Abstract
The PIK3CA gene encoding the p110α subunit of Class IA phosphatidylinositol 3-kinases (PI3Ks) is frequently mutated in human tumors. Mutations in the PIK3CB gene encoding p110β, the only other widely expressed Class IA PI3K, have not been reported. We compared the biochemical activity and transforming potential of mutant forms of p110α and p110β in a human mammary epithelial cell system. The two most common tumor-derived alleles of p110α, H1047R and E545K, potently activated PI3K signaling. Human mammary epithelial cells expressing these alleles grew efficiently in soft agar and as orthotopic tumors in nude mice. We also examined a third class of mutations in p110α, those in the p85-binding domain. A representative tumor-derived p85-binding-domain mutant R38H showed modestly reduced p85 binding and weakly activated PI3K/Akt signaling. In contrast, a deletion mutant lacking the entire p85-binding domain efficiently activated PI3K signaling. When we constructed in p110β a mutation homologous to the E545K allele of p110α, the resulting p110β mutant was only weakly activated and allowed minimal soft-agar growth. However, a gene fusion of p110β with the membrane anchor from c-Src was highly active and transforming in both soft-agar and orthotopic nude mouse assays. Thus, although introduction of activating mutations from p110α at the corresponding sites in p110β failed to render the enzyme oncogenic in human cells, the possibility remains that other mutations might activate the β isoform.
Keywords: orthotopical tumor, PIK3CA, PIK3CB, Akt, oncogene
The phosphoinositide 3-kinases (PI3Ks) are a family of lipid kinases defined by their ability to phosphorylate the 3′-OH of phosphoinositides. Three classes of mammalian PI3Ks have been characterized and shown to differ in their expression patterns, activation mechanisms, and substrate specificities (1, 2). The Class I PI3Ks have been most widely investigated, because it is these isoforms that are generally coupled to extracellular stimuli. This class is further divided into Class IA enzymes activated by receptor tyrosine kinases (RTKs) and Class IB enzymes regulated by G protein-coupled receptors. Although PI3Ks have been implicated in the regulation of a wide variety of cellular processes, only Class IA enzymes are clearly implicated in human cancers (3).
Class IA PI3Ks consist of a p110 catalytic subunit and a regulatory subunit. Currently, three isoforms (α, β, and δ) exist for the p110 subunit, whereas there are seven known proteins for the regulatory subunit: p85α, p85β, and p55γ and their splicing variants. The p85 subunit (or the shorter isoform) keeps the p110 subunit in a stable but low activity state in quiescent cells (4). Upon stimulation by growth factors, the p85 subunit recruits the catalytic subunit to the membrane through the interaction of its SH2 domains and the phosphotyrosine motifs on activated RTKs. Subsequently, the activated p110 catalytic subunit converts phosphatidylinoditol-4, 5-biphosphate to phosphatidylinoditol-3, 4, 5-triphosphate at the membrane, providing docking sites for signaling proteins with pleckstrin-homology domains. Among these proteins are the protein Ser-Thr kinase Akt (also called protein kinase B or PKB) and the phosphoinositide-dependent kinase 1 (PDK1), which come into proximity with each other through PIP3 association. PDK1 phosphorylates and activates Akt, which subsequently phosphorylates a series of other signaling proteins that affect cell growth, proliferation, survival, and transformation.
Deregulation of the Class IA PI3K–Akt pathway is common in human tumors. PTEN, a lipid phosphatase which dephosphorylates the 3′-OH of phosphoinositides and, thus, counteracts the action of PI3Ks, is mutated or silenced in many human malignancies, including breast cancer, prostate cancer, and glioblastoma (5–7). Akt, the major downstream effector of PI3K, has been found amplified or overexpressed in breast, ovarian, and thyroid cancers (8, 9). Recently, mutations have been identified at a high frequency in the PIK3CA gene encoding the p110α subunit in many types of human cancers (10–13). So far, these revealed mutations are exclusively somatic missense mutations and mostly clustered at hot spots within the helical and catalytic domains. Less frequent mutations within the p85-binding domain have also been identified but not characterized. Interestingly, mutations in the PIK3CB gene, encoding p110β, the only other widely expressed catalytic subunit of Class IA PI3Ks, have not yet been reported. However, the mutational analysis of p110β has been performed only on a very limited number of tumors.
In this article, we compare the biochemical activity and transforming potential for several mutant forms of p110α and p110β by using a genetically engineered human mammary epithelial cell (HMEC) transformation system. Our data suggest that p110α is potently activated by the “hotspot” mutations found in human tumors, whereas tumor mutations in the p85-binding domain are only weakly activating. Furthermore, we demonstrate that it may be more difficult to activate p110β than p110α by missense mutation, but p110β, nonetheless, possesses considerable tumorigenic potential.
Materials and Methods
Plasmid Construction. To construct hemagglutinin (HA)-tagged alleles of p110α and p110β, cDNA clones of human PIK3CA and PIK3CB (obtained from the FLEXGene Repository of the Harvard Institute of Proteomics) were subcloned into pBabepuro by PCR with primers containing HA-epitope coding sequence with subsequent restriction and ligation. To make amino-terminal myristoylated alleles of p110 isoforms, a coding sequence for the myristoylation domain of c-Src was fused in-frame with a HA-tagged PIK3CA or PIK3CB sequence. Other mutations were made by PCR-mediated deletion or by using the QuickChange site-directed mutagenesis kit (Stratagene). Primers used are described in Supporting Materials and Methods, which is published as supporting information on the PNAS web site. All of the constructs were verified by sequencing.
Immunoprecipitation and Immunoblotting. Cells were starved in DMEM/F-12 (Invitrogen) without growth factors for 1.5 h, washed twice with cold PBS, and lysed in Nonidet P-40 lysis buffer as described in ref. 14. For HA-IPs, an aliquot (≈450 μg of protein) from each lysate was mixed with anti-HA affinity matrix (Cat. 11815016, Roche Diagnostics) overnight at 4°C and washed four times with cold 1% Nonidet P-40 in PBS. The precipitated proteins were subsequently separated by 6% SDS/PAGE. In parallel, aliquots of the same cell lysates (≈20 μg of protein) were separated by 8% SDS/PAGE. Proteins were then transferred to nitrocellulose membranes (Cat. 10402580, Schleicher & Schuell). After the membranes were blocked with 5% milk of TBS/T [TBST for film exposure; TBS for Odyssey Scanner (Li-Cor, Lincoln, NE)], they were incubated with primary antibodies overnight at 4°C. Anti-p110α, anti-phospho-Akt (Ser-473), and anti-Akt antibodies were obtained from Cell Signaling Technology (Beverly, MA). Anti-pan-p85 antibody was obtained from Upstate Biotechnology (Lake Placid, NY). Anti-HA antibody (HA11) was obtained from Covance (Princeton, NJ). Anti-vinculin antibody was obtained from Sigma. After rinsing with TBS-0.2% Tween 20, the membranes were incubated with secondary antibodies diluted in 3% milk of TBST for 50 min at room temperature. The bands were detected by using chemiluminescence (PerkinElmer Life Sciences) or Odyssey Scanner according to the manufacturer's instructions.
Stable Cell Lines. HMEC cell lines stably expressing various alleles of p110α and p110β were generated by infection of retroviruses carrying these p110 isoforms encoding genes or other specific genes that were generated as described in ref. 14. After infection, successfully transduced polyclonal cell populations were obtained by selection with puromycin (0.5 μg/ml) or other appropriate drug to pools of stable clones. The culture conditions of HMECs were as described in ref. 14.
Soft-Agar Colony-Formation Assays. For soft-agar colony-formation assays, 5 × 104 cells were plated into each 60-mm plate with a bottom layer of 0.6% agar and a top layer of 0.3% agar. Colonies were counted after 3 weeks of incubation and analyzed as described in ref. 14.
s.c. and Orthotopic Tumorigenicity Assays. Six- to eight-week-old immunocompromised mice (BALB/cAnNTac-Foxn1nuN9, Taconic Farms, and CanN.Cg-Foxn1nu/Crl, Charles River Laboratories) were γ-irradiated with one dose of 400 rad before injections. For s.c. tumor-formation assay, cells (4 × 106) were resuspended in 50 μl of PBS and 50 μl of Matrigel (Becton Dickinson) and injected by using a 25-gauge needle s.c. into nude mice anesthetized by inhalation of isoflurane. For orthotopic tumorigenicity assays, mice were anesthetized with avertin i.p., and the axial mammary fat pad regions were exposed by incisions for injection. Cells (4 × 106) were resuspended in 50 μl of PBS and 50 μl of Matrigel (Becton Dickinson) and injected by using insulin syringes with 28-gauge needles. The condition of the mice and tumor development were monitored daily. Mice were killed when a single tumor reached 1.5 cm in diameter or after 2 months of monitoring. Tumors were fixed in 10% buffered formalin (SF100–4, Fisher) for histological examinations.
Results
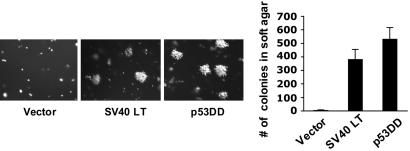
Inactivation of p53 Is Sufficient to Replace SV40 Large T Antigen (LT) in the Transformation of HMECs. In our prior work, we found that the PI3K pathway is the major target of SV40 small t antigen in HMEC transformation. As a result, we were able to effect the transformation of HMECs through the activation of PI3K in the presence of hTERT, SV 40 LT, and an elevated myc level (14). Before studying tumor-derived mutations in PI3K, we wished to create a human epithelial transformation system that was independent of viral oncogenes. We chose to use our previously characterized high-passage HMEC cells population-doubling 130 (14) for our study, because these cells exhibit elevated levels of endogenous c-myc and, thus, require one less genetic manipulation. SV40 LT is known to bind and inactivate, among other target proteins, the retinoblastoma (pRB) and p53 tumor suppressors (15). It has been demonstrated that inactivation of both p53 and pRB pathways is required for human cell transformation (16). Consistent with prior observations (17, 18), the hTERT-immortalized HMECs used in our study have lost the expression of p16INK4 (data not shown). Thus, one immediate question raised was whether loss of p53 could replace SV 40 LT to transform these hTERT-immortalized HMECs lacking functional p16INK4A. To inactivate p53, we introduced into HMEC-hTERT cells a dominant negative mutant allele of p53 (p53DD) that is known to disrupt the sequence-specific DNA-binding activity of wild-type p53 (19). We next introduced the activated allele of bovine p110α, myr-Flag-p110α, used in our previous study (14) and tested the resulting cells for their ability to form colonies in soft agar. Satisfyingly, p53DD could fully replace SV40 LT, in combination with hTERT, high myc level, and myr-Flag-p110α, to efficiently transform HMEC, as measured by the anchorage-independent (AI) growth (Fig. 1).
Fig. 1.
Substitution of SV40 LT with a dominant negative allele of p53 in the transformation of HMECs. p53DD sufficed to replace SV40 LT and promoted soft-agar colony growth of HMEC in combination with hTERT, elevated c-myc, and myristoylated bovine p110α. The assay was carried out as described in Materials and Methods, and the means (±SD) for three experiments are shown.
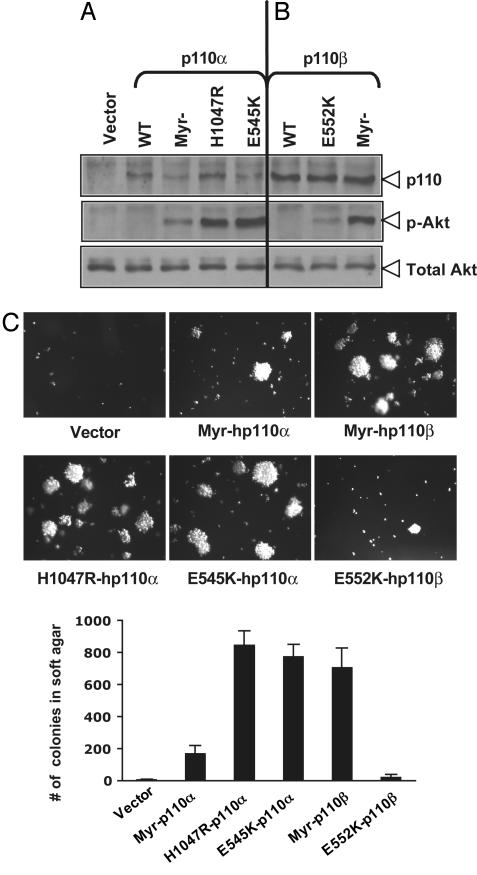
Expression of the Two Most Frequently Occurring Tumor Mutant Alleles of p110α, E545K and H1047R, Induces Oncogenic Transformation in HMECs. The HMEC transformation system devoid of viral oncogenes described above still requires PI3K activation for transformation (Fig. 1), thus providing us with an ideal model system to evaluate the oncogenic potential of mutant alleles of PIK3CA found in human cancers. To begin characterization of PIK3CA mutants, we first introduced the E545K and H1047R mutations, the two most commonly found mutations in human cancer, into the wild-type human PIK3CA gene. We also generated a constitutively activated allele of human p110α (hp110α) by the addition of a myristoylation motif at the amino terminus of hp110α, termed myr-hp110α. All three alleles of hp110α were stably introduced into HMECs in the presence of hTERT, p53DD, and a high level of myc. Protein expression levels were determined by immunoblotting with anti-HA antibody, because all three alleles of hp110α are HA-epitope tagged (Fig. 2A). Both the E545K and H1047R hotspot mutants have been described as activating alleles of p110α in colon cancer cells (10, 20) or avian cells expressing the avian version of p110α mutated in the corresponding sites of these two hotspots (21). To evaluate the activities of mutant hp110α alleles, HMEC cells expressing these various alleles of hp110α were starved in basal medium free of serum and growth factors for 1.5 h, and the levels of phosphorylated Akt in each cell type were determined by using immunoblotting with phospho-Akt-specific antibodies. The myristoylated hp110α induced constitutive and growth-factor-independent activation of Akt signaling in HMECs (Fig. 2A), although the activation level is not as high as that of myristoylated bovine p110α, probably because of its reduced expression level (data not shown). Notably, although all hp110α alleles expressed at relatively low levels, both E545K and H1047R hotspot mutants induced much higher phosphorylation of Akt than that of myr-hp110α (Fig. 2A).
Fig. 2.
Soft-agar colony formation of HMECs expressing mutant alleles of human p110 isoforms. (A) Relative expression levels of hp110α mutant alleles and their activation on Akt signaling in HMECs. Cells were starved in basal medium, and lysates were prepared as described in Materials and Methods for immunoblot analysis with an anti-HA antibody and anti-phospho-Akt(Ser-473) or total Akt antibodies. (B) Expression of hp110β mutant alleles and their activation on Akt signaling in HMECs were performed as described in A. (C) Soft-agar colony growth of HMEC-hTERT-p53DD expressing various alleles of hp110α or hp110β. The assay was carried out as described in Materials and Methods, and the means (±SD) for three experiments are shown.
To examine the transforming activities of these tumor mutants p110α, we tested these HMEC cells for their ability to grow in an anchorage-independent manner. Cells expressing E545K and H1047R hotspot tumor mutants grew robustly in soft agar as colonies, whereas cells expressing myr-hp110α were less potent in AI growth (Fig. 2C). Thus, the transforming potencies of these cells expressing different activating mutant alleles of p110α seem to correlate well with their kinase activities, as measured by phospho-akt levels (Fig. 2 A and C). Taken together, our results demonstrated that the naturally occurring activating mutations in hp110α are highly active and transforming.
HMECs Expressing Both E545K and H1047R Generate Invasive Orthotopic Tumors in Animal Hosts. We and others have reported that the combination of hTERT, SV40 LT, oncogenic Ras, and SV40 small t antigen (st) (or myr-p110α) confers on HMEC cells the ability to form s.c. tumors in nude mice (14, 22). We have now transformed HMECs with exclusively cellular genes, because we have replaced SV40 LT with p53DD and SV40 st with activating alleles of p110α. Because the addition of oncogenic Ras, in addition to an active PI3K allele, was shown to be required for tumor formation in HMECs (14, 22), we also introduced H-rasV12 into HMECs expressing hTERT and p53DD in combination with myr-hp110α, E545K, or H1047R, to assess whether the mutant alleles of p110α could induce tumor formation in animals. We initially injected these cells s.c. into the flank regions of nude mice. Tumors were formed from all of the injections of HMECs expressing E545K or H1047R, but only one tumor was found from injections of HMECs expressing myr-hp110α at 8 weeks after injection (Table 1). This result, again, suggests that the level of PI3K activity displayed by any given mutant appears to correlate its ability to induce the tumors in HMECs.
Table 1. Formation of tumors in nude mice.
| HMEC expressing hTERT, p53DD, RasV12
|
No. of tumors/injections
|
|
|---|---|---|
| s.c. | Fat pad | |
| Vector | 0/6 | 0/6 |
| Myr-p110α | 1/6 | N/D |
| E545K | 6/6 | 8/8 |
| H1047R | 6/6 | 8/8 |
| Myr-p110β | N/D | 6/6 |
For each injection, 4 × 106 of indicated cell populations mixed with an equal volume of matrigel in a final volume of 100 μl were injected s.c. or orthotopically into axial mammary fat pads of nude mice. Mice were killed when tumor size reached 1.0-1.5 cm in diameter or after 2 months of monitoring. N/D, not determined.
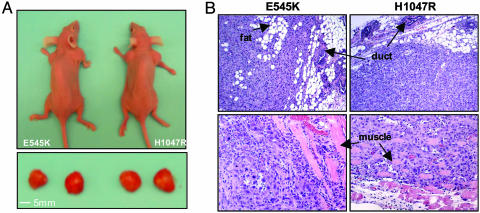
To examine breast tumor formation driven by mammary epithelial cells in their physiologic environment, we repeated the tumor-formation experiments by injecting HMECs expressing E545K or H1047R orthotopically in axial mammary fat pads. Tumors developed in 100% of cases (eight of eight) for HMEC lines expressing either E545K or H1047R and reached a size of 1.0–1.2 cm in diameter within 8 weeks after injection (Fig. 3A and Table 1), consistent with the findings in the s.c. tumor experiments. Whereas the s.c. tumors were confined to the site of injection, both E545K and H1047R mutant alleles of p110α promoted the formation of high-grade orthotopic tumors that infiltrated into neighboring fat and muscle tissue layers (Fig. 3B). However, metastatic spread to other sites, such as lung, bone, and ovaries, was not observed.
Fig. 3.
Orthotopic tumor formation of HMECs expressing mutant alleles of human p110α.(A) HMECs expressing E545K or H1047R hp110α in combination with hTERT, p53DD, and HrasV12 were injected into axial mammary fat pads as described in Materials and Methods. Orthotopic tumors developed and reached a 1-cm diameter within 8 wks after injection. (B) Histology of orthotopic tumors derived from HMECs expressing E545K or H1047R mutant hp110α. All tissues were stained with hematoxylin and eosin. The tumors were poorly differentiated and infiltrated through adipocytes and muscle tissue layers (indicated by arrows).
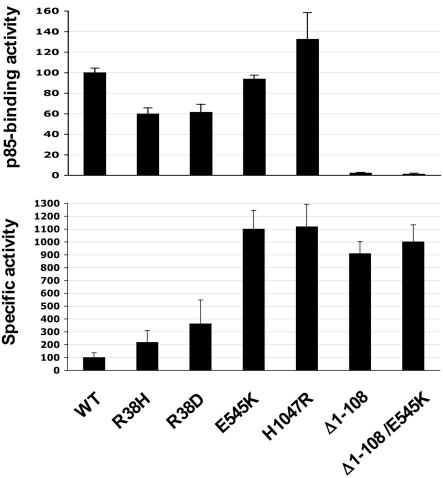
Effects of p85-Binding Domain Mutations on p110α Activity. As noted above, both H1047R and E545K tumor mutant alleles of p110α exhibited much higher constitutive activities than that of the myristoylated allele of hp110α, suggesting that mechanism(s) other than stable membrane localization may be involved in the oncogenic activation of p110α. To gain further insight into the mechanism of the activation of oncogenic p110α, we analyzed a third class of hp110α mutants carrying mutations in the p85-binding domain. The p85 regulatory subunit has been reported both to stabilize the p110 subunit and to inhibit its catalytic activity (4). In all, three mutations in the p85-binding domain were analyzed: R38H, which was identified in two independent human tumors (10), arose through a single base conversion in a conserved residue in all Class IA p110 isoforms (see Fig. 6, which is published as supporting information on the PNAS web site); R38D, constructed to reverse the charge at the R38 residue, in the hope of making a mutant with a more exaggerated phenotype than R38H; and Δ1–108, a deletion mutant lacking the entire p85-binding domain, constructed to analyze the effect of total loss of p85 binding. All these constructs were stably expressed in HMEC cells along with p53DD and hTERT, as described above. The expression levels of R38H, R38D, and the two hotspot mutant alleles of hp110α were reduced moderately when compared with that of wild-type hp110α, as measured by antibodies specific to p110α after immunoprecipitation with anti-HA antibodies (see Fig. 7A, which is published as supporting information on the PNAS web site). The level of Δ1–108 was reduced to a greater extent, presumably because of the total loss of binding and stabilization by the p85 subunit (Fig. 7A). We also assessed the p85 binding and akt activation of these mutants (Fig. 4; and see Fig. 7). We found that the two p85-binding domain mutants, R38H and R38D, displayed modest reductions in their p85-binding abilities, whereas both of the hotspot mutants, E545K and H1047R, retained full p85-binding capacity (Figs. 4 and 7). Moreover, both R38H and R38D mutants showed only slightly enhanced phospho-Akt levels (Figs. 4 and 7A) and failed to induce HMEC to grow as colonies in soft agar (data not shown). However, the Δ1–108 deletion mutant of p110α consistently exhibited an elevated phospho-Akt level in HMEC (Fig. 7), probably because Δ1–108 deletion relieved p110α from the p85's inhibitory effects. Interestingly, the Δ1–108 deletion mutant showed roughly the same level of “specific activity” as the E545K and H1047R alleles (Figs. 4 and 7A).
Fig. 4.
Effects of p85-binding-domain mutations on p110α activity. (Upper) The p85-binding activities of various alleles of hp110α, as indicated. The normalized level of p85-binding activity of each allele was determined by dividing the amount of p85 bound by the amount of each hp110α allele expressed. (Lower) The specific activity of each hp110α allele, determined by normalizing the level of phospho-Akt to the expression level of each hp110α allele, as indicated. Each column reflects the average of three independent experiments, with error bars indicating SD.
These results have two implications: First, the tumor mutants in the R38 residue seem to have little effect on p110α's activity, as measured here. Further work would be required to test their tumorgenicity. Second, the fact that the Δ1–108 allele has the same specific activity as the E545K and H1047R alleles suggests that the effect of the E545K and H1047R mutations was to mitigate or eliminate the negative effects of p85 binding on p110α activity. To further examine this hypothesis, we constructed an additional hp110α mutant, Δ1–108/E545K, that carries a double mutation in which the Δ1-108 deletion was combined with the E545K mutation in p110α, allowing us to directly compare the specific activities of the wild-type and the mutant alleles of hp110α when both lack p85-binding domains. Notably, both deletion mutants were expressed at much lower levels than the full-length alleles, presumably because of lowered stability and, yet, are highly effective in Akt activation (Figs. 4 and 7B). Indeed, the two deletion constructs show similar specific activity (Fig. 4).
Activation of p110βby Myristoylation Is Oncogenic in HMECs. Whereas somatic mutations of p110α occur frequently in human cancers, p110β, the other ubiquitously expressed Class IA PI3K, has not yet been found to be mutant in human cancers, raising the question whether p110β can potentially be oncogenic. In an attempt to activate p110β, we fused human p110β (hp110β) with the amino terminus myristoylation domain to provide constitutive membrane anchorage. In addition, we made a mutation within the helical region of hp110β, E552K, corresponding to the E545K hotspot mutation within hp110α, because this site is conserved among the Class 1A p110 isoforms (Fig. 6). We then stably introduced wild-type, myristoylated, and E552K mutant alleles of hp110β into HMEC in the presence of hTERT and p53DD. Interestingly, protein expression levels of all hp110β alleles in HMECs were consistently higher when compared with that of hp110α alleles, as measured by immunoblot with anti-HA antibodies (see Fig. 2 A and B). The levels of phospho-Akt of cells expressing different alleles of p110β were examined to evaluate the activities of these various p110β alleles. Myr-hp110β induced considerably higher levels of Akt phosphorylation than myr-hp110α (Fig. 2 A and B), perhaps because of its higher expression level. However, the E552K allele of hp110β exhibited only minimum activity, despite its high expression level (Fig. 2B). Transforming activities were determined by measuring the AI growth of cells expressing myr-hp110β or E552K. Consistent with the findings from hp110α alleles that mutants with higher PI3K activity exhibited higher transforming potency, Myr-hp110β induced robust colony formation in HMEC cells plated in soft agar, whereas E552K displayed relatively weak activity (Fig. 2C).
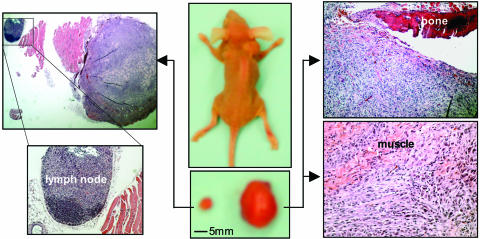
To test whether myr-hp110β can be tumorigenic, we performed orthotopic injections of HMECs expressing myr-hp110β in combination with hTERT, p53DD, and H-rasV12 into mammary fat pads of nude mice. Tumors were formed from all injected sites (six of six) within 8 weeks of implantation (Table 1). Two of the six tumors developed quickly and reached 1.5 cm in diameter in 6 weeks, and the mice bearing the large tumors showed deformed backbones and emaciation (Fig. 5). Both large and small tumors infiltrated into the adjacent scapular bones and muscle layers (Fig. 5). A metastatic spread was identified in a lymph node proximal to one of the small tumors, as shown in Fig. 5.
Fig. 5.
Orthotopic tumor formation by HMECs expressing myristoylated human p110β. HMECs expressing myr-hp110β in combination with hTERT, p53DD, and HrasV12 were injected into axial mammary fat pads as described in Materials and Methods. The tumors were examined as described in Fig. 4. The tumors were also poorly differentiated but showed considerable size heterogeneity. The large tumor showed infiltration through bone and adipocyte muscle tissue layers, and a lymphoma was identified adjacent to the small tumor located in the contralateral axial mammary fat pad, as indicated.
Discussion
One of the major goals of developing defined human cancer models is to achieve oncogenic transformation of normal human cells through genetic changes characteristic of the tumor type being studied (24). Introduction of SV40 LT or coexpression of E6 and E7 papillomaviral oncoproteins has been used to inactivate both pRB and p53 tumor-suppressor pathways in human cell transformation (16, 25). In ridding the HMEC transformation system of viral oncogenes, we found that expression of p53DD in combination with hTERT, high myc, and activation of PI3K is sufficient to transform HMEC cells to grow colonies in soft agar. The need to inactivate p53 and not pRb is presumably because of the loss of p16INK4 expression, which is common in HMEC cells. The dependency of HMEC on the activation of PI3K for the transformation provides us with an excellent system to study the effects of mutational activation of PI3Ks.
Consistent with results for the H1047R and E545K mutants of p110α evaluated in colon (20) and avian (21) cells, we found that, in HMEC cells, these two most commonly mutated alleles of hp110α greatly enhanced Akt phosphorylation and promoted both robust colony formation in soft agar and efficient tumor growth in animal hosts. Furthermore, the orthotopic breast tumors derived from HMECs expressing H1047R or E545K displayed similar phenotypes, with high-grade malignancy and invasion of adjacent tissue layers. However, distant metastatic spread was not observed.
We have shown that both H1047R and E545K mutations drive constitutive, growth-factor-independent PI3K activity. However, the mechanism(s) of their activation is not clear. The p110 catalytic subunit is normally activated when it is recruited to the plasma membrane through docking of its regulatory subunit p85 to the activated growth-factor receptors. But constitutive membrane association of p110α is unlikely to explain the activation of these two tumor mutant alleles of p110α, because membrane anchoring of p110α by myristoylation resulted in a much lower level of PI3K activity compared with the H1047R and E545K mutations. We also investigated an alternative mechanism for the oncogenic activity of the p110α mutants. The commonly accepted model for p110 regulation supposes that most regulatory signals come through its associated p85 subunit, which both stabilizes p110 and inhibits its activity before receptor activation. Indeed, we found that the Δ1–108 mutant of p110α lacking the entire p85-binding domain is poorly expressed but shows a relatively high activity, as judged by Akt phosphorylation. Interestingly, when we normalize Akt phosphorylation for the level of expression of the various p110α alleles, the specific activity of Δ1–108 is similar to that of the two hotspot tumor p110α alleles. Our finding suggests that H1047R and E545K mutations in p110α may have managed to relieve the negative effects of p85 on p110α but not affect the stabilization afforded by p85 binding. Consistent with this hypothesis, we found that activities of the wild-type and E545K mutant p110α alleles are roughly equivalent when we delete the p85-binding domain from each. This idea is supported by a recent study describing a truncated murine p85 mutant that binds but does not inhibit p110, leading to constitutively increased p110 activity (26). Further experimentation will be required to prove this model.
The point mutants in the p85-binding domain of p110α remain something of a puzzle, because the tumor-derived mutation within the p85-binding domain displayed a modest effect on its p85 binding and a small effect on p110α's activity. We are unable to explain what role this mutation might play in the genesis of the tumors. It remains possible that the minimal increase in activity seen in the mutant has some small selective advantage or that the mutation might activate a pathway other than the Akt pathway, which is of importance in another cell type. A similar situation is seen with B-Raf mutations in tumors, where the hotspot mutations are clearly activating, but other sites of mutation can have little effect on activity (27).
We also wished to assess the oncogenic potential of p110β, although no mutations in p110β have been reported in human tumors to date. Our data on the E552K point mutation in p110β showed relatively weak transforming activity in HMECs. However, the E552K point mutant in p110β did, indeed, generate an increase in p110β activity, as judged by Akt phosphorylation. It is possible that this allele could be selected for in the context of partial loss of PTEN function or HER2 overexpression. The relatively high transformation potential of the myr-p110β allele, as measured in soft-agar growth and in orthotopic tumor formation, clearly demonstrates the tumorigenic potential of p110β. In separate work, we have expressed a myristoylated murine p110β allele in mouse prostate and observed hyperplastic prostate growth in the resulting transgenic animals (S. H. Lee and T.M.R., unpublished results). Taken together, our data suggest that the PIK3CB gene should be examined for both point mutations and gene fusions in a larger number of human tumors before it is abandoned as a potential human oncogene.
Supplementary Material
Acknowledgments
We thank L. Cantley, W. Sellers, J. D. Iglehart, and A. Miron for helpful discussions; Q. Yu for help on the animal experiments; and Harvard Institute of Proteomics for reagents. This work was supported by National Institutes of Health Grants P01-CA50661 and CA30002 (to T.M.R.), CA089021 (to T.M.R. and M.F.L.), and Specialized Programs of Research Excellence Grant 5P50CA090381-05 (to J.J.Z. and M.F.L.); a Claudia Barr Award and the Friends Woman's Cancer Program Award (to J.J.Z.); and the Prostate Cancer Foundation (M.F.L.). In compliance with Harvard Medical School guidelines, we disclose the consulting relationships: Novartis Pharmaceuticals, Inc. (T.M.R. and M.F.L.) and Upstate Biotechnology (T.M.R.).
Author contributions: J.J.Z., Z.L., M.F.L., and T.M.R. designed research; J.J.Z., Z.L., L.W., and E.S. performed research; J.J.Z. and Z.L. contributed new reagents/analytic tools; J.J.Z., Z.L., M.F.L., and T.M.R. analyzed data; and J.J.Z., Z.L., M.F.L., and T.M.R. wrote the paper.
Conflict of interest statement: No conflicts declared.
Abbreviations: HA, hemagglutinin; HMEC, human mammary epithelial cell; LT, large T antigen; PI3K, phosphatidylinositol 3-kinase.
References
- 1.Fruman, D. A., Meyers, R. E. & Cantley, L. C. (1998) Annu. Rev. Biochem. 67, 481–507. [DOI] [PubMed] [Google Scholar]
- 2.Parekh, D. B., Katso, R. M., Leslie, N. R., Downes, C. P., Procyk, K. J., Waterfield, M. D. & Parker, P. J. (2000) Biochem. J. 352, 425–433. [PMC free article] [PubMed] [Google Scholar]
- 3.Vivanco, I. & Sawyers, C. L. (2002) Nat. Rev. Cancer 2, 489–501. [DOI] [PubMed] [Google Scholar]
- 4.Yu, J., Zhang, Y., McIlroy, J., Rordorf-Nikolic, T., Orr, G. A. & Backer, J. M. (1998) Mol. Cell. Biol. 18, 1379–1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li, J., Yen, C., Liaw, D., Podsypanina, K., Bose, S., Wang, S. I., Puc, J., Miliaresis, C., Rodgers, L., McCombie, R., et al. (1997) Science 275, 1943–1947. [DOI] [PubMed] [Google Scholar]
- 6.Sansal, I. & Sellers, W. R. (2004) J. Clin. Oncol. 22, 2954–2963. [DOI] [PubMed] [Google Scholar]
- 7.Steck, P. A., Pershouse, M. A., Jasser, S. A., Yung, W. K., Lin, H., Ligon, A. H., Langford, L. A., Baumgard, M. L., Hattier, T., Davis, T., et al. (1997) Nat. Genet. 15, 356–362. [DOI] [PubMed] [Google Scholar]
- 8.Ringel, M. D., Hayre, N., Saito, J., Saunier, B., Schuppert, F., Burch, H., Bernet, V., Burman, K. D., Kohn, L. D. & Saji, M. (2001) Cancer Res. 61, 6105–6111. [PubMed] [Google Scholar]
- 9.Bellacosa, A., de Feo, D., Godwin, A. K., Bell, D. W., Cheng, J. Q., Altomare, D. A., Wan, M., Dubeau, L., Scambia, G., Masciullo, V., et al. (1995) Int. J. Cancer 64, 280–285. [DOI] [PubMed] [Google Scholar]
- 10.Samuels, Y., Wang, Z., Bardelli, A., Silliman, N., Ptak, J., Szabo, S., Yan, H., Gazdar, A., Powell, S. M., Riggins, G. J., et al. (2004) Science 304, 554. [DOI] [PubMed] [Google Scholar]
- 11.Bachman, K. E., Argani, P., Samuels, Y., Silliman, N., Ptak, J., Szabo, S., Konishi, H., Karakas, B., Blair, B. G., Lin, C., et al. (2004) Cancer Biol. Ther. 3, 772–775. [DOI] [PubMed] [Google Scholar]
- 12.Broderick, D. K., Di, C., Parrett, T. J., Samuels, Y. R., Cummins, J. M., McLendon, R. E., Fults, D. W., Velculescu, V. E., Bigner, D. D. & Yan, H. (2004) Cancer Res. 64, 5048–5050. [DOI] [PubMed] [Google Scholar]
- 13.Campbell, I. G., Russell, S. E., Choong, D. Y., Montgomery, K. G., Ciavarella, M. L., Hooi, C. S., Cristiano, B. E., Pearson, R. B. & Phillips, W. A. (2004) Cancer Res. 64, 7678–7681. [DOI] [PubMed] [Google Scholar]
- 14.Zhao, J. J., Gjoerup, O. V., Subramanian, R. R., Cheng, Y., Chen, W., Roberts, T. M. & Hahn, W. C. (2003) Cancer Cell 3, 483–495. [DOI] [PubMed] [Google Scholar]
- 15.Ali, S. H. & DeCaprio, J. A. (2001) Semin. Cancer Biol. 11, 15–23. [DOI] [PubMed] [Google Scholar]
- 16.Hahn, W. C., Dessain, S. K., Brooks, M. W., King, J. E., Elenbaas, B., Sabatini, D. M., DeCaprio, J. A. & Weinberg, R. A. (2002) Mol. Cell. Biol. 22, 2111–2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Foster, S. A., Wong, D. J., Barrett, M. T. & Galloway, D. A. (1998) Mol. Cell. Biol. 18, 1793–1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Romanov, S. R., Kozakiewicz, B. K., Holst, C. R., Stampfer, M. R., Haupt, L. M. & Tlsty, T. D. (2001) Nature 409, 633–637. [DOI] [PubMed] [Google Scholar]
- 19.Shaulian, E., Zauberman, A., Ginsberg, D. & Oren, M. (1992) Mol. Cell. Biol. 12, 5581–5592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Samuels, Y., Diaz, L. A., Jr., Schmidt-Kittler, O., Cummins, J. M., Delong, L., Cheong, I., Rago, C., Huso, D. L., Lengauer, C., Kinzler, K. W., et al. (2005) Cancer Cell 7, 561–573. [DOI] [PubMed] [Google Scholar]
- 21.Kang, S., Bader, A. G. & Vogt, P. K. (2005) Proc. Natl. Acad. Sci. USA 102, 802–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Elenbaas, B., Spirio, L., Koerner, F., Fleming, M. D., Zimonjic, D. B., Donaher, J. L., Popescu, N. C., Hahn, W. C. & Weinberg, R. A. (2001) Genes Dev. 15, 50–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Davies, H., Bignell, G. R., Cox, C., Stephens, P., Edkins, S., Clegg, S., Teague, J., Woffendin, H., Garnett, M. J., Bottomley, W., et al. (2002) Nature 417, 949–954. [DOI] [PubMed] [Google Scholar]
- 24.Zhao, J. J., Roberts, T. M. & Hahn, W. C. (2004) Trends Mol. Med. 10, 344–350. [DOI] [PubMed] [Google Scholar]
- 25.Yu, J., Boyapati, A. & Rundell, K. (2001) Virology 290, 192–198. [DOI] [PubMed] [Google Scholar]
- 26.Shekar, S. C., Wu, H., Fu, Z., Yip, S. C., Nagajyothi, Cahill, S. M., Girvin, M. E. & Backer, J. M. (2005) J. Biol. Chem. 280, 27850–27855. [DOI] [PubMed] [Google Scholar]
- 27.Wan, P. T., Garnett, M. J., Roe, S. M., Lee, S., Niculescu-Duvaz, D., Good, V. M., Jones, C. M., Marshall, C. J., Springer, C. J., Barford, D. & Marais, R. (2004) Cell 116, 855–867. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.