Abstract
By using the 1.6 million single-nucleotide polymorphism (SNP) genotype data set from Perlegen Sciences [Hinds, D. A., Stuve, L. L., Nilsen, G. B., Halperin, E., Eskin, E., Ballinger, D. G., Frazer, K. A. & Cox, D. R. (2005) Science 307, 1072–1079], a probabilistic search for the landscape exhibited by positive Darwinian selection was conducted. By sorting each high-frequency allele by homozygosity, we search for the expected decay of adjacent SNP linkage disequilibrium (LD) at recently selected alleles, eliminating the need for inferring haplotype. We designate this approach the LD decay (LDD) test. By these criteria, 1.6% of Perlegen SNPs were found to exhibit the genetic architecture of selection. These results were confirmed on an independently generated data set of 1.0 million SNP genotypes (International Human Haplotype Map Phase I freeze). Simulation studies indicate that the LDD test, at the megabase scale used, effectively distinguishes selection from other causes of extensive LD, such as inversions, population bottlenecks, and admixture. The ≈1,800 genes identified by the LDD test were clustered according to Gene Ontology (GO) categories. Based on overrepresentation analysis, several predominant biological themes are common in these selected alleles, including host–pathogen interactions, reproduction, DNA metabolism/cell cycle, protein metabolism, and neuronal function.
Keywords: balancing selection, Bayesian probabilistic modeling, common disorders, human evolution, single nucleotide polymorphism
Human genotype information on an unprecedented scale is now available for analysis, because of both privately and publicly funded research efforts [Perlegen Sciences (1) and the International Haplotype Map (HapMap) project (2), respectively]. These data sets provide a global map of human variability by using single-nucleotide polymorphisms (SNPs) as markers. The major stated reason for generating such data is to define a core set of haplotypes useful for genome association studies (1, 2). Driving this haplotype search is the hypothesis that common DNA variants underlie many common disorders and that these high frequency variants can be identified either directly or by means of linkage disequilibrium (LD) to nearby SNPs (3, 4). It has been proposed that these “disease” variants reached polymorphic frequency either by chance (5) or through selection for related phenotypes (6–8) and now predispose to disease because of recent environmental/genetic factors (4, 6). Defining “functional” SNP variants among the millions present in human DNA, however, remains an ongoing challenge.
The contemporary human genome is likely the result of a complex history of many different genomic/population events, including ancient and recent selection. Uncovering evidence for selection is one approach to defining functional human DNA variation. To date, studies have focused largely on specific genomic regions (6, 9–13) and suggested that recent selection among humans may be common (14). Extending such studies to the entire genome is a challenging undertaking. Most traditional population genetics tests for selection (15, 16) rely predominately on observing either deviations in local heterozygosity (17) or unusually high singleton pairwise differences within a given sample (18). For example, in Tajima's D and Fu and Li's D and F test statistics, positive scores are indicative of unusually high heterozygosity within the data set. Additionally, these tests usually do not take distance between variable sites into consideration and rely heavily on statistics obtained from rare mutational events. The selection criterion for the Perlegen and HapMap genotyping efforts, however, was the high heterozygosity and equal spacing of SNPs (1, 2). Hence, these data sets have high ascertainment bias. Using tests that rely on heterozygosity and frequency of rare mutations to infer selection on such biased data sets should be largely meaningless.
Although most tests are insensitive to all but extreme examples of selection (1, 2, 8, 10, 19), relatively sensitive tests for positive selection have been developed recently, based on the probability of seeing two random chromosomes from the sample that share the same haplotype (6, 8, 9, 20–22). For example, in the first example of this type of analysis, Serre et al. (20) surveyed polymorphic sites closely linked to the ΔF508 allele of CFTR. This analysis calculated an allele age of 3,000 years for the ΔF508 mutation, suggesting that the high frequency of this allele in European ancestry populations might be explained by heterozygote advantage. Recently, Sabeti and colleagues (10) used this approach to confirm selection at two well characterized genes (G6PD and TNFSF5). Their method uses computationally estimated haplotypes, however, making global chromosomal scans a daunting computational task. In addition, this approach does not consider the expected decay of LD surrounding a selected allele, in contrast to the method presented in this work.
Here, we construct a probabilistic model, based on our prior experimental approach (6, 8), designated the LD decay (LDD) test. The method relies only on high-heterozygosity SNPs for analysis, exactly the type of data obtained in the Perlegen and HapMap efforts (1, 2). This “first-pass” analysis uncovers a surprising number of alleles with the fingerprint of recent positive selection, in contrast to other global approaches using less-sensitive methods (1, 2). We outline several predominant biological themes among genes detected with this strategy and suggest that selection for alleles in these categories accompanied the major “out of Africa” population expansion of humankind and/or the radical shift from hunter–gatherer to agricultural societies (23–26).
Materials and Methods
The G6PD V202M data set was obtained from Sabeti et al. (10). The Perlegen data set was obtained from Hinds et al. (1), and the HapMap data set (2) was obtained from the Phase 1 freeze. Details of our computational approach are described in Results, with additional information in Supporting Text and Figs. 6–8, which are published as supporting information on the PNAS web site.
Results
We developed a simple computational approach to distinguish large differences in LD surrounding a given SNP pair based on our prior experimental approach (6, 8). By examining individuals homozygous for a given SNP, the fraction of inferred recombinant chromosomes (FRC) at adjacent polymorphisms can be directly computed without the need to infer haplotype (8). We use the expected increase with distance in FRC surrounding a selected allele to identify such alleles. Importantly, the method is insensitive to local recombination rate, because local rate will influence the extent of LD surrounding both alleles, while the method looks for LD differences between alleles.
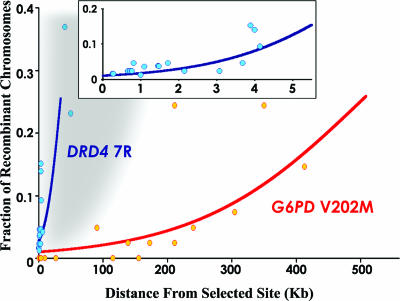
As two well characterized examples, the patterns of FRCs surrounding the selected alleles DRD4 7R, a dopamine receptor (6, 8), and G6PD V202M, a variant conferring malaria resistance in African populations (9, 10), are strong indicators of selection (Fig. 1). The new allele attained a high population frequency yet still retained a strong local LD block in comparison with the alternative allele. More importantly, the progressive decay of this strong LD with distance from the selected allele is further evidence of selection acting on such sites. One observes this pattern because the number of possible meiotic recombinations not eliminating the advantageous allele increases as a function of distance from the selected site. The overall “rate” of LDD is influenced by the intraallelic coalescence time of the inferred selection and local recombination rate. For example, the G6PD V202M variant exhibits LDD similar to DRD4 7R, although the decay is 14 times slower (Fig. 1). This result is consistent, however, with the calculated 5- to 10-fold younger allele age of G6PD V202M and the 2- to 4-fold increase in recombination rate at the DRD4 locus (6, 8–10).
Fig. 1.
LD patterns surrounding DRD4 7R and G6PD V202M. The observed FRC, associated with a minor allele under selection (DRD4 7R and G6PD V202M), are plotted vs. distance. FRC is calculated assuming the selected variant arose on a single chromosome (haplotype) (8). The indicated logistic function curves are approximated as sigmoidal, indicating the increasing decay of LD with distance with maximum assumed value of 0.5. Only sites in one direction from the selected allele are shown. The proximal region of the DRD4 7R data are shown at increased resolution in Inset. The approximate current Perlegen (1) data set detection limit (gray) is indicated.
Although the analytical concept of defining recent allele age (and hence implied selection) based on this predicted exponential decay of LD is well established (6, 8, 20–22), little mention of the approach is found in the population genetics literature (16). Although this expected LDD can be approximated by various linear or exponential curves (depending on the assumptions made regarding recombination), we used a standard sigmoidal curve, consistent with prior work on allele age calculations (6, 8, 20–22) and the acknowledgment that inferred recombination has a maximum value of 0.5 (Fig. 1). Given the current SNP database depth, however, any reasonable approximation to the expected LDD yields comparable results (data not shown). Obviously, further work can refine this analytical approach if warranted, as experimental data (and the accuracy of inferred FRC) increases.
A simplified example of our computational approach is shown in Fig. 2, and details are presented in Supporting Text and Figs. 6–8. First, each SNP (S) is sorted by homozygous common and minor alleles, and heterozygous individuals are discarded. This method allows the direct measure of adjacent inferred FRC without the need to infer haplotype (8). All adjacent SNP markers within ±500 Kb are binned according to the separation distance from site S (Fig. 2 A, arrowhead). For each neighboring site, we then compute its inferred FRC (8), assuming the S variant arose on a single chromosome (haplotype). The distance away from S and each associated FRC is then recorded as a value pair into a list for S (Fig. 2B). From this list, average log likelihood (ALnLH) is computed based on the sum of the square of the differences between the input model and the actual data, with uniform prior and a deviation function to account for experimental and recombination variation. This process is then repeated for all sites, using a “sliding-window” of 1 Mb (containing 150–300 SNPs).
Fig. 2.
Probabilistic method for finding unusual genetic architectures. (A) Binning on major/minor alleles. Each individual is sorted based on homozygosity at the major or minor allele at site S (arrowhead). (B) Compute fraction of adjacent recombinant chromosomes. The distance (d1–d3) and FRC for each neighboring SNP is then computed and stored. This list is then used to compute the ALnLH for each site (see text). Using only homozygous individuals for the computation eliminates the need to infer haplotypes.
On average, the distance between each SNP in the Perlegen data set is 2 kb (1). These data were generated by genotyping 71 unrelated individuals from 3 populations: 24 European Americans, 23 African Americans, and 24 Han Chinese from the Los Angeles area. Because there are relatively few individuals in each population, the total Perlegen data set was initially analyzed. Approximately 68% of the 1,586,383 Perlegen sites have minor allele frequency of >10%. Approximately 49% of the total sites have homozygous minor allele individuals of >5%, which we take as our cut-off for analysis (≈0.22 allele frequency). If the homozygous minor allele is population specific in the Perlegen data set, this cut-off represents an allele frequency of >38% in that population.
The LDD test can be used with many a priori combinations of inferred recombination/coalescence parameters. For this initial analysis, we broadly sampled the range of potential selected allele LDD defined by the nongray area in Fig. 1, ≈1 SD from the genome average (see Supporting Text and Figs. 6–8). This cut-off excludes some well documented selected alleles such as the telomeric gene DRD4 7R, which have allelic frequencies below our cut-off and/or are in regions with too few neighboring SNPs currently typed in the database to stringently distinguish such alleles from background. The LDD test can be applied in such regions by high density SNP-typing/resequencing (6, 8).
For the purpose of this analysis, then, we define “recently” selected alleles, which include a number of loci such as PTC (12) and LCT (13) in addition to G6PD (Fig. 1), as ones that can be distinguished given the Perlegen resolution, coalescent time, and local recombination frequency, as well as their high (>0.22) allele frequency. Although “hotspots” for recombination likely occur in human DNA, the large-scale (megabase) variation in recombination frequency in most nontelomeric euchromatic regions does not vary beyond 2- to 4-fold (1, 2, 27). Selected alleles detected with our approach, therefore, should have estimated coalescent times up to 10,000 years in areas of high recombination to >40,000 years (the upper Paleolithic; ref. 23) in areas of low recombination (21).
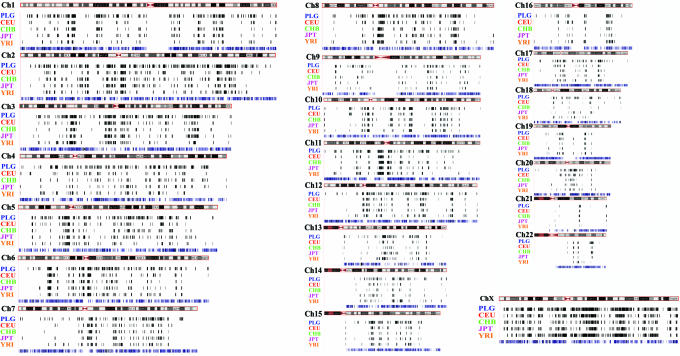
We set a detection threshold at an ALnLH of >2.6SD(>99.5th percentile) from the genome average, or 0.61 for the Perlegen data set (see Supporting Text and Figs. 6–8). The calculated genomewide Perlegen ALnLH scores exhibit an average of 0.043, but with a SD of 0.22. Hence, an ALnLH of 0.61 represents a highly unusual genetic architecture. In total, 25,386 (1.6%) of the 1.6 million Perlegen SNPs met these criteria. Because of the extensive LD detected by this analysis, many adjacent SNPs are calculated with high ALnLH values. Interestingly, ≈29% of these SNP clusters show signs of selection in individuals from all three Perlegen populations (European, African, and Asian American) and ≈78% in at least two populations. It is important to note that “absence” in a particular population using the LDD test is only a reflection that homozygous minor allele individuals are not observed in the sample. A display of regions of inferred selection along all chromosomes for the Perlegen (PLG) data set is shown in Fig. 3; see also Fig. 9, which is published as supporting information on the PNAS web site.
Fig. 3.
Darwin's fingerprint. The global landscape (black lines) of recent inferred Darwinian selection for the Perlegen (PLG) and HapMap (CEU, CHB, JPT, and YRI) data sets is shown, aligned along chromosomes and genes (blue lines). A larger version of this figure is available as Fig. 9, and higher-resolution analysis can be obtained from the authors for display on the University of California at Santa Cruz Genome Browser (28).
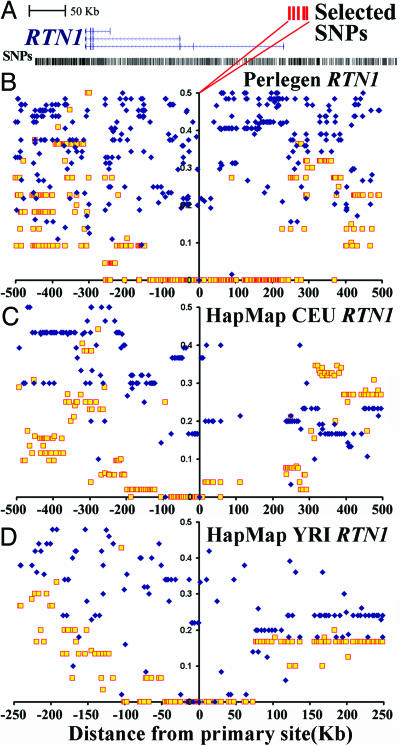
As an example of representative data, Fig. 4 shows the local genetic architecture centered at a 25-kb region defining the promoter of the Reticulon gene (RTN1) on chromosome 14 (Online Mendelian Inheritance in Man accession no. 600865) (29). This gene encodes a neuroendocrine-specific protein thought to affect cellular amyloid-β and the formation of amyloid plaques in Alzheimer's disease (30). The randomness for neighboring recombinant chromosomes for the major RTN1 allele at this site exemplifies the genome average, with little long-range LD. In contrast, the minor RTN1 allele at this site closely matches the LDD model (Fig. 4). The large LD block around Perlegen SNP rs9323357 and its disproportionately high allelic frequency (35%) suggests a possible recent selective event at the RTN1 locus.
Fig. 4.
Example of inferred selection at the Reticulon gene (RTN1), which encodes a neuroendocrine-specific protein thought to affect the formation of amyloid plaques in Alzheimer's disease (29, 30). (A) Inferred selected SNPs in the promoter region (red) are shown along with all annotated SNPs (black). (B–D) The randomness for neighboring recombinant chromosomes for the major RTN1 allele (blue) at this site exemplifies the genome average, with little long-range LD. In contrast, the minor RTN1 allele (yellow) at this site closely matches the LDD model for selection. The horizontal axis labels distance away from each centered SNP, and the vertical axis is FRC (Fig. 1). (B) Perlegen data set. (C) CEU HapMap data set. (D) African ancestry (YRI) HapMap data set. The Asian HapMap data sets resemble the CEU architecture (data not shown). Note the twofold horizontal axis scale change for the YRI display, reflecting the more rapid LDD at this site in this population.
Although the Perlegen data set (1) has high SNP resolution, population depth is limited. The recently released HapMap data set (Phase I freeze) (2), conversely, has fewer SNPs (1.0 million) but deeper population coverage: 90 European ancestry (CEU), 90 African (Yoruba) ancestry (YRI), 45 Han Chinese (CHB), and 45 Japanese (JPT) individuals. This data set allows for an independent confirmation of our results. In addition, the greater depth of the HapMap data set allows better definition of potential population-specific selective events, which account for only 22% of the Perlegen clusters.
Calculations of ALnLH were conducted separately on all four HapMap populations, again using a cut-off of >2.6 SD (>99.5th percentile) from the genome average (ranging from 0.51 to 0.71 for YRI and CEU populations, respectively). Merging all four HapMap populations yielded a total of 20,786 SNPs with evidence of selection, similar to the Perlegen data set. Inferred selection for the four HapMap populations is shown in Figs. 3 and 9. Because there is only partial overlap between SNPs used by the Perlegen and HapMap efforts, both data sets were aligned along the Human Genome (hg17) sequence (28), using a 10-kb window for assigning regions. Encouragingly, there is a 77% (YRI) to 96% [Han Chinese (CHB)] overlap between the inferred selected regions identified by the Perlegen and HapMap data sets. For example, the RTN1 promoter region originally identified in the Perlegen data set shows evidence for selection in all four HapMap populations (Fig. 4). Interestingly, the LDD at this locus is greater in the YRI population, as expected for an older population that has not undergone the severe recent bottlenecks (31, 32) inferred for Asian and European populations. In general, regions of inferred selection that are found in all populations exhibit this African-specific faster LDD.
The genomic distribution of inferred selection using the LDD test is in general random, with no bias toward or against other unusual genomic regions such as segmental duplications or inversions (33, 34) (Fig. 3). Although inversions can suppress recombination and produce large LD blocks, large (>100 kb) inversions are not common in human DNA, do not produce a gradual LDD as observed for selected alleles, and would not eliminate recombination at the high frequency of alleles reported in this work. For example, a recently reported large chromosome 17 inversion (34) produces a distinct pattern of flat LD clearly distinguishable from the alleles identified in this work (data not shown). The few inferred inversions detected by our analysis are not excluded, because their high frequency implies that selection may be maintaining them in the population (34).
There is a slight underrepresentation of detected selection in high-recombination areas such as telomeric regions, as expected given the particular parameters used for this initial screen. One strikingly nonrandom distribution, however, is an ≈2-fold overrepresentation of such alleles on the X chromosome (Fig. 3; P ≪ 0.00001). Given that overall population recombination frequency on the X chromosome is not significantly lower than the genome average (27, 35), this result is consistent with the hypothesis that alleles on the “haploid” X chromosome will be under stronger selective pressure than those on diploid autosomes.
In addition to selection, are there other mechanisms that could produce these unusual long-range genetic architectures? It is commonly assumed that one summary statistic is often insufficient to unambiguously detect recent selection from other population events (16, 36). Many population-genetics tests, indeed, cannot distinguish selection from bottlenecks/admixture. This lack of discrimination is because of both a lack of acknowledgment of LD structure in these tests (as discussed above) and the usual examination of small (≪1 Mb) genomic regions (16). Supporting Text describes permutations/simulations of admixture and bottleneck models, using actual Perlegen or HapMap data sets. These simulations were conducted because prior population genetics simulations and coalescence models of population structure cannot be compared directly with the highly biased Perlegen and HapMap SNP data set, consisting largely of high-heterozygosity SNPs. These simulations indicate that the LDD test, at the megabase scale used, appears to effectively distinguish between effects due to selection vs. demographic history. We conclude that inferred recent Darwinian selection is the most likely explanation for these unusual genomic architectures (Figs. 3 and 4).
The inferred selected SNPs were queried into the National Center for Biotechnology Information SNP Database (dbSNP; www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=snp) Build 123 for associated genes/exons within a 100-kb radius. A 100-kb radius was chosen as a reasonable first-pass distance in which a SNP could influence a gene's expression/function, given current knowledge of gene organization and regulation (33, 37). Approximately 35% of the inferred selected SNPs were not within a 100-kb radius of known genes and were not analyzed further. Whether this fraction represents selection at noncoding regions or the inability to identify all potential gene regions in the current HGP assembly is unclear. In the Perlegen data set, inferred selected SNPs clustered in 1,799 genes (Fig. 3). Similar results were obtained with the HapMap data sets. A total of 112 annotated genes showed evidence for selection in both the Perlegen data set and all four HapMap populations (see Table 1, which is published as supporting information on the PNAS web site).
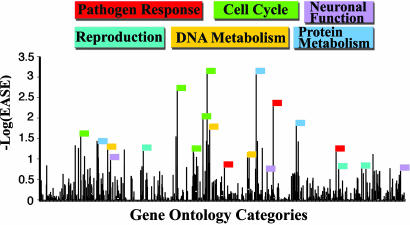
We examined whether there are predominant biological themes represented among these selected genes, using ease for the analysis of overrepresentation (26). Similar ease results were obtained for all populations. As an example, Fig. 5 shows ease values determined for the 407 HapMap CEU selected genes classifiable under Gene Ontology (GO) Biological Process categories. These 870 overrepresented categories are <1% of the total currently annotated GO categories.
Fig. 5.
Overrepresented GO categories are not random and represent six biological themes. A total of 407 HapMap CEU selected genes are classifiable under Biological Process GO categories. For these classified genes, 870 biological themes with positive ease values were identified, as indicated. Six functional categories constitute 82% of the –log(ease) scores of >0.65, indicated by colored flags. Each flag is color-coded for one of these specific categories, namely pathogen–host interaction, reproduction, DNA metabolism (including putative transcription factors), cell cycle, protein metabolism, and neuronal function.
Overall, the observed genes in overrepresented GO categories are not random. For example, six functional categories constitute 82% of the HapMap CEU –log(ease) scores of >0.65, represented by color flags in Fig. 5. We have defined these more general functional categories to include a number of individual GO categories associated with pathogen–host interaction, reproduction, DNA metabolism/cell cycle, protein metabolism, and neuronal function. We emphasize that many genes appear in multiple GO categories, and hence exact classification is not possible. Nevertheless, the clustering of most high-scoring GO categories into one of these generally defined functional categories is striking (Fig. 5). In the 112 genes with evidence for selection in all populations (Table 1) the proportion of genes in each of these categories is as follows: reproduction, 7%; host–pathogen interaction, 10%; cell cycle, 13%; protein metabolism, 15%; neuronal function, 17%; and DNA metabolism (including putative transcription factors), 21%.
Selection for alleles in some of these categories might be anticipated, such as host–pathogen interaction and reproduction, given prior selection studies in humans and other organisms (15, 38, 39). Pathogen defense has long been suspected to be under constant evolutionary pressure. The beginning of agriculture and animal domestication 10,000 years ago not only brought domesticated animals close to humans but also established permanent human settlements (24). Such shifts from a hunter–gatherer nomadic lifestyle to agrarian societies likely facilitated the wide spread of infectious agents (38, 40). Our results suggest that human populations may have encountered many selective events associated with pathogen–host interaction. Examples of genes identified under host–pathogen interaction include CSF2, CCNT2, DEFB118, STAB1, SP1, and Zap70, and under reproduction, BIRC6, CUGBP1, DLG3, HMGCR, STS, and XRN2.
The other overrepresented GO categories contain a number of unexpected genes. For example, it has been suggested that changes in organic compound metabolism may have been influenced by increases in meat consumption by early humans (41). Overrepresented genes in protein metabolism could be the result of this shift in dietary composition and/or the profound changes associated with a restricted agrarian diet (40). The large number of selected genes under DNA metabolism is also unexpected. We suggest that many of these selected alleles may be involved in the recent inferred increase in longevity of humans (42). Modifications to our immune system, increases in tumor suppression, and enhanced DNA repair (Fig. 5) are likely molecular components of our unique primate longevity. Some examples of selected genes in protein metabolism include ADAMTS19–20, APEH, PLAU, HDAC8, UBR1, and USP26, and under DNA metabolism CKN1, FANCC, RAD51C, HDAC8, PDCD8, and SMC1L1.
One of the more intriguing categories overrepresented in inferred selective events is neuronal function. We define this category to include a diverse assortment of genes, including the serotonin transporter (SLC6A4), glutamate and glycine receptors (GRM3, GRM1, and GLRA2), olfactory receptors (OR4C13 and OR2B6), synapse-associated proteins (RAPSN), and a number of brain-expressed genes with largely unknown function (ASPM, RNT1; see Fig. 4).
Discussion
It is well established that new mutations have a very high likelihood of being lost within a few generations (43). This principle is the basis of our probabilistic approach, which asserts that a high-frequency allele with large LD is almost impossible to achieve by chance (Figs. 1, 2, 3, 4). Further, by requiring that the pattern of LD exhibit an expected decay with distance, other possible causes of long-range LD (inversions, population admixture, and small bottlenecks) are less likely to account for this anomalous genetic architecture (see Supporting Text and Figs. 6–8). Finally, the clustering of identified alleles into a few specific GO categories is distinctly nonrandom (Fig. 5). Together, the evidence of this first-pass study shows many promising regions with inferred recent Darwinian selection, which undoubtedly will guide numerous future studies. Specifically, our method identifies the region likely responsible for the inferred selection, at or near the minima of calculated recombination (Figs. 3 and 4).
Although our approach searches for the fingerprint of directional selection, it is important to note that many of the identified alleles may be part of a balancing selection allelic system (8, 14). Although it is common to distinguish between directional and balancing selection, in actuality all balancing selection alleles, unless they are ancient, must also have the fingerprint of a directional component [i.e., one of the alleles must be a “younger” variant (8)]. Even modestly selected alleles (s = 0.05) will become fixed in as little as 200 generations. Such selection would not be detected with the LDD test, because the locus is no longer polymorphic. Indeed, the observation that many of these alleles are found in most examined populations (Fig. 4), yet have not reached fixation, argues for balanced selection. Obviously, only further work directed at specific alleles can clarify the nature and mode of the selection acting at the loci described in this work.
It is intriguing that a significant fraction of inferred selected alleles are found in most of the examined populations (Fig. 4). Although the calculated intraallelic coalescence time for many of these alleles in European and Asian ancestry populations is similar, the same allele exhibits a more rapid LDD, and hence a longer coalescence time in African ancestry populations. The model that best explains these data is the ongoing balanced selection for these alleles for at least the last 40,000–50,000 years after the out-of-Africa expansion (14). In African populations, these alleles exhibit faster LDD, reflecting the original coalescence. In European and Asian populations, ongoing selection “reset” the LD “clock” on the few chromosomes containing the selected variant during the inferred bottlenecks generating these populations.
Although our studies have confirmed previously known selected alleles (such as LCT; ref. 13) and have uncovered hundreds of putative newly selected alleles (Fig. 3), there are a few related caveats that deserve mention. First, although the populations used likely represent a reasonable sample of humankind, they are not without bias. As one example, many of the Centre d'Etude du Polymorphisme Humain (CEPH) families used in the HapMap Project (2) originated from Utah, where polygamous marriages were common a number of generations ago. Significant homozygous regions in CEPH families have been observed (44), although these haplotype blocks, as expected, are significantly larger than detected in this study (see Supporting Text and Figs. 6–8).
A second concern is that although ancient (>50,000 years) population bottlenecks or admixture would produce LD blocks much smaller than those observed in this study (32), more recent undetected admixture might contribute to these results. The extreme bottleneck/admixture models simulated in this study (see Supporting Text and Figs. 6–8), however, indicate that this concern is unlikely, again because of our probabilistic model's reliance on the distinct pattern of LD loss with distance. Further, for the alleles found across all populations (Table 1), it is difficult to see what type of recent admixture could produce such a result. We conclude that few of the observed anomalous architectures could be the result of undetected population stratification. Nevertheless, we caution that any particular site identified in this study not be assumed to be unambiguously the result of selection without further confirmatory studies. Likewise, although the number of inferred selected alleles uncovered in this study is large (Fig. 3), it should not be considered a complete list, but rather a first-pass analysis given the current data sets (1, 2) and the particular method of analysis.
In conclusion, we have introduced a simple probabilistic method to detect unusual genetic architectures associated with recent selection that does not require haplotype information. It is, therefore, suitable for large chromosomal scans with large population samples. Homo sapiens have undoubtedly undergone strong recent selection for many different phenotypes, including but certainly not limited to the general categories we have defined in this work (Fig. 5). Such inferred selective events are not rare (Fig. 3). The numbers obtained, however, are similar to estimated numbers obtained for artificial selection (by humans) on the maize genome (45). Given that most of these selective events likely occurred in the last 10,000–40,000 years, a time of major population expansion out of Africa followed by regional shifts from hunter–gatherer to agrarian societies, it is tempting to speculate that gene–culture interactions directly or indirectly shaped our genomic architecture (46, 47). As such, we suggest that such recently selected alleles may provide useful “markers” for investigating the evolutionary migrations of our species, as an adjunct to studies using neutral markers. We also propose that many of these alleles, because of their high prevalence and recent selection, should be considered likely “functional candidates” for association with human variability and the common disorders afflicting humankind.
Supplementary Material
Acknowledgments
We thank Pardis Sabeti (Whitehead Institute, Massachusetts Institute of Technology, Cambridge, MA) for providing the G6PD data set and Brad Sherman for updating the ease LocusLink population library. This work was supported by grants from the U.S. Department of Energy, the National Institute of Mental Health, and the National Institute of Aging (to R.K.M.) and a National Library of Medicine National Research Service Award (to E.T.W.).
Author contributions: E.T.W., G.K., P.B., and R.K.M. designed research; E.T.W. performed research; E.T.W., P.B., and R.K.M. contributed new reagents/analytic tools; E.T.W. and R.K.M. analyzed data; and E.T.W., P.B., and R.K.M. wrote the paper.
Conflict of interest statement: No conflicts declared.
Abbreviations: ALnLH, average log likelihood; CEU, European ancestry; CHB, Han Chinese ancestry; FRC, fraction of inferred recombinant chromosomes; GO, Gene Ontology; HapMap, International Haplotype Map; JPT, Japanese ancestry; LD, linkage disequilibrium; LDD, LD decay; SNP, single-nucleotide polymorphism; YRI, African (Yoruba) ancestry.
References
- 1.Hinds, D. A., Stuve, L. L., Nilsen, G. B., Halperin, E., Eskin, E., Ballinger, D. G., Frazer, K. A. & Cox, D. R. (2005) Science 307, 1072–1079. [DOI] [PubMed] [Google Scholar]
- 2.The International HapMap Consortium (2005) Nature 437, 1299–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Risch, N. & Merikangas, K. (1996) Science 273, 1516–1517. [DOI] [PubMed] [Google Scholar]
- 4.Zwick, M. E., Cutler, D. J. & Chakravarti, A. (2000) Annu. Rev. Genomics Hum. Genet. 1, 387–407. [DOI] [PubMed] [Google Scholar]
- 5.Reich, D. E. & Lander, E. S. (2001) Trends Genet. 17, 502–510. [DOI] [PubMed] [Google Scholar]
- 6.Ding, Y. C., Chi, H. C., Grady, D. L., Morishima, A., Kidd, J. R., Kidd, K. K., Flodman, P., Spence, M. A., Schuck, S., Swanson, J. M., et al. (2002) Proc. Natl. Acad. Sci. USA 99, 309–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grady, D. L., Chi, H. C., Ding, Y. C., Smith, M., Wang, E., Schuck, S., Flodman, P., Spence, M. A., Swanson, J. M. & Moyzis, R. K. (2003) Mol. Psychiatry 8, 536–545. [DOI] [PubMed] [Google Scholar]
- 8.Wang, E., Ding, Y. C., Flodman, P., Kidd, J. R., Kidd, K. K., Grady, D. L., Ryder, O. A., Spence, M. A., Swanson, J. M. & Moyzis, R. K. (2004) Am. J. Hum. Genet. 74, 931–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tishkoff, S. A., Varkonyi, R., Cahinhinan, N., Abbes, S., Argyropoulos, G., Destro-Bisol, G., Drousiotou, A., Dangerfield, B., Lefranc, G., Loiselet, J., et al. (2001) Science 293, 455–462. [DOI] [PubMed] [Google Scholar]
- 10.Sabeti, P. C., Reich, D. E., Higgins, J. M., Levine, H. Z., Richter, D. J., Schaffner, S. F., Gabriel, S. B., Platko, J. V., Patterson, N. J., McDonald, G. J., et al. (2002) Nature 419, 832–837. [DOI] [PubMed] [Google Scholar]
- 11.Stephens, J. C., Reich, D. E., Goldstein, D. B., Shin, H. D., Smith, M. W., Carrington, M., Winkler, C., Huttley, G. A., Allikmets, R., Schriml, L., et al. (1998) Am. J. Hum. Genet. 62, 1507–1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wooding, S., Kim, U. K., Bamshad, M. J., Larsen, J., Jorde, L. B. & Drayna, D. (2004) Am. J. Hum. Genet. 74, 637–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bersaglieri, T., Sabeti, P. C., Patterson, N., Vanderploeg, T., Schaffner, S. F., Drake, J. A., Rhodes, M., Reich, D. E. & Hirschhorn, J. N. (2004) Am. J. Hum. Genet. 74, 1111–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harpending, H. & Rogers, A. (2000) Annu. Rev. Genomics Hum. Genet. 1, 361–385. [DOI] [PubMed] [Google Scholar]
- 15.Vallender, E. J. & Lahn, B. T. (2004) Hum. Mol. Genet. 13, Spec. 2, R245–R254. [DOI] [PubMed] [Google Scholar]
- 16.Kreitman, M. (2000) Annu. Rev. Genomics Hum. Genet. 1, 539–559. [DOI] [PubMed] [Google Scholar]
- 17.Tajima, F. (1989) Genetics 123, 585–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fu, Y. X. & Li, W. H. (1993) Genetics 133, 693–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Simonsen, K. L., Churchill, G. A. & Aquadro, C. F. (1995) Genetics 141, 413–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Serre, J. L., Simon-Bouy, B., Mornet, E., Jaume-Roig, B., Balassopoulou, A., Schwartz, M., Taillandier, A., Boue, J. & Boue, A. (1990) Hum. Genet. 84, 449–454. [DOI] [PubMed] [Google Scholar]
- 21.Slatkin, M. & Rannala, B. (2000) Annu. Rev. Genomics Hum. Genet. 1, 225–249. [DOI] [PubMed] [Google Scholar]
- 22.Fay, J. C. & Wu, C. I. (2000) Genetics 155, 1405–1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bar-Yosef, O. (2002) Annu. Rev. Anthropol. 31, 363–393. [Google Scholar]
- 24.Flannery, K. V. (1972) Annu. Rev. Ecol. Syst. 3, 399–426. [Google Scholar]
- 25.Cowen, C. W. & Watson, P. J. (1992) The Origins of Agriculture (Smithsonian Institution Press, Washington, DC).
- 26.Hosack, D. A., Dennis, G., Jr., Sherman, B. T., Lane, H. C. & Lempicki, R. A. (2003) Genome Biol. 4, R70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kong, A., Gudbjartsson, D. F., Sainz, J., Jonsdottir, G. M., Gudjonsson, S. A., Richardsson, B., Sigurdardottir, S., Barnard, J., Hallbeck, B., Masson, G., et al. (2002) Nat. Genet. 31, 241–247. [DOI] [PubMed] [Google Scholar]
- 28.Kent, W. J., Sugnet, C. W., Furey, T. S., Roskin, K. M., Pringle, T. H., Zahler, A. M. & Haussler, D. (2002) Genome Res. 12, 996–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oertle, T., Klinger, M., Stuermer, C. A. & Schwab, M. E. (2003) FASEB J. 17, 1238–1247. [DOI] [PubMed] [Google Scholar]
- 30.He, W., Lu, Y., Qahwash, I., Hu, X. Y., Chang, A. & Yan, R. (2004) Nat. Med. 10, 959–965. [DOI] [PubMed] [Google Scholar]
- 31.Reich, D. E., Cargill, M., Bolk, S., Ireland, J., Sabeti, P. C., Richter, D. J., Lavery, T., Kouyoumjian, R., Farhadian, S. F., Ward, R. & Lander, E. S. (2001) Nature 411, 199–204. [DOI] [PubMed] [Google Scholar]
- 32.Kruglyak, L. (1999) Nat. Genet. 22, 139–144. [DOI] [PubMed] [Google Scholar]
- 33.The International Human Genome Sequencing Consortium (2004) Nature 431, 931–945. [DOI] [PubMed] [Google Scholar]
- 34.Stefansson, H., Helgason, A., Thorleifsson, G., Steinthorsdottir, V., Masson, G., Barnard, J., Baker, A., Jonasdottir, A., Ingason, A., Gudnadottir, V. G., et al. (2005) Nat. Genet. 37, 129–137. [DOI] [PubMed] [Google Scholar]
- 35.Ross, M. T., Grafham, D. V., Coffey, A. J., Scherer, S., McLay, K., Muzny, D., Platzer, M., Howell, G. R., Burrows, C., Bird, C. P., et al. (2005) Nature 434, 325–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Charlesworth, B., Charlesworth, D. & Barton, N. H. (2003) Annu. Rev. Ecol. Evol. Syst. 34, 99–125. [Google Scholar]
- 37.Wasserman, W. W. & Sandelin, A. (2004) Nat. Rev. Genet. 5, 276–287. [DOI] [PubMed] [Google Scholar]
- 38.Williams, G. C. & Nesse, R. M. (1991) Q. Rev. Biol. 66, 1–22. [DOI] [PubMed] [Google Scholar]
- 39.Swanson, W. J. & Vacquier, V. D. (2002) Annu. Rev. Ecol. Syst. 33, 161–179. [Google Scholar]
- 40.Larsen, C. S. (1995) Annu. Rev. Anthropol. 24, 185–213. [Google Scholar]
- 41.Finch, C. E. & Stanford, C. B. (2004) Q. Rev. Biol. 79, 3–50. [DOI] [PubMed] [Google Scholar]
- 42.Caspari, R. & Lee, S.-H. (2004) Proc. Natl. Acad. Sci. USA 101, 10895–10900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kimura, M. & Ohta, T. (1971) Theoretical Aspects of Population Genetics (Princeton Univ. Press, Princeton). [PubMed]
- 44.Broman, K. W. & Weber, J. L. (1999) Am. J. Hum. Genet. 65, 1493–1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wright, S. I., Bi, I. V., Schroeder, S. G., Yamasaki, M., Doebley, J. F., McMullen, M. D. & Gaut, B. S. (2005) Science 308, 1310–1314. [DOI] [PubMed] [Google Scholar]
- 46.Cavalli-Sforza, L. L., Menozzi, P. & Piazza, A. (1994) The History and Geography of Human Genes (Princeton Univ. Press, Princeton).
- 47.Darwin, C. (1871) The Descent of Man and Selection in Relation to Sex (Murray, London).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.