in S. cerevisiae (61).
We confirmed the functional relevance of the SAGE data by demonstrating that acid phosphatase activity was elevated and polyphosphate accumulation was reduced in the ubc1 mutant. Analysis of the SAGE data indicated that the acid phosphatase gene that we identified was specifically transcribed in the ubc1 mutant. S. cerevisiae has three repressible acid phosphatases, one of which is Pho5p; a search of the genome sequence of U. maydis (MIPS) with the Pho5p sequence revealed five genes encoding probable acid phosphatases and phytases. Given the number of genes encoding putative phosphatases, their genetic analysis via gene disruption would be a significant undertaking. One of these genes (Um06428) is related to a thiamine-repressible acid phosphatase precursor. In S. pombe, thiamine is not essential for growth but does play a role in the regulation of mating (18). Thiamine represses the expression of a gene encoding an acid phosphatase that dephosphorylates thiamine phosphates present in growth medium. In this context, we noted that SAGE tags for two genes implicated in thiamine metabolism (thiazole biosynthetic enzyme and thiamine biosynthesis protein) had altered levels in the PKA mutants. These observations suggest a further linkage between phosphate metabolism and PKA in U. maydis.
Phosphate, PKA, and morphogenesis.
Inorganic phosphate in S. cerevisiae, as sensed by the Pho84 and Pho87 permeases, has recently been identified as a nutrient signal that activates that PKA pathway (21). The readdition of phosphate to starved cells influenced the expression and activities of PKA targets, including the RP genes, although a specific cAMP signal was not triggered. This independence of cAMP signaling and PKA activation may be relevant to the observed differences between our SAGE data collected with the adr1 mutant and the SSH data collected with the uac1 mutant (discussed above). The coincident relationships between phosphate acquisition, RP gene expression, and PKA revealed by the SAGE data suggest that phosphate may also act as a nutrient signal in U. maydis. To begin to examine this possibility, we tested whether phosphate levels influence the dimorphic transition that U. maydis displays in response to lipids as the sole carbon source (36). We found that an elevated phosphate concentration promoted filamentation in response to lipids, thus suggesting that phosphate perception is part of the nutritional influence on morphogenesis in U. maydis. The influence of phosphate levels on fungal morphogenesis may be a general phenomenon because Hornby et al. (29) have demonstrated that high phosphate levels promote pseudohyphal growth in Candida albicans. It will be important to follow up these observations with experiments to determine whether phosphate perception plays a role during filamentous growth of the fungus in host tissue.
The SAGE data indicated other possible connections between cAMP signaling and morphogenesis in U. maydis. For example, we identified a tag for a septin with reduced abundance in the ubc1 mutant compared to that in the wild type, a result that is interesting because of the multiple-bud phenotype of this mutant. Our analysis of the SAGE data is consistent with the hypothesis that the multiple-bud phenotype may be related to an influence of the cAMP pathway on the expression of the septin gene. Consistent with this idea, we recently found that disruption of the septin gene results in a morphological defect (9). Additional tags that represented genes with potential morphological or cell wall functions were identified, and these included genes for glucanases and a GTP-binding protein. It is reasonable to expect changes in cell wall functions as a function of PKA activity, given that ubc1 mutants display a wet colony phenotype that may be related to the production of an extracellular matrix (22, 37). In addition, precedent for this observation comes from microarray experiments with a yeast mutant defective in a cAMP phosphodiesterase (encoded by PDE2); a number of genes encoding cell wall functions had altered expression in the mutant (31).
Many of the tags that show different levels of abundance in the mutants match genes encoding hypothetical proteins. The expression patterns of these genes in the morphological mutants suggest that future evaluation of these genes may contribute to a deeper understanding of the role of cAMP signaling in the morphogenesis of U. maydis and other fungal pathogens. In particular, some of the tags for transcripts that are elevated in the filamentous adr1 mutant may identify important functions for morphogenesis, a key aspect of pathogenesis in U. maydis. This suggestion is supported by the SAGE results that identified genes whose expression is regulated both by PKA activity and by the bE/bW mating-type regulator. Coregulation of specific genes by mating type and cAMP signaling has been described previously (37). For example, the transcription of the bW and bE genes depends on the phosphorylation of the transcription factor Prf1 by both the PKA and MAP kinase signaling pathways (33), and targets of direct or indirect regulation by the bE/bW transcription factor are also regulated by cAMP (10, 65, 75). The completion and annotation of the genomic sequence for U. maydis will allow a more detailed exploration of the functions of the genes whose expression is influenced by PKA and mating.
Regulation of transcription.
Analysis of the SAGE data suggests that PKA activity may have general and specific influences on the transcription of genes in U. maydis. For example, we noted that two of the genes identified in the SAGE data encoded histone 4 and histone H2B; the tags for these genes were found at lower levels in the libraries from the mutants than in the wild-type library, although the extent of reduced expression was not dramatic. It is possible that part of the influence of PKA on the transcriptome may be an indirect effect of changes in histone levels. In yeast, a reduction in nucleosome content by the depletion of histone H4 causes increased expression of 15% of genes and reduced expression of 10% of genes (76). Also, the depletion of histone H4 activates the PHO5 gene that is part of the yeast phosphate (PHO) regulatory pathway (25). Consistent with this idea, we find that the tag for histone H4 had a reduced level in the ubc1 mutant and that acid phosphatase activity is elevated in this strain. Furthermore, modification of histone H3 is known to influence the expression of the yeast genes encoding functions that we also observe to be regulated in U. maydis, including inositol 1-phosphate synthase, acid phosphatase, and phosphate permease. The observation that a tag for the gene encoding inositol 1-phosphate synthase was reduced in the ubc1 mutant is interesting in light of the known regulation of this gene by PKA via the Opi1 transcription factor in yeast. Specifically, PKA phosphorylation of Opi1 stimulates its negative regulatory influence on the expression of the INO1 gene, which encodes inositol 1-phosphate synthase (68). Opi1 is also maintained in an inactive state by phosphatidic acid, and inositol addition relieves this control to allow the transcription factor to regulate phospholipid biosynthesis (42). In general, the regulatory mechanisms for lipid signaling and phospholipid metabolism in yeast may serve as a paradigm for exploring lipid-related functions that influence morphogenesis in U. maydis.
Acknowledgments
We thank Barry Saville for generously providing access to the EST data for U. maydis. We also thank Aliya Hasham and Scott Zuyderdyn for assistance with preliminary tag to gene matching.
This work was supported by the Canadian Institutes of Health Research (J.W.K.). J.W.K. is a Burroughs Wellcome Fund Scholar in Molecular Pathogenic Mycology. M.M. and S.J. are scholars of the Michael Smith Foundation for Health Research, and M.M. is a Terry Fox/NCIC Young Investigator.
REFERENCES
- 1.Andrews, D. L., J. D. Egan, M. E. Mayorga, and S. E. Gold. 2000. The Ustilago maydis ubc4 and ubc5 genes encode members of a MAP kinase cascade required for filamentous growth. Mol. Plant-Microbe Interact. 13:781-786. [DOI] [PubMed] [Google Scholar]
- 2.Andrews, D. L., M. D. Garcia-Pedrajas, and S. E. Gold. 2004. Fungal dimorphism regulated gene expression in Ustilago maydis: I. Filament up-regulated genes. Mol. Plant Pathol. 5:281-293. [DOI] [PubMed] [Google Scholar]
- 3.Angelastro, J. M., B. Torocsik, and L. A. Greene. 2002. Nerve growth factor selectively regulates expression of transcripts encoding ribosomal proteins. BMC Neurosci. 3:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Audic, S., and J. M. Claverie. 1997. The significance of digital gene expression profiles. Genome Res. 7:986-995. [DOI] [PubMed] [Google Scholar]
- 5.Banuett, F., and I. Herskowitz. 1994. Identification of fuz7, a Ustilago maydis MEK/MAPKK homolog required for a-locus-dependent and -independent steps in the fungal life cycle. Genes Dev. 8:1367-1378. [DOI] [PubMed] [Google Scholar]
- 6.Barrett, K. J., S. E. Gold, and J. W. Kronstad. 1993. Identification and complementation of a mutation to constitutive filamentous growth in Ustilago maydis. Mol. Plant-Microbe Interact. 6:274-283. [DOI] [PubMed] [Google Scholar]
- 7.Bölker, M., S. Genin, C. Lehmler, and R. Kahmann. 1995. Genetic regulation of mating and dimorphism in Ustilago maydis. Can. J. Bot. 73:320-325. [Google Scholar]
- 8.Bölker, M., M. Urban, and R. Kahmann. 1992. The a mating type locus of U. maydis specifies cell signaling components. Cell 68:441-450. [DOI] [PubMed] [Google Scholar]
- 9.Boyce, K. J., H. Chang, C. A. D'Souza, and J. W. Kronstad. 2005. An Ustilago maydis septin is required for filamentous growth in culture and for full symptom development in maize. Eukaryot. Cell 4:2053-2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brachmann, A., G. Weinzierl, J. Kamper, and R. Kahmann. 2001. Identification of genes in the bW/bE regulatory cascade in Ustilago maydis. Mol. Microbiol. 42:1047-1063. [DOI] [PubMed] [Google Scholar]
- 11.Cardenas, M. E., N. S. Cutler, M. C. Lorenz, C. J. Di Como, and J. Heitman. 1999. The TOR signaling cascade regulates gene expression in response to nutrients. Genes Dev. 13:3271-3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cohen, A., N. Perzov, H. Nelson, and N. Nelson. 1999. A novel family of yeast chaperones involved in the distribution of V-ATPase and other membrane proteins. J. Biol. Chem. 274:26885-26893. [DOI] [PubMed] [Google Scholar]
- 13.D'Souza, C. A., and J. Heitman. 2001. Conserved cAMP signaling cascades regulate fungal development and virulence. FEMS Microbiol. Rev. 25:349-364. [DOI] [PubMed] [Google Scholar]
- 14.Dürrenberger, F., R. D. Laidlaw, and J. W. Kronstad. 2001. The hgl1 gene is required for dimorphism and teliospore formation in the fungal pathogen Ustilago maydis. Mol. Microbiol. 41:337-348. [DOI] [PubMed] [Google Scholar]
- 15.Dürrenberger, F., K. Wong, and J. W. Kronstad. 1998. Identification of a cAMP-dependent protein kinase catalytic subunit required for virulence and morphogenesis in Ustilago maydis. Proc. Natl. Acad. Sci. USA 95:5684-5689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ewing, B., and P. Green. 1998. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 8:186-194. [PubMed] [Google Scholar]
- 17.Ewing, B., L. Hillier, M. C. Wendl, and P. Green. 1998. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 8:175-185. [DOI] [PubMed] [Google Scholar]
- 18.Fankhauser, H., A. Zurlinden, A. M. Schweingruber, E. Edenharter, and M. E. Schweingruber. 1995. Schizosaccharomyces pombe thiamine pyrophosphokinase is encoded by gene tnr3 and is a regulator of thiamine metabolism, phosphate metabolism, mating, and growth. J. Biol. Chem. 270:28457-28462. [DOI] [PubMed] [Google Scholar]
- 19.Garcia-Pedrajas, M. D., and S. E. Gold. 2004. Fungal dimorphism regulated gene expression in Ustilago maydis: II. Filament down-regulated genes. Mol. Plant Pathol. 5:295-307. [DOI] [PubMed] [Google Scholar]
- 20.Gillissen, B., J. Bergemann, C. Sandmann, B. Schroeer, M. Bolker, and R. Kahmann. 1992. A two-component regulatory system for self/non-self recognition in Ustilago maydis. Cell 68:647-657. [DOI] [PubMed] [Google Scholar]
- 21.Giots, F., M. C. Donaton, and J. M. Thevelein. 2003. Inorganic phosphate is sensed by specific phosphate carriers and acts in concert with glucose as a nutrient signal for activation of the protein kinase A pathway in the yeast Saccharomyces cerevisiae. Mol. Microbiol. 47:1163-1181. [DOI] [PubMed] [Google Scholar]
- 22.Gold, S., G. Duncan, K. Barrett, and J. Kronstad. 1994. cAMP regulates morphogenesis in the fungal pathogen Ustilago maydis. Genes Dev. 8:2805-2816. [DOI] [PubMed] [Google Scholar]
- 23.Gordon, D., C. Abajian, and P. Green. 1998. Consed: a graphical tool for sequence finishing. Genome Res. 8:195-202. [DOI] [PubMed] [Google Scholar]
- 24.Gstaiger, M., B. Luke, D. Hess, E. J. Oakeley, C. Wirbelauer, M. Blondel, M. Vigneron, M. Peter, and W. Krek. 2003. Control of nutrient-sensitive transcription programs by the unconventional prefoldin URI. Science 302:1208-1212. [DOI] [PubMed] [Google Scholar]
- 25.Han, M., U. J. Kim, P. Kayne, and M. Grunstein. 1988. Depletion of histone H4 and nucleosomes activates the PHO5 gene in Saccharomyces cerevisiae. EMBO J. 7:2221-2228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hartmann, H. A., R. Kahmann, and M. Bolker. 1996. The pheromone response factor coordinates filamentous growth and pathogenicity in Ustilago maydis. EMBO J. 15:1632-1641. [PMC free article] [PubMed] [Google Scholar]
- 27.Hartmann, H. A., J. Kruger, F. Lottspeich, and R. Kahmann. 1999. Environmental signals controlling sexual development of the corn smut fungus Ustilago maydis through the transcriptional regulator Prf1. Plant Cell 11:1293-1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hoffman, C. S. 2005. Glucose sensing via the protein kinase A pathway in Schizosaccharomyces pombe. Biochem. Soc. Trans. 33:257-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hornby, J. M., R. Dumitru, and K. W. Nickerson. 2004. High phosphate (up to 600 mM) induces pseudohyphal development in five wild type Candida albicans. J. Microbiol. Methods 56:119-124. [DOI] [PubMed] [Google Scholar]
- 30.Jensen, L. T., M. Ajua-Alemanji, and V. Cizewski Culotta. 2003. The Saccharomyces cerevisiae high affinity phosphate transporter encoded by PHO84 also functions in manganese homeostasis. J. Biol. Chem. 278:42036-42040. [DOI] [PubMed] [Google Scholar]
- 31.Jones, D. L., J. Petty, D. C. Hoyle, A. Hayes, E. Ragni, L. Popolo, S. G. Oliver, and L. I. Stateva. 2003. Transcriptome profiling of a Saccharomyces cerevisiae mutant with a constitutively activated Ras/cAMP pathway. Physiol. Genomics 16:107-118. [DOI] [PubMed] [Google Scholar]
- 32.Jorgensen, P., I. Rupes, J. R. Sharom, L. Schneper, J. R. Broach, and M. Tyers. 2004. A dynamic transcriptional network communicates growth potential to ribosome synthesis and critical cell size. Genes Dev. 18:2491-2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kaffarnik, F., P. Muller, M. Leibundgut, R. Kahmann, and M. Feldbrugge. 2003. PKA and MAPK phosphorylation of Prf1 allows promoter discrimination in Ustilago maydis. EMBO J. 22:5817-5826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kamper, J., M. Reichmann, T. Romeis, M. Bolker, and R. Kahmann. 1995. Multiallelic recognition: nonself-dependent dimerization of the bE and bW homeodomain proteins in Ustilago maydis. Cell 81:73-83. [DOI] [PubMed] [Google Scholar]
- 35.Klein, C., and K. Struhl. 1994. Protein kinase A mediates growth-regulated expression of yeast ribosomal protein genes by modulating RAP1 transcriptional activity. Mol. Cell. Biol. 14:1920-1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Klose, J., M. M. de Sá, and J. W. Kronstad. 2004. Lipid-induced filamentous growth in Ustilago maydis. Mol. Microbiol. 52:823-835. [DOI] [PubMed] [Google Scholar]
- 37.Kruger, J., G. Loubradou, E. Regenfelder, A. Hartmann, and R. Kahmann. 1998. Crosstalk between cAMP and pheromone signalling pathways in Ustilago maydis. Mol. Gen. Genet. 260:193-198. [DOI] [PubMed] [Google Scholar]
- 38.Lee, N., C. A. D'Souza, and J. W. Kronstad. 2003. Of smuts, blasts, mildews, and blights: cAMP signaling in phytopathogenic fungi. Annu. Rev. Phytopathol. 41:399-427. [DOI] [PubMed] [Google Scholar]
- 39.Li, Y., R. D. Moir, I. K. Sethy-Coraci, J. R. Warner, and I. M. Willis. 2000. Repression of ribosome and tRNA synthesis in secretion-defective cells is signaled by a novel branch of the cell integrity pathway. Mol. Cell. Biol. 20:3843-3851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lian, T. S., M. I. Simmer, C. A. D'Souza, B. R. Steen, S. D. Zuyderduyn, S. J. M. Jones, M. A. Marra, and J. W. Kronstad. 2005. Iron-regulated transcription and capsule formation in the fungal pathogen Cryptococcus neoformans. Mol. Microbiol. 55:1452-1472. [DOI] [PubMed] [Google Scholar]
- 41.Lichter, A., and D. Mills. 1998. Control of pigmentation of Ustilago hordei: the effect of pH, thiamine, and involvement of the cAMP cascade. Fungal Genet. Biol. 25:63-74. [DOI] [PubMed] [Google Scholar]
- 42.Loewen, C. J., M. L. Gaspar, S. A. Jesch, C. Delon, N. T. Ktistakis, S. A. Henry, and T. P. Levine. 2004. Phospholipid metabolism regulated by a transcription factor sensing phosphatidic acid. Science 304:1644-1647. [DOI] [PubMed] [Google Scholar]
- 43.Longtine, M. S., and E. Bi. 2003. Regulation of septin organization and function in yeast. Trends Cell Biol. 13:403-409. [DOI] [PubMed] [Google Scholar]
- 44.Lubkowitz, M. A., D. Barnes, M. Breslav, A. Burchfield, F. Naider, and J. M. Becker. 1998. Schizosaccharomyces pombe isp4 encodes a transporter representing a novel family of oligopeptide transporters. Mol. Microbiol. 28:729-741. [DOI] [PubMed] [Google Scholar]
- 45.Martin, D. E., A. Soulard, and M. N. Hall. 2004. TOR regulates ribosomal protein gene expression via PKA and the Forkhead transcription factor FHL1. Cell 119:969-979. [DOI] [PubMed] [Google Scholar]
- 46.Martinez-Espinoza, A. D., M. D. Garcia-Pedrajas, and S. E. Gold. 2002. The Ustilaginales as plant pests and model systems. Fungal Genet. Biol. 35:1-20. [DOI] [PubMed] [Google Scholar]
- 47.Mayorga, M. E., and S. E. Gold. 1999. A MAP kinase encoded by the ubc3 gene of Ustilago maydis is required for filamentous growth and full virulence. Mol. Microbiol. 34:485-497. [DOI] [PubMed] [Google Scholar]
- 48.Mayorga, M. E., and S. E. Gold. 2001. The ubc2 gene of Ustilago maydis encodes a putative novel adaptor protein required for filamentous growth, pheromone response and virulence. Mol. Microbiol. 41:1365-1379. [DOI] [PubMed] [Google Scholar]
- 49.McGrath, J. W., and J. P. Quinn. 2000. Intracellular accumulation of polyphosphate by the yeast Candida humicola G-1 in response to acid pH. Appl. Environ. Microbiol. 66:4068-4073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Muller, P., C. Aichinger, M. Feldbrugge, and R. Kahmann. 1999. The MAP kinase kpp2 regulates mating and pathogenic development in Ustilago maydis. Mol. Microbiol. 34:1007-1017. [DOI] [PubMed] [Google Scholar]
- 51.Neuman-Silberberg, F. S., S. Bhattacharya, and J. R. Broach. 1995. Nutrient availability and the RAS/cyclic AMP pathway both induce expression of ribosomal protein genes in Saccharomyces cerevisiae but by different mechanisms. Mol. Cell. Biol. 15:3187-3196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nugent, K. G., K. Choffe, and B. J. Saville. 2004. Gene expression during Ustilago maydis diploid filamentous growth: EST library creation and analyses. Fungal Genet. Biol. 41:349-360. [DOI] [PubMed] [Google Scholar]
- 53.Ogawa, N., J. DeRisi, and P. O. Brown. 2000. New components of a system for phosphate accumulation and polyphosphate metabolism in Saccharomyces cerevisiae revealed by genomic expression analysis. Mol. Biol. Cell 11:4309-4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Orth, A. B., M. Rzhetskaya, E. J. Pell, and M. Tien. 1995. A serine (threonine) protein kinase confers fungicide resistance in the phytopathogenic fungus Ustilago maydis. Appl. Environ. Microbiol. 61:2341-2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Oshima, Y. 1997. The phosphatase system in Saccharomyces cerevisiae. Genes Genet. Syst. 72:323-334. [DOI] [PubMed] [Google Scholar]
- 56.Parle-McDermott, A., P. McWilliam, O. Tighe, D. Dunican, and D. T. Croke. 2000. Serial analysis of gene expression identifies putative metastasis-associated transcripts in colon tumour cell lines. Br. J. Cancer 83:725-728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Persson, B. L., J. O. Lagerstedt, J. R. Pratt, J. Pattison-Granberg, K. Lundh, S. Shokrollahzadeh, and F. Lundh. 2003. Regulation of phosphate acquisition in Saccharomyces cerevisiae. Curr. Genet. 43:225-244. [DOI] [PubMed] [Google Scholar]
- 58.Potapova, O., S. V. Anisimov, M. Gorospe, R. H. Dougherty, W. A. Gaarde, K. R. Boheler, and N. J. Holbrook. 2002. Targets of c-Jun NH(2)-terminal kinase 2-mediated tumor growth regulation revealed by serial analysis of gene expression. Cancer Res. 62:3257-3263. [PubMed] [Google Scholar]
- 59.Ramesh, M., D. Laidlaw, F. Dürrenberger, A. Orth, and J. W. Kronstad. 2001. The cAMP signal transduction pathway mediates resistance to dicarboximide and aromatic hydrocarbon fungicides in Ustilago maydis. Fungal Genet. Biol. 32:183-193. [DOI] [PubMed] [Google Scholar]
- 60.Robertson, L. S., H. C. Causton, R. A. Young, and G. R. Fink. 2000. The yeast A kinases differentially regulate iron uptake and respiratory function. Proc. Natl. Acad. Sci. USA 97:5984-5988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ruis, H., and C. Schuller. 1995. Stress signaling in yeast. Bioessays 17:959-965. [DOI] [PubMed] [Google Scholar]
- 62.Sacadura, N. T., and B. J. Saville. 2003. Gene expression and EST analysis of Ustilago maydis germinating teliospores. Fungal Genet. Biol. 40:47-64. [DOI] [PubMed] [Google Scholar]
- 63.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 64.Sato, S., H. Suzuki, U. Widyastuti, Y. Hotta, and S. Tabata. 1994. Identification and characterization of genes induced during sexual differentiation in Schizosaccharomyces pombe. Curr. Genet. 26:31-37. [DOI] [PubMed] [Google Scholar]
- 65.Schauwecker, F., G. Wanner, and R. Kahmann. 1995. Filament-specific expression of a cellulase gene in the dimorphic fungus Ustilago maydis. Biol. Chem. Hoppe-Seyler 376:617-625. [DOI] [PubMed] [Google Scholar]
- 66.Schmelzle, T., T. Beck, D. E. Martin, and M. N. Hall. 2004. Activation of the RAS/cyclic AMP pathway suppresses a TOR deficiency in yeast. Mol. Cell. Biol. 24:338-351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Serafim, L. S., P. C. Lemos, C. Levantesi, V. Tandoi, H. Santos, and M. A. Reis. 2002. Methods for detection and visualization of intracellular polymers stored by polyphosphate-accumulating microorganisms. J. Microbiol. Methods 51:1-18. [DOI] [PubMed] [Google Scholar]
- 68.Sreenivas, A., and G. M. Carman. 2003. Phosphorylation of the yeast phospholipid synthesis regulatory protein Opi1p by protein kinase A. J. Biol. Chem. 278:20673-20680. [DOI] [PubMed] [Google Scholar]
- 69.Steen, B. R., T. Lian, S. Zuyderduyn, W. K. MacDonald, M. Marra, S. J. Jones, and J. W. Kronstad. 2002. Temperature-regulated transcription in the pathogenic fungus Cryptococcus neoformans. Genome Res. 12:386-400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Steen, B. R., S. Zuyderduyn, D. L. Toffaletti, M. Marra, S. J. M. Jones, J. R. Perfect, and J. Kronstad. 2003. Cryptococcus neoformans gene expression during experimental cryptococcal meningitis. Eukaryot. Cell 2:1336-1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Thevelein, J. M., R. Gelade, I. Holsbeeks, O. Lagatie, Y. Popova, F. Rolland, F. Stolz, S. Van de Velde, P. Van Dijck, P. Vandormael, A. Van Nuland, K. Van Roey, G. Van Zeebroeck, and B. Yan. 2005. Nutrient sensing systems for rapid activation of the protein kinase A pathway in yeast. Biochem. Soc. Trans. 33:253-256. [DOI] [PubMed] [Google Scholar]
- 72.Velculescu, V. E., L. Zhang, B. Vogelstein, and K. W. Kinzler. 1995. Serial analysis of gene expression. Science 270:484-487. [DOI] [PubMed] [Google Scholar]
- 73.Velculescu, V. E., L. Zhang, W. Zhou, J. Vogelstein, M. A. Basrai, D. E. Bassett, Jr., P. Hieter, B. Vogelstein, and K. W. Kinzler. 1997. Characterization of the yeast transcriptome. Cell 88:243-251. [DOI] [PubMed] [Google Scholar]
- 74.Wang, Y., M. Pierce, L. Schneper, C. G. Guldal, X. Zhang, S. Tavazoie, and J. R. Broach. 2004. Ras and Gpa2 mediate one branch of a redundant glucose signaling pathway in yeast. PLoS Biol. 2:610-622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wosten, H. A., R. Bohlmann, C. Eckerskorn, F. Lottspeich, M. Bolker, and R. Kahmann. 1996. A novel class of small amphipathic peptides affect aerial hyphal growth and surface hydrophobicity in Ustilago maydis. EMBO J. 15:4274-4281. [PMC free article] [PubMed] [Google Scholar]
- 76.Wyrick, J. J., F. C. Holstege, E. G. Jennings, H. C. Causton, D. Shore, M. Grunstein, E. S. Lander, and R. A. Young. 1999. Chromosomal landscape of nucleosome-dependent gene expression and silencing in yeast. Nature 402:418-421. [DOI] [PubMed] [Google Scholar]
- 77.Zhang, L., W. Zhou, V. E. Velculescu, S. E. Kern, R. H. Hruban, S. R. Hamilton, B. Vogelstein, and K. W. Kinzler. 1997. Gene expression profiles in normal and cancer cells. Science 276:1268-1272. [DOI] [PubMed] [Google Scholar]
- 78.Zurita-Martinez, S. A., and M. E. Cardenas. 2005. Tor and cyclic AMP-protein kinase A: two parallel pathways regulating expression of genes required for cell growth. Eukaryot. Cell 4:63-71. [DOI] [PMC free article] [PubMed] [Google Scholar]