Abstract
Nutritional excess and/or obesity represent well-known predisposition factors for the development of non-insulin-dependent diabetes mellitus (NIDDM). However, molecular links between obesity and NIDDM are only beginning to emerge. Here, we demonstrate that nutrients suppress phosphatidylinositol 3 (PI3)-kinase/Akt signaling via Raptor-dependent mTOR (mammalian target of rapamycin)-mediated phosphorylation of insulin receptor substrate 1 (IRS-1). Raptor directly binds to and serves as a scaffold for mTOR-mediated phosphorylation of IRS-1 on Ser636/639. These serines lie close to the Y632MPM motif that is implicated in the binding of p85α/p110α PI3-kinase to IRS-1 upon insulin stimulation. Phosphomimicking mutations of these serines block insulin-stimulated activation of IRS-1-associated PI3-kinase. Knockdown of Raptor as well as activators of the LKB1/AMPK pathway, such as the widely used antidiabetic compound metformin, suppress IRS-1 Ser636/639 phosphorylation and reverse mTOR-mediated inhibition on PI3-kinase/Akt signaling. Thus, diabetes-related hyperglycemia hyperactivates the mTOR pathway and may lead to insulin resistance due to suppression of IRS-1-dependent PI3-kinase/Akt signaling.
mTOR (mammalian target of rapamycin) is a Ser/Thr kinase that belongs to the phosphatidylinositol (PI) kinase-related protein kinase family (17). In the budding yeast Saccharomyces cerevisiae, TOR functions as a nutrient-dependent mediator of cell autonomous growth (17). In metazoans, TOR participates in both nutrient- and hormone-dependent signaling pathways (17). mTOR partitions between two scaffold proteins, Raptor (14, 27) and Rictor (25, 33, 52). The rapamycin-sensitive Raptor/mTOR complex (TORC1) regulates growth via S6K1 and 4EBP1/PHAS (14, 27). The rapamycin-insensitive Rictor/mTOR (TORC2) complex regulates cellular proliferation via Akt (51) and cytoskeleton organization via protein kinase Cα (52) and small GTPases Rho and Rac (25).
AMP-activated protein kinase (AMPK) acts as an intracellular energy sensor that controls glucose and lipid metabolism in peripheral tissues (6). Its activation leads to a homeostatic response by down-regulation of anabolic pathways and up-regulation of catabolic pathways (6). AMPK- and PI3-kinase/Akt-dependent signaling pathways converge at the level of tuberus sclerosis complex 2 (TSC2 or tuberin) which functions as a GTPase-activating protein for the small GTPase Rheb (20-22). Genetic studies in Drosophila melanogaster place Rheb downstream of insulin/insulin-like growth factor I and nutrient-dependent signaling pathways (53). Rheb is negatively regulated by AMPK/TSC (20, 22) and functions as an upstream activator of mTOR (20, 34, 53). Normal cell growth and homeostasis rely on a proper balance between nutrient- and hormone-dependent regulation of the AMPK/mTOR axis, while its deregulation could lead to metabolic and growth-related diseases (11, 20, 22,67).
Several in vitro and in vivo studies show a beneficial metabolic effect of AMPK activation on glucose homeostasis and peripheral insulin sensitivity (37, 68, 75). In contrast, hyperactivation of the mTOR-mediated pathway(s) is involved in several desensitizing events downstream of the insulin receptor. The inhibitory effect of mTOR/S6K1 activation correlates with increased Ser/Thr phosphorylation of insulin receptor substrate 1 (IRS-1). For example, mTOR/S6K1-dependent phosphorylation of IRS-1 on Ser307 uncouples its interaction with the insulin receptor (15, 72). On the contrary, knockout of S6K1 increases insulin sensitivity and protects mice against age- and diet-induced obesity (67).
In this study, we show that hyperglycemia, the main metabolic abnormality in non-insulin-dependent diabetes mellitus (NIDDM) subjects, up-regulates activity of the mTOR/S6K1 axis. This may lead not only to previously characterized inhibitory phosphorylation of IRS-1 on Ser307 by S6K1 (15, 72) but also to phosphorylation of IRS-1 on Ser636/639 (human isoform) directly by mTOR. These serine residues lie at the +4/+7 position of the Y632MPM motif that, along with the Y612MPM, is implicated in binding of the two Src homology 2 domains of the regulatory p85α subunit of PI3-kinase (13, 38). Point mutations of Ser636/639 residues to aspartic acid abolish insulin-stimulated activation of IRS-1-associated PI3-kinase. Our data show that IRS-1 coimmunoprecipitates with the mTOR scaffold protein, Raptor, and that the Raptor/mTOR complex exhibits kinase activity toward Ser636/639 of recombinant IRS-1 in vitro. Deletion of the carboxy terminus of Raptor significantly decreases Raptor-IRS-1 interaction and attenuates IRS-1 Ser636/639 phosphorylation, suggesting that mTOR phosphorylates IRS-1 in a Raptor-dependent fashion. Similarly, knockdown of Raptor attenuates nutrient-, insulin-, and Rheb-stimulated IRS-1 Ser636/639 phosphorylation and reverses the inhibitory role of mTOR on PI3-kinase/Akt signaling. Moreover, long-term activation of mTOR by Rheb overexpression leads to IRS-1 degradation. Activators of AMPK, such as LKB1 (32), 2-deoxyglucose (22), and the antidiabetic drug metformin (75) suppress IRS-1 Ser636/639 phosphorylation and enhance PI3-kinase/Akt signaling. This suggests that some of the antidiabetic effects of metformin could be mediated by AMPK-dependent inhibition of mTOR.
MATERIALS AND METHODS
Materials.
Anti-Raptor antibody was purchased from Bethyl Laboratories. Antibodies pY20 and HA.11 were from BD Transduction Laboratories and Covance, respectively. All other antibodies were from Cell Signaling. Insulin, IBMX, dexamethasone, protein A, protein G, horseradish peroxidase-conjugated secondary antibodies, protease and phosphatase inhibitor cocktails, and metformin (1,1-dimethyl biguanide hydrochloride) were from Sigma. Rapamycin and fatty acid-free bovine serum albumin (catalog no. 126575) were obtained from Calbiochem. Tumor necrosis factor alpha (TNF-α) was from Research Diagnostics, lipids were from Avandi Polar Lipids, and Raptor small interfering RNA (siRNA) was from MWG-Biotech.
Plasmid construction.
Rat wild-type pCMVhis-IRS-1 was a gift from M. White. For Myc-tagged IRS-1, the first 211 nucleotides of IRS-1 were PCR amplified with 5′-ATATGAATTCGCCGCCACCATGGAACAAAAACTTATTTCTGAAGAAGATCTGGCGAGCCCTCCGGATACCGATGGC-3′ (sense, includes the Myc epitope as well as a Kozak sequence for efficient translational initiation) and 5′-TTGAAACAGCTCTCGAGGGGG-3′ (antisense). The amplified fragment was pasted in the EcoRI and XhoI sites of pCMV-MCS vector (Stratagene). The full-length IRS-1 tagged with the Myc epitope generated by subcloning the XhoI/SalI fragment of wild-type (WT) IRS-1 in the XhoI site of the intermediate plasmid from the previous step to obtain the full-length pCMV-Myc-IRS-1 WT.
Site-specific mutagenesis of pCMVhis-IRS-1 was carried out with the help of the overlap extension method. Amino acid numbering refers to the human isoform. The following sense primers have been used: (i) for IRS-1 S636/639D, 5′-TATATGCCAATGGACCCCAAGGATGTATCTGCC-3′; (ii) for IRS-1 S636/639A, 5′-TATATGCCAATGGCTCCCAAGGCTGTATCTGCC-3′; (iii) for IRS-1 S616D, 5′-TACATGCCAATGGATCCCGGAGTG-3′; (iv) for IRS-1 S616/636D, 5′-CCAATGGATCCCGGAGTGGCTCCAGTGCCCAGCAAC CGCAAAGGAAATG GGGACTATATGCCAATGGAC-3′. Mutated nucleotides are underlined; silent mutagenized nucleotides are in boldface type. Outer primers spanned two NcoI restriction enzyme sites in the 5′ and 3′ ends of the aforementioned mutagenized sites.
Human LKB1/STK11 was amplified with the primers 5′-GAATTCGAGGTGGTGGACCCGCAGCAGCTG-3′ (sense) and 5′-TCTAGAAAATCACTGCTGCTTGCAGGCCGACAG-3′ (antisense) from the clone MGC-16254 (ATCC) and subcloned in frame in the EcoRI and XbaI sites of pCMV5-Myc vector to generate pCMV5-Myc-LKB1.
Wild-type pRK5-HA-Raptor was a gift from D. Sabatini. For the carboxy-terminal deletions, the following primers have been used: (i) for ΔWD40 (Δ1011 to 1335), 5′-TTGCCTTCTGGCCGGCCCGGC-3′ (sense) and 5′-GCGGCCGCACTACGTAATGCCCTTCTGAATGACTTG-3′ (antisense); (ii) for ΔWD40/Hinge (Δ678 to 1335), 5′-TGGCAGCCAAGGGCCCGGTACAAG-3′ (sense) and 5′-GCGGCCGCACTAATTGCTTTCATACTGAACCACAAG-3′ (antisense). PCR-amplified fragments were subcloned in the FseI and NotI and ApaI and NotI sites of digested pRK5-HA-Raptor, respectively.
Wild-type rat pCGN-HA-Rheb was a gift from J. Der and J. Clark. For the C181S point mutation, the following primers have been used: 5′-ATGGCTTCTAGCTATCCTTATGAC-3′ (sense) and 5′-ATGGATCCTCACATCACCGAGCTCGAAGACTTCCC-3′ (antisense).
Kinase-deficient rat pRK7-HA-S6K1 αII was a gift from J. Blenis. Wild-type rat pMT2-HA-S6K1 αI WT and the rapamycin-resistant mutant pMT2-HA-S6K1-Δ2-46/ΔCT104 were a gift from J. Avruch.
Cell culture.
Murine 3T3-L1 cells were grown in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% calf serum until confluence. Two days later, cells were transferred to the differentiation medium (DMEM containing 10% fetal bovine serum [FBS], 0.5 mM 3-isobutyl-1-methylxanthine, 1 μM dexamethasone, and 1.7 μM insulin). After 48 h, the differentiation medium was replaced with DMEM supplemented with 10% FBS. Cells were used after 6 to 8 days of differentiation. The myoblast line L6 was grown under subconfluent conditions in DMEM containing 20% FBS. Cells at ∼80 to 90 confluence were maintained in DMEM supplemented with 2% calf serum for 5 to 7 days for differentiation into myotubes. HepG2, NIH 3T3, and 293HEK cells were grown in DMEM supplemented with 10% FBS. C2C12 fibroblasts were grown similar to L6 myoblasts. For stable expression of hemagglutinin (HA)-tagged Rheb, C2C12 fibroblasts were transfected with pCGN-HA-Rheb, and several clones were selected for 7 to 10 days in the medium containing 300 μg/ml hygromycin B (Invitrogen).
Subcellular fractionation.
Cells were washed twice in ice-cold phosphate-buffered saline (PBS) and homogenized in buffer A (20 mM HEPES, pH 7.4, 1 mM EDTA, 1 mM phenylmethylsulfonyl fluoride [PMSF], 2 mM Na3VO4, 30 mM NaF, supplemented with a mixture of protease and phosphatase inhibitors) by 16 strokes in a ball-bearing homogenizer (Isobiotec, Heidelberg, Germany) with a 12-μm clearance. The homogenate was centrifuged at 14,000 × g for 20 min at 4°C, and the supernatant was recentrifuged at 200,000 × g for 1 h at 4°C in a Beckman type Ti42.2 rotor. The cytosol and high-speed pellet were analyzed by Western blotting.
Cell lysis and immunoprecipitations.
Cells were washed twice in ice-cold PBS and solubilized in the lysis buffer (50 mM Tris, pH 7.5, 150 mM NaCl, 1% Triton X-100, 10 mM Na3VO4, 50 mM NaF, 1 mM β-glycerophosphate, 1 mM sodium pyrophosphate, 1 mM EDTA, 1 mM EGTA, 1 mM PMSF, supplemented with a mixture of protease and phosphatase inhibitors). The lysate was cleared by centrifugation for 10 min at 14,000 × g at 4°C. For immunoprecipitation, 0.5 to 1 mg of the lysate was incubated overnight with 1 to 2 μg of the specific and nonspecific antibody coupled to protein A- or G-Sepharose beads under gentle agitation. After extensive washing with lysis buffer, beads were resuspended in sodium dodecyl suflate sample buffer supplemented with 5% β-mercaptoethanol, boiled for 5 min, and subjected to Western blot analysis.
In vitro measurement of IRS-1-associated PI3-kinase activity. IRS-1 was immunoprecipitated from cell lysates, and immune complexes were washed 3 times with lysis buffer and then with wash buffer A (100 mM Tris, pH 7.4, 500 mM LiCl, 2 mM Na3VO4). Then, immunoprecipitates were washed once with buffer B (10 mM Tris, pH 7.4, 100 mM NaCl, 1 mM EDTA, 2 mM Na3VO4), and twice with the kinase buffer (10 mM MgCl2 and 50 mM Tris, pH 7.4). After that, 45 μl of kinase buffer, 5 μl of sonicated lipid substrate (phosphatidylinositol and phosphatidylserine (1:1) in a concentration of 1 mg/ml in 25 mM HEPES, pH 7.4, and 1 mM EDTA), and 20 μCi of [γ-32P]ATP were added to washed beads for 20 min at room temperature. Reactions were stopped by the addition of 100 μl of 1 N HCl. Lipids were extracted and spotted on potassium oxalate-treated silica gel thin-layer chromatography (TLC) plates (250-μm layer, catalog no. 4420221; Whatman) and separated in chloroform, methanol, acetone, acetic acid, and H2O (60:20:23:18:12, vol/vol/vol/vol/vol). The plate was dried and exposed either to the X-ray film with an intensifying screen at −80°C or in Instant Imager (Packard) for quantification of radioactivity. Radioactive spots were identified by comigration with lipid standards detected by iodine vapors.
In vitro kinase assay.
293HEK cells transiently transfected with Myc-mTOR and Myc-Raptor were harvested in lysis buffer which contained 0.3% 3-[(3-cholamidylpropyl)-dimethylammonio]-1-propanesulfonate (CHAPS) instead of Triton X-100. Ectopically expressed proteins were immunoprecipitated with anti-Myc antibody as described in the previous section. Beads were washed twice with mTOR kinase buffer (10 mM HEPES, pH 7.4, 50 mM NaCl, and 10 mM MnCl2), resuspended in 30 μl of kinase buffer containing 200 μM ATP with or without 20 μCi of [γ-32P]ATP, and recombinant IRS-1 (600 ng; catalog no. 13-124; Upstate) was immediately added to the kinase mixture. The phosphorylation reaction was carried out at 30°C for 30 min and terminated by the addition of an equal volume of 4× sample buffer. Phosphorylated proteins were separated by polyacrylamide gel electrophoresis and analyzed by Western blotting with phosphospecific antibody and/or autoradiography.
siRNA transfection.
siRNA against human Raptor was transfected into 293HEK cells with the help of Lipofectamine 2000 as described previously by Kim and colleagues (27). Transfected cells were further grown for 48 to 60 h before the experiment.
Measurements of GDP-bound Rheb.
293 HEK cells transiently transfected with pCGN-HA-Rheb were washed twice in ice-cold PBS and lysed in Rheb lysis buffer (50 mM Tris, pH 7.5, 150 mM NaCl, 1% Triton X-100, 1 mM PMSF, 20 mM MgCl2 supplemented with the mixture of protease inhibitors). The lysate was cleared by centrifugation for 10 min at 14,000 × g at 4°C, and the NaCl concentration was adjusted to 500 mM to inhibit GTPase-activating protein activity in the lysate. HA-Rheb was immunoprecipitated with anti-HA antibody coupled to protein G-Sepharose beads under gentle agitation at 4°C. The beads were washed sequentially with high-salt buffer (50 mM Tris, pH 8, 500 mM NaCl, 10 mM MgCl2, and 0.5% Triton X-100) and with low-salt buffer (50 mM Tris, pH 8, 100 mM NaCl, 10 mM MgCl2, and 0.1% Triton X-100). The beads were resuspended in 30 μl elution buffer (5 mM Tris phosphate, pH 7.8, 2 mM EDTA, and 4 mM dithiothreitol), heated for 3 min at 100°C, cooled on ice, and pelleted for 3 min at 14,000 × g. The supernatant containing guanine nucleotides dissociated from bead-bound Rheb was collected and either analyzed immediately or stored at −80°C. The beads were resuspended in sodium dodecyl sulfate sample buffer, supplemented with 5% β-mercaptoethanol, boiled for 5 min, and subjected to Western blot analysis. For the measurement of GDP-bound Rheb, eluted GDP was converted to [γ-32P]GTP using nucleoside diphosphate (NDP) kinase (catalog no. N-2635; Sigma) and 1 μCi/μl [γ-32P]ATP for 90 min at 37°C in NDP reaction buffer (50 mM Tris-HCl, pH 7.4, 10 mM MgCl2 in the presence of 4 mU/μl of NDP kinase) in the following reaction: GDP + [γ-32P]ATP → [γ-32P]GTP + ADP. The labeled nucleotides were separated by chromatography on plastic-backed cellulose TLC plates (catalog no. 10089; Selecto Scientific) in saturated ammonium sulfate (53.1 g/100 ml H2O), H2O, 3 M sodium acetate, 10 N sodium hydroxide, isopropanol (80/10/6/2/2, vol/vol/vol/vol/vol). The Rf of GTP was 0.71, and that of ATP was 0.5. The proper migration was assessed by comigration of [α-32P]GTP and [γ-32P]ATP. The reaction was linear in the range of 1 to 50 pM of the GDP standard. Data were presented as the amount of GDP-bound Rheb normalized by the amount of immunoprecipitated HA-Rheb as determined by Western blotting.
Statistics.
Results are expressed as means ± standard errors of the means (SEM). Significance was analyzed with the help of Student's t test.
Experimental protocol.
In all experiments, cells were incubated in either high-glucose (25 mM) serum-free DMEM (catalog no. 11995-065; Invitrogen) or in glucose-free Krebs-Ringer phosphate (KRP) buffer (12 mM HEPES, 120 mM NaCl, 6 mM KCl, 1.2 mM MgSO4, 1 mM CaCl2, 0.6 mM Na2HPO4, 0.4 mM NaH2PO4, and 0.1% fatty acid-free bovine serum albumin, pH 7.4) for 6 h unless indicated otherwise. Rapamycin, insulin, 2-deoxyglucose, leucine, glucose, and TNF-α were added to cells for 30 min unless indicated otherwise. In transfection experiments, the corresponding empty vector was used to balance the amounts of transfected DNA. Shifts in electrophoretic mobility of IRS-1 were analyzed in 6% gels.
RESULTS
Rapamycin inhibits IRS-1 Ser636/639 phosphorylation and enhances insulin signaling via the PI3-kinase/Akt pathway.
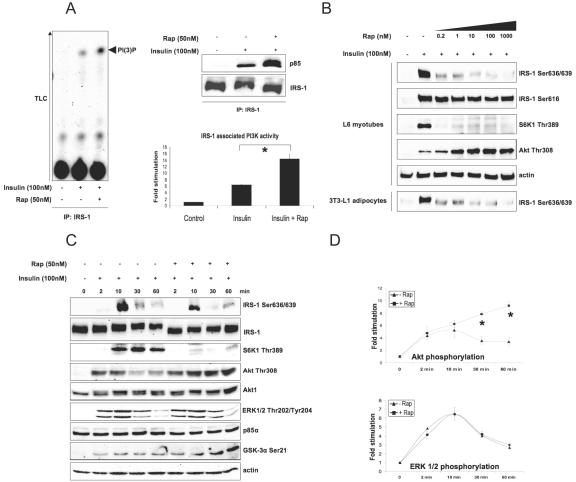
Rapamycin forms a complex with the immunophilin FKBP12 and inhibits mTOR activity with a 50% inhibitory concentration in vitro ∼50 pM by inducing the dissociation of the Raptor/mTOR complex (41). Figure 1A demonstrates that upon insulin or insulin-like growth factor I (not shown) stimulation, rapamycin increases the amount of p85 that coimmunoprecipitates with IRS-1 as well as the IRS-1-associated PI3-kinase activity by a factor of 2.4. In addition, rapamycin completely reverses the shift in the electrophoretic mobility of IRS-1 that is attributed to Ser/Thr phosphorylation and enhances insulin-stimulated association of PI3-kinase with IRS-1 within 10 to 60 min of insulin administration (not shown).
FIG. 1.
Rapamycin inhibits phosphorylation of IRS-1 on Ser636/639 and enhances PI3-kinase/Akt signaling. (A) L6 myotubes were serum starved for 6 h, pretreated with rapamycin (Rap) for 30 min, and then treated with insulin for another 30 min as described in Materials and Methods, and endogenous IRS-1 was immunoprecipitated (IP). The presence of PI3-kinase in the immunoprecipitated material was determined by Western blotting (top right panel) and its activity was measured by TLC (left panel). The bottom right panel shows normalized IRS-1-associated PI3-kinase activity (mean values ± SEM) of the results from three independent experiments. (B and C) Total cell lysates treated as described for panel A were analyzed by Western blotting. (D) Normalized mean values ± SEM of results from three independent experiments shown in panel C; asterisks indicate that P values of <0.01. +, present; −, absent.
Figure 1B shows that, in both L6 myotubes and 3T3-L1 adipocytes, insulin-induced phosphorylation of IRS-1 on Ser636/639 is sensitive to picomolar concentrations of rapamycin similar to phosphorylation of S6K1 on Thr389. In contrast, phosphorylation of IRS-1 on Ser616 is not sensitive to even micromolar concentrations of the drug. Increasing concentrations of rapamycin enhance insulin-stimulated Akt phosphorylation in a way antiparallel to that of IRS-1 Ser636/639 phosphorylation, suggesting a potential role for these two serine residues in regulating IRS-1-associated PI3-kinase activity.
The peak of mTOR activity takes place after 10 to 15 min of insulin stimulation (5), following Akt and ERK1/2 activation (1 to 2 min), and coincides with IRS-1 Ser636/639 and S6K1 Thr389 phosphorylation (Fig. 1C). Rapamycin specifically enhances insulin-stimulated phosphorylation of Akt (2.3 and 2.8 times after 30 and 60 min, respectively) (Fig. 1D) and glycogen synthetase kinase 3α without affecting ERK1/2 phosphorylation (Fig. 1C and 1D).
Phosphomimicking mutations of S636/639 suppresses IRS-1-associated PI3-kinase activity and may control intracellular localization of IRS-1.
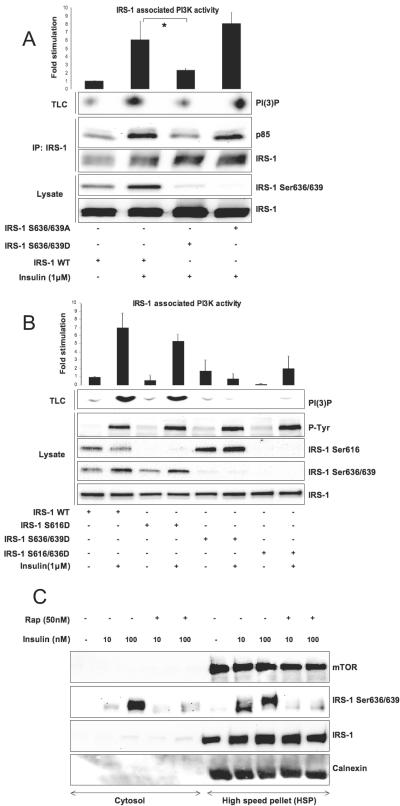
To address the role of IRS-1 Ser636/639 phosphorylation in PI3-kinase activation, we created a double IRS-1 mutant by replacing these two serine residues with either aspartic acid (S636/639D) or alanine (S636/639A). Figure 2A shows that insulin stimulation significantly increases the amount of p85 and PI3-kinase activity that coimmunoprecipitates with wild-type IRS-1. On the contrary, the S636/639D mutant does not mediate insulin-dependent activation of the enzyme. Mutation of Ser636/639 to Ala completely restores insulin-stimulated IRS-1-associated PI3-kinase activity, suggesting that Ser636/639 phosphorylation negatively regulates PI3-kinase.
FIG. 2.
Phosphorylation of IRS-1 on Ser 636/639 but not on Ser616 uncouples insulin signaling. (A) 293HEK cells transiently transfected with specified IRS-1 mutants (4 μg of each cDNA per 10-cm plate) were serum starved for 6 h followed by 5 min of insulin stimulation and immunoprecipitation (IP) of IRS-1. The presence of PI3-kinase in the immunoprecipitated material was determined by Western blotting, and its activity was measured by TLC. The top panel shows normalized PI3-kinase activity (mean values ± SEM) of three independent experiments. The bottom panel shows Western blot analysis of total cell lysates. (B) Same as described for panel A with different IRS-1 mutants. Note that in panels A and B, phosphospecific antibodies do not recognize mutated serine residues in IRS-1, verifying that mutations have been introduced correctly. (C) L6 myotubes were homogenized and fractionated as described in Materials and Methods into cytosol and high-speed pellet. Each fraction was analyzed by Western blotting. The same amount of protein (30 μg) was loaded on each lane; asterisks indicate P values of < 0.01. +, present; −, absent.
Both Y612MPM and Y632MPM motifs are involved in PI3-kinase recruitment (13, 38) and have a serine residue at the +4 position from the phosphorylated tyrosine. Phosphorylation of both these serines correlates with inhibition of PI3-kinase/Akt signaling (Ser636/639 are discussed in this study) (see references 26 and 62 for Ser616). However, phosphorylation of IRS-1 on Ser616 is not sensitive to rapamycin (Fig. 1B). To analyze the relative impact of these two serine residues on activation of PI3-kinase, we generated two more mutants, S616D and S616/636D. Figure 2B shows that insulin-stimulated PI3-kinase activity associated with the S616D mutant is decreased by ∼30%. However, the S616/636D and Ser636/639D mutants repress PI3-kinase activity to a similar extent, suggesting a prominent negative role of Ser636 in the regulation of IRS-1-associated PI3-kinase activity. Lastly, neither mutation visibly interferes with tyrosine phosphorylation of IRS-1 (Fig. 2B). Although our findings, in agreement with previous reports (29, 62), do not support the role of Ser616 phosphorylation in regulation of PI3-kinase activity, we do not rule out the possibility that phosphorylation of Ser616 may be implicated in suppression of PI3-kinase/Akt signaling promoted by other factors, such as TNF-α and/or metabolites (60, 64), by acting synergistically with phosphorylation of another serine residue(s).
Upon insulin stimulation, phosphorylation-dependent redistribution of IRS-1 from microsomes to cytosol coincides with cessation of PI3-kinase signaling and development of insulin resistance (9, 23). Figure 2C shows that, upon insulin treatment, IRS-1 phosphorylated on Ser636/639 is enriched in the cytosol, while the majority of IRS-1 stays in tight association with the microsomal fraction recovered in the high-speed pellet. Therefore, mTOR/S6K1-mediated IRS-1 Ser636/639 phosphorylation may have an additional inhibitory effect on the PI3-kinase/Akt pathway by disrupting the spatial organization of the signaling components. To what extent Ser636/639 phosphorylation regulates IRS-1 compartmentalization is unknown. Interestingly, in silico analysis of the IRS-1 primary sequence reveals a putative 14-3-3 binding motif KSVS641AP, similar to the R/KSXS*XP motif described by Yaffe et al. (73), localized close to the Ser636/639 sites. Our preliminary studies (unpublished data) suggest that IRS-1 Ser641 phosphorylation is rapamycin sensitive and that 14-3-3 proteins may be coimmunoprecipitated with IRS-1. The 14-3-3 family of proteins is known to regulate compartmentalization of various signal transduction pathways. Further studies are needed to identify the specific isoform(s) of 14-3-3 that binds to IRS-1 and to characterize molecular mechanisms that may regulate this interaction.
Rheb antagonizes insulin signaling via IRS-1 Ser636/639 phosphorylation and degradation.
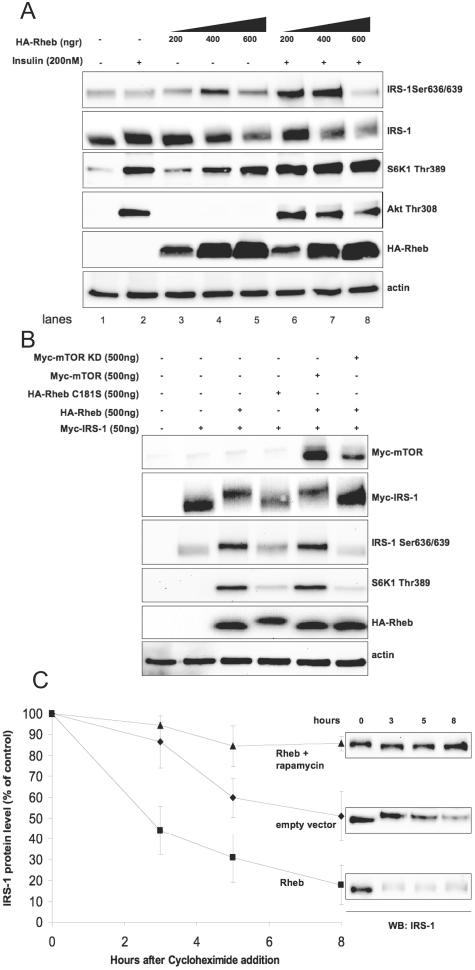
TSC2 integrates nutrient- and hormone-dependent signals and regulates mTOR in a Rheb-dependent fashion (20-22, 34, 35). Figure 3A shows that increasing doses of Rheb lead to activation of mTOR, as shown by S6K1 Thr389 and IRS-1 Ser636/639 phosphorylation even in the absence of insulin. Importantly, overexpression of Rheb antagonizes insulin-stimulated Akt phosphorylation (compare lanes 2 and 8). In these experiments, we noticed that high levels of overexpressed Rheb decrease the amount of endogenous IRS-1, probably by stimulating its degradation (Fig. 3A, compare lanes 1 and 2 with lanes 5 and 8). To test this hypothesis and also to avoid any effect of Rheb overexpression on endogenous IRS-1 at the transcriptional level (15), we ectopically expressed Myc-tagged IRS-1 in 293HEK cells. We studied the involvement of the mTOR pathway in degradation of ectopically expressed Myc-IRS-1 by three independent approaches: (i) by overexpression of wild-type Rheb and a farnesylation-defective mutant, Rheb C181S, (ii) by overexpression of wild-type and kinase-deficient mTOR along with Rheb, and (iii) by using the farnesyl transferase inhibitor FTI-277 that blocks membrane association of Rheb. Figure 3B shows that the levels of Myc-IRS-1 are regulated by Rheb in an mTOR-dependent manner. Overexpression of Rheb alone or together with wild-type mTOR leads to Ser636/639 phosphorylation and degradation of Myc-IRS-1. Kinase-deficient mTOR and, to a lesser extent, FTI-277 (not shown) reverse IRS-1 Ser636/639 phosphorylation as well as the electrophoretic mobility shift of IRS-1 and block its degradation. Rheb C181S has only a limited effect, since this mutant is not dominant negative and partially preserves its signaling capacity (30).
FIG. 3.
Activation of mTOR/S6K1 by Rheb suppresses insulin signaling. (A) 293HEK cells were transfected with different amounts of Rheb cDNA for 36 h, serum starved for 6 h, and then stimulated with insulin for 30 min. Total cell lysates were analyzed by Western blotting (WB). (B) 293HEK cells were transfected as indicated on the figure. Cells were serum starved for 6 h, and total cell lysates were analyzed by Western blotting. (C) C2C12 fibroblasts stably transfected with Rheb or empty vector were grown in DMEM supplemented with 20% FBS and treated with cycloheximide (40 μg/ml) in the presence or absence of rapamycin (50 nM) for different periods of time. Levels of endogenous IRS-1 in cell lysates were determined by Western blotting. The graph shows the IRS-1 protein level (as percentage of control) normalized to actin of three independent experiments (mean values ± SEM). +, present; −, absent.
To further establish the role of mTOR signaling in IRS-1 turnover, we generated several lines of C2C12 fibroblasts stably transfected with Rheb and studied the half-life of endogenous IRS-1. Figure 3C shows that, in Rheb-overexpressing cells, the IRS-1 half-life is reduced from ∼8 h to ∼2.5 h. Importantly, rapamycin inhibits Rheb-stimulated degradation of IRS-1 and increases its half-life over control values. This result is consistent with recent reports showing that IRS-1 is virtually undetectable in TSC1/2−/− fibroblasts (15, 57). The S636/639A mutant exhibits turnover times similar to those of wild-type IRS-1 (not shown), suggesting that IRS-1 degradation is independent of its phosphorylation on these serine residues. This question, however, deserves further investigation. Lastly, insulin-stimulated IRS-1 phosphorylation on Ser307 is preserved in the S636/639A mutant (not shown).
Nutrients stimulate IRS-1 Ser636/639 phosphorylation and suppress IRS-1-associated PI3-kinase/Akt signaling.
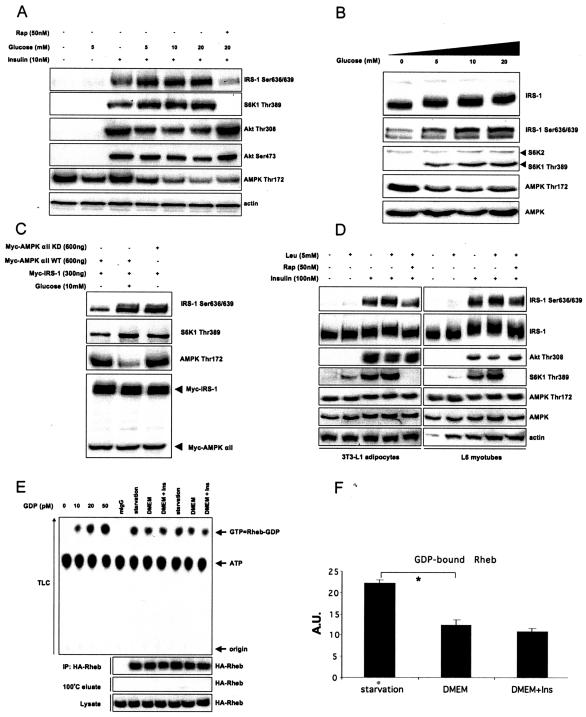
L6 myotubes were starved in glucose- and amino acid-free KRP buffer and treated with increasing concentrations of glucose that mimic either normoglycemic (5 mM) or hyperglycemic conditions (10 and 20 mM). In the presence of insulin, glucose potently stimulates IRS-1 Ser636/639 phosphorylation and inhibits Akt and AMPK phosphorylation in a dose-dependent fashion (Fig. 4A). Consistently, Ser636/639 is hyperphosphorylated in the skeletal muscle of NIDDM subjects (2). Rapamycin abrogates mTOR-mediated IRS-1 Ser636/639 phosphorylation and fully reverses the inhibitory effect of glucose on insulin stimulated PI3-kinase/Akt activation. Moreover, when we increase the sensitivity of our detection system by either using super enhanced chemiluminescence (Fig. 4B) or by overexpressing IRS-1 (see Fig. S1 in the supplemental material), we observe that glucose stimulates phosphorylation of Ser636/639 in an insulin-independent manner. Lastly, Fig. 4C shows that the glucose effect on Ser636/639 phosphorylation is dependent on the kinase activity of AMPK.
FIG. 4.
Nutrients stimulate phosphorylation of IRS-1 on Ser636/639 and inhibit insulin signaling. (A) L6 myotubes were starved in glucose-free KRP for 6 h, pretreated with glucose for 30 min, and stimulated with insulin for another 30 min as indicated. (B) L6 myotubes were starved in glucose-free KRP for 6 h and stimulated with the indicated concentrations of glucose for 30 min. (C) 293HEK cells were transfected with the indicated constructs and were starved in glucose-free KRP for 6 h following stimulation with glucose for 30 min. Total cell lysates were analyzed by Western blotting. (D) 3T3-L1 adipocytes and L6 myotubes were starved in glucose- and amino acid-free KRP for 6 h, treated with rapamycin as indicated for 30 min, and stimulated with leucine and insulin for another 30 min. Total cell lysates were analyzed by Western blotting. (E) 293HEK cells were transfected with HA-Rheb and were starved in glucose-free KRP or in serum-free DMEM following insulin stimulation for 30 min as indicated on the figure. HA-Rheb-bound GDP was measured as described in Materials and Methods. Results of two independent experiments are presented. The first four lanes of the TLC panel show the calibration curve. (F) Normalized results (mean values ± SEM) of three independent experiments shown in panel E. The asterisk indicates a P value of < 0.01. +, present; −, absent.
Branched-chain amino acids (especially leucine) inhibit insulin signaling in vitro and in vivo (43, 65, 66). Figure 4D demonstrates that, in both 3T3-L1 adipocytes and L6 myotubes, leucine stimulates Ser636/639 phosphorylation in a rapamycin-sensitive but AMPK-independent manner, thus providing a common theme in the nutrient-dependent regulation of PI3-kinase/Akt signaling.
To further explore the role of nutrients in insulin signaling, we measured the guanine nucleotide load of Rheb under different nutritional conditions. Rheb is an unusual GTPase in a sense that, under basal conditions (i.e., in serum-free media) when less than 5% of Ras is present in its active GTP-bound conformation (30, 56), ∼40% of Rheb is associated with GTP (30; data not shown). Thus, assuming that GTP-bound Rheb is active and GDP-bound Rheb is not, the major Rheb-related regulatory event may be not GDP/GTP exchange, as is the case with Ras, but rather hydrolysis of Rheb-bound GTP.
First, we studied the GTP/GDP load of Rheb in nutrient-depleted cells using in vivo cell labeling with 32P. This, however, led to extensive cell death that we attribute to the lack of nutrients (including phosphorus) in the medium together with radioactivity-induced cellular stress. Moreover, previous studies suggest that incubation of cells in phosphate-free medium markedly decreases the intracellular ATP concentration (56), which can potentially interfere with AMPK activity and cause misinterpretation of the results. Therefore, we measured the amount of GDP-bound Rheb in nonlabeled cells as described previously (56). Using this approach, we demonstrate that nutrient deprivation increases the amount of GDP-bound Rheb ∼1.8 times compared to cells maintained in serum-free DMEM supplied with 25 mM glucose (Fig. 4E and F). Insulin stimulation has a marginal effect on GDP-bound Rheb (Fig. 4E and F). In summary, our data suggest that nutrients may use both AMPK-dependent and -independent mechanisms to uncouple PI3-kinase/Akt signaling at the level of IRS-1 via the mTOR pathway.
AMPK effectors suppress IRS-1 Ser636/639 phosphorylation and activate PI3-kinase/Akt signaling.
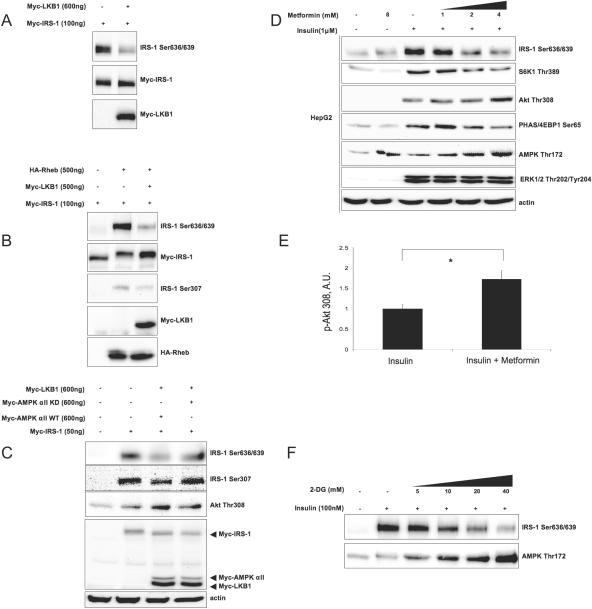
Metformin activates AMPK in vitro (75) and in vivo (37) in an LKB1-dependent manner (32). To this end, we found that ectopically expressed LKB1 inhibits basal (Fig. 5A) as well as Rheb-stimulated (Fig. 5B) phosphorylation of IRS-1 on Ser636/639 and Ser307 in an AMPK-dependent fashion (Fig. 5C). Overexpression of LKB1 enhances insulin-stimulated Akt phosphorylation (Fig. 5C and results not shown), in agreement with previous findings showing that LKB1−/− mouse embryonic fibroblasts exhibit reduced Akt phosphorylation (58). Furthermore, metformin, in HepG2 cells (Fig. 5D) and less so in L6 myotubes (see Fig. S2 in the supplemental material), suppresses insulin-stimulated mTOR activity toward IRS-1 Ser636/639 and S6K1 in a dose-dependent manner. This leads to the enhancement of insulin-stimulated Akt phosphorylation (by a factor of 1.7 in HepG2 cells) (Fig. 5E) and activation (see the panel with phosphorylated glycogen synthetase kinase 3β in Fig. S3 in the supplemental material) without interfering with ERK1/2 activity (Fig. 5D). Since the effect of metformin on AMPK is LKB1 dependent (32, 50), the differential sensitivity to the drug observed in vivo (63) and in vitro (this study) could be attributed to different expression levels of endogenous LKB1 (74). Finally, Fig. 5F shows that another activator of AMPK, 2-deoxyglucose, previously shown to suppress mTOR signaling to S6K1 and PHAS/4E-BP1 (22), also inhibits insulin-stimulated IRS-1 Ser636/639 phosphorylation in a reverse fashion to activation of AMPK. Therefore, we conclude that AMPK-dependent suppression of the mTOR/S6K1 pathway positively regulates PI3-kinase/Akt signaling.
FIG. 5.
LKB1, metformin, and 2-deoxyglucose suppress phosphorylation of IRS-1 on Ser636/639 and enhance Akt phosphorylation. (A to C) 293HEK cells were transfected with indicated constructs. Cells were either serum starved for 6 h (A and B) or grown in 20% FBS (C). Total cell lysates were analyzed by Western blotting. (D) HepG2 cells were serum starved for 6 h, pretreated for 60 min with indicated doses of metformin, and stimulated with insulin for another 30 min. Total cell lysates were analyzed by Western blotting. (E) Insulin-induced Akt Thr308 phosphorylation in HepG2 cells in the absence and in the presence of 5 mM metformin was normalized by actin. The panel shows mean values ± SEM of the results from three independent experiments. The asterisk indicates a P value of <0.03. A.U., arbitrary units. (F) 3T3-L1 adipocytes were pretreated with increasing concentrations of 2-deoxyglucose (2-DG) for 30 min and then stimulated with insulin for another 30 min. Total cell lysates were analyzed by Western blotting. +, present; −, absent.
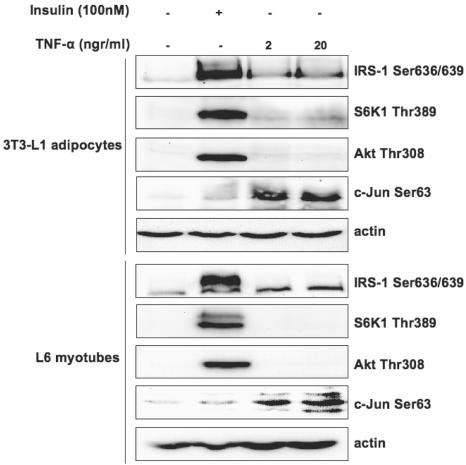
Several in vitro and in vivo studies have described the involvement of TNF-α in the development of insulin resistance secondary to obesity and proinflammatory conditions (42, 60, 64). Figure 6 shows that, in contrast to insulin, short-term TNF-α treatment does not activate mTOR-dependent S6K1 Thr389 and IRS-1 Ser636/639 phosphorylation in L6 myotubes and 3T3-L1 adipocytes, although it induces activation of JNK kinase. Moreover, pharmacological inhibition of JNK and Ras/Raf/ERK1/2 pathways with SP600128 and PD98059, respectively, does not enhance insulin-stimulated IRS-1-associated PI3-kinase activity (not shown), rendering their involvement in regulation of Ser636/639 phosphorylation unlikely.
FIG. 6.
TNF-α does not stimulate the mTOR/S6K1 pathway and IRS-1 Ser636/639 phosphorylation. L6 myotubes and 3T3-L1 adipocytes were serum starved for 6 h and stimulated with the indicated doses of either TNF-α or insulin for another 30 min. Total cell lysates were analyzed by Western blotting. +, present; −, absent.
Phosphorylation of IRS-1 on Ser636/639 is Raptor dependent.
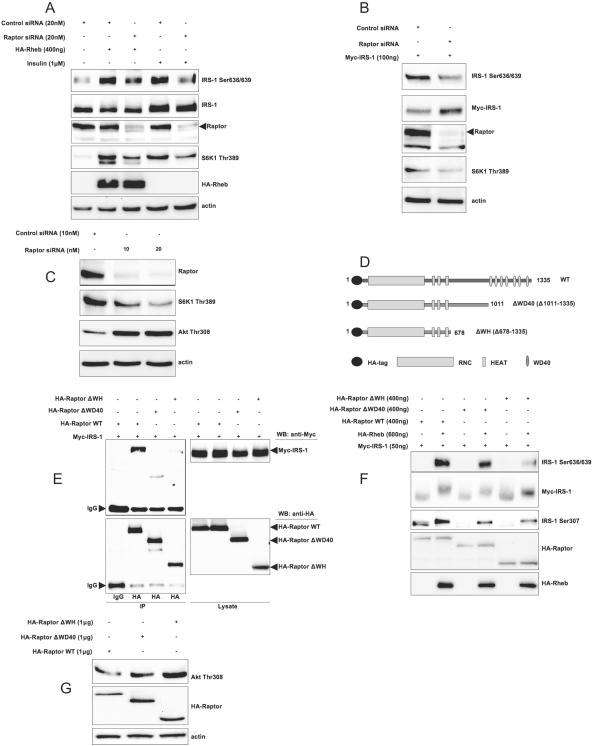
Since Ser636/639 phosphorylation is inhibited by low concentrations of rapamycin (Fig. 1B), we have thought that this phosphorylation can happen in a Raptor-dependent manner. Indeed, Fig. 7A shows that Raptor knockdown with the help of siRNA not only decreases Rheb- and insulin-dependent phosphorylation of the bona fide mTOR substrate S6K1 but also reduces phosphorylation of IRS-1 on Ser636/639. In addition, Raptor knockdown suppresses basal (i.e., nutrient dependent) IRS-1 Ser636/639 phosphorylation (Fig. 7B) and enhances PI3-kinase/Akt signaling (Fig. 7C) in cells growing in serum-rich media.
FIG. 7.
Raptor is essential for Rheb-, insulin-, and nutrient-dependent phosphorylation of IRS-1 on Ser636/639 and attenuation of PI3-kinase/Akt signaling. (A) 293HEK cells were transfected with Rheb, siRNA against Raptor, or control siRNA as indicated on the figure. Cells that were serum starved for 6 h were stimulated or not with insulin, and total cell lysates were analyzed by Western blotting (WB). (B) 293HEK cells were transfected with Myc-IRS-1 and the indicated siRNAs. Cells were incubated in serum-free media for 6 h, and total cell lysates were analyzed by Western blotting. (C) 293HEK cells were transfected with indicated amounts of siRNA against Raptor or with control siRNA. Cells were grown in 20% FBS, and total cell lysates were analyzed by Western blotting. (D) The domain structure of wild-type Raptor and mutants (not to scale). (E) 293HEK cells were transfected with the indicated constructs (2 μg each per 10-cm plate). HA-tagged Raptor mutants were immunoprecipitated (IP) with anti-HA antibody; immunoprecipitates (left) and total cell lysates (right) were analyzed by Western blotting. (F and G) 293HEK cells were transfected with indicated constructs. In panel F, cells were serum starved for 6 h, and in panel G, cells were grown in 20% FBS. +, present; −, absent; IgG, immunoglobulin G.
Raptor is an ∼149-kDa protein with a poorly characterized conserved N-terminal domain followed by three HEAT repeats, hinge domain, and seven WD-40 repeats at the carboxy terminus (27) (Fig. 7D). In agreement with data shown in Fig. 7A, B, and C, we found that IRS-1 may be coimmunoprecipitated with Raptor even in the presence of 1% Triton X-100. Deletion of the seven C-terminal WD-40 repeats and the adjacent hinge domain (amino acids 678 to 1011 for the human isoform) dramatically decreases interaction of Raptor with IRS-1 (Fig. 7E). In addition, these deletions attenuate Rheb-stimulated phosphorylation of IRS-1 on Ser636/639 and on Ser307 (Fig. 7F) and enhance PI3-kinase/Akt signaling in 293HEK cells growing in serum-rich conditions (Fig. 7G). We conclude that the C terminus of Raptor, including the WD-40 repeats and, possibly, the hinge region, may be important for Raptor-IRS-1 interaction and for mTOR/S6K1-mediated phosphorylation of IRS-1. WD-40 domains serve as a stable propeller-like platform involved in protein-protein interactions. Further studies are needed for the precise mapping of Raptor-IRS-1 interaction as well as the identification of a potential Raptor-binding motif in IRS-1.
Raptor/mTOR complex phosphorylates IRS-1 on Ser636/639 in vitro.
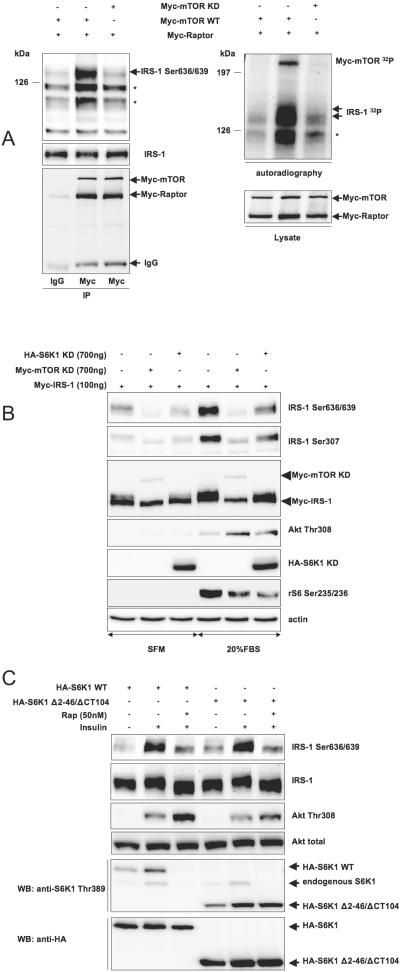
Figure 8A demonstrates that Myc-tagged Raptor/mTOR complex immunoprecipitated form 293HEK cells in the presence of 0.3% CHAPS can phosphorylate recombinant IRS-1 on Ser636/639 in vitro, whereas the same complex with kinase-deficient mTOR is virtually inactive.
FIG. 8.
Raptor/mTOR/S6K1 complex phosphorylates in vitro IRS-1 on Ser636/639. (A) 293HEK cells were transfected with indicated cDNAs. Cells were harvested in 0.3% CHAPS, and Raptor/mTOR complexes were immunoprecipitated (IP) with anti-Myc antibody. In vitro phosphorylation with recombinant IRS-1 as a substrate was performed as described in Materials and Methods. Phosphorylation products were analyzed by Western blotting with phosphospecific antibody (top left panel) or by autoradiography (top right panel). Asterisks indicate degradation products of recombinant IRS-1. (B) 293HEK cells were transfected as indicated and incubated in serum-free medium (SFM) or in 20% FBS as shown on the figure. Total cell lysates were analyzed by Western blotting (WB). (C) NIH 3T3 cells were transfected as indicated, incubated in serum-free media for 6 h, pretreated with rapamycin (Rap) for 30 min, and stimulated with insulin for another 30 min. Total cell lysates were analyzed by Western blotting. +, present; −, absent; IgG, immunoglobulin G.
Raptor may form a complex with both mTOR and S6K1 (14). Thus, we sought to determine which protein kinase phosphorylates IRS-1 in our system by overexpressing kinase-deficient mutants of mTOR and S6K1 along with IRS-1 in 293HEK cells. Figure 8B shows that kinase-deficient mTOR (i) completely abolishes phosphorylation of IRS-1 on Ser636/639, (ii) eliminates the shift in the electrophoretic mobility of IRS-1, and (iii) up-regulates serum-induced Akt phosphorylation. Kinase-deficient S6K1 only partly recapitulates all these effects, suggesting that mTOR per se significantly contributes to the phosphorylation of Ser636/639 under basal (nutrient dependent) and serum-rich conditions. Consistently, overexpression of wild-type S6K1 does not interfere with basal or insulin-stimulated phosphorylation of IRS-1 on Ser636/639 (see Fig. S4 in the supplemental material).
To confirm this result, we used the rapamycin-resistant mutant of S6K1, Δ2 to 46/ΔCT104, that cannot interact with TORC1 (8, 12, 40, 54, 55, 69-71) but can still be phosphorylated by TORC2 (1). Figure 8C shows that, in NIH 3T3 cells expressing this mutant and wild-type S6K1, rapamycin still inhibits insulin-dependent phosphorylation of Ser636/639. Therefore, phosphorylation of Ser636/639 can be attributed to activation of mTOR, rather than S6K1.
Long and colleagues have shown that Rheb directly binds and activates mTOR in a GTP-dependent manner (34, 35). We have overexpressed Rheb together with wild-type S6K1 and its rapamycin-resistant mutant Δ2 to 46/ΔCT104 in 293HEK cells. In serum-starved cells, Rheb stimulates phosphorylation of wild-type S6K1, but not the rapamycin-resistant mutant (see Fig. S5 in the supplemental material). Nevertheless, Rheb triggers phosphorylation of endogenous IRS-1 on Ser636/639 in both cases, suggesting that S6K1 may not be involved in this phosphorylation.
DISCUSSION
Epidemiological studies suggest that obesity represents a causative factor for the development of insulin resistance that precedes and predicts NIDDM (31, 45, 60). The primary molecular defect in NIDDM subjects is impaired insulin-stimulated glucose transport and decreased glycogen synthesis in skeletal muscle (10, 31, 61), the tissue which accounts for 90% of nonoxidative glucose disposal and represents ∼70% of whole-body glucose metabolism (61). The resulting hyperglycemia antagonizes metabolic and prosurvival effects of PI3-kinase/Akt signaling in vivo (28) and in vitro (18, 19), a phenomenon known as glucotoxicity. The molecular basis of this effect is not yet known.
An important finding of this study is that nutrients regulate PI3-kinase/Akt signaling via Raptor-dependent mTOR-mediated IRS-1 phosphorylation. More precisely, glucose, by suppressing AMPK, and leucine, by an as yet unknown mechanism, activate mTOR that leads to Raptor-dependent phosphorylation of IRS-1 on Ser636/639. This and other mTOR/S6K1-mediated phosphorylation events (15, 72) abolish the adaptor abilities of IRS-1 by suppressing its associated insulin-stimulated PI3-kinase activity (this study) (Fig. 2A and B and 9) and its interaction with the insulin receptor (15, 72) and eventually may lead to degradation of IRS-1 (this study) (15).
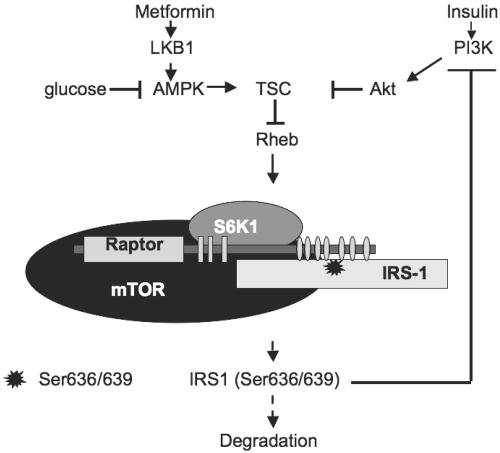
FIG. 9.
Diabetic milieu (hyperglycemia and hyperinsulinemia) down-regulates PI3-kinase/Akt signaling via Raptor-dependent mTOR/S6K1 mediated phosphorylation of IRS-1 on Ser636/639 (not to scale).
We suggest that mTOR/S6K1-mediated phosphorylation of IRS-1 acts as a homeostatic negative feedback loop between nutrient- and growth factor-dependent signaling networks. Under normal conditions, assessment of the cell nutritional status via AMPK and other mechanisms constantly adjusts metabolic, prosurvival, and growth decisions by reversible fine-tuning of PI3-kinase/Akt signaling. In obesity and obesity-related NIDDM, nutritional excess and compensatory hyperinsulinemia may synergistically and permanently hyperactivate the mTOR/S6K1 pathway. This leads not only to attenuated PI3-kinase/Akt signaling due to phosphorylation of IRS-1 on Ser636/639 but also to IRS-1 degradation (Fig. 3A to C and 9) (44). This model may provide a mechanistic explanation for the earlier findings showing that decreased IRS-1 protein levels predict the development of insulin resistance (7) and that IRS-1 is reduced in NIDDM subjects (48).
Several lines of evidence suggest that Raptor directly binds to and serves as a scaffold for mTOR/S6K1-mediated phosphorylation of IRS-1: (i) low doses of rapamycin cause dissociation of the Raptor-mTOR complex (41) and inhibit phosphorylation of IRS-1 on Ser636/639 (Fig. 1B), (ii) Raptor is required for nutrient-, Rheb-, and insulin-stimulated phosphorylation of Ser636/639 (Fig. 7 A to C), (iii) Raptor and IRS-1 are coimmunoprecipitated even in the presence of 1% Triton X-100 (Fig. 7E), (iv) the WD-40 repeats and, less so, the hinge domain of Raptor are required for both interaction with IRS-1 and Rheb-induced phosphorylation on Ser636/639 (Fig. 7E and F), (v) functionally, knockdown of Raptor as well as Raptor carboxy-terminal deletions exhibit the same positive effect on Akt phosphorylation as kinase-deficient mTOR (compare Fig. 7C and G and 8B).
Raptor/mTOR complex that may also contain S6K1 (14) exhibits kinase activity toward recombinant IRS-1 in vitro (Fig. 8A) (14, 16, 42). mTOR catalyzes phosphorylation of Ser/Thr-Pro motifs, such as T37PGGTLFSTT46P in 4EBP1/PHAS (3-5) and PVDS371P in S6K1 (36, 49). In addition, mTOR can phosphorylate Phe-Ser/Thr-Phe/Tyr motifs, such as LGFT389Y and EKFS404F in S6K1 (5, 24, 36, 46) and PQFS473Y in Akt (51). On the contrary, S6K1 preferentially phosphorylates RXRXXS motifs followed by distal serines, such as those present in rS6 protein (RRRLS235S236), SKAR protein (RRVNS383AS385) (47), and IRS-1 (RRSRTES307) (15). The S636PKS639V motif belongs to the Ser/Thr-Pro prototype which suggests that it may represent a better substrate for mTOR than for S6K1. Consistently, Harrington and colleagues showed that S6K1 does not phosphorylate in vitro a glutathione S-transferase-fused IRS-1 516 to 896 fragment (15). Moreover, the sensitivity of Ser636/639 phosphorylation to insulin, Rheb, and rapamycin (Fig. 8C; see also Fig. S4 and S5 in the supplemental material) is unaffected by overexpression (at least 10 times over the basal S6K1 level) (data not shown) of a rapamycin-resistant mutant of S6K1 (Δ2 to 46/ΔCT104) deficient in Raptor binding, suggesting that mTOR per se phosphorylates IRS-1. Consistently, kinase-deficient mTOR completely eliminates Ser636/639 phosphorylation, while kinase-deficient S6K1 has only a partial effect (Fig. 8B). We cannot, however, exclude a possibility that in vitro phosphorylation of IRS-1 is mediated by an unknown protein kinase(s) that may copurify with the Raptor/mTOR complex (for an example, see reference 39). Further studies are needed to characterize the molecular topography of TORC1 and its ability to signal to downstream targets, including IRS-1.
When the manuscript was under review, Sarbassov and colleagues showed that Raptor/mTOR and Rictor/mTOR complexes regulate Akt phosphorylation in a reverse manner (51). In agreement with our results, they found that TORC1 complex suppressed Akt phosphorylation. Here, we demonstrate that this inhibition takes place at least in part via Raptor-dependent mTOR-mediated phosphorylation of IRS-1. Thus, mTOR signaling toward Akt is scaffold dependent.
Rheb integrates nutrient- and hormone-dependent inputs downstream of the TSC complex and activates the mTOR pathway (17). Here, we show that the Rheb effect on mTOR signaling is Raptor dependent (Fig. 7A) and that nutrients regulate changes in the GTP/GDP ratio (Fig. 4E and F). Consistently, Long and colleagues showed that Rheb directly binds Raptor and mTOR (34) and that its binding to mTOR is amino acid dependent (35).
Another significant finding of this study is that metformin, the most commonly used oral antidiabetic agent (75), suppresses mTOR/S6K1-mediated IRS-1 phosphorylation in an AMPK-dependent fashion. Metformin enhances insulin sensitivity in liver and, to a lesser extent, in muscle (63). At the molecular level, metformin, via AMPK-dependent activation, inhibits acetyl-coenzyme A carboxylase (75), the rate-limiting enzyme of de novo fatty acid biosynthesis, and down-regulates the hepatic expression of SREBP-1 as well as other lipogenic enzymes (75), thus targeting diabetes-related dyslipidemia (45, 75). In addition, metformin, synergistically with insulin, induces skeletal muscle glucose uptake (50, 75). Our study with HepG2 and L6 cells suggests that the observed beneficial effect of metformin on insulin sensitivity in liver and skeletal muscle takes place at least partially via AMPK-dependent suppression of the mTOR/S6K1 pathway. Overexpression of LKB1, an upstream activator of AMPK (32), recapitulates the positive effect of metformin on PI3-kinase/Akt signaling. Noteworthy, LKB1 regulates 13 different kinases of the AMPK subfamily, while metformin specifically activates AMPK in an LKB1-dependent fashion (32). Recently, Sakamoto and colleagues reported the first genetic evidence that LKB1 is a major regulator of the AMPK αII isoform in skeletal muscle (50). Importantly, they showed that LKB1 is required for phenformin (a metformin analog)-induced activation of AMPK αII. Our study extends these in vivo findings and shows that the LKB1/AMPK pathway increases PI3-kinase/Akt signaling via down-regulation of IRS-1 Ser636/639 phosphorylation by the Raptor/mTOR complex (11, 59). Therefore, identification of activators of LKB1 and/or AMPK could lead to the development of new antidiabetic compounds targeting insulin resistance.
Supplementary Material
Acknowledgments
We are indebted to J. Avruch, M. Birnbaumn, J. Blenis, G. Clark, C. Der, P. Pilch, D. Sabatini, and M. White for providing useful reagents. A.T. acknowledges A. Papavassiliou, D. Synetos, A. Charonis, J. Dresios, G. Maniatis, D. Drainas, D. Kalpaxis, C. Petrovas, G. Thoidis, J. Jimenez, K. Ravid, and A. Phinikaridou for stimulation and encouragement in different aspects of this work.
This work was supported by research grants DK52057 and DK56736 from the NIH and by a research grant from the American Diabetes Association to K.V.K.
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Ali, S. M., and D. M. Sabatini. 2005. Structure of S6 kinase 1 determines whether raptor-mTOR or rictor-mTOR phosphorylates its hydrophobic motif site. J. Biol. Chem. 280:19445-19448. [DOI] [PubMed] [Google Scholar]
- 2.Bouzakri, K., M. Roques, P. Gual, S. Espinosa, F. Guebre-Egziabher, J. P. Riou, M. Laville, Y. Le Marchand-Brustel, J. F. Tanti, and H. Vidal. 2003. Reduced activation of phosphatidylinositol-3 kinase and increased serine 636 phosphorylation of insulin receptor substrate-1 in primary culture of skeletal muscle cells from patients with type 2 diabetes. Diabetes 52:1319-1325. [DOI] [PubMed] [Google Scholar]
- 3.Brunn, G. J., P. Fadden, T. A. Haystead, and J. C. Lawrence, Jr. 1997. The mammalian target of rapamycin phosphorylates sites having a (Ser/Thr)-Pro motif and is activated by antibodies to a region near its COOH terminus. J. Biol. Chem. 272:32547-32550. [DOI] [PubMed] [Google Scholar]
- 4.Brunn, G. J., C. C. Hudson, A. Sekulic, J. M. Williams, H. Hosoi, P. J. Houghton, J. C. Lawrence, Jr., and R. T. Abraham. 1997. Phosphorylation of the translational repressor PHAS-I by the mammalian target of rapamycin. Science 277:99-101. [DOI] [PubMed] [Google Scholar]
- 5.Burnett, P. E., R. K. Barrow, N. A. Cohen, S. H. Snyder, and D. M. Sabatini. 1998. RAFT1 phosphorylation of the translational regulators p70 S6 kinase and 4E-BP1. Proc. Natl. Acad. Sci. USA 95:1432-1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carling, D. 2004. The AMP-activated protein kinase cascade—a unifying system for energy control. Trends Biochem. Sci. 29:18-24. [DOI] [PubMed] [Google Scholar]
- 7.Carvalho, E., P. A. Jansson, M. Axelsen, J. W. Eriksson, X. Huang, L. Groop, C. Rondinone, L. Sjostrom, and U. Smith. 1999. Low cellular IRS 1 gene and protein expression predict insulin resistance and NIDDM. FASEB J. 13:2173-2178. [DOI] [PubMed] [Google Scholar]
- 8.Cheatham, L., M. Monfar, M. M. Chou, and J. Blenis. 1995. Structural and functional analysis of pp70S6k. Proc. Natl. Acad. Sci. USA 92:11696-11700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clark, S. F., J. C. Molero, and D. E. James. 2000. Release of insulin receptor substrate proteins from an intracellular complex coincides with the development of insulin resistance. J. Biol. Chem. 275:3819-3826. [DOI] [PubMed] [Google Scholar]
- 10.Cline, G. W., K. F. Petersen, M. Krssak, J. Shen, R. S. Hundal, Z. Trajanoski, S. Inzucchi, A. Dresner, D. L. Rothman, and G. I. Shulman. 1999. Impaired glucose transport as a cause of decreased insulin-stimulated muscle glycogen synthesis in type 2 diabetes. N. Engl. J. Med. 341:240-246. [DOI] [PubMed] [Google Scholar]
- 11.Corradetti, M. N., K. Inoki, N. Bardeesy, R. A. DePinho, and K. L. Guan. 2004. Regulation of the TSC pathway by LKB1: evidence of a molecular link between tuberous sclerosis complex and Peutz-Jeghers syndrome. Genes Dev. 18:1533-1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dennis, P. B., N. Pullen, S. C. Kozma, and G. Thomas. 1996. The principal rapamycin-sensitive p70(s6k) phosphorylation sites, T-229 and T-389, are differentially regulated by rapamycin-insensitive kinase kinases. Mol. Cell. Biol. 16:6242-6251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Esposito, D. L., Y. Li, A. Cama, and M. J. Quon. 2001. Tyr(612) and Tyr(632) in human insulin receptor substrate-1 are important for full activation of insulin-stimulated phosphatidylinositol 3-kinase activity and translocation of GLUT4 in adipose cells. Endocrinology 142:2833-2840. [DOI] [PubMed] [Google Scholar]
- 14.Hara, K., Y. Maruki, X. Long, K. Yoshino, N. Oshiro, S. Hidayat, C. Tokunaga, J. Avruch, and K. Yonezawa. 2002. Raptor, a binding partner of target of rapamycin (TOR), mediates TOR action. Cell 110:177-189. [DOI] [PubMed] [Google Scholar]
- 15.Harrington, L. S., G. M. Findlay, A. Gray, T. Tolkacheva, S. Wigfield, H. Rebholz, J. Barnett, N. R. Leslie, S. Cheng, P. R. Shepherd, I. Gout, C. P. Downes, and R. F. Lamb. 2004. The TSC1-2 tumor suppressor controls insulin-PI3K signaling via regulation of IRS proteins. J. Cell Biol. 166:213-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hartman, M. E., M. Villela-Bach, J. Chen, and G. G. Freund. 2001. Frap-dependent serine phosphorylation of IRS-1 inhibits IRS-1 tyrosine phosphorylation. Biochem. Biophys. Res. Commun. 280:776-781. [DOI] [PubMed] [Google Scholar]
- 17.Hay, N., and N. Sonenberg. 2004. Upstream and downstream of mTOR. Genes Dev. 18:1926-1945. [DOI] [PubMed] [Google Scholar]
- 18.Huang, C., R. Somwar, N. Patel, W. Niu, D. Torok, and A. Klip. 2002. Sustained exposure of L6 myotubes to high glucose and insulin decreases insulin-stimulated GLUT4 translocation but upregulates GLUT4 activity. Diabetes 51:2090-2098. [DOI] [PubMed] [Google Scholar]
- 19.Ido, Y., D. Carling, and N. Ruderman. 2002. Hyperglycemia-induced apoptosis in human umbilical vein endothelial cells: inhibition by the AMP-activated protein kinase activation. Diabetes 51:159-167. [DOI] [PubMed] [Google Scholar]
- 20.Inoki, K., Y. Li, T. Xu, and K. L. Guan. 2003. Rheb GTPase is a direct target of TSC2 GAP activity and regulates mTOR signaling. Genes Dev. 17:1829-1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Inoki, K., Y. Li, T. Zhu, J. Wu, and K. L. Guan. 2002. TSC2 is phosphorylated and inhibited by Akt and suppresses mTOR signalling. Nat. Cell Biol. 4:648-657. [DOI] [PubMed] [Google Scholar]
- 22.Inoki, K., T. Zhu, and K. L. Guan. 2003. TSC2 mediates cellular energy response to control cell growth and survival. Cell 115:577-590. [DOI] [PubMed] [Google Scholar]
- 23.Inoue, G., B. Cheatham, R. Emkey, and C. R. Kahn. 1998. Dynamics of insulin signaling in 3T3-L1 adipocytes. Differential compartmentalization and trafficking of insulin receptor substrate (IRS)-1 and IRS-2. J. Biol. Chem. 273:11548-11555. [DOI] [PubMed] [Google Scholar]
- 24.Isotani, S., K. Hara, C. Tokunaga, H. Inoue, J. Avruch, and K. Yonezawa. 1999. Immunopurified mammalian target of rapamycin phosphorylates and activates p70 S6 kinase alpha in vitro. J. Biol. Chem. 274:34493-34498. [DOI] [PubMed] [Google Scholar]
- 25.Jacinto, E., R. Loewith, A. Schmidt, S. Lin, M. A. Ruegg, A. Hall, and M. N. Hall. 2004. Mammalian TOR complex 2 controls the actin cytoskeleton and is rapamycin insensitive. Nat. Cell Biol. 6:1122-1128. [DOI] [PubMed] [Google Scholar]
- 26.Jiang, G., Q. Dallas-Yang, S. Biswas, Z. Li, and B. B. Zhang. 2004. Rosiglitazone, an agonist of peroxisome-proliferator-activated receptor gamma (PPARgamma), decreases inhibitory serine phosphorylation of IRS1 in vitro and in vivo. Biochem. J. 377:339-346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim, D. H., D. D. Sarbassov, S. M. Ali, J. E. King, R. R. Latek, H. Erdjument-Bromage, P. Tempst, and D. M. Sabatini. 2002. mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell 110:163-175. [DOI] [PubMed] [Google Scholar]
- 28.Kim, J. K., A. Zisman, J. J. Fillmore, O. D. Peroni, K. Kotani, P. Perret, H. Zong, J. Dong, C. R. Kahn, B. B. Kahn, and G. I. Shulman. 2001. Glucose toxicity and the development of diabetes in mice with muscle-specific inactivation of GLUT4. J. Clin. Investig. 108:153-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li, J., K. DeFea, and R. A. Roth. 1999. Modulation of insulin receptor substrate-1 tyrosine phosphorylation by an Akt/phosphatidylinositol 3-kinase pathway. J. Biol. Chem. 274:9351-9356. [DOI] [PubMed] [Google Scholar]
- 30.Li, Y., K. Inoki, and K. L. Guan. 2004. Biochemical and functional characterizations of small GTPase Rheb and TSC2 GAP activity. Mol. Cell. Biol. 24:7965-7975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lillioja, S., D. M. Mott, B. V. Howard, P. H. Bennett, H. Yki-Jarvinen, D. Freymond, B. L. Nyomba, F. Zurlo, B. Swinburn, and C. Bogardus. 1988. Impaired glucose tolerance as a disorder of insulin action. Longitudinal and cross-sectional studies in Pima Indians. N. Engl. J. Med. 318:1217-1225. [DOI] [PubMed] [Google Scholar]
- 32.Lizcano, J. M., O. Goransson, R. Toth, M. Deak, N. A. Morrice, J. Boudeau, S. A. Hawley, L. Udd, T. P. Makela, D. G. Hardie, and D. R. Alessi. 2004. LKB1 is a master kinase that activates 13 kinases of the AMPK subfamily, including MARK/PAR-1. EMBO J. 23:833-843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Loewith, R., E. Jacinto, S. Wullschleger, A. Lorberg, J. L. Crespo, D. Bonenfant, W. Oppliger, P. Jenoe, and M. N. Hall. 2002. Two TOR complexes, only one of which is rapamycin sensitive, have distinct roles in cell growth control. Mol. Cell 10:457-468. [DOI] [PubMed] [Google Scholar]
- 34.Long, X., Y. Lin, S. Ortiz-Vega, K. Yonezawa, and J. Avruch. 2005. Rheb binds and regulates the mTOR kinase. Curr. Biol. 15:702-713. [DOI] [PubMed] [Google Scholar]
- 35.Long, X., S. Ortiz-Vega, Y. Lin, and J. Avruch. 2005. Rheb binding to mammalian target of rapamycin (mTOR) is regulated by amino acid sufficiency. J. Biol. Chem. 280:23433-23436. [DOI] [PubMed] [Google Scholar]
- 36.Moser, B. A., P. B. Dennis, N. Pullen, R. B. Pearson, N. A. Williamson, R. E. Wettenhall, S. C. Kozma, and G. Thomas. 1997. Dual requirement for a newly identified phosphorylation site in p70s6k. Mol. Cell. Biol. 17:5648-5655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Musi, N., M. F. Hirshman, J. Nygren, M. Svanfeldt, P. Bavenholm, O. Rooyackers, G. Zhou, J. M. Williamson, O. Ljunqvist, S. Efendic, D. E. Moller, A. Thorell, and L. J. Goodyear. 2002. Metformin increases AMP-activated protein kinase activity in skeletal muscle of subjects with type 2 diabetes. Diabetes 51:2074-2081. [DOI] [PubMed] [Google Scholar]
- 38.Myers, M. G., Jr., Y. Zhang, G. A. Aldaz, T. Grammer, E. M. Glasheen, L. Yenush, L. M. Wang, X. J. Sun, J. Blenis, J. H. Pierce, and M. F. White. 1996. YMXM motifs and signaling by an insulin receptor substrate 1 molecule without tyrosine phosphorylation sites. Mol. Cell. Biol. 16:4147-4155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nishiuma, T., K. Hara, Y. Tsujishita, K. Kaneko, K. Shii, and K. Yonezawa. 1998. Characterization of the phosphoproteins and protein kinase activity in mTOR immunoprecipitates. Biochem. Biophys. Res. Commun. 252:440-444. [DOI] [PubMed] [Google Scholar]
- 40.Nojima, H., C. Tokunaga, S. Eguchi, N. Oshiro, S. Hidayat, K. Yoshino, K. Hara, N. Tanaka, J. Avruch, and K. Yonezawa. 2003. The mammalian target of rapamycin (mTOR) partner, raptor, binds the mTOR substrates p70 S6 kinase and 4E-BP1 through their TOR signaling (TOS) motif. J. Biol. Chem. 278:15461-15464. [DOI] [PubMed] [Google Scholar]
- 41.Oshiro, N., K. Yoshino, S. Hidayat, C. Tokunaga, K. Hara, S. Eguchi, J. Avruch, and K. Yonezawa. 2004. Dissociation of raptor from mTOR is a mechanism of rapamycin-induced inhibition of mTOR function. Genes Cells 9:359-366. [DOI] [PubMed] [Google Scholar]
- 42.Ozes, O. N., H. Akca, L. D. Mayo, J. A. Gustin, T. Maehama, J. E. Dixon, and D. B. Donner. 2001. A phosphatidylinositol 3-kinase/Akt/mTOR pathway mediates and PTEN antagonizes tumor necrosis factor inhibition of insulin signaling through insulin receptor substrate-1. Proc. Natl. Acad. Sci. USA 98:4640-4645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Patti, M. E., E. Brambilla, L. Luzi, E. J. Landaker, and C. R. Kahn. 1998. Bidirectional modulation of insulin action by amino acids. J. Clin. Investig. 101:1519-1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pirola, L., S. Bonnafous, A. M. Johnston, C. Chaussade, F. Portis, and E. Van Obberghen. 2003. Phosphoinositide 3-kinase-mediated reduction of insulin receptor substrate-1/2 protein expression via different mechanisms contributes to the insulin-induced desensitization of its signaling pathways in L6 muscle cells. J. Biol. Chem. 278:15641-15651. [DOI] [PubMed] [Google Scholar]
- 45.Powers, A. C. 2001. Diabetes mellitus, p. 2109-2137. In E. Braunwald, S. A. Fauci, L. D. Karsper, L. S. Hauser, L. D. Longo, and J. L. Jameson (ed.), Harrison's principles of internal medicine, 15th ed. McGraw-Hill Book Co., New York, N.Y.
- 46.Pullen, N., P. B. Dennis, M. Andjelkovic, A. Dufner, S. C. Kozma, B. A. Hemmings, and G. Thomas. 1998. Phosphorylation and activation of p70s6k by PDK1. Science 279:707-710. [DOI] [PubMed] [Google Scholar]
- 47.Richardson, C. J., M. Broenstrup, D. C. Fingar, K. Julich, B. A. Ballif, S. Gygi, and J. Blenis. 2004. SKAR is a specific target of S6 kinase 1 in cell growth control. Curr. Biol. 14:1540-1549. [DOI] [PubMed] [Google Scholar]
- 48.Rondinone, C. M., L. M. Wang, P. Lonnroth, C. Wesslau, J. H. Pierce, and U. Smith. 1997. Insulin receptor substrate (IRS) 1 is reduced and IRS-2 is the main docking protein for phosphatidylinositol 3-kinase in adipocytes from subjects with non-insulin-dependent diabetes mellitus. Proc. Natl. Acad. Sci. USA 94:4171-4175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Saitoh, M., N. Pullen, P. Brennan, D. Cantrell, P. B. Dennis, and G. Thomas. 2002. Regulation of an activated S6 kinase 1 variant reveals a novel mammalian target of rapamycin phosphorylation site. J. Biol. Chem. 277:20104-20112. [DOI] [PubMed] [Google Scholar]
- 50.Sakamoto, K., A. McCarthy, D. Smith, K. A. Green, D. Grahame Hardie, A. Ashworth, and D. R. Alessi. 2005. Deficiency of LKB1 in skeletal muscle prevents AMPK activation and glucose uptake during contraction. EMBO J. 24:1810-1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sarbassov, D. D., D. A. Guertin, S. M. Ali, and D. M. Sabatini. 2005. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science 307:1098-1101. [DOI] [PubMed] [Google Scholar]
- 52.Sarbassov, D. D., S. M. Ali, D. H. Kim, D. A. Guertin, R. R. Latek, H. Erdjument-Bromage, P. Tempst, and D. M. Sabatini. 2004. Rictor, a novel binding partner of mTOR, defines a rapamycin-insensitive and raptor-independent pathway that regulates the cytoskeleton. Curr. Biol. 14:1296-1302. [DOI] [PubMed] [Google Scholar]
- 53.Saucedo, L. J., X. Gao, D. A. Chiarelli, L. Li, D. Pan, and B. A. Edgar. 2003. Rheb promotes cell growth as a component of the insulin/TOR signalling network. Nat. Cell Biol. 5:566-571. [DOI] [PubMed] [Google Scholar]
- 54.Schalm, S. S., and J. Blenis. 2002. Identification of a conserved motif required for mTOR signaling. Curr. Biol. 12:632-639. [DOI] [PubMed] [Google Scholar]
- 55.Schalm, S. S., D. C. Fingar, D. M. Sabatini, and J. Blenis. 2003. TOS motif-mediated raptor binding regulates 4E-BP1 multisite phosphorylation and function. Curr. Biol. 13:797-806. [DOI] [PubMed] [Google Scholar]
- 56.Scheele, J. S., J. M. Rhee, and G. R. Boss. 1995. Determination of absolute amounts of GDP and GTP bound to Ras in mammalian cells: comparison of parental and Ras-overproducing NIH 3T3 fibroblasts. Proc. Natl. Acad. Sci. USA 92:1097-1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shah, O. J., Z. Wang, and T. Hunter. 2004. Inappropriate activation of the TSC/Rheb/mTOR/S6K cassette induces IRS1/2 depletion, insulin resistance, and cell survival deficiencies. Curr. Biol. 14:1650-1656. [DOI] [PubMed] [Google Scholar]
- 58.Shaw, R. J., N. Bardeesy, B. D. Manning, L. Lopez, M. Kosmatka, R. A. DePinho, and L. C. Cantley. 2004. The LKB1 tumor suppressor negatively regulates mTOR signaling. Cancer Cell 6:91-99. [DOI] [PubMed] [Google Scholar]
- 59.Shaw, R. J., M. Kosmatka, N. Bardeesy, R. L. Hurley, L. A. Witters, R. A. DePinho, and L. C. Cantley. 2004. The tumor suppressor LKB1 kinase directly activates AMP-activated kinase and regulates apoptosis in response to energy stress. Proc. Natl. Acad. Sci. USA 101:3329-3335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shulman, G. I. 2000. Cellular mechanisms of insulin resistance. J. Clin. Investig. 106:171-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shulman, G. I., D. L. Rothman, T. Jue, P. Stein, R. A. DeFronzo, and R. G. Shulman. 1990. Quantitation of muscle glycogen synthesis in normal subjects and subjects with non-insulin-dependent diabetes by 13C nuclear magnetic resonance spectroscopy. N. Engl. J. Med. 322:223-228. [DOI] [PubMed] [Google Scholar]
- 62.Sommerfeld, M. R., S. Metzger, M. Stosik, N. Tennagels, and J. Eckel. 2004. In vitro phosphorylation of insulin receptor substrate 1 by protein kinase C-zeta: functional analysis and identification of novel phosphorylation sites. Biochemistry 43:5888-5901. [DOI] [PubMed] [Google Scholar]
- 63.Stumvoll, M., N. Nurjhan, G. Perriello, G. Dailey, and J. E. Gerich. 1995. Metabolic effects of metformin in non-insulin-dependent diabetes mellitus. N. Engl. J. Med. 333:550-554. [DOI] [PubMed] [Google Scholar]
- 64.Sykiotis, G. P., and A. G. Papavassiliou. 2001. Serine phosphorylation of insulin receptor substrate-1: a novel target for the reversal of insulin resistance. Mol. Endocrinol. 15:1864-1869. [DOI] [PubMed] [Google Scholar]
- 65.Tremblay, F., A. Gagnon, A. Veilleux, A. Sorisky, and A. Marette. 2005. Activation of the mammalian target of rapamycin pathway acutely inhibits insulin signaling to Akt and glucose transport in 3T3-L1 and human adipocytes. Endocrinology 146:1328-1337. [DOI] [PubMed] [Google Scholar]
- 66.Tremblay, F., and A. Marette. 2001. Amino acid and insulin signaling via the mTOR/p70 S6 kinase pathway. A negative feedback mechanism leading to insulin resistance in skeletal muscle cells. J. Biol. Chem. 276:38052-38060. [DOI] [PubMed] [Google Scholar]
- 67.Um, S. H., F. Frigerio, M. Watanabe, F. Picard, M. Joaquin, M. Sticker, S. Fumagalli, P. R. Allegrini, S. C. Kozma, J. Auwerx, and G. Thomas. 2004. Absence of S6K1 protects against age- and diet-induced obesity while enhancing insulin sensitivity. Nature 431:200-205. [DOI] [PubMed] [Google Scholar]
- 68.Viollet, B., F. Andreelli, S. B. Jorgensen, C. Perrin, A. Geloen, D. Flamez, J. Mu, C. Lenzner, O. Baud, M. Bennoun, E. Gomas, G. Nicolas, J. F. Wojtaszewski, A. Kahn, D. Carling, F. C. Schuit, M. J. Birnbaum, E. A. Richter, R. Burcelin, and S. Vaulont. 2003. The AMP-activated protein kinase alpha2 catalytic subunit controls whole-body insulin sensitivity. J. Clin. Investig. 111:91-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Weng, Q. P., K. Andrabi, A. Klippel, M. T. Kozlowski, L. T. Williams, and J. Avruch. 1995. Phosphatidylinositol 3-kinase signals activation of p70 S6 kinase in situ through site-specific p70 phosphorylation. Proc. Natl. Acad. Sci. USA 92:5744-5748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Weng, Q. P., K. Andrabi, M. T. Kozlowski, J. R. Grove, and J. Avruch. 1995. Multiple independent inputs are required for activation of the p70 S6 kinase. Mol. Cell. Biol. 15:2333-2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Weng, Q. P., M. Kozlowski, C. Belham, A. Zhang, M. J. Comb, and J. Avruch. 1998. Regulation of the p70 S6 kinase by phosphorylation in vivo. Analysis using site-specific anti-phosphopeptide antibodies. J. Biol. Chem. 273:16621-16629. [DOI] [PubMed] [Google Scholar]
- 72.Werner, E. D., J. Lee, L. Hansen, M. Yuan, and S. E. Shoelson. 2004. Insulin resistance due to phosphorylation of insulin receptor substrate-1 at serine 302. J. Biol. Chem. 279:35298-35305. [DOI] [PubMed] [Google Scholar]
- 73.Yaffe, M. B., K. Rittinger, S. Volinia, P. R. Caron, A. Aitken, H. Leffers, S. J. Gamblin, S. J. Smerdon, and L. C. Cantley. 1997. The structural basis for 14-3-3:phosphopeptide binding specificity. Cell 91:961-971. [DOI] [PubMed] [Google Scholar]
- 74.Yoo, L. I., D. C. Chung, and J. Yuan. 2002. LKB1—a master tumour suppressor of the small intestine and beyond. Nat. Rev. Cancer 2:529-535. [DOI] [PubMed] [Google Scholar]
- 75.Zhou, G., R. Myers, Y. Li, Y. Chen, X. Shen, J. Fenyk-Melody, M. Wu, J. Ventre, T. Doebber, N. Fujii, N. Musi, M. F. Hirshman, L. J. Goodyear, and D. E. Moller. 2001. Role of AMP-activated protein kinase in mechanism of metformin action. J. Clin. Investig. 108:1167-1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.