Abstract
The structure of a cell surface enzyme from a Gram-positive pathogen has been determined to 2-Å resolution. Gram-positive pathogens have a thick cell wall to which proteins and carbohydrate are covalently attached. Streptococcal C5a peptidase (SCP), is a highly specific protease and adhesin/invasin. Structural analysis of a 949-residue fragment of the [D130A,S512A] mutant of SCP from group B Streptococcus (S. agalactiae, SCPB) revealed SCPB is composed of five distinct domains. The N-terminal subtilisin-like protease domain has a 134-residue protease-associated domain inserted into a loop between two β-strands. This domain also contains one of two Arg-Gly-Asp (RGD) sequences found in SCPB. At the C terminus are three fibronectin type III (Fn) domains. The second RGD sequence is located between Fn1 and Fn2. Our analysis suggests that SCP binding to integrins by the RGD motifs may stabilize conformational changes required for substrate binding.
Keywords: bacterial adhesins, endopeptidases, molecular models, Streptococcus agalactiae, x-ray crystallography
The complement cascade is a key mediator of the innate immune response (1, 2). The 74-residue complement factor C5a is a chemotactic peptide that attracts polymorphonuclear neutrophils (PMNs) and an anaphylatoxin, promoting histamine release and increasing capillary permeability. PMNs are critical for the phagocytosis and clearance of microorganisms upon entry into the host. Two important species of β-hemolytic streptococci, Streptococcus pyogenes (group A Streptococcus, GAS) and Streptococcus agalactiae (group B Streptococcus, GBS), which cause a variety of serious human infections that range from mild cases of pharyngitis and impetigo to serious invasive diseases such as necrotizing fasciitis (GAS) and neonatal sepsis (GBS) have developed a way to defeat this immune response. All human isolates of β-hemolytic streptococci, including GAS and GBS, produce a highly conserved cell-wall protein SCP that specifically inactivates C5a by releasing a seven-residue carboxyl-terminal fragment that contains the PMN binding site (3). This proteolysis abolishes C5a-induced recruitment of PMNs to the site of infection. It has also been demonstrated that streptococcal C5a peptidase (SCP) from GBS acts as an adhesin to fibronectin type III (Fn) and as an invasin of epithelial cells (4, 5).
The scp genes from GAS and GBS encode a polypeptide containing between 1,134 and 1,181 amino acids (6). The first 31 residues are the export signal presequence and are removed upon passing through the cytoplasmic membrane (see Fig. 1B). The next 68 residues serve as a pro-sequence and must be removed to produce active SCP from GBS (SCPB) (7). The next 10 residues can be removed without loss of protease activity (unpublished observation). At the other end, starting with Lys-1034, are four consecutive 17-residue motifs followed by a cell sorting and cell-wall attachment signal (8, 9). This combined signal is composed of a 20-residue hydrophilic sequence containing an LPTTND sequence, a 17-residue hydrophobic sequence, and a short basic carboxyl terminus.
Fig. 1.
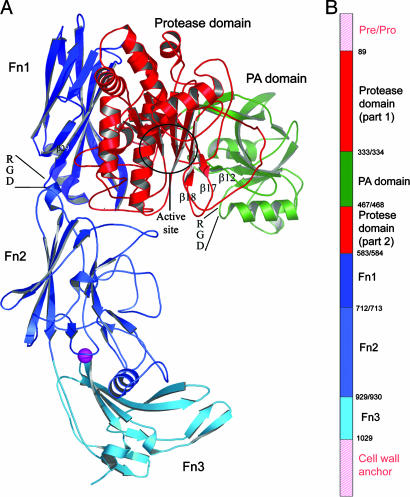
Domain structure of SCPBw. (A) Ribbon drawing of SCPBw monomer. Protease domain is red, PA domain is green, and three Fn domains are navy, sky blue, and cyan. Maroon sphere is calcium ion. The β12–β17–β18 sheet, the two RGD sequences, and the active-site region are indicated. (B) Schematic representation of SCPB domain organization. Domains are colored as in A. The domains not in SCPBw are indicated by pink labels and pink hatching. Numbers indicate domain boundaries in SCPB.
A construct was produced that encoded the 949-residue fragment corresponding to residues 89 (numbers correspond to the prepro-protein as transcribed) through 1037 of SCPB. On the basis of sequence homology with the subtilisin family of serine proteases, Asp-130, His-193, and Ser-512 had been suggested to form the active-site catalytic triad (10, 11). Site-directed mutagenesis supported this hypothesis. Studies by Anderson et al. (7) reported that recombinant SCPB was unstable as it was subject to autoproteolysis. Mutating Asp-130 and Ser-512 to alanines produced a stable molecule suitable for crystallization trials. In addition, immunization with the [D130A/S512A]89–1037 peptide, hereafter designated SCPBw, enhanced clearance of streptococci from the lungs of intranasally infected mice, consistent with the absence of any large-scale structural changes as a result of these mutations (13). Finally, CD analysis of SCPBw and WT SCPB show identical spectra and thermal denaturation behavior (data not shown). Thus the structure of SCPBw was determined by using multiple wavelength anomalous diffraction (14) from selenomethionine-substituted crystals.
Methods
Protein Production and Crystallization. The nucleotide region corresponding to bases 265-3112 was amplified from a serotype II strain of S. agalactiae (GBS) by PCR and cloned into the arabinose-inducible plasmid pBAD18. Mutations were introduced that result in amino acid changes from Asp-130 and Ser-512 to alanines (pLP676). The plasmid pLP676 was transformed into Escherichia coli B834(DE3) and cultured in M9 minimal media supplemented with selenomethionine plus the other 19 aa at 4 mg/ml and containing 30 mg/ml chloramphenicol. For expression, an overnight culture was diluted 1:100 in fresh media and grown while shaking at 37°C to an OD600 of 0.8. The culture was subsequently induced with 0.4 mM arabinose for 3 h, and the cells were collected by centrifugation. Recombinant SCPBw was purified from the soluble fraction of bacterial lysate as described (7). Briefly, the soluble fraction was subjected to sequential fractionation with 50% and then 70% ammonium sulfate. This fractionation was followed by ion-exchange chromatography on Q-Sepharose (Amersham Pharmacia). Bound SCP was eluted by using a linear gradient of NaCl, dialyzed against TBS, aliquoted, and stored frozen at –20°C. All purification steps were performed under reducing conditions. Crystals were grown from 10 mg/ml solutions of SCPBw by using the hanging drop method with 2 + 2-μl drops with 10–15% PEG 4000, 0.2 M sodium acetate in 100 mM citrate buffer (pH 3–5) in the reservoir.
Data Collection and Processing. Three wavelength anomalous diffraction data for SCPBw were collected to 2-Å resolution at the X25 beamline at the National Synchrotron Light Source at Brookhaven National Laboratory, Upton, NY. The crystals belong to space group P21, with unit cell dimensions of a = 114.7 Å, b = 75.1 Å, c = 132.4 Å, and β = 105°. There are two SCPBw polypeptide chains per asymmetric unit, referred to as chain A and chain B. However, SCPBw is not a dimer in solution. Diffraction data were processed and reduced with the hkl package (15). Data reduction statistics are summarized in Table 1.
Table 1. Data collection and refinement statistics.
| Data collection | |
| Space group | P21 |
| Temperature | -170°C |
| Unit cell dimensions | a = 114.7 Å, b = 75.1 Å, |
| c = 132.4 Å, β = 105° | |
| Data resolution, Å | 50-2 |
| Completeness, % | 99.8 (99.2) |
| Redundancy | 5.5 (1.0) |
| Rsym | 0.086 |
| 〈(I)/σ(I) 〉 | 45.1 (2.6) |
| Molecules/asymmetric unit | 2 |
| Refinement statistics | |
| Rfactor | 0.229 |
| Rfree | 0.269 |
| No. of non-H protein atoms | 14,364 |
| No. of solvent molecules | 1,057 |
| No. of citrates | 8 |
| No. of acetates | 2 |
| No. of calciums | 2 |
| Average B, Å2 | |
| Chain A | 48.8 |
| Chain B | 53.3 |
| Solvents | 50.3 |
| Other | 51.9 |
| Ramachandran regions | |
| % in core | 80.4 |
| % in allowed | 15.7 |
| % in generously allowed | 2.9 |
| % in disallowed | 1.1 |
| RMSD from ideal geometry | |
| Bond distance, Å | 0.007 |
| Bond angles, ° | 1.6 |
| Dihedral angles, ° | 24.4 |
Values in parentheses are for highest-resolution shell of reflections.
Structure Determination and Refinement. The locations of the 32 Se atoms in the crystal were determined with the program solve (16), and initial phases to 2.3-Å resolution were calculated. resolve (16) and maid (17) were used to automatically trace 40–50% of the model. The remainder of the model was built into the electron density with o (18). Structure refinement was carried out with cns (19). The final model contained 14,364 protein atoms, 2 calcium ions, 6 citrates, and 1,056 solvent molecules and produced an Rwork of 0.23 and an Rfree of 0.27. Refinement statistics are summarized in Table 1.
Results
SCPBw is an L-shaped molecule containing 50 β-strands and 19 helices divided among the major five domains of SCPBw. These domains are the protease domain (residues 109–333 and 468–582), the protease-associated domain (PA domain) (residues 334–467), and three Fn domains (Fn1, residues 584–712; Fn2, residues 713–928; Fn3, residues 930-1031). The positions of the secondary structural elements in the primary and tertiary structures are given in, respectively, Figs. 5 and 6, which are published as supporting information on the PNAS web site.
With the exception of residues 185–205 all of SCPBw is well ordered with unambiguous electron density. Residues 185–205 are part of a 112-residue fragment of SCPBw that Beckmann et al. (5) showed binds fibronectin. It is to the left side of the active site (see Fig. 1) and includes His-193 of the active-site triad (7, 12).
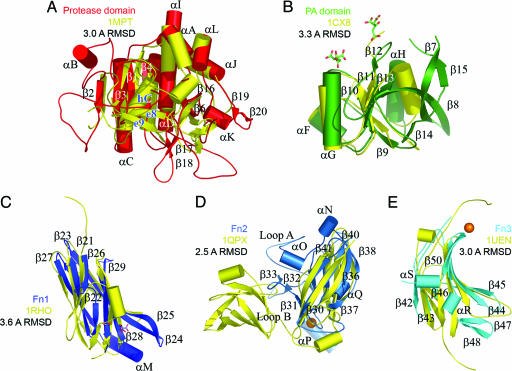
Protease Domain. blast searches using the sequence of the protease domain reveal homologies of 25–42% with other subtilisins. A structural search with dali (20) aligns the Cαs of 200 residues with the structure of a subtilisin (M-protease) from Bacillus sp. KSM-K16 (Protein Data Bank ID code 1MPT) with an overall rms deviation (RMSD) of 3.0 Å (see Fig. 5). There are only 25% of the amino acid residues identical between this subtilisin and the protease domain. Many of the loops connecting the core subtilisin secondary structure elements are more elaborate in SCPBw and are often used to interact with adjacent domains (see Fig. 2A). To discuss these differences it is convenient to use the nomenclature of Siezen et al. (11).
Fig. 2.
Ribbon drawings of individual SCPBw domains (colored as in Fig. 1) superimposed with nearest structural homolog (yellow, Protein Data Bank ID code and RMSD between Cαs are indicated). Secondary structural elements are labeled as in Fig. 3. Calcium binding sites are shown as spheres. Bound citrates and acetate are shown as sticks. (A) Protease domain with Bacillus M-protease. (B) PA domain with the apical domain from TfR (26). (C) Fn1 with human Rho guanine nucleotide dissociation inhibitor (37). (D) Fn2 with PapD from E. coli (38). (E) Fn3 with third Fn from human Kiaa0343 protein.
The loop between strands e1 and e2 (strands β1 and β2 in Figs. 5 and 6) has an additional 33 residues that form the three-turn helix αB and a short extended segment that runs antiparallel to the final strand (β2) of the seven-strand central parallel β-sheet. The additional five residues between strand e4 (β4) and helix hE (αD) fold into a crevice at the top of Fn2 with Tyr-268 OH forming a hydrogen bond with Val-315 O in the segment preceding strand e6 (β6). Just before Val-315 is an insertion of 12 residues forming a loop that is inserted between Fn1 and Fn2. Between strands e6 (β6) and e7 (β16) are two insertions. The first is 134 residues and forms the PA domain discussed below. The second is six residues and forms a loop that interacts with Fn1 and Fn2. Eight residues are inserted between helices hF (αI) and hG (αJ) that add an additional turn to the former helix. Similarly eight residues with sequence YDEDEKAY are inserted between helices hG (αJ) and hH (αK) to form a short β-hairpin (strands β19 and β20, on back of protease domain as positioned in Fig. 2 A). This very acidic hairpin inserts into a basic pocket formed by Lys-421 from the PA domain, Lys-470 and Arg-479 of the protease domain, and Lys-632 of Fn1.
PA Domain. The PA domain is inserted between strands e6 and e7 (strands β6 and β16 in the protease domain). Based on sequence analysis PA domains (21, 22) are reported in several other subtilases including cucumisin (23), lactococcal cell wall protease (24), some zinc endopeptidases, e.g., yeast aminopeptidase Y (25), and the human transferrin receptor (TfR, Protein Data Bank ID code 1CX8) (26). The only structural data for a PA domain is from TfR. Although that protein has a metalloprotease fold, it has no enzymatic activity unlike SCP.
Comparison of the structures of the apical domain of TfR (26) and the SCPBw PA domain is shown in Fig. 2B. The Cαs of these proteins can be superimposed over 116 residues with an RMSD of 3.3 Å despite only 23% sequence identity. The PA domain has more extensive regular secondary structure than the apical domain of TfR with loops in TfR becoming β-strands in the PA domain forming a mixed topology seven-strand barrel. Inserted between adjacent barrel parallel strands β11 and β13 is a loop containing the short β12 strand. This strand forms main-chain hydrogen bonds with the twisted hairpin formed by strands β17 and β18 of the protease domain (see Fig. 1 A). As discussed later, we hypothesize that it is the release of this hairpin that is important in forming a functioning active site.
There are three helices in the PA domain. The nine-residue αH helix is across the end of the barrel nearest the active site in the protease domain. Against the adjacent parallel β-strands are the very short αF helix and the 10-residue αG helix. Just before the αG helix is one of two Arg-Gly-Asp (RGD) sequences in SCPB.
Fn Domains. Fn1, Fn2, and Fn3, like all Fn domains (27), have a core structure of a sandwich formed by a four-strand and a three-strand antiparallel β sheet. Looking at the common core, the Cαs of these domains all can be superimposed on other examples of this fold with RMSDs of 2.5–3.6 Å despite sequence identities <15%. Similar structural and sequence agreement are seen in superimposing Fn1, Fn2, and Fn3 on each other. Drawings of Fn1, Fn2, and Fn3 superimposed on their nearest structural homologs are presented in Fig. 2 C–E.
Fn1 and Fn3 are the smaller Fn domains, having 128 and 101 residues, respectively; Fn2 is the largest with 215 residues. The overall shape of an Fn domain is a 25-Å-diameter cylinder with connecting polypeptide loops emerging from opposite ends. Both Fn1 and Fn3 have long loops extending from the carboxyl-terminal ends of the domains; Fn2 has extremely small loops at the carboxyl-terminal end of the domain. For the amino-terminal end of the domains, Fn3 has significantly smaller loops than either Fn1 or Fn2. Thus in the region of the active site of the protease domain, the ends of both Fn1 and Fn2 are more elaborate, facilitating interdomain interactions. For example, the loop between strand β38 and β39 of Fn2 (at the top of Fig. 2D) inserts into a groove at the bottom of the protease domain and forms a number of hydrogen bonds. The hydrophilic interface between Fn2 and the protease domain contrasts with the interface between Fn1 and the protease domain. That interface is primarily hydrophobic with the notable exception of Asp-631 and Glu-687 of Fn1 and Arg-568 of the protease domain, which are buried.
Significant features of Fn2 are the three loops extending from the side of the domain below the entrance to the active site (see Fig. 1). The first side loop (loop A) is from Tyr-733 through Asn-757. This 28-residue Ω-shaped loop forms a concave hydrophilic cylindrical surface 23 Å wide and 14 Å high just below the active site of the protease domain, suggesting a possible role in binding substrate. The largest of these side loops (loop B, 55 residues from Glu-764 through Tyr-818) can be divided into two halves. The second half of loop B (Thr-797 through Tyr-818) forms a narrow twisted Ω-shaped loop perpendicular to the bottom of loop A and makes a number of ionic and hydrogen-bond interactions with strands β44 and β45 of Fn3. The first half of loop B (Glu-674 through Ile-793) forms a J-shaped loop the bend of which makes extensive hydrophobic interactions with strands β48, β49, and β50 of Fn3. Between loops A and B is a 14-Å × 7-Å cavity whose floor is formed by strands β44, β49, and β50 of Fn3. The cavity is lined with hydrophilic groups with Arg-961 and Arg-963 at the base. It is tempting to speculate that this cavity participates in binding carbohydrate. Behind the upper corner of this cavity under loop B is the third side loop, loop C. This small loop (Asn-824 through Asn-828) forms part of a calcium binding site (see Fig. 2D) with Asp-826 and Asp-830 serving as counterions. This calcium-binding site is not an EF-hand site but rather is between two adjacent β-strands. Such sites most frequently occur in cell adhesion, receptors, and membrane proteins (28, 29).
There are four other small molecule binding sites in SCPBw (see Fig. 2). A citrate from the crystallization solution is found at the interface between Fn2 and the protease domain. The one-carboxylate is exposed; the two-carboxylate interacts with Lys-868; and the three-carboxylate interacts with Asp-312, which in turn interacts with Arg-840. This finding suggests that the three-carboxylate is protonated, which is consistent with the pH of the crystallization solution. Two other citrates are found between the protease and PA domains under the β12-β17-β18 mixed sheet. An acetate is found between the protease domain and Fn1. The stabilizing effect of these small molecules may be key in crystallization.
Discussion
As mentioned above, residues 185–205 are disordered in the crystal structure of SCPBw, which is important because they include His-193 of the active-site triad (7, 12). On the basis of structural homology His-193 should form hydrogen bonds with Ser-512 and Asp-130. The mutation of these latter residues to alanines could contribute to the mobility of this segment. In addition, the right side of the protease domain is twisted almost 17° when compared with Bacillus mesentericus subtilisin. This twist results in the Cα of the active-site serine (Ala-512) sliding 3.8 Å away from the central β-sheet. This motion rotates the path of the active site cleft of the protease domain by ≈60° when compared with the structures of other subtilisins.
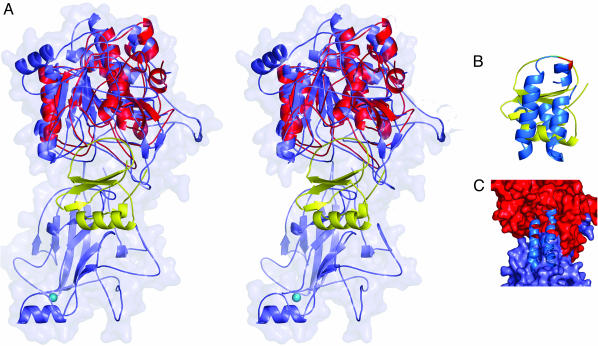
A key question is how does SCPB recognize and cleave C5a. It had been postulated that PA domains are involved in substrate recognition (22). This is not the case in SCPB because of its position relative to the active site. Analysis using the structures of C5a (Protein Data Bank ID code 1KJS) (30–32) and the B. mesentericus subtilisin–eglin C complex (Protein Data Bank ID code 1MEE) (33, 34) indicated that it was not possible to dock C5a into the SCPBw active-site cleft without significant structural changes by one or both molecules (Fig. 3). Fig. 3A shows the result of superimposing the subtilisin–eglin C complex onto the protease domain. In Fig. 3B the structure of C5a is superimposed on that of eglin C by using the region immediately surrounding the cleavage site. Using this chain of superpositions one can model the SCPB–C5a complex shown in Fig. 3C, which has serious steric clashes. The most significant of these occur with Fn2 and the β17 (e8)–β18 (e9) hairpin of the protease domain.
Fig. 3.
Modeling of the SCPB–C5a complex. (A) Stereo ribbon drawing of superposition of the protease domain and Fn2 of SCPBw (light blue with van der Waals surface) with B. mesentericus subtilisin (red)–eglin C (yellow) complex. (B) Superposition of C5a (green) onto eglin C (yellow) using residues around the cleavage site to determine orientation. (C) Model of the SCPB–C5a complex using results in A and B. Protease domain and Fn2 are shown as van der Waals surface in red and light blue, respectively; C5a is blue.
In subtilisin the e8–e9 hairpin is rotated nearly 90° and held in place by several hydrogen bonds with residues in the hC helix, which contains the active-site histidine (see Fig. 1). In SCPBw the β17–β18 hairpin is held in place by hydrogen bonds between the peptide backbones of β17 and β12 of the PA domain. The PA domain is mobile. For the B chain, which has the fewer intermolecular contacts, the average Cα thermal parameter for the PA domain is 32 Å2 more that that of the A chain. A similar mobility was reported for the homologous apical domain of TfR (35). If the stabilizing hydrogen bonds between β12 of the PA domain and β17 of the protease domain were to break, the β17–β18 hairpin could pop up and be able to help the disordered 21-residue loop of SCPBw containing the active-site His-193 to order.
We hypothesize that there is a conformational trigger in the interaction of SCP from GAS and GBS with a cellular receptor. Near the active site are two RGD sequences, which in other proteins have been shown to bind integrins (see Fig. 1) (36). The role of the RGD sequences in binding to Hep2 epithelial cells was evaluated by using mutant forms of full-length SCPA protein (1,164 residues). Single site mutations in which the aspartates in these sequences were replaced by alanines individually and jointly were constructed in the WT scpA gene. Conversion of either aspartate to alanine reduced binding to epithelial cells by >50%; conversion of both aspartates to alanines eliminated binding to Hep2 cells (see Fig. 7, which is published as supporting information on the PNAS web site). Interaction of these mutant proteins to fibronectin was also reduced relative to the WT protein but differences were not statistically significant.
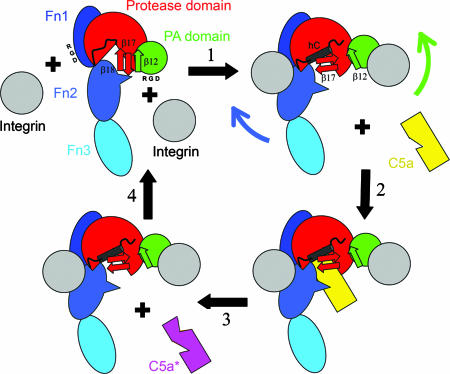
One RGD is at the end of the loop between strand β10 and helix αG of the PA domain (see Figs. 1, 2, 5, and 6). At the nearest point this loop is 8 Å from the β17–β18 hairpin. Integrin binding (step 1 in Fig. 4) to this RGD sequence would disrupt the hydrogen bonds between strands β12 and β17, freeing the β17–β18 hairpin. The second RGD sequence is at the very end of Fn1. Integrin binding to this site would force the C-terminal end of Fn1 away from the protease domain, which would pull Fn2 with its loop A away from the active site, opening it up for C5a binding. Because SCPB is active in the absence of other proteins these domain motions must be able to occur spontaneously. However, we hypothesize that binding to cellular receptors alters this equilibrium and increases the activity of SCPB.
Fig. 4.
Cartoon showing hypothesized effect of integrins (gray) on SCPB structure. Domains of SCPW are indicated and colored as in Fig. 1. Positions of RGD sequences in PA domain and Fn1 are indicated as are the β12, β17, and β18 strands. In step 1, binding of integrins to RGD sequences disrupts the β12–β17 interactions, allowing the β17–β18 hairpin to rotate and promote formation of helix hC. Fn2 is pulled back, making space for C5a binding in step 2. In step 3, C5a is proteolyzed. In step 4, the integrins dissociate regenerating free SCPB.
The surface of streptococci, what the host sees, is far from smooth. Supercoiled helices of the M protein extend out 600 Å; teichoic acids extend up to 200 Å. SCPBw is ≈110 Å in length plus the four 17-residue Pro, Gly-rich hydrophilic repeats at the C-terminal end of SCPB. The structure of these 68 residues is unknown. If these residues are helical they could push the N-terminal 100 Å farther from the cell surface (see Fig. 1 A). If the segments are extended they can push the N-terminal twice as far. It appears likely that the stalk formed by these repeats places the RGD sequences and active site at the outer edge or beyond the carpet of antigenic carbohydrates where they can interact with the host.
Supplementary Material
Acknowledgments
We thank Edward Hoeffner (University of Minnesota) and Brian O'Mara (Wyeth Research) for superb technical assistance. This work was supported by National Institutes of Health Grants R01 AI57585 (to C.A.E.), R01 AI50607 (to D.H.O.), and R01 AI20016 (to P.P.C.). Diffraction data were collected by using the facilities of beamline X25 of the National Synchrotron Light Source at Brookhaven National Laboratory. Computational facilities were provided by the Basic Sciences Computer Laboratory of the Minnesota Supercomputing Institute at the University of Minnesota.
Author contributions: P.P.C., D.H.O., and C.A.E. designed research; C.K.B., Z.-Y.G., S.S.P., L.A.W., and C.A.E. performed research; Z.-Y.G., Y.V.M., P.P.C., S.B.O., and C.A.E. contributed new reagents/analytic tools; C.K.B., D.H.O., and C.A.E. analyzed data; and C.K.B., P.P.C., D.H.O., and C.A.E. wrote the paper.
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: Fn, fibronectin type III; GAS, group A Streptococcus; GBS, group B Streptococcus; PA domain, protease-associated domain; RGD, Arg-Gly-Asp; RMSD, rms deviation; SCP, streptococcal C5a peptidase; SCPB, SCP from GBS; TfR, human transferrin receptor.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 1XF1).
References
- 1.Walport, M. J. (2001) N. Engl. J. Med. 344, 1140–1144. [DOI] [PubMed] [Google Scholar]
- 2.Walport, M. J. (2001) N. Engl. J. Med. 344, 1058–1066. [DOI] [PubMed] [Google Scholar]
- 3.Hill, H. R., Bohnsack, J. F., Morris, E. Z., Augustine, N. H., Parker, C. J., Cleary, P. P. & Wu, J. T. (1988) J. Immunol. 141, 3551–3556. [PubMed] [Google Scholar]
- 4.Cheng, Q., Stafslien, D., Purushothaman, S. S. & Cleary, P. (2002) Infect. Immun. 70, 2408–2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beckmann, C., Waggoner, J. D., Harris, T. O., Tamura, G. S. & Rubens, C. E. (2002) Infect. Immun. 70, 2869–2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cleary, P. (1998) in Handbook of Proteolytic Enzymes, eds. Barrett, A. J., Rawlings, N. D. & Woessner, J. F. (Academic, New York), pp. 308–310.
- 7.Anderson, E. T., Wetherell, M. G., Winter, L. A., Olmsted, S. B., Cleary, P. P. & Matsuka, Y. V. (2002) Eur. J. Biochem. 269, 4839–4851. [DOI] [PubMed] [Google Scholar]
- 8.Janulczyk, R. & Rasmussen, M. (2001) Infect. Immun. 69, 4019–4026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schneewind, O., Fowler, A. & Faull, K. F. (1995) Science 268, 103–106. [DOI] [PubMed] [Google Scholar]
- 10.Chen, C. C. & Cleary, P. P. (1990) J. Biol. Chem. 265, 3161–3167. [PubMed] [Google Scholar]
- 11.Siezen, R., de Vos, W., Leunissen, J. & Dijkstra, B. (1991) Protein Eng. 4, 719–737. [DOI] [PubMed] [Google Scholar]
- 12.Stafslien, D. K. & Cleary, P. P. (2000) J. Bacteriol. 182, 3254–3258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cleary, P. P., Matsuka, Y. V., Huynh, T., Lam, H. & Olmsted, S. B. (2004) Vaccine 22, 5332–5341. [DOI] [PubMed] [Google Scholar]
- 14.Hendrickson, W. A., Horton, J. R. & LeMaster, D. M. (1990) EMBO J. 9, 1665–1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Otwinowski, Z. & Minor, W. (1997) Methods Enzymol. 276, 307–326. [DOI] [PubMed] [Google Scholar]
- 16.Terwilliger, T. (2004) J. Sync. Rad. 11, 49–52. [DOI] [PubMed] [Google Scholar]
- 17.Levitt, D. G. & Banaszak, L. J. (1993) J. Appl. Crystallogr. 26, 736–745. [Google Scholar]
- 18.Jones, T. A. (1978) J. Appl. Crystallogr. 11, 268–272. [Google Scholar]
- 19.Brünger, A. T., Kuriyan, J. & Karplus, M. (1987) Science 235, 458–460. [DOI] [PubMed] [Google Scholar]
- 20.Holm, L. & Sander, C. (1993) J. Mol. Biol. 233, 123–186. [DOI] [PubMed] [Google Scholar]
- 21.Luo, X. & Hofmann, K. (2001) Trends Biochem. Sci. 26, 147–148. [DOI] [PubMed] [Google Scholar]
- 22.Mahon, P. & Bateman, A. (2000) Protein Sci. 9, 1930–1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yamagata, H., Masuzawa, T., Nagaoka, Y., Ohnishi, T. & Iwasaki, T. (1994) J. Biol. Chem. 269, 32725–32731. [PubMed] [Google Scholar]
- 24.Siezen, R. J. (1999) Antonie Van Leeuwenhoek 76, 139–155. [PubMed] [Google Scholar]
- 25.Nishizawa, M., Yasuhara, T., Nakai, T., Fujiki, Y. & Ohashi, A. (1994) J. Biol. Chem. 269, 13651–13655. [PubMed] [Google Scholar]
- 26.Lawrence, C. M., Ray, S., Babyonyshev, M., BGalluser, R., Borhani, D. W. & Harrison, S. C. (1999) Science 286, 779–782. [DOI] [PubMed] [Google Scholar]
- 27.Bork, P., Downing, A. K., Kieffer, B. & Campbell, I. D. (1996) Q. Rev. Biophys. 29, 119–167. [DOI] [PubMed] [Google Scholar]
- 28.Glusker, J. P. (1991) Adv. Protein Chem. 42, 1–76. [DOI] [PubMed] [Google Scholar]
- 29.McPhalen, C. A., Strynadka, N. C. & James, M. N. (1991) Adv. Protein Chem. 42, 77–144. [DOI] [PubMed] [Google Scholar]
- 30.Zhang, X., Boyar, W., Toth, M. J., Wennogle, L. & Gonnella, N. C. (1997) Proteins 28, 261–267. [DOI] [PubMed] [Google Scholar]
- 31.Zuiderweg, E., Nettesheim, D., Mollison, K. & Carter, G. (1989) Biochemistry 28, 172–185. [DOI] [PubMed] [Google Scholar]
- 32.Williamson, M. & Madison, V. (1990) Biochemistry 29, 2895–2905. [DOI] [PubMed] [Google Scholar]
- 33.Dauter, Z., Betzel, C., Genov, N., Pipon, N. & Wilson, K. S. (1991) Acta Crystallogr. B 47, 707–730. [DOI] [PubMed] [Google Scholar]
- 34.Bode, W., Papamokos, E. & Musil, D. (1987) Eur. J. Biochem. 166, 673–692. [DOI] [PubMed] [Google Scholar]
- 35.Cheng, Y., Zak, O., Aisen, P., Harrison, S. C. & Walz, T. (2004) Cell 116, 565–576. [DOI] [PubMed] [Google Scholar]
- 36.Ruoslahti, E. & Pierschbacher, M. D. (1986) Cell 44, 517–518. [DOI] [PubMed] [Google Scholar]
- 37.Keep, N. H., Barnes, M., Barsukov, I., Badii, R., Lian, L. Y., Segal, A. W., Moody, P. C. & Roberts, G. C. (1997) Structure (London) 5, 623–633. [DOI] [PubMed] [Google Scholar]
- 38.Hung, D., Knight, S. & Hultgren, S. J. (1999) Mol. Microbiol. 31, 773–783. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.