Abstract
Background
Long alpha-helical coiled-coil proteins are involved in diverse organizational and regulatory processes in eukaryotic cells. They provide cables and networks in the cyto- and nucleoskeleton, molecular scaffolds that organize membrane systems and tissues, motors, levers, rotating arms, and possibly springs. Mutations in long coiled-coil proteins have been implemented in a growing number of human diseases. Using the coiled-coil prediction program MultiCoil, we have previously identified all long coiled-coil proteins from the model plant Arabidopsis thaliana and have established a searchable Arabidopsis coiled-coil protein database.
Results
Here, we have identified all proteins with long coiled-coil domains from 21 additional fully sequenced genomes. Because regions predicted to form coiled-coils interfere with sequence homology determination, we have developed a sequence comparison and clustering strategy based on masking predicted coiled-coil domains. Comparing and grouping all long coiled-coil proteins from 22 genomes, the kingdom-specificity of coiled-coil protein families was determined. At the same time, a number of proteins with unknown function could be grouped with already characterized proteins from other organisms.
Conclusion
MultiCoil predicts proteins with extended coiled-coil domains (more than 250 amino acids) to be largely absent from bacterial genomes, but present in archaea and eukaryotes. The structural maintenance of chromosomes proteins and their relatives are the only long coiled-coil protein family clearly conserved throughout all kingdoms, indicating their ancient nature. Motor proteins, membrane tethering and vesicle transport proteins are the dominant eukaryote-specific long coiled-coil proteins, suggesting that coiled-coil proteins have gained functions in the increasingly complex processes of subcellular infrastructure maintenance and trafficking control of the eukaryotic cell.
Background
The coiled-coil was one of the earliest protein structures described and first discovered in the two-stranded coiled-coil protein alpha-keratin [1]. Coiled-coils consist of two or more alpha-helices winding around each other in a supercoil, a simple yet versatile protein fold [2]. Mutations in coiled-coil proteins have been implicated in a large variety of human diseases such as severe skin fragility, muscular dystrophies, neurodegenerative diseases, progeria, and cancer [3-10]. Spurred by medical interest, the number of investigated long coiled-coil proteins in yeast and animals has rapidly grown in recent years. Recently, a database of all long coiled-coil proteins in the model plant Arabidopsis was established to facilitate the identification and characterization of long coiled-coil proteins in plants [11]. In contrast to eukaryotic organisms, only few long coiled-coil proteins have been characterized in prokaryotes. Examples include chaperonins and nucleases, secretion proteins, and cytadherence factors [12-15].
The foremost feature of coiled-coil domains appears to be their ability to act as "cellular velcro" to hold together molecules, subcellular structures, and even tissues. They can act as protein-protein interaction motifs, for examples as dimerization domains in transcription factors and receptor kinases [16-18]. They function as "zippers" in membrane fusion proteins [19], and as adapters between molecules and solid state cellular structures, such as in microtubule organizing centers, the nuclear pores and lamina, actin- and microtubule-associated proteins and cytoskeleton-associated E3 ubiquitin ligases [20-24]. Extracellular coiled-coil proteins include cell adherence factors and surface receptors, vertebrate blood components such as apolipoproteins and fibrinogen-like clotting factors, and extracellular matrix components such as laminins and cartilage matrix proteins forming tissue scaffolds in metazoa [25,26].
Besides associating with and interconnecting other molecules and macromolecular structures, long coiled-coil domains exhibit a number of structural and mechanical functions [27]. Typically, long coiled-coil domains form rod-like tertiary structures [2] and assemble to dynamic fibers, meshworks and scaffolds. Examples are the intermediate filaments of the cytoskeleton and nuclear lamina [28]. Recent evidence suggests an important role for the dynamic properties of cytoplasmic intermediate filaments in neurodegenerative diseases [29]. Other coiled-coils act as spacers, for example in the yeast spindle pole body where the distance between the plaques is determined by the length of the coiled-coil domain in the connecting proteins [30,31]. Membrane-bound coiled-coil proteins such as the spectrins and golgins form scaffolds for membrane structures within the cell [32,33]. In combination with other functional domains, coiled-coil domains are an integral part of molecular motors, such as the actin motor myosin and the microtubule motors kinesin and dynein [34]. Other coiled-coil proteins with ATPase and GTPase domains often function in folding and repair, e.g. as chaperonins in protein folding, and topoisomerases and nucleases in DNA remodeling [35-37].
On a primary structure level, amino acid sequences with the capacity to form left-handed alpha-helical coiled-coils are characterized by a heptad repeat pattern in which residues in the first and fourth position are hydrophobic, and residues in the fifth and seventh position are predominantly charged or polar [38]. This pattern of hydrophobic and polar residues interferes with sequence comparison algorithms, which often lead to false predictions of homology between long coiled-coil proteins based on the low complexity and repeat nature of the underlying sequence motif. On the other hand, this repeat pattern can also be used to predict coiled-coil domains in amino acid sequences by computational means [39-42].
In the post-genomics era, such structure-prediction algorithms can now be applied to whole proteomes. Based on the prediction algorithm COILS, roughly 10% of all proteins encoded by eukaryotic genomes contain coiled-coil domains whereas prokaryotic genomes contain only 4–5% [43]. Using the MultiCoil program, one in every 11 proteins in yeast was predicted to contain a coiled-coil sequence [44]. However, these studies did not use a cut-off for domain length to determine coiled-coils. A minimum length of three to four heptad repeats is required for the formation of a stable coiled-coil using synthetic peptides [45-47]. Using this minimum domain length of 20 amino acids (or about three heptad repeats), 5.6% of the predicted ORFs in the fully sequenced Arabidopsis genome were found to encode coiled-coil proteins [11].
In a comparative genomics approach, we determined the coiled-coil content of 22 predicted whole proteomes using the prediction pipeline and processing software developed to create the ARABI-COIL database [11]. The 22 genomes analyzed included four archaeal genomes, ten bacterial genomes (three gram-positive and seven gram-negative species), and eight eukaryotic genomes (two each for yeasts, invertebrates, mammals, and plants).
Results
Prediction and selection of coiled-coil proteins was performed using the MultiCoil algorithm [42] and the ExtractProp processing software [11]. For the purpose of this study, "long coiled-coil" proteins were defined according to the parameters used to establish the ARABI-COIL database and included all sequences with at least one coiled-coil domain and minimum domain length of 70, two domains and minimum domain length of 50, and three or more domains and minimum domain length of 30 [11].
Eukaryotic genomes contain higher percentages of long coiled-coil proteins than prokaryotic genomes
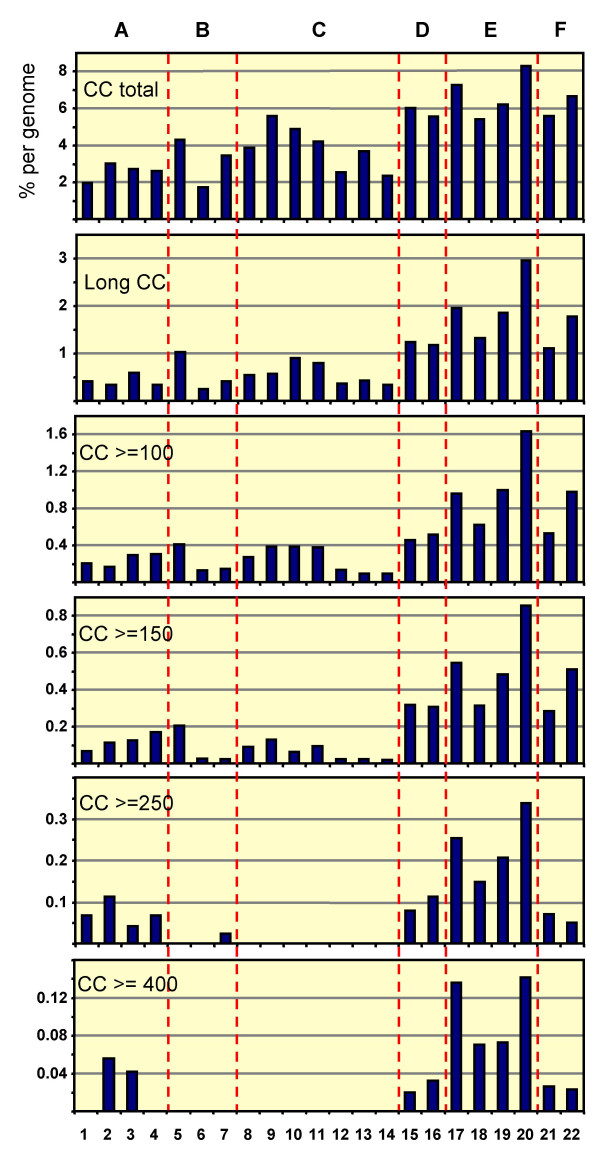
Proteins predicted to form coiled-coil domains were present in all genomes analyzed (Table 1, Figure 1) and comprised between 2% and 8% of the total proteomes. The most pronounced difference between prokaryotic and eukaryotic genomes was in the percentage of genes per genome predicted to encode long or multiple coiled-coil domains. With increasing coiled-coil domain length cut-off, lower percentages of proteins were identified in bacterial genomes. With the exception of Bacillus subtilis, MultiCoil predicted no coiled-coil proteins with domains longer than 250 amino acids in the bacterial genomes analyzed. However, archaeal and eukaryotic genomes contain proteins predicted to form coiled-coils of this length. Strikingly, prediction of coiled-coil domains over 400 amino acids in length was completely absent in bacterial genomes, but present in eukaryotes as well as two archaea, Sulfolobus solfataricus and Archeoglobus fulgidus. These numbers however do not take discontinuous coiled-coil prediction into account, as evident in the case of prokaryotic SMC proteins (Figure 2).
Table 1.
Proteome sequence data sets downloaded for MultiCoil analysis
| Organism | Number of proteins | Date of download | Comments (info as provided by EBI and TIGR) |
| Archaea (extremophiles): | |||
| Archaeoglobus fulgidus (A.f.) | 2400 | 10-Jun-2004 | hyperthermophilic, organoheterotrophic-lithoautotrophic, sulfur-metabolizing; glycoprotein envelope, flagellated |
| Methanococcus jannaschii (M.j.) | 1782 | 10-Jun-2004 | thermophilic, methanogenic, autotrophic, strict anaerobic; grows under high pressure in deep sea, flagellated |
| Sulfolobus solfataricus (S.s.) | 2939 | 28-Jan-2004 | thermophilic, sulfuric acid-producing, aerobic; no flagella, but pilus-like and pseudopodium-like structures |
| Thermoplasma acidophilum (T.a.) | 1479 | 28-Jan-2004 | thermoacidophilic; flagellated, no cell wall |
| Gram-positive bacteria: | |||
| Actinobacteria: Mycobacterium tuberculosis (M.t.) | 3995 | 10-Jun-2004 | animal pathogen (tuberculosis); no flagella |
| Bacilli: Bacillus subtilis (B.s.) | 4167 | 28-Jan-2004 | capable of producing endospores, flagellated |
| Mollicutes: Mycoplasma genitalium (M.g.) | 486 | 10-Jun-2004 | animal pathogen (surface parasite), smallest known self-replication cell & genome; no flagella |
| Gram-negative bacteria: | |||
| Alphaproteobacteria: Agrobacterium tumefaciens (A.tu.) | 5393 | 10-Jun-2004 | plant pathogen (crown gall); flagellated |
| Betaproteobacteria: Chromobacterium violaceum (C.v.) | 4400 | 10-Jun-2004 | subtropical/tropical; produces antimicrobial violacein, flagellated |
| Gammaproteobacteria: Escherichia coli K12 (E.c.) | 4356 | 28-Jan-2004 | enterobacterium, laboratory strain, flagellated |
| Epsilonproteobacteria: Heliobacter pylori (H.p.) | 1556 | 10-Jun-2004 | animal pathogen; micro-aerophilic, spiral-shaped, flagellated |
| Chlamydiae: Chlamydia pneumoniae (C.p.) | 1110 | 10-Jun-2004 | animal pathogen (obligate intracellular parasite), no flagella |
| Spirochaetes: Borrelia burgdorferi (B.b.) | 1558 | 10-Jun-2004 | animal pathogen (Lime disease); spiral-shaped, flagellated |
| Cyanobacteria: Synechocystis sp. PCC6803 (S.sp.) | 3164 | 28-Jan-2004 | photosynthetic (oxygenic), no flagella |
| Yeast: | |||
| Saccharomyces cerevisiae (S.c.) | 6191 | 28-Jan-2004 | baker's yeast |
| Schizosaccaromyces pombe (S.p.) | 5037 | 28-Jan-2004 | fission yeast |
| Metazoa: | |||
| Caenorhabditis elegans (C.e.) | 22873 | 28-Jan-2004 | nematode |
| Drosophila melanogaster (D.m.) | 16196 | 28-Jan-2004 | insect (fruitfly) |
| Mus musculus (M.m.) | 27577 | 28-Jan-2004 | mammal (mouse) |
| Homo sapiens (H.s.) | 29024 | 28-Jan-2004 | mammal (human) |
| Plants: | |||
| Arabidopsis thaliana (A.t.) | 26945 | 28-Jan-2004 | dicot |
| Oryza sativa ssp. Japonica (O.s.) | 56056 | 9-Jan-2004 | monocot (rice) |
Proteome sequence sets were downloaded from the European Bioinformatics Institute (EBI) [106] or The Institute for Genome Research (TIGR) [107]. The number of protein sequence entries reflects the annotation of ORFs at the time of download.
Figure 1.
Percentages of long coiled-coil proteins per genome. CC, coiled-coil length in amino acids, "CC total" includes all sequences predicted to contain a minimum stretch of 20 amino acids predicted to form a coiled-coil, "Long CC" includes all sequences with at least one coiled-coil domain and minimum domain length of 70, two domains and minimum domain length of 50, and three or more domains and minimum domain length of 30. A, archaea; B, Gram+ bacteria; C, Gram- bacteria; D, yeasts; E, metazoa; F, plants. 1, Thermoplasma acidophilum; 2, Methanococcus jannaschii; 3, Archaeoglobus fulgidus; 4, Sulfolobus solfataricus; 5, Mycoplasma genitalium; 6, Mycobacterium tuberculosis; 7, Bacillus subtilis; 8, Clamydia pneumoniae; 9, Heliobacter pylori; 10, Borrelia burgdorferi; 11, Synechocystis sp. PCC6803; 12, Escherichia coli; 13, Chromobacterium violaceum; 14, Agrobacterium tumefaciens; 15, Schizosaccharomyces pombe; 16, Saccharomyces cerevisiae; 17, Drosophila melanogaster; 18, Caenorhabditis elegans; 19, Mus musculus; 20, Homo sapiens; 21, Arabidopsis thaliana; 22, Oryza sativa.
Figure 2.
ABC-ATPases in archaea and bacteria. Phylogenetic tree and schematic representation of domain structures of ABC-ATPases and related sequences found in the prokaryotic genomes analyzed. Conserved domains shown as identified in CDD [49]. aa, amino acids. For species name abbreviations, see Table 1.
Prokaryotic long coiled-coil proteins
Archaea
Four archaeal genomes were included in this study and tables with coiled-coil protein details are available in additional file 1 (Archeoglobus fulgidus, Table S1; Methanococcus jannaschii, Table S2; Sulfolobus solfataricus, Table S3; and Thermoplasma acidophilum, Table S4). 2–3% of the genes in these archaea were found to code for coiled-coil proteins. In contrast to eubacteria, all of the coiled-coil size-classes analyzed are represented in this group, with proteins predicted to form coiled-coils longer than 400 residues present in Methanococcus jannaschii and Archeoglobus fulgidus proteomes (see Figure 1).
Eubacteria
Bacterial genomes for this study were chosen from different families to represent a wide range of prokaryotic species. Three gram-positive bacterial genomes (additional file 1; Mycobacterium tuberculosis, Table S5; Bacillus subtilis, Table S6; and Mycoplasma genitalium, Table S7), and seven gram-negative bacterial genomes (Agrobacterium tumefaciens, Table S8; Chromobacterium violaceum, Table S9; Escherichia coli, Table S10; Heliobacter pylori, Table S11; Chlamydia pneumoniae, Table S12; Borrelia burgdorferi, Table S13; and the cyanobacterium Synechocystis, Table S14) were analyzed.
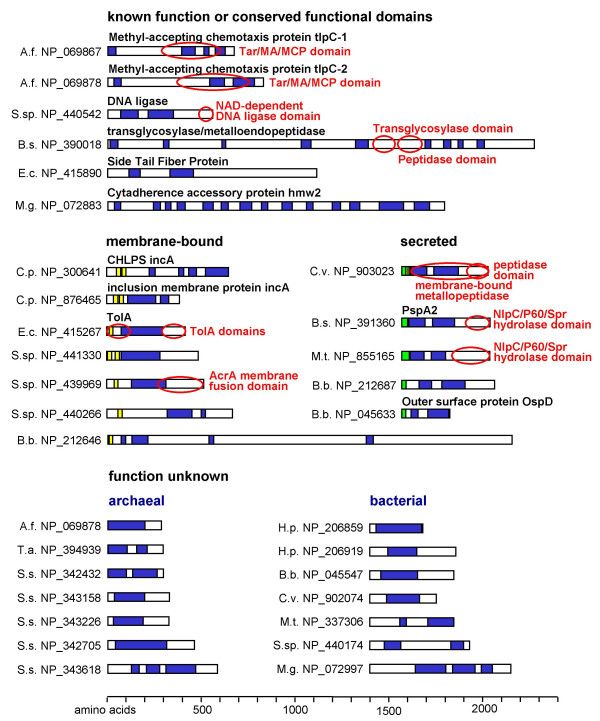
The largest prokaryotic coiled-coil domains were identified in proteins of the SMC, Rad50, SbcC and MukB families. These proteins contain globular head and tail domains separated by a coiled-coil rod with a hinge [48]. Figure 2 summarizes schematic diagrams of the domain structures of the prokaryotic SMC and SMC-like proteins identified in this study based on our coiled-coil prediction data and conserved domains as identified through Conserved Domain Database (CDD) searches [49]. Figure 3 shows a summary of additional long coiled-coil proteins with domains of at least 150 amino acids in length present in prokaryotic genomes. A number of these proteins are involved in membrane events, such as chemosensing via methyl-accepting chemotaxis proteins [50] and membrane fusion and vesicle formation mediated by AcrA, TolA, and incA proteins [51-53]. Others function as adhesion proteins, for example the lambda phage side tail fiber protein [54] and the hmw2 protein of the attachment organelle of Mycoplasma pneumoniae [15], or as enzymes of the cell wall such as the NlpC/P60 proteins [55].
Figure 3.
Prokaryotic long coiled-coil proteins. Schematic representation of prokaryotic long coiled-coil proteins not belonging to the ABC-ATPase family. Only proteins with at least 150 amino acids predicted to be in a coiled-coil are shown. Blue, coiled-coil domain; green, signal peptide; yellow, transmembrane domain. Functional domains as identified in the CDD [49] are circled in red. tlpC-1, tlpC-2, methyl-accepting chemotaxis proteins homologous to B.s. tlpC [111]; hmw2, cytadherence protein [15], CHLPS incA, incA, inclusion membrane proteins [53]; TolA, [52]; OspD, outer surface protein D [112], [113]. For species name abbreviations, see Table 1.
Long coiled-coil domains cause clustering of unrelated coiled-coil sequences
Sequences predicted to form long coiled-coil domains were analyzed for family relationships and conservation across species in an all-against-all approach using the Smith-Waterman sequence comparison algorithm followed by clustering based on an adaptation of Kruskal's minimum cost spanning tree algorithm [56,57].
In a pilot analysis to test the feasibility of the clustering approach, all prokaryotic sequences meeting the aforementioned criteria for "long coiled-coil" proteins were included in the clustering. Due to the larger number of qualified sequences in the eukaryotic species, only the longest domains (at least 250 residues in length) or sequences largely covered by coiled-coil (at least 60% of the sequence) were included in the combined pilot sequence set comprising 527 unique sequences. A maximum P-score of 1.0e-20 was used as the critical threshold when selecting only the most prominent sequence similarities in this test group. In all, 12,013 pair-wise P-score values were selected, defining as many unique relationships from the 277,729 possible pair-wise relationships. Sequences were then grouped using Kruskal's minimum cost spanning tree algorithm using the P-score value as the edge weight for the selected P-score values. 166 independent non-overlapping sequence subsets (subtrees) were defined in this manner. The largest grouping consisted of 270 sequences, representing over half of the sequences in the pilot sequence set and including functionally distinct families such as for example myosins, golgins, and SMC proteins. Distinct clusters of long coiled-coil proteins besides this large, heterogeneous group were formed by the animal and yeast tropomyosins (two separate clusters), the laminins, the CASP/CDP-family and the nuclear lamins.
Masking of coiled-coil domains before clustering
To prevent clustering based on the inherent coiled-coil repeat similarities, amino acids predicted to form coiled-coil domains were computationally masked out before being subjected to sequence similarity comparison (Figure 4). The clustering of the sequences with masked coiled-coil domains yielded a much more accurate grouping of known long coiled-coil protein families such as the myosins, golgins, and SMC proteins (Table 2). The largest group of long coiled-coil proteins with 58 sequences comprised the myosin motor proteins. The laminins, CASP/CDP, and nuclear lamins still exhibited the prior cluster profile, however the tropomyosin clusters did not appear after masking the coiled-coil domains. The coiled-coil coverage for many of the tropomyosins was predicted as 100% in our analysis, effectively excluding this protein family from the sequence comparison after masking.
Figure 4.
Flowchart of sequence comparison and clustering. Coiled-coil prediction data was generated using the program MultiCoil [42] and output processing and coiled-coil domain selection were performed as described for the ARABI-COIL database [11]. Coiled-coil prediction data was used to generate a set of sequences with coiled-coil domains masked out. The masked sequences were used as a query against unmasked sequences in an all-against-all Smith-Waterman sequence comparison (SW Search). The P-scores from this comparison were used for clustering of the output.
Table 2.
Clustering results
| Annotation | # sequences | Species |
| Myosins | 56 | A.t., C.e., D.m., H.s., M.m., O.s., S.c., S.p. |
| SMCs | 13 | A.f., A.tu., B.b., B.s., C.v., H.s., M.g., M.j., M.m., M.t., S.c., S.sp., T.a. |
| Laminins | 10 | D.m., H.s., M.m. |
| ROCK | 9 | C.e., D.m., H.s., M.m. |
| ELKS/ERC1 | 7 | H.s., M.m. |
| SLAP | 5 | H.s., M.m. |
| Kinectin | 5 | H.s., M.m. |
| Periplakin | 5 | H.s., M.m. |
| DOC1/FILIP | 5 | H.s. |
| C-Nap | 5 | D.m., H.s., M.m. |
| CASP/CDP | 4 | H.s., M.m. |
| CENP-F | 4 | H.s., M.m. |
| Lamins | 4 | H.s., M.m. |
| Hypothetical | 4 | C.e. |
| Unknown | 4 | A.t., O.s., S.p. |
| Unknown | 4 | A.t., O.s. |
Largest clusters identified in a pilot analysis using all prokaryotic long coiled-coil proteins and eukaryotic proteins with coiled-coil domains longer than 250 amino acids or more than 60% coverage. Only clusters with four or more members are listed. ROCK, Rho-associated coiled-coil containing kinase [114]; ELKS/ERC1, Rab6-interacting protein [115], SLAP, sarcolemmal-associated protein [116]; DOC1, downregulated in ovarian cancer 1 [117]; CENP-F, centromer protein F [118]. For species name abbreviations, see Table 1.
Clustering analysis with masked coiled-coil domains
After determining the consistency of clusters formed after masking coiled-coil domains with well-known coiled-coil protein families such as the SMC proteins, myosins and kinesins, we proceeded to cluster all 3576 predicted long coiled-coil sequences from the 22 genomes. The clustering algorithm was further improved to first preclude transitively similar sequences by requiring all sequences in each cluster to satisfy the P-score threshold for all pair-wise relationships within the cluster and secondly to identify "bridge" sequences meeting these criteria for multiple clusters (see Material and Methods for details). A P-score threshold of 10e-06 was selected as the appropriate balance of sequence coverage and cluster discrimination. Table 3 gives an overview of the sequences from each species contributing to the clustering analysis using the 1.0e-06 P-score cut-off. The high number of species-specific sequences found in rice is caused by retrotransposon repeats in the rice genome containing predicted coiled-coil domains within a putative transposase ORF. Figure 5 shows the distribution of clusters among the different kingdoms. Sequence annotation including species origin provided further insight into functions and relationships among sequences in each cluster. Additional information was obtained using Conserved Domain Database searches, multiple sequence alignments, and phylogenetic tree analysis of selected clusters (see Materials and Methods).
Table 3.
Contribution to clusters
| species | ORFs total | CCs total | long CCs | species-specific | in cross-species clusters |
| Archaea | |||||
| T.a. | 1479 | 29 | 6 | 1 | 5 |
| M.j. | 1782 | 54 | 6 | 2 | 4 |
| A.f. | 2400 | 65 | 14 | 6 | 8 |
| S.s. | 2939 | 77 | 10 | 8 | 2 |
| gram + bacteria | |||||
| M.g. | 486 | 21 | 5 | 3 | 2 |
| M.t. | 3995 | 70 | 10 | 7 | 3 |
| B.s. | 4167 | 144 | 17 | 10 | 7 |
| gram - bacteria | |||||
| C.p. | 1110 | 43 | 6 | 5 | 1 |
| H.p. | 1556 | 87 | 9 | 6 | 3 |
| B.b. | 1558 | 76 | 14 | 13 | 1 |
| S. sp. | 3164 | 133 | 25 | 16 | 9 |
| E.c. | 4356 | 111 | 16 | 6 | 10 |
| C.v. | 4400 | 161 | 19 | 9 | 10 |
| A.tu. | 5393 | 161 | 18 | 10 | 8 |
| yeast | |||||
| S.p. | 5037 | 303 | 62 | 25 | 37 |
| S.c. | 6191 | 344 | 73 | 35 | 38 |
| plants | |||||
| A.t. | 26945 | 1518 | 284 | 59 | 225 |
| O.s. | 56056 | 3740 | 997 | 795 | 202 |
| animals | |||||
| D.m. | 16196 | 1174 | 317 | 117 | 200 |
| C.e. | 22873 | 1234 | 304 | 144 | 160 |
| M.m. | 27577 | 1709 | 512 | 56 | 456 |
| H.s. | 29024 | 2400 | 855 | 189 | 666 |
Contribution of genomes to cross-species clusters (based on clustering using a P-score cut-off of 1.0e-06). For species name abbreviations, see Table 1.
Figure 5.
Cluster distribution. Clustering after Smith-Waterman comparison of sequences with coiled-coil domains masked. Numbers within the circles and overlapping sections represent numbers of clusters containing sequences from the respective kingdoms. For kingdom-specific clusters, only clusters with sequences from at least two different species were counted.
Coiled-coil proteins conserved between prokaryotes and eukaryotes
The SMC proteins were identified as the single major cluster of long coiled-coil proteins containing sequences from eukaryotic as well as prokaryotic genomes (see Table 4). Another group of conserved proteins with long coiled-coils comprised a number of eukaryotic Ser/Thr-kinases and a homolog from the cyanobacterium Synechocystis (sll0776 in Figure S1, additional file 2). However, proteins belonging to this cluster could not be found in any other prokaryotic genome.
Table 4.
Clusters with sequences from prokaryotes and eukaryotes
|
Cluster size (# of sequences) |
Max. edge (P-score) |
protein family | putative function/site of action | organisms represented |
| 45 | 4.6E-7 | Structural maintenance of chromosomes 1–4 | condensin, cohesin (chromatin) | A.f., A.t., A.tu., B.b., B.s., C.e., C.v., D.m., H.s., M.g., M.j., M.m., M.t., O.s., S.c., S.p, S.sp., T.a. |
| 26 | 1.6E-10 | Ser/Thr-kinases (DAP, DMK, GIN4, ROCK) | signal transduction | C.e., D.m., H.s., M.m., O.s., S.c., S.sp. |
Numbers include "bridge" sequences qualifying for more than one cluster. Only clusters represented by at least 10 sequences are listed. DAP, death-associated protein kinase [119]; DMK, myotonic dystrophy kinase [120]; GIN4, growth inhibitory gene 4 [121]; ROCK, Rho-associated coiled-coil containing kinase [114]. For species name abbreviations, see Table 1.
A number of smaller cluster were formed containing proteins with shorter coiled-coil domains close to the cut-off for our analysis. One cluster comprised the translation initiation factor IF-2, containing the respective sequences from Drosophila, E. coli, mouse, rice and yeast. Another cluster with sequences conserved in prokaryotes as well as eukaryotes contained the AAA+ family ATPase ClpB/Hsp104 represented by plant, yeast and bacterial sequences. This protein functions as a protease/chaperonin in eubacteria, plants and mitochondria [35]. Two small clusters combined sequences from prokaryotes and plant genomes. One cluster comprised mitochondrial seryl-tRNA synthetases conserved in plant mitochondria as well as archaea while the second cluster comprised the PspA-like VIPP1 protein from plastids and the cyanobacterium Synechocystis. VIPP1 is involved in thylakoid biosynthesis in both chloroplasts as well as cyanobacteria, possibly acting in thylakoid membrane trafficking [58,59].
Prokaryotic coiled-coil protein clusters
Prokaryotic clusters comprised membrane-bound proteins and signal transducers, as well as membrane-spanning transporters and secretion proteins such as the HlyD family [60]. The only cluster specific to prokaryotes represented by more than ten sequences in this study comprised the methyl-accepting chemotaxis proteins (MCPs; Table 5; [50]). Smaller prokaryotic clusters contained the aforementioned ABC-ATPases RAD50 and SbcC involved in DNA repair and a highly conserved group of archaeal proteins of unknown function (COG1340, represented by NP_394939 in Figure 3).
Table 5.
Prokaryotic clusters
|
Cluster size (# of sequences) |
Max. edge (P-score) |
protein family | putative function/site of action | organism |
| 12 | <E-40 | Methyl-accepting chemotaxis proteins | chemotactic sensor/signal transducer (bacterial envelope membrane) | A.f., A.tu., B.s., C.v., E.c., S.sp. |
Numbers include "bridge" sequences qualifying for more than one cluster. Only clusters represented by at least 10 sequences are listed. For species name abbreviations, see Table 1.
Eukaryotic coiled-coil protein clusters
The main clusters formed by eukaryotic sequences only (Table 6) were the eukaryotic motor proteins: the actin motor myosin and the microtubule motor kinesin and the related kinesin-like calmodulin-binding protein KCBP [34,61,62]. The proteins of the SMC5 and SMC6 families formed a eukaryotic cluster instead of clustering together with the condensin/cohesin SMCs 1–4 and the prokaryotic SMC proteins in our analysis (Figure 6B). Eukaryotic RAD50 proteins clustered separately from prokaryotic RAD50s as well, indicating a higher convergence of the non-coiled-coil RAD50 ATPase domains as compared to the SMC 1–4 head and tail domains. Additional larger clusters included eukaryotic Ser/Thr-kinases and a family comprised of the Retinoblastoma-associated protein RBP95, Ring Finger Proteins 20 and 40, and yeast Bre1p [63,64,23] (Figure S2, additional file 2, and Table S15, additional file 3). Formin-related proteins associated with growing actin fibers [65,66] were found in animal/yeast and animal/plant cluster combinations. Smaller conserved eukaryotic clusters included a number of proteins involved in vesicle transport, such as a Rab6 GTPase-activating protein involved in retrograde transport [67], the golgin CASP [68] and the vesicular transport proteins P115 (see Figure S3, additional file 2), autophagy protein APG6 [69,70], and early endosome antigen (EEA1, [71]) homologs (see Figure S4, additional file 2).
Table 6.
Eukaryotic clusters
|
Cluster size (# of sequences) |
Max. edge (P-score) |
protein family | putative function/site of action | organisms represented |
| 94 | 5.4E-10 | Myosin heavy chain | actin motor protein (muscle, cytoskeleton) | A.t., C.e., D.m., H.s., M.m., O.s., S.c., S.p. |
| 27 | 1.5E-36 | Kinesin heavy chain (KCBP, KIFC) | MT motor protein (cytoskeleton) | A.t., C.e., D.m., H.s., M.m., O.s., S.c., S.p. |
| 21 | 1.1E-35 | Kinesin heavy chain (Cmet/Cana, MKRPs, NACK/HINKEL) | MT motor protein (cytoskeleton) | A.t., D.m., H.s., M.m., O.s., S.p. |
| 17 | 1.2E-7 | Structural maintenance of chromosomes 5–6/RAD18 | DNA repair (chromatin) | A.t., C.e., D.m., H.s., M.m., O.s., S.c., S.p. |
| 12 | 5.0E-7 | Kinases (GIN4, MET) | signal transduction | A.t., D.m., H.s., O.s., S.c. |
| 11 | <E-40 | RAD50 (eukaryotic) | DNA repair | A.t., C.e., D.m., H.s., M.m., O.s., S.c., S.p. |
| 11 | 1.2E-7 | Retinoblastoma-associated protein, RING finger protein 20 | E3 Ubi. ligase for H2B histone modification (nuclear)? | A.t., C.e., D.m., H.s., M.m., O.s., S.c, S.p. |
Numbers include "bridge" sequences qualifying for more than one cluster. Only clusters represented by at least 10 sequences are listed. MKRPs, mitochondrial kinesin-related proteins [122]; NACK/HINKEL, NPK1-activating kinesin [123]; MET, [124]. For species name abbreviations, see Table 1.
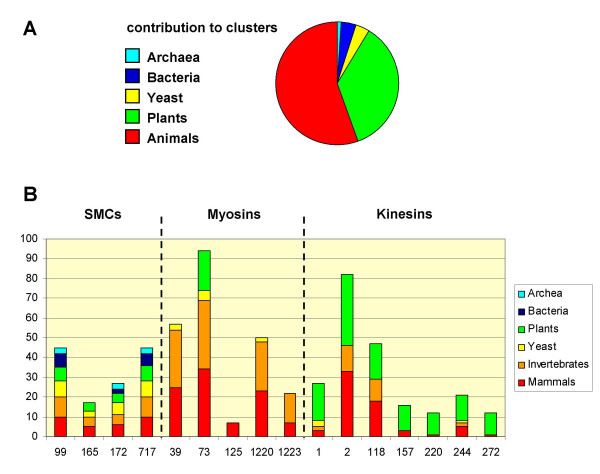
Figure 6.
Contribution to clusters. Contribution of the different kingdoms to the complete sequence pool analyzed (A) and to SMC, myosin and kinesin clusters (B). Y-axis, number of sequences; X-axis, cluster IDs. Examples for characteristic protein families represented in clusters are as follows: clusters 99, 172, 717, SMC 1–4; cluster 165, SMC 5–6; clusters 39, 1220, type II myosins; cluster 125, type X myosins; cluster 1223, non-muscle myosins; clusters 1, 272, KIFCs; clusters 157, 220, PAKRPs; cluster 244, NACK, MKRPs. Proteins may qualify for two or more overlapping cluster, e.g. resulting in prokaryotic SMCs clustering with different types of diverged eukaryotic SMC proteins.
Yeast, yeast-plant, and yeast-animal coiled-coil protein clusters
Eukaryotic genomes included the baker's yeast (Saccharomyces cerevisiae) and fission yeast (Schizosaccharomyces pombe) as eukaryotic, unicellular organisms. Protein clusters found to be specific for yeast were typically small (one sequence from each yeast genome, see additional file 4) and comprised proteins involved in RNA export, such as Gle1, [72] and Mlp1 [73], the spindle assembly checkpoint protein Mad1 [74], and GRIP-domain golgins [75,76]. These proteins have known homologs in other eukaryotic proteomes, which did not cluster together with the yeast proteins, likely due to a high overall coverage with coiled-coil sequences (e.g. up to 70% coiled-coil coverage for Mlp1/Tpr, up to 74% for MAD1, and up to 75% for GRIP-golgins). Another functional group of yeast proteins were cell polarity proteins such as Spa2 and Tea1 [77,78]. Tea1 clustered together with a number of plant sequences of unknown function containing Kelch repeats [79] in combination with coiled-coil domains. Proteins that were found in clusters specific to yeasts and animals (Table 7) included the microtubule motor dynein as well as proteins involved in endocytosis and microtubule dynamics, such as intersectin, restin and cytoplasmic linker proteins (CLIP) [80]. A number of myosin subclusters, for example myosin type II, was represented only by yeast and animal but not plant sequences, consistent with previous findings [81] (see Table 7 and Figure 6B).
Table 7.
Animal and yeast clusters
|
Cluster size (# of sequences) |
Max. edge (P-score) |
protein family | putative function/site of action | organism |
| 57 | 3.3E-7 | Myosin heavy chain XVIII | non-muscle and smooth muscle myosins | C.e., D.m., H.s., M.m., S.p. |
| 50 | 6.5E-8 | Myosin heavy chain type 2 | actin motor protein (cytoskeleton) | C.e., D.m., H.s., M.m., S.c., S.p. |
| 27 | <E-40 | Dynein heavy chain | MT motor (cytoskeleton, flagella) | C.e., D.m., H.s., M.m., S.c. |
| 17 | 3.1E-9 | Intersectins, Eps15 | endocytosis | C.e., D.m., H.s., M.m., S.c., S.p. |
| 16 | 9.5E-7 | Png-1, IF-2, Neurofilament triplet L, Troponin T | D.m., H.s., M.m., O.s., S.c. | |
| 14 | <E-40 | Restin, Dynactin, CLIP proteins | linking endocytic vesicles to MTs (IF cytoskeleton), dynein activator (MTs in neurons), MT/IF associated (cytoskeleton) | C.e., D.m., H.s., M.m., S.c., S.p. |
| 14 | 1.6E-15 | Myosin heavy chain V | unusual myosin | C.e., D.m., H.s., M.m., S.c. |
| 14 | <E-40 | DRFs | binds Rho-GTP and profilin, promotes actin polymerization (membrane cytoskeleton) | D.m., H.s., M.m., S.c. |
Numbers include "bridge" sequences qualifying for more than one cluster. Only clusters represented by at least 10 sequences are listed. CLIP, cytoplasmic linker protein [80]; DRFs, diaphanous-related formins [66]; Eps15, epidermal growth factor receptor substrate 15 [125]; Png-1, postmitotic neural gene-1 [126]. For species name abbreviations, see Table 1.
Animal coiled-coil protein clusters
From the metazoan kingdom, genomes from nematodes (Caenorhabditis elegans), flies (Drosophila melanogaster), and mammals (Mus musculus and Homo sapiens) were analyzed. Clusters that appeared to be specific to animals (Table 8) comprised a variety of proteins crosslinking cytoskeletal components with membranes, such as spectrin- and periplakin-like membrane-actin and membrane-IF crosslinkers [32,82], the plasmamembrane-scaffolding Liprins [83], the family of Merlin and Ezrin/Radixin/Moesin (ERM) proteins [84,85], and a number of Golgi- and vesicle-associated proteins. Other groups comprised centrosome-associated and mitotic spindle checkpoint proteins. Type X myosins grouped together in a metazoan cluster without plant or yeast sequences. Another animal-specific group contained coiled-coil proteins involved in structural integrity such as the extracellular scaffolding protein Laminin [26] and intermediate filament proteins including the nuclear lamins and neurofilaments [86,87]. Smaller animal-specific clusters contained protein sequences involved in cell attachment and motility, embryogenesis, spermatogenesis, and immune cell movement.
Table 8.
Animal-specific clusters
|
Cluster size (# of sequences) |
Max. edge (P-score) |
protein family | putative function/site of action | organism |
| 39 | 9.9E-7 | Spectrins, Dystrophin, Nesprins | membrane/actin/MT crosslinkers (cytoskeleton) | C.e., D.m., H.s., M.m. |
| 32 | <E-40 | Laminins | scaffold protein (extracellular matrix) | C.e., D.m., H.s., M.m. |
| 25 | 6.7E-8 | Plectin/Desmoplakin | actin/MT crosslinkers (cytoskeleton) | C.e., D.m., H.s., M.m. |
| 22 | 6.5E-8 | Myosin heavy chain (muscle) | actin motor protein, muscle fibers | C.e., D.m., H.s., M.m. |
| 18 | 9.3E-9 | Lamins | nuclear IFs | C.e., D.m., H.s., M.m. |
| 17 | 5.8E-7 | Neurofilament triplet L, M, Death inducer, Troponin T | C.e., D.m., M.m., H.s. | |
| 14 | 7.5E-7 | Neurofilament triplet H, M | C.e., D.m., H.s., M.m. | |
| 14 | 7.5E-7 | Neurofilaments (Desmin, Vimentin) | IFs | C.e., H.s., M.m. |
| 13 | 2.6E-7 | PP1, ASPP | apoptosis stimulating | D.m., H.s., M.m. |
| 12 | <E-40 | Moesin, Ezrin, Radixin | membrane organization and stabilization (membrane cytoskeleton, cytovilli) | C.e., D.m, H.s., M.m. |
| 11 | 1.7E-43 | RUFY | possible role in vesicle trafficking (endosomes?) | D.m., H.s., M.m. |
| 11 | 2.2E-11 | Lamins | nuclear IFs | D.m., H.s., M.m. |
| 11 | 3.4E-7 | Restin, Dynactin, CLIP proteins | linking endocytic vesicles to MTs (IF cytoskeleton), dynein activator (MTs in neurons), MT/IF associated (cytoskeleton) | C.e., D.m., H.s., M.m. |
| 11 | 3.9E-7 | Png-1, Neurofilament triplet M, Troponin T | C.e., D.m., H.s., M.m. | |
| 10 | <E-40 | Dystrophins | C.e., D.m., H.s., M.m. | |
| 10 | 2.7E-8 | prion-like protein | C.e., D.m. |
Numbers include "bridge" sequences qualifying for more than one cluster. Only clusters represented by at least 10 sequences are listed. ASPP, apoptosis stimulating of p53 protein [127]; PP1, protein phosphatase 1 [128]; RUFY, RUN and FYVE domain containing proteins [129]. For species name abbreviations, see Table 1.
A number of the clusters containing animal sequences were limited to mammalian sequences only (Table 9). The hair fiber protein keratin was found to form the largest group of proteins specific to mammals. Other mammlian clusters comprised neurofilament proteins and crosslinkers of the actin cytoskeleton and were found to overlap with clusters containing invertebrate sequences as well. A number of smaller mammalian clusters (see additional file 5, Table S17) contained sequences of unknown function which have so far only been characterized as autoantigens or cancer antigens. Smaller clusters included the centrosomal protein Ninein, which is involved in anchoring microtubule minus ends [88], and a number of other centrosomal proteins including TACCs, C-NAP1, and Centriolin [89-91]. Other clusters included mammalian reproductive organ-specific proteins, such as sperm tail-associated proteins and mammary gland-specific proteins, vertebrate-specific transcription factors and coactivators such as the SOX proteins [92], and regulators of endothelial cell motility and clotting factors in blood vessels.
Table 9.
Clusters with mammalian sequences only
|
Cluster size (# of sequences) |
Max. edge (P-score) |
protein family | putative function/site of action | organism |
| 51 | <E-40 | Keratin type II | IF, cytoskeletal (cytokeratin), hair | H.s., M.m. |
| 45 | 5.0E-7 | Keratin type I (hair keratin) | IF, cuticular/hair | H.s., M.m. |
| 32 | 4.2E-7 | Keratin type I | IF, cytoskeletal (cytokeratin), root sheeth | H.s., M.m. |
| 31 | 9.2E-7 | Keratin type II | IF, cytoskeletal (cytokeratin), hair | H.s., M.m. |
| 24 | 3.9E-7 | Keratin type I (hair keratin) | IF, cuticular/hair | H.s., M.m. |
| 18 | 1.4E-14 | Neurofilaments (Desmin, Internexin, Peripherin, Vimentin) | IFs | H.s., M.m. |
| 13 | 1.2E-25 | Interferon-induced guanylate-binding proteins | H.s., M.m. | |
| 10 | 1.9E-8 | Plectin, Desmoplakin, Periplakin, Envoplakin | H.s., M.m. |
Numbers include "bridge" sequences qualifying for more than one cluster. Only clusters represented by at least 10 sequences are listed. See Table S16 for protein details on clusters smaller than 10. For species name abbreviations, see Table 1.
Plant coiled-coil protein clusters
As representatives for the plant kingdom, a dicot (Arabidopsis thaliana) and a monocot (Oryza sativa) plant genome were analyzed. Clusters of long coiled-coil proteins specific to Arabidopsis and rice contained mostly sequences of so far unknown function (Table 10). The rice genome contains a large number of transposon-derived ORFs which are predicted to contain coiled-coil domains, therefore a large number of plant-specific clusters was represented by rice sequences only. These have been omitted from Table 10. Plant-specific clusters represented by both plant species analyzed included kinase interacting protein 1 (KIP1) and its relatives [93], the family of filament-like plant proteins, FPPs [94], and a cluster of putative Zinc finger transcription factors homologous to the x1 gene of maize [95]. Smaller clusters (see additional file 6, Table S18) included nuclear matrix constituent protein 1 (NMPC1) and relatives [96], and the chloroplast unusual positioning 1 (CHUP1) actin-interacting protein [97]. Several clusters showed overlap between the plant and animal kingdoms (Table 11). These included a number of kinesin subclusters, vesicle trafficking proteins, and Guanylate-binding proteins (Figure S5).
Table 10.
Plant-specific clusters
|
Cluster size (# of sequences) |
Max. edge (P-score) |
protein family | putative function/site of action | organism |
| 21 | 1.2E-7 | Kinase-interacting protein 1-like | signal transduction | A.t., O.s. |
| 13 | 7.6E-12 | expressed proteins | unknown | A.t., O.s. |
| 12 | 2.4E-9 | FPPs | unknown | A.t., O.s. |
| 11 | 2.9E-7 | putative receptor kinases | signal transduction | A.t., O.s. |
| 10 | 5.9E-18 | Transcription factor X1-like proteins | transcription | A.t., O.s. |
Numbers include "bridge" sequences qualifying for more than one cluster. Only clusters represented by at least 10 sequences are listed. See Table S17 for protein details on clusters smaller than 10. FPPs, filament-like plant proteins [94]. For species name abbreviations, see Table 1.
Table 11.
Animal and plant clusters
|
Cluster size (# of sequences) |
Max. edge (P-score) |
protein family | putative function/site of action | organism |
| 83 | 1.6E-38 | Kinesin heavy chain (Chromokinesin, KIF3, 4) | MT motor protein (cytoskeleton), nuclear | A.t., C.e., D.m., H.s., M.m., O.s. |
| 47 | 4.6E-39 | Kinesin heavy chain (KIF2-4, NACK, FRA) | MT motor protein (cytoskeleton) | A.t., C.e., D.m., H.s., M.m., O.s. |
| 29 | 3.0E-32 | Kinesin heavy chain (KIF1, 13, 14, 16, 17) | MT motor protein (cytoskeleton), axonal transporter of synaptic vesicles | A.t., C.e., D.m., H.s., M.m., O.s. |
| 16 | 5.9E-38 | Kinesin heavy chain (PAKRP) | MT motor protein (cytoskeleton) | A.t., H.s., O.s. |
| 14 | 2.1E-9 | Plexin, Rab6 GTPase activating protein | vesicle trafficking | A.t., C.e., D.m., H.s., M.m., O.s. |
| 13 | 9.4E-7 | Guanylate-binding protein | A.t., H.s., M.m., O.s. | |
| 12 | 4.3E-27 | Kinesin heavy chain (KIFC1, TH65) | MT motor protein (cytoskeleton) | A.t., H.s., O.s. |
| 12 | 9.6E-35 | Kinesin heavy chain (PAKRP, MKRP) | MT motor protein (cytoskeleton) | A.t., H.s., O.s. |
| 11 | 3.4E-7 | DRFs | binds Rho-GTP and profilin, promotes actin polymerization (membrane cytoskeleton) | A.t., C.e., H.s., M.m., O.s. |
Numbers include "bridge" sequences qualifying for more than one cluster. Only clusters represented by at least 10 sequences are listed. FRA, fragile fiber [130]; PAKRP, phragmoplast-associated kinesin-related protein [131], [132]. For species name abbreviations, see Table 1.
Discussion
The SMC proteins are the most widely conserved coiled-coil proteins
The most widely conserved family of long coiled-coil proteins found in our study comprised the SMC proteins. Representatives from almost all species analyzed were found in this cluster, with a few exceptions such as the gram-negative bacterium E. coli. This is consistent with previous findings that SMC proteins are present in eukaryotes as well as all gram-positive bacteria and nearly all archaea, but only less than half of the gram-negative bacteria. It has been proposed that eukaryotic smc genes evolved from archaeal precursors by two consecutive gene duplications [48]. Bacteria without SMC proteins often contain an SMC-related long coiled-coil protein involved in chromosome segregation or DNA repair, such as MukB or SbcC [98,13].
Prokaryotic coiled-coil filament proteins
While prokaryotic genomes contained less long coiled-coil proteins than eukaryotes, we found a number of so far uncharacterized long coiled-coil proteins as candidates for filament-forming prokaryotic coiled-coils. These included Heliobacter pylori proteins previously suggested as candidates for bacterial filament proteins [99].
Metazoan mitotic motor proteins lack homologs in plants
The presence of a nucleus in eukaryotic cells is closely linked with the presence of a motile cytoskeleton, in particular the mitotic structures necessary to orchestrate nuclear division, and the endocytic pathway. Dolan et al. [100] proposed a list of motility proteins involved in mitotic processes as candidates for homology searches in prokaryotes to determine their evolutionary origin. We found 70% of the suggested proteins (Astrin, CENP-E, Centrin, Dynein, Dynactin, Kinesin, Kinectin, MAD, NuMA, Pericentrin) among the long coiled-coil proteins identified in our analysis, however none of them clustered together with sequences from archaea or bacteria. Interestingly, with the exception of the kinesins, we also could not find any of these proteins clustering with plant sequences. With the exception of dynein, kinesin and MAD proteins, we could not find clustering of these mitotic motility proteins with yeast sequences either.
The organization of mitotic microtubule nucleation and the composition of the nuclear envelope in plant cells differ significantly from metazoan cells [101]. One hypothesis to explain these differences is the separate development of specialized mechanisms to orchestrate open mitosis in metazoan and plant lineages, leading to the evolution of different nuclear envelope compositions, targeting mechanisms, and mitotic spindle nucleation in the plant and animal kingdoms. This model explains the absence of many metazoan mitotic motility proteins in plants as well as yeast, which undergoes closed mitosis, and suggests that this group of proteins evolved after the occurrence of open mitosis.
We could not find any plant-specific classes of coiled-coil motor proteins, but noted kinesin subclusters largely represented by plant sequences only, indicating an expansion of this group of motor proteins during plant evolution (see Figure 6B). It has been noted before that Arabidopsis contains a surprisingly large number of kinesins [102], and it has been suggested that plant-specific kinesin subfamilies might be involved in stress responses or pathogen defenses [103].
Differences and similarities in cytoskeletal and membrane infrastructure between plants and animals
Besides the motor proteins (myosins, kinesins, dyneins), membrane tethering and vesicle transport proteins appear to be specific for eukaryotes in our clustering analysis, indicating another major class of specialized coiled-coil proteins that evolved after the formation of eukaryotic cells. It has been previously suggested that the higher content of long coiled-coil domains in metazoa compared to plants and protists indicates the presence of extensive coiled-coil matrices in animal cells and tissues [25]. One of the groups of coiled-coil proteins apparently absent in plants and yeasts are the nuclear matrix and intermediate filament proteins. No lamin sequences could be identified from the plant genomes. Other differences we noted between the plant and animal kingdoms are the lack of membrane-cytoskeleton crosslinkers and scaffolding proteins, such as spectrin-like proteins and many actin- and microtubule-associated proteins, in plant proteomes. This might indicate differences in the overall organization and networking of membrane systems and the actin and microtubule cytoskeleton in plant and animal cells.
Differences in coiled-coil content between genomes
Earlier surveys of coiled-coil sequences in GenBank had suggested that invertebrate genomes contain more coiled-coils than vertebrates, and that animal genomes contain four times more "extended" coiled-coils (>75 amino acids) than plant genomes [25]. While we could not find such a difference for the overall coiled-coil content or the group of proteins defined as "long" coiled-coils in this study, we did note a significantly lower percentage of coiled-coils longer than 250 amino acids in yeast as well as plants compared to the animal genomes (see Figure 1). On average, the yeasts contained one third of the percentage of coiled-coils present in vertebrate genomes with domains longer than 100 and longer than 250 residues (37% and 35%, respectively), whereas invertebrates contained about two thirds (60% and 73%, respectively). The plant genomes, however, contained on average 57% of the percentage of proteins with coiled-coil domains longer than 100 amino acids, but only 22% of the coiled-coils with 250 amino acids and longer when compared to vertebrates. An interesting observation is that the human genome appears to contain more extended coiled-coil proteins than the mouse genome. Our data suggests that this is caused by the human proteome sequence set containing more unique long coiled-coil proteins without homologs in other species (see Table 3), as well as more redundant sequences in clusters (e.g. comparing counts of human versus mouse sequences in clusters listed in additional file 5, Table S17).
Comparison with other genome-wide coiled-coil predictions
Comparable with the Arabidopsis coiled-coil protein database ARABI-COIL, this study takes a more restrictive approach to identifying coiled-coil proteins than previous genome-wide approaches to predict coiled-coil proteins [44,43]. In contrast to the older studies, our prediction criteria included a minimum coiled-coil domain length corresponding to about three heptad repeats to eliminate sequences with short stretches of predicted coiled-coils unlikely to form stable structures [11]. Using these parameters, on average about 6.4% of all proteins in the eukaryotic proteomes and about 3.5% in the prokaryotic proteomes (2.6% in archaea, 3.7% in bacteria) contained coiled-coil domains. Our results were consistent with the study of Liu and Rost [43] in that most eukaryotic genomes contained more coiled-coil proteins than prokaryotic genomes, and most bacterial genomes more than archaea. The more restrictive parameters used here resulted in predicting on average about 65–70% of the number of proteins found in those previous studies. Liu and Rost [43] further found an exceptionally high coiled-coil content in Heliobacter pylori with a higher percentage than C. elegans, and an exceptionally low coiled-coil content in Mycobacterium tuberculosis. Our analysis was consistent with these previous observations and resulted in 5.6% coiled-coil for Heliobacter pylori versus 5.4% in C. elegans and only 1.8% in Mycobacterium tuberculosis, the lowest percentage for all 22 genomes analyzed here.
Limitations of the prediction and clustering analysis
Discontinuous coiled-coil domain predictions
MultiCoil provides a more stringent coiled-coil prediction than other programs such as COILS, resulting in less false positive predictions. In tests on the PDB database of solved protein structures, two-thirds of the sequences predicted by COILS did not contain coiled-coils [104]. By comparison, the programs PAIRCOIL and MultiCoil perform significantly better [42]. Occasionally, however, the increased stringency might lead to prediction of fragmented domains where continuous domains have been experimentally verified, as evident in the case of the SMC proteins (see Figure 2).
Selection of long coiled-coil proteins only
In this study, we focused on proteins potentially involved in structural functions. As the emphasis was placed on proteins with long or multiple coiled-coil domains, it is possible that our selection criteria resulted in the exclusion of homologs of proteins with short stretches of coiled-coil that barely qualified for the analysis. The selection criteria applied in this study have been shown to exclude 97% of the known bZIP proteins from Arabidopsis [11]. Other examples we noted are the translation initiation factor IF-2, mitochondrial and prokaryotic seryl-tRNA synthetases, and the ClpB/HSP104 family of heatshock proteins. Members of these protein families failed to meet the selection criteria for long coiled-coil domains, making it difficult to draw conclusions for these protein families from our clustering analysis. We therefore focused our attention on clusters with mainly proteins containing longer coiled-coils (>150 amino acids).
Effect of coiled-coil masking in the clustering analysis
When clustering sequences with long coiled-coil domain in the pilot analysis, the majority of proteins with long coiled-coil domains was grouped together in one large cluster. Many of the proteins with unknown functions in this group were annotated as "myosin-like", however only about 20% of the proteins in the cluster actually contained a myosin motor domain. In the other cases, the only similarity to myosin was the presence of a long coiled-coil domain similar to the myosin coiled-coil tail. This illustrates the ease with which long coiled-coil domains can lead to misannotations in databases with annotations based on sequence similarity searches.
Masking the coiled-coil domains before sequence comparison and clustering significantly increased the specificity of the clustering analysis, however protein sequences with high coiled-coil coverage were lost in the subsequent clustering as the masking left little to no sequence for comparison. Examples are the animal and yeast tropomyosins, many of which were predicted to contain 100% coiled-coil coverage, paramyosin, and the plant cytoskeletal protein CIP1 with more than 80% coiled-coil coverage [105].
Conclusion
Our genome-wide identification of coiled-coil proteins and subsequent clustering provides data suggesting evolutionary conservation or uniqueness of coiled-coil proteins among 22 fully sequenced genomes. We found SMC, MukB, SbcC and Rad50 proteins to be the proteins with the longest coiled-coil domains occurring in prokaryotes, whereas eukaryotic proteomes also contained proteins with stretches of coiled-coil longer than the SMC rod domains. The high conservation of the SMC proteins and their structural relatives involved in chromosome maintenance and repair demonstrates the universal importance and conservation of DNA housekeeping mechanisms.
Long coiled-coil proteins specific to eukaryotes are predominantly involved in subcellular infrastructure maintenance and trafficking control. Table 12 gives an overview of the functional classes of long coiled-coil proteins found in our analysis and their representation in different kingdoms. The genomes of higher plants lack sequences coding for intermediate filament proteins. Many of the known mitotic spindle associated coiled-coil motor proteins in animals lack homologs in plants, consistent with the absence of a centrosomal microtubule organization center in plant cells. However, the kinesin family of microtubule motor proteins appears to have expanded during the evolution of higher plants.
Table 12.
Summary of coiled-coil protein functions
| Functional groups of coiled-coil proteins | Examples | Species represented |
| Chromatin organization and maintenance, chromosome segregation and DNA repair | SMCs, Rad50, SbcC, MukB, MutS | all kingdoms |
| Transcription and translation | Transcription and translation initiation factors, reverse transcriptase | all kingdoms |
| Protein trafficking and quality control | Chaperonins, secretion proteins | prokaryotes and organelles |
| Membrane sensors, channels and regulation of influx/export | MCPs, ion channels | prokaryotes |
| Sensor mechanisms and signal transduction | Receptor kinases, GTPases | eukaryotes – conserved, as well as plant and animal specific |
| Compartmentalization, stabilization and dynamics of membrane systems | Golgins, SNAREs, endocytic proteins | eukaryotes |
| Adherence | Cell adherence, extracellular matrix, intracellular adapters | eukaryotes and parasitic prokaryotes |
| Mechanical fiber and meshwork formation | Keratin, intermediate filaments, flagellar (e.g. sperm tail) proteins | eukaryotes, keratin only in mammals |
| Motility | Muscle fibers, cell motility, actin and microtubule motors | eukaryotes |
| Organization, stabilization and dynamics of the cytoskeleton | Actin and microtubule crosslinkers | eukaryotes, predominantly metazoa |
| Mitotic spindle assembly and checkpoint control | Centrosome, kinetochore and spindle pole body proteins | metazoa and yeast |
The repeat nature of the coiled-coil motif makes it difficult to clearly determine sequence homology relationships between long coiled-coil proteins. Functional studies will have to reveal whether so far uncharacterized prokaryotic and plant coiled-coil proteins fulfill similar functions to metazoan counterparts.
Methods
Sequence data and pre-processing
Proteome sequence sets of fully sequenced genomes were downloaded from the European Bioinformatics Institute (EBI) [106] for organisms listed in Table 1, with the exception of rice. The rice proteome set was downloaded from The Institute for Genome Research (TIGR) [107]. An initial preprocessing of the FASTA files was conducted to standardize identifiers for the sequences for easier incorporation into a MySQL database.
Coiled-coil prediction and post-processing
Prediction and selection of coiled-coil proteins was performed using the underlying schema and software systems developed to create the ARABI-COIL database [11]. In summary, the modified FASTA files were used as input for the MultiCoil application installed on the Linux Cluster of the Ohio Supercomputer Center (OSC, Columbus, OH). The MultiCoil output was post-processed using the previously described Java-based ExtractProp Suite [11] and used to establish a database of coiled-coil prediction data for each organism. The same coiled-coil selectivity criteria applied to ARABI-COIL were used to select sequences predicted to contain long or multiple coiled-coil domains. These criteria impose a minimum coiled-coil domain of 30 residues if at least three domains are present in the translated reading frame, a minimum of 50 residues if at least two domains are present, and a minimum coiled-coil length of 70 residues if only a single domain is present. Intra-domain gaps of less than 20 residues were considered contiguous for purposes of establishing domain length. The resulting data was converted to XML and used to populate MySQL databases for each genome.
Masking of coiled-coil domains
To eliminate interference of the coiled-coil repeat motif with sequence homology analysis, coiled-coil domains were "masked" before subjecting the sequences to Smith-Waterman sequence similarity searches. Mask information was created based on the processed MultiCoil prediction data generated to populate the MySQL databases for each genome. A Java-based program was applied to the FASTA sequences selected for Smith-Waterman comparison to replace all amino acids predicted to be contained in coiled-coil domains with the letter X, effectively masking coiled-coil domains.
Sequence similarity comparison
Smith-Waterman comparison was conducted using the TimeLogic Smith-Waterman implementation at OSC and the Blosum62 scoring matrix on all unique sequences in the combined sequences set. Sequences with masked coiled-coil domains were used as query on unmasked sequence sets as target. A P-score cut-off of 1.0e-03 was used as a threshold for selecting sequence similarity relationships. For sequences to be characterized as pair-wise similar and recovered for use in the clustering analysis, the P-score value must be less than this threshold based on the query-target Smith-Waterman comparison.
Clustering analysis
After completing the pair-wise similarity calculation using the Smith-Waterman algorithm and extracting sequence pairs and associated P-scores, sequences were grouped using a modified version of Kruskal's minimum cost spanning tree algorithm [57]. The algorithm creates and progressively merges sub-trees of a graph in building a minimum cost spanning tree. In the algorithm, the weights of edges in the directed graph were determined by the pair-wise P-score similarity value for the sequence as a query relative to the related sequence as a target. An effective clustering can be achieved by using only P-score similarity values which are below a specified threshold, effectively creating a disconnected series of related sequences.
The clustering was tested in a pilot analysis on a combined sequence set including 527 prokaryotic long coiled-coil proteins and eukaryotic proteins containing extended coiled-coil domains of at least 250 amino acids in length or at least 60% of the protein sequence in a coiled-coil. Edges with P-scores greater than 1.0e-03 to 1.0e-15 were ignored when combining sub-trees in the algorithm. The success of the clustering was estimated by observing the clustering behavior of well-known coiled-coil protein families, such as SMC proteins and myosins. After testing the effects of masking the coiled-coil domains and optimizing cut-offs for P-scores during clustering, the complete coiled-coil sequence set containing 3576 long coiled-coil proteins from the 22 genomes was processed similarly. Different P-score thresholds were explored in efforts to increase specificity in the multi-genome sequence set while preserving comprehensive coverage. Employing Kruskal's algorithm, the 3576 sequence set resulted in 156 clusters covering 3567 sequences using a threshold of 1.0e-03, 467 clusters covering 3551 sequences using a threshold of 1.0e-6 and 850 clusters covering 3520 sequences using a threshold of 1.0e-15. (For comparison, the same algorithm yielded 490 clusters for the unmasked sequence set).
Even with the improved selectivity of the clustering demonstrated in the pilot investigation using masked coiled-coil sequences, the overall effectiveness of the resulting clustering still required refinement to achieve sufficient specificity. The use of Kruskal's algorithm for subset selection enabled transitively similar sequences to be included in specific clusters. (Transitively similar sequences are sequences in which sequence A is similar to sequence B and sequence B is similar to sequence C thereby clustering sequence A and C which would otherwise not belong to the same cluster.) One drawback of this simplified clustering is that a given sequence need only be similar to at least one other sequence in the cluster. This limitation resulted in clusters containing sequences which, while closely related to at least one other sequence in a cluster, were not closely related to every sequence within the cluster.
The algorithm was consequently improved to specifically preclude transitively similar sequences by requiring all sequences in a given cluster to satisfy the P-score threshold for all pair-wise relationships in the cluster. The new algorithm dramatically improved specificity, with the same 3576 masked sequence set generating 1213 non-overlapping clusters covering 3567 sequences, 1263 non-overlapping clusters covering 3551 sequences, and 1384 non-overlapping clusters covering 3520 sequences with the improved algorithm for the same corresponding P-score threshold values. The P-score threshold of 1.0e-06 was selected as the appropriate balance of sequence coverage and cluster discrimination required.
The interest in identifying sequences which qualified for more than one cluster and bridged multiple clusters of protein families drove a second modification of the clustering algorithm. By design, the modified Kruskal's algorithm created mutually orthogonal, non-overlapping clusters while precluding transitively similar sequences from populating the same cluster. The 'greedy' algorithm was modified to specifically identify transitively similar sequences between clusters, enabling a unique ability to identify "bridge" sequences which satisfy participation criteria in multiple clusters or protein families. The modification amounted to simply validating each sequence's individual ability to satisfy participation criteria for a cluster based on the non-overlapping cluster partitioning.
The software used to conduct the actual cluster analysis in the study is available for download at the Ohio Bioscience Library [108].
Cluster alignments and phylogenetic tree generation
Multiple sequence alignments and phylogenetic trees were generated for clusters of interest using sequences with masked coiled-coil domains and ClustalW version 1.82 incorporating the Blossum scoring matrix [109]. Phylogenetic trees were generated using the ClustalW program with a bootstrap parameter of 10,000 and displayed using the program TreeView v.1.6.6 [110].
List of abbreviations
CASP, CDP/cut alternatively spliced product
CC, coiled-coil
CDD, conserved domain database
CDP, CCAAT displacement protein
CENP, centromer protein
CIP1, COP1-interactive protein 1
CHUP1, chloroplast unusual positioning 1
CLIP, cytoplasmic linker protein
DAM, disheveled associated activator of morphogenesis
DIA1, Diaphanous-related formin 1
DOC1, downregulated in ovarian cancer 1
EBI, European Bioinformatics Institute
ERM, ezrin/radixin/moesin
FPPs, filament-like plant proteins
Hsp, heat shock protein
IF, intermediate filament
KCBP, kinesin-like calmodulin-binding protein
KIP1, kinase interacting protein 1
KLP, kinesin-like protein
MCP, methyl-accepting chemotaxis protein
MKRP, mitochondrial kinesin-related protein
MLP, myosin-like protein
NuMA, nuclear mitotic apparatus
ORF, open reading frame
OSC, Ohio Supercomputer Center
PAKRP, phragmoplast-associated kinesin-related protein
PP1, protein phosphatase 1
RBP, Retinoblastoma-binding protein
ROCK, Rho-associated coiled-coil containing kinase
SLAP, sarcolemmal-associated protein
SMC, structural maintenance of chromosomes
S/W, Smith-Waterman sequence comparison
TACC, transforming acidic coiled-coil
Tpr, translocated promoter region
VIPP1, vesicle-inducing plastid protein 1
XML, extensible markup language
Authors' contributions
AR coordinated this study, analyzed the data, and prepared the manuscript. SJS participated in MultiCoil and Smith-Waterman output processing and ClustalW analysis. EAS generated MultiCoil and Smith-Waterman outputs, developed software for pre- and post-processing and coiled-coil masking, and wrote the code for the clustering algorithm. IM proposed and supervised the study and edited the manuscript. All authors read and approved the final manuscript.
Supplementary Material
Prokaryotic coiled-coil proteins Tables S1-S14: Protein details of all long coiled-coil proteins predicted in the prokaryotic genomes analyzed in this study. Open file with Acrobat Reader.
Eukaryotic clusters of interest Figures S1-S6: Phylogenetic trees based on ClustalW alignments of the sequences, displayed using TreeView v.1.6.6. Open file with Acrobat Reader.
Sequence details for Figures S1-S6, supplement to additional file 2. Table S15: Protein information and prediction data for sequences contained in Figures S1-S6. AGI locus numbers from TAIR are used as sequence IDs for Arabidopsis, TIGR sequence IDs are used for rice and Synechocystis. All other sequence IDs correspond to the EBI identifiers in the downloaded FASTA files. Max. Coil Length, longest coiled-coil domain in the protein sequence; Coil Coverage, percent of sequence predicted to be in a coiled-coil. Open file with Microsoft Excel.
Yeast clusters Table S18: Protein information and prediction data for sequences in yeast clusters with two species (Saccharomyces cerevisiae and Schizosaccharomyces pombe) represented. Sequence IDs correspond to the EBI identifiers in the downloaded FASTA files. Max. Coil Length, longest coiled-coil domain in the protein sequence; Coil Coverage, percent of sequence predicted to be in a coiled-coil. Open file with Microsoft Excel.
Small mammalian clusters; supplement to Table 9. Table S16: Protein information and prediction data for sequences in mammalian clusters with two species (mouse, human) represented and less than 10 sequences per cluster. Sequence IDs correspond to the EBI identifiers in the downloaded FASTA files. Max. Coil Length, longest coiled-coil domain in the protein sequence; Coil Coverage, percent of sequence predicted to be in a coiled-coil. Open file with Microsoft Excel.
Small plant clusters; supplement to Table 10. Table S17: Protein information and prediction data for sequences in plant clusters with two species (Arabidopsis, rice) represents and less than 10 sequences per cluster. AGI locus numbers from TAIR or NCBI RefSeq numbers are used as sequence IDs for Arabidopsis, TIGR sequence IDs are used for rice. Max. Coil Length, longest coiled-coil domain in the protein sequence; Coil Coverage, percent of sequence predicted to be in a coiled-coil. Open file with Microsoft Excel.
Acknowledgments
Acknowledgements
We thank the Ohio Supercomputer Center for providing usage time for this analysis. This work was supported by the National Science Foundation 2010 Project (grant no. NSF 0209339 to I.M.).
Contributor Information
Annkatrin Rose, Email: rose.273@osu.edu.
Shannon J Schraegle, Email: shannon@osc.edu.
Eric A Stahlberg, Email: eas@osc.edu.
Iris Meier, Email: meier.56@osu.edu.
References
- Crick FH. The packing of alpha-helices: Simple coiled-coils. Acta Cryst. 1953;6:689–697. doi: 10.1107/S0365110X53001964. [DOI] [Google Scholar]
- Burkhard P, Stetefeld J, Strelkov SV. Coiled coils: a highly versatile protein folding motif. Trends Cell Biol. 2001;11:82–88. doi: 10.1016/S0962-8924(00)01898-5. [DOI] [PubMed] [Google Scholar]
- Magin TM, Reichelt J, Hatzfeld M. Emerging functions: diseases and animal models reshape our view of the cytoskeleton. Exp Cell Res. 2004;301:91–102. doi: 10.1016/j.yexcr.2004.08.018. [DOI] [PubMed] [Google Scholar]
- Mounkes L, Kozlov S, Burke B, Stewart CL. The laminopathies: nuclear structure meets disease. Curr Opin Genet Dev. 2003;13:223–230. doi: 10.1016/S0959-437X(03)00058-3. [DOI] [PubMed] [Google Scholar]
- Puls I, Jonnakuty C, LaMonte BH, Holzbaur ELF, Tokito M, Mann E, Floeter MK, Bidus K, Drayna D, Oh SJ, Brown RH, Jr, Ludlow CL, Fischbeck KH. Mutant dynactin in motor neuron disease. Nat Genet. 2003;33:455–456. doi: 10.1038/ng1123. [DOI] [PubMed] [Google Scholar]
- Hirokawa N, Takemura R. Molecular motors in neuronal development, intracellular transport and diseases. Curr Opin Neurobiol. 2004;14:564–573. doi: 10.1016/j.conb.2004.08.011. [DOI] [PubMed] [Google Scholar]
- Chigira S, Sugita K, Kita K, Sugaya S, Arase Y, Ichinose M, Shirasawa H, Suzuki N. Increased expression of the Huntingtin interacting protein-1 gene in cells from Hutchinson Gilford Syndrome (progeria) patients and aged donors. J Gerontol A Biol Sci Med Sci. 2003;58:B873–878. doi: 10.1093/gerona/58.10.b873. [DOI] [PubMed] [Google Scholar]
- Mounkes LC, Stewart CL. Aging and nuclear organization: lamins and progeria. Curr Opin Cell Biol. 2004;16:322–327. doi: 10.1016/j.ceb.2004.03.009. [DOI] [PubMed] [Google Scholar]
- Raff JW. Centrosomes and cancer: lessons from a TACC. Trends Cell Biol. 2002;12:222–225. doi: 10.1016/S0962-8924(02)02268-7. [DOI] [PubMed] [Google Scholar]
- McClatchey AI. Merlin and ERM proteins: unappreciated roles in cancer development? Nat Rev Cancer. 2003;3:877–883. doi: 10.1038/nrc1213. [DOI] [PubMed] [Google Scholar]
- Rose A, Manikantan S, Schraegle SJ, Maloy MA, Stahlberg EA, Meier I. Genome-wide identification of Arabidopsis coiled-coil proteins and establishment of the ARABI-COIL database. Plant Physiol. 2004;134:927–939. doi: 10.1104/pp.103.035626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundin VF, Stirling PC, Gomez-Reino J, Mwenifumbo JC, Obst JM, Valpuesta JM, Leroux MR. Molecular clamp mechanism of substrate binding by hydrophobic coiled-coil residues of the archaeal chaperone prefoldin. Proc Natl Acad Sci USA. 2004;101:4367–4372. doi: 10.1073/pnas.0306276101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connelly JC, Kirkham LA, Leach DRF. The SbcCD nuclease of Escherichia coli is a structural maintenance of chromosomes (SMC) family protein that cleaves hairpin DNA. Proc Natl Acad Sci USA. 1998;95:7969–7974. doi: 10.1073/pnas.95.14.7969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delahay RM, Frankel G. Coiled-coil proteins associated with type III secretion systems: a versatile domain revisited. Mol Microbiol. 2002;45:905–916. doi: 10.1046/j.1365-2958.2002.03083.x. [DOI] [PubMed] [Google Scholar]
- Balish MF, Ross SM, Fisseha M, Krause DC. Deletion analysis identifies key functional domains of the cytadherence-associated protein HMW2 of Mycoplasma pneumoniae. Mol Microbiol. 2003;50:1507–1516. doi: 10.1046/j.1365-2958.2003.03807.x. [DOI] [PubMed] [Google Scholar]
- Jakoby M, Weisshaar B, Droge-Laser W, Vincente-Carbajosa J, Tiedemann J, Kroi T, Parcy F. bZIP transcription factors in Arabidopsis. Trends Plant Sci. 2002;7:106–111. doi: 10.1016/S1360-1385(01)02223-3. [DOI] [PubMed] [Google Scholar]
- Vinson C, Myakishev M, Acharya A, Mir AA, Moll JR, Bonovich M. Classification of human B-ZIP proteins based on dimerization properties. Mol Cell Biol. 2002;22:6321–6335. doi: 10.1128/MCB.22.18.6321-6335.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Gao R, Lynn DG. Racheting up vir gene expression in Agrobacterium tumefaciens: coiled coils in histidine kinase signal transduction. ChemBioChem. 2002;3:311–317. doi: 10.1002/1439-7633(20020402)3:4<311::AID-CBIC311>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- Blumenthal R, Clague MJ, Durell SR, Epand RM. Membrane fusion. Chem Rev. 2003;103:53–69. doi: 10.1021/cr000036+. [DOI] [PubMed] [Google Scholar]
- Sillibourne JE, Milne DM, Takahashi M, Ono Y, Meek DW. Centrosomal anchoring of the protein kinase CK1delta mediated by attachment to the large, coiled-coil scaffolding protein CG-NAP/AKAP450. J Mol Biol. 2002;322:785–797. doi: 10.1016/S0022-2836(02)00857-4. [DOI] [PubMed] [Google Scholar]
- Zhao X, Wu CY, Blobel G. Mlp-dependent anchorage and stabilization of a desumoylating enzyme is required to prevent clonal lethality. J Cell Biol. 2004;167:605–11. doi: 10.1083/jcb.200405168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhen YY, Libotte T, Munck M, Noegel AA, Korenbaum E. NUANCE, a giant protein connecting the nucleus and actin cytoskeleton. J Cell Sci. 2002;115:3207–3222. doi: 10.1242/jcs.115.15.3207. [DOI] [PubMed] [Google Scholar]
- Wood A, Krogan NJ, Dover J, Schneider J, Heidt J, Boateng MA, Dean K, Golshani A, Zhang Y, Greenblatt JF, Johnston M, Shilatifard A. Bre1, an E3 ubiquitin ligase required for recruitment and substrate selection of Rad6 at a promoter. Mol Cell. 2003;11:267–274. doi: 10.1016/S1097-2765(02)00802-X. [DOI] [PubMed] [Google Scholar]
- Hardtke CS, Deng XW. The cell biology of the COP/DET/FUS proteins. Regulating proteolysis in photomorphogenesis and beyond? Plant Physiol. 2000;124:1548–1557. doi: 10.1104/pp.124.4.1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odgren PR, Harvie LW, Jr, Fey EG. Phylogenetic occurrence of coiled coil proteins: implications for tissue structure in metazoa via a coiled coil tissue matrix. Proteins. 1996;24:467–484. doi: 10.1002/(SICI)1097-0134(199604)24:4<467::AID-PROT6>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- Kammerer RA. alpha-Helical coiled-coil oligomerization domains in extracellular proteins. Matrix Biol. 1997;15:555–565. doi: 10.1016/S0945-053X(97)90031-7. [DOI] [PubMed] [Google Scholar]
- Rose A, Meier I. Scaffolds, levers, rods and springs: diverse cellular functions of long coiled-coil proteins. Cell Mol Life Sci. 2004;61:1996–2009. doi: 10.1007/s00018-004-4039-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strelkov SV, Herrmann H, Aebi U. Molecular architecture of intermediate filaments. BioEssays. 2003;25:243–251. doi: 10.1002/bies.10246. [DOI] [PubMed] [Google Scholar]
- Helfand BT, Chang L, Goldman RD. Intermediate filaments are dynamic and motile elements of cellular architecture. J Cell Sci. 2004;117:133–141. doi: 10.1242/jcs.00936. [DOI] [PubMed] [Google Scholar]
- Schaerer F, Morgan G, Winey M, Philippsen P. Cnm67p is a spacer protein of the Saccharomyces cerevisiae spindle pole body outer plaque. Mol Biol Cell. 2001;12:2519–2533. doi: 10.1091/mbc.12.8.2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilmartin JV, Dyos SL, Kershaw S, Finch JT. A spacer protein in the Saccharomyces cerevisiae spindle pole body whose transcript is cell cycle-regulated. J Cell Biol. 1993;123:1175–1184. doi: 10.1083/jcb.123.5.1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Matteis MA, Morrow JS. Spectrin tethers and mesh in the biosynthetic pathway. J Cell Sci. 2000;113:2331–2343. doi: 10.1242/jcs.113.13.2331. [DOI] [PubMed] [Google Scholar]
- Barr FA, Short B. Golgins in the structure and dynamics of the Golgi apparatus. Curr Opin Cell Biol. 2003;15:405–413. doi: 10.1016/S0955-0674(03)00054-1. [DOI] [PubMed] [Google Scholar]
- Schliwa M, Woehlke G. Molecular motors. Nature. 2003;422:759–765. doi: 10.1038/nature01601. [DOI] [PubMed] [Google Scholar]
- Mogk A, Bukau B. Molecular chaperones: structure of a protein disaggregase. Curr Biol. 2004;14:R78–R80. doi: 10.1016/j.cub.2003.12.051. [DOI] [PubMed] [Google Scholar]
- Graumann PL. SMC proteins in bacteria: condensation motors for chromosome segregation? Biochimie. 2001;83:53–59. doi: 10.1016/S0300-9084(00)01218-9. [DOI] [PubMed] [Google Scholar]
- Cromie GA, Leach DRF. Recombinational repair of chromosomal DNA double-strand breaks generated by a restriction endonuclease. Mol Microbiol. 2001;41:873–883. doi: 10.1046/j.1365-2958.2001.02553.x. [DOI] [PubMed] [Google Scholar]
- Mason JM, Arndt KM. Coiled coil domains: stability, specificity, and biological implications. ChemBioChem. 2004;5:170–176. doi: 10.1002/cbic.200300781. [DOI] [PubMed] [Google Scholar]
- Berger B, Wilson DB, Wolf E, Tonchev T, Milla M, Kim PS. Predicting coiled coils by use of pairwise residue correlations. Proc Natl Acad Sci USA. 1995;92:8259–8263. doi: 10.1073/pnas.92.18.8259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parry DA. Coiled-coils in alpha-helix-containing proteins: analysis of the residue types within the heptad repeat and the use of these data in the prediction of coiled-coils in other proteins. Biosci Rep. 1982;2:1017–1024. doi: 10.1007/BF01122170. [DOI] [PubMed] [Google Scholar]
- Lupas A. Predicting coiled-coil regions in proteins. Curr Opin Struct Biol. 1997;7:388–393. doi: 10.1016/S0959-440X(97)80056-5. [DOI] [PubMed] [Google Scholar]
- Wolf E, Kim PS, Berger B. MultiCoil: a program for predicting two- and three-stranded coiled coils. Protein Sci. 1997;6:1179–1189. doi: 10.1002/pro.5560060606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Rost B. Comparing function and structure between entire genomes. Protein Sci. 2001;10:1970–1979. doi: 10.1110/ps.10101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman JRS, Wolf E, Kim PS. A computationally directed screen identifying interacting coiled coils from Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 2000;97:13203–13208. doi: 10.1073/pnas.97.24.13203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lumb KJ, Carr CM, Kim PS. Subdomain folding of the coiled coil leucine zipper from the bZIP transcriptional activator GCN4. Biochemistry. 1994;33:7361–7367. doi: 10.1021/bi00189a042. [DOI] [PubMed] [Google Scholar]
- Su JY, Hodges RS, Kay CM. Effect of chain length on the formation and stability of synthetic alpha-helical coiled-coils. Biochemistry. 1994;33:15501–15510. doi: 10.1021/bi00255a032. [DOI] [PubMed] [Google Scholar]
- Litowski JR, Hodges RS. Designing heterodimeric two-stranded alpha-helical coiled-coils: the effect of chain length on protein folding, stability and specificity. J Pept Res. 2001;58:477–492. doi: 10.1034/j.1399-3011.2001.10972.x. [DOI] [PubMed] [Google Scholar]
- Soppa J. Prokaryotic structural maintenance of chromosomes (SMC) proteins: distribution, phylogeny, and comparison with MukBs and additional prokaryotic and eukaryotic coiled-coil proteins. Gene. 2001;278:253–264. doi: 10.1016/S0378-1119(01)00733-8. [DOI] [PubMed] [Google Scholar]
- Marchler-Bauer A, Anderson JB, Cherukuri PF, DeWeese-Scott C, Geer LY, Gwadz M, He S, Hurwitz DI, Jackson JD, Ke Z, Lanczycki CJ, Liebert CA, Liu C, Lu F, Marchler GH, Mullokandov M, Shoemaker BA, Simonyan V, Song JS, Thiessen PA, Yamashita RA, Yin JJ, Zhang D, Bryant SH. CDD: a conserved domain database for protein classification. Nucl Acids Res. 2005;33:D192–D196. doi: 10.1093/nar/gki069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira M, Parente JA, Bataus LA, Cardoso DD, Soares RB, Soares CM. Chemotaxis and flagellar genes of Chromobacterium violaceum. Genet Mol Res. 2004;3:92–101. [PubMed] [Google Scholar]
- Ip H, Stratton K, Zgurskaya H, Liu J. pH-induced conformational changes of AcrA, the membrane fusion protein of Escherichia coli multidrug efflux system. J Biol Chem. 2003;278:50474–50482. doi: 10.1074/jbc.M305152200. [DOI] [PubMed] [Google Scholar]
- Henry T, Pommier S, Journet L, Bernadac A, Gorvel JP, Lloubes R. Improved methods for producing outer membrane vesicles in Gram-negative bacteria. Res Microbiol. 2004;155:437–446. doi: 10.1016/j.resmic.2004.04.007. [DOI] [PubMed] [Google Scholar]
- Hackstadt T, Scidmore-Carlson MA, Shaw EI, Fischer ER. The Chlamydia trachomatis IncA protein is required for homotypic vesicle fusion. Cell Microbiol. 1999;1:119–130. doi: 10.1046/j.1462-5822.1999.00012.x. [DOI] [PubMed] [Google Scholar]
- Montag D, Schwarz H, Henning U. A component of the side tail fiber of Escherichia coli bacteriophage lambda can functionally replace the receptor-recognizing part of a long tail fiber protein of the unrelated bacteriophage T4. J Bacteriol. 1989;171:4378–4384. doi: 10.1128/jb.171.8.4378-4384.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anantharaman V, Aravind L. Evolutionary history, structural features and biochemical diversity of the NlpC/P60 superfamily of enzymes. Genome Biol. 2003;4:R11. doi: 10.1186/gb-2003-4-2-r11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith TF, Waterman MS. Identification of common molecular subsequences. J Mol Biol. 1981;147:195–197. doi: 10.1016/0022-2836(81)90087-5. [DOI] [PubMed] [Google Scholar]
- Kruskal JB. On the shortest spanning subtree of a graph and the traveling salesman problem. Proc Amer Math Soc. 1956;7:48–50. [Google Scholar]
- Kroll D, Meierhoff K, Bechtold N, Kinoshita M, Westphal S, Vothknecht UC, Soll J, Westhoff P. VIPP1, a nuclear gene of Arabidopsis thaliana essential for thylakoid membrane formation. Proc Natl Acad Sci USA. 2001;98:4238–4242. doi: 10.1073/pnas.061500998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westphal S, Heins L, Soll J, Vothknecht UC. Vipp1 deletion mutant of Synechocystis: a connection between bacterial phage shock and thylakoid biogenesis? Proc Natl Acad Sci USA. 2001;98:4243–4248. doi: 10.1073/pnas.061501198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentschev I, Dietrich G, Goebel W. The E. coli alpha-hemolysin secretion system and its use in vaccine development. Trends Microbiol. 2002;10:39–45. doi: 10.1016/S0966-842X(01)02259-4. [DOI] [PubMed] [Google Scholar]
- Vos JW, Safida F, Reddy ASN, Hepler PK. The kinesin-like calmodulin binding protein is differentially involved in cell division. Plant Cell. 2000;12:979–990. doi: 10.1105/tpc.12.6.979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirokawa N, Takemura R. Kinesin superfamily and their various functions and dynamics. Exp Cell Res. 2004;301:50–59. doi: 10.1016/j.yexcr.2004.08.010. [DOI] [PubMed] [Google Scholar]
- Wen H, Ao S. RBP95, a novel leucine zipper protein, binds to retinoblastoma protein. Biochem Biophys Res Comm. 2000;275:141–148. doi: 10.1006/bbrc.2000.3242. [DOI] [PubMed] [Google Scholar]
- Hwang WW, Venkatasubrahmanyam S, Ianculescu AG, Tong A, Boone A, Madhani HD. A conserved RING finger protein required for histone H2B monoubiquitination and cell size control. Mol Cell. 2003;11:261–266. doi: 10.1016/S1097-2765(02)00826-2. [DOI] [PubMed] [Google Scholar]
- Watanabe N, Higashida C. Formins: processive cappers of growing actin filaments. Exp Cell Res. 2004;301:16–22. doi: 10.1016/j.yexcr.2004.08.020. [DOI] [PubMed] [Google Scholar]
- Madrid R, Gasteier JE, Bouchet J, Schröder S, Geyer M, Benichou S, Fackler OT. Oligomerization of the diaphanous-related formin FHOD1 requires a coiled-coil motif critical for its cytoskeletal and transcriptional activities. FEBS Letters. 2005;579:441–448. doi: 10.1016/j.febslet.2004.12.009. [DOI] [PubMed] [Google Scholar]
- Cuif MH, Possmayer F, Zander H, Bordes N, Jollivet F, Couedel-Courteille A, Janoueix-Lerosey I, Langsley G, Bornens M, Goud B. Characterization of GAPCenA, a GTPase activating protein for Rab6, part of which associates with the centrosome. EMBO J. 1999;18:1772–1782. doi: 10.1093/emboj/18.7.1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillingham AK, Pfeifer AC, Munro S. CASP, the alternatively spliced product of the gene encoding the CCAAT-displacement protein transcription factor, is a Golgi membrane protein related to Giantin. Mol Biol Cell. 2002;13:3761–3774. doi: 10.1091/mbc.E02-06-0349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puthenveedu M, Linstedt AD. Gene replacement reveals that p115/SNARE interactions are essential for Golgi biogenesis. Proc Natl Acad Sci USA. 2004;101:1253–1256. doi: 10.1073/pnas.0306373101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kametaka S, Okano T, Ohsumi M, Ohsumi Y. Apg14p and Apg6/Vps30p form a protein complex essential for autophagy in the yeast, Saccharomyces cerevisiae. J Biol Chem. 1998;273:22284–22291. doi: 10.1074/jbc.273.35.22284. [DOI] [PubMed] [Google Scholar]
- Mu FT, Callaghan JM, Steele-Mortimer O, Stenmark H, Parton RG, Campbell PL, McCluskey J, Yeo JP, Tock EPC, Toh BH. EEA1, and early endosome-associated protein. EEA1 is a conserved alpha-helical peripheral membrane protein flanked by cysteine "fingers" and contains a calmodulin-binding IQ motif. J Biol Chem. 1995;270:13503–13511. doi: 10.1074/jbc.270.22.13503. [DOI] [PubMed] [Google Scholar]
- Suntharalingam M, Alcazar-Roman AR, Wente SR. Nuclear export of the yeast mRNA-binding protein Nab2 is linked to a direct interaction with Gfd1 and to Gle1 function. J Biol Chem. 2004;279:35384–35391. doi: 10.1074/jbc.M402044200. [DOI] [PubMed] [Google Scholar]
- Green DM, Johnson CP, Hagan H, Corbett AH. The C-terminal domain of myosin-like protein 1 (Mlp1p) is a docking site for heterogenous nuclear ribonucleoproteins that are required for mRNA export. Proc Natl Acad Sci USA. 2003;100:1010–1015. doi: 10.1073/pnas.0336594100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillet ES, Espelin CW, Sorger PK. Spindle checkpoint proteins and chromosome-microtubule attachment in budding yeast. J Cell Biol. 2004;164:535–546. doi: 10.1083/jcb.200308100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barr FA. A novel Rab6-interacting domain defines a family of Golgi-targeted coiled-coil proteins. Curr Biol. 1999;9:381–384. doi: 10.1016/S0960-9822(99)80167-5. [DOI] [PubMed] [Google Scholar]
- Munro S, Nichols BJ. The GRIP domain – a novel Golgi-targeting domain found in several coiled-coil proteins. Curr Biol. 1999;9:377–380. doi: 10.1016/S0960-9822(99)80166-3. [DOI] [PubMed] [Google Scholar]
- van Drogen F, Peter M. Spa2p functions as a scaffold-like protein to recruit the Mpk1p MAP kinase module to sites of polarized growth. Curr Biol. 2002;12:1698–1703. doi: 10.1016/S0960-9822(02)01186-7. [DOI] [PubMed] [Google Scholar]
- Behrens R, Nurse P. Roles of fission yeast tea1p in the localization of polarity factors and in organizing the microtubular cytoskeleton. J Cell Biol. 2002;157:783–793. doi: 10.1083/jcb.200112027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams J, Kelso R, Cooley L. The kelch repeat superfamily of proteins: propellers of cell function. Trends Cell Biol. 2000;10:17–24. doi: 10.1016/S0962-8924(99)01673-6. [DOI] [PubMed] [Google Scholar]
- Arnal I, Heichette C, Diamantopoulos GS, Chretien D. CLIP-170/tubilin-curved oligomers coassemble at microtubule ends and promote rescues. Curr Biol. 2004;14:2086–2095. doi: 10.1016/j.cub.2004.11.055. [DOI] [PubMed] [Google Scholar]
- Reddy ASN, Day IS. Analysis of the myosins encoded in the recently completed Arabidopsis thaliana genome sequence. Genome Biol. 2001;2:RESEARCH0024. doi: 10.1186/gb-2001-2-7-research0024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karashima T, Watt FM. Interaction of periplakin and envoplakin with intermediate filaments. J Cell Sci. 2002;115:5027–5037. doi: 10.1242/jcs.00191. [DOI] [PubMed] [Google Scholar]
- Serra-Pages C, Medley QG, Tang M, Hart A, Streuli M. Liprins, a family of LAR transmembrane protein-tyrosine phosphatase-interacting proteins. J Biol Chem. 1998;273:15611–15620. doi: 10.1074/jbc.273.25.15611. [DOI] [PubMed] [Google Scholar]
- Bretscher A, Edwards K, Fehon RG. ERM proteins and merlin: integrators at the cell cortex. Nat Rev Mol Cell Biol. 2002;3:586–599. doi: 10.1038/nrm882. [DOI] [PubMed] [Google Scholar]
- Ivetic A, Ridley AJ. Ezrin/radixin/moesin proteins and Rho GTPase signalling in leucocytes. Immunology. 2004;112:165–176. doi: 10.1111/j.1365-2567.2004.01882.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePianto D, Coulombe PA. Intermediate filaments and tissue repair. Exp Cell Res. 2004;301:68–76. doi: 10.1016/j.yexcr.2004.08.007. [DOI] [PubMed] [Google Scholar]
- Goldman RD, Gruenbaum Y, Moir RD, Shumaker DK, Spann TP. Nuclear lamins: building blocks of nuclear architecture. Genes Dev. 2002;16:533–547. doi: 10.1101/gad.960502. [DOI] [PubMed] [Google Scholar]
- Baird DH, Myers KA, Mogensen M, Moss D, Baas PW. Distribution of the microtubule-related protein ninein in developing neurons. Neuropharmacology. 2004;47:677–683. doi: 10.1016/j.neuropharm.2004.07.016. [DOI] [PubMed] [Google Scholar]
- Gergely F. Centrosomal TACCtics. Bioessays. 2002;24:915–925. doi: 10.1002/bies.10162. [DOI] [PubMed] [Google Scholar]
- Mayor T, Stierhof YD, Tanaka K, Fry AM, Nigg EA. The centrosomal protein C-Nap1 is required for cell cycle-regulated centrosome cohesion. J Cell Biol. 2000;151:837–846. doi: 10.1083/jcb.151.4.837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gromley A, Jurczyk A, Sillibourne J, Halilovic E, Mogensen M, Groisman I, Blomberg M, Doxsey S. A novel human protein of the maternal centriole is required for the final stages of cytokinesis and entry into S phase. J Cell Biol. 2003;161:535–545. doi: 10.1083/jcb.200301105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson M, Koopman P. Matching SOX: partner proteins and co-factors of the SOX family of transcriptional regulators. Curr Opin Genet Dev. 2002;12:441–446. doi: 10.1016/S0959-437X(02)00323-4. [DOI] [PubMed] [Google Scholar]
- Skirpan AL, McCubbin AG, Ishimizu T, Wang X, Hu Y, Dowd PE, Ma H, Kao TH. Isolation and characterization of kinase interacting protein 1, a pollen protein that interacts with the kinase domain of PRK1, a receptor-like kinase of petunia. Plant Physiol. 2001;126:1480–1492. doi: 10.1104/pp.126.4.1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gindullis F, Rose A, Patel S, Meier I. Four signature motifs define the first class of structurally related large coiled-coil proteins in plants. BMC Genomics. 2002;3:9. doi: 10.1186/1471-2164-3-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao H, Zhou Q, Li J, Smith H, Yandeau M, Nikolau BJ, Schnable PS. Molecular characterization of meiotic recombination across the 140-kb multigenic a1-sh2 interval of maize. Proc Natl Acad Sci USA. 2002;99:6157–6162. doi: 10.1073/pnas.082562199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda K, Xu ZJ, Takahashi S, Ito A, Ono M, Nomura K, Inoue M. Peripheral framework of carrot cell nucleus contains a novel protein predicted to exhibit a long alpha-helical domain. Exp Cell Res. 1997;232:173–181. doi: 10.1006/excr.1997.3531. [DOI] [PubMed] [Google Scholar]
- Oikawa K, Kasahara M, Kiyosue T, Kagawa T, Suetsugu N, Takahashi F, Kanegae T, Niwa Y, Kadota A, Wada M. CHLOROPLAST UNUSUAL POSITIONING1 is essential for proper chloroplast positioning. Plant Cell. 2003;15:2805–2815. doi: 10.1105/tpc.016428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasgupta S, Maisnier-Patin S, Nordström K. New genes with old modus operandi. EMBO Rep. 2000;1:323–327. doi: 10.1093/embo-reports/kvd077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausmess N, Kuhn JR, Jacobs-Wagner C. The bacterial cytoskeleton: an intermediate filament-like function in cell shape. Cell. 2003;115:705–713. doi: 10.1016/S0092-8674(03)00935-8. [DOI] [PubMed] [Google Scholar]
- Dolan MF, Melnitsky H, Margulis L, Kolnicki R. Motility proteins and the origin of the nucleus. Anat Rec. 2002;268:290–301. doi: 10.1002/ar.10161. [DOI] [PubMed] [Google Scholar]
- Rose A, Patel S, Meier I. The plant nuclear envelope. Planta. 2004;218:327–336. doi: 10.1007/s00425-003-1132-2. [DOI] [PubMed] [Google Scholar]
- Reddy ASN, Day IS. Kinesins in the Arabidopsis genome: a comparative analysis among eukaryotes. BMC Genomics. 2001;2:2. doi: 10.1186/1471-2164-2-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YRJ, Liu B. Cytoskeletal motors in Arabidopsis. Sixty-one kinesins and seventeen myosins. Plant Physiol. 2004;136:3877–3883. doi: 10.1104/pp.104.052621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger B, Singh M. An iterative method for improved protein structural motif recognition. J Comput Biol. 1997;4:261–273. doi: 10.1089/cmb.1997.4.261. [DOI] [PubMed] [Google Scholar]
- Matsui M, Stoop CD, von Armin AG, Wei N, Deng XW. Arabidopsis COP1 protein specifically interacts in vitro with a cytoskeleton-associated protein, CIP1. Proc Natl Acad Sci USA. 1995;92:4239–4243. doi: 10.1073/pnas.92.10.4239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- European Bioinformatics Institute (EBI) http://www.ebi.ac.uk/
- The Institute for Genome Research (TIGR) http://www.tigr.org/
- Ohio Bioscience Library http://bioinformatics.osc.edu/obl/
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page RDM, TreeView, 2001 http://taxonomy.zoology.gla.ac.uk/rod/treeview.html
- Hanlon DW, Rosario MM, Ordal GW, Venema G, Van Sinderen D. Identification of TlpC, a novel 62 kDa MCP-like protein from Bacillus subtilis. Microbiology. 1994;140:1847–1854. doi: 10.1099/13500872-140-8-1847. [DOI] [PubMed] [Google Scholar]
- Marconi RT, Samuels DS, Landry RK, Garon CF. Analysis of the distribution and molecular heterogeneity of the ospD gene among the lyme disease spirochetes: evidence for lateral gene exchange. J Bacteriol. 1994;176:4572–4582. doi: 10.1128/jb.176.15.4572-4582.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norris SJ, Carter CJ, Howell JK, Barbour AG. Low-passage-associated proteins of Borrelia burgdorferi B31: characterization and molecular cloning of OspD, a surface-exposed, plasmid-encoded lipoprotein. Infect Immun. 1992;60:4662–4672. doi: 10.1128/iai.60.11.4662-4672.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amano M, Fukata Y, Kaibuchi K. Regulation and functions of Rho-associated kinase. Exp Cell Res. 2000;261:44–51. doi: 10.1006/excr.2000.5046. [DOI] [PubMed] [Google Scholar]
- Ko J, Na M, Kim S, Lee JR, Kim E. Interaction of the ERC family of RIM-binding proteins with the liprin-alpha family of multidomain proteins. J Biol Chem. 2003;278:42377–42385. doi: 10.1074/jbc.M307561200. [DOI] [PubMed] [Google Scholar]
- Wielowieyski PA, Sevinc S, Guzzo R, Salih M, Wigle JT, Tuana BS. Alternative splicing, expression, and genomic structure of the 3' region of the gene encoding the sarcolemmal-associated proteins (SLAPs) defines a novel class of coiled-coil tail-anchored membrane proteins. J Biol Chem. 2000;275:38474–38481. doi: 10.1074/jbc.M007682200. [DOI] [PubMed] [Google Scholar]
- Mok SC, Wong KK, Chan RK, Lau CC, Tsao SW, Knapp RC, Berkowitz RS. Molecular cloning of differentially expressed genes in human epithelian ovarian cancer. Gynecol Oncol. 1994;52:247–252. doi: 10.1006/gyno.1994.1040. [DOI] [PubMed] [Google Scholar]
- Liao H, Winkfein RJ, Mack G, Rattner JB, Yen TJ. CENP-F is a protein of the nuclear matrix that assembles onto kinetochores at late G2 and is rapidly degraded after mitosis. J Cell Biol. 1995;130:507–518. doi: 10.1083/jcb.130.3.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng MH. Death associated protein kinase: from regulation of apoptosis to tumor suppressive functions and B cell malignancies. Apoptosis. 2002;7:261–270. doi: 10.1023/A:1015364104672. [DOI] [PubMed] [Google Scholar]
- Zhang R, Epstein HF. Homodimerization through coiled-coil regions enhance activity of the myotonic dystrophy protein kinase. FEBS Lett. 2003;546:281–287. doi: 10.1016/S0014-5793(03)00601-X. [DOI] [PubMed] [Google Scholar]
- Altman R, Kellogg D. Control of mitotic events by Nap1 and he Gin4 kinase. J Cell Biol. 1997;138:119–130. doi: 10.1083/jcb.138.1.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh R, Fujiwara M, Yoshida S. Kinesin-related proteins with a mitochondrial targeting signal. Plant Physiol. 2001;127:724–726. doi: 10.1104/pp.127.3.724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka H, Ishikawa M, Kitamura S, Takahashi Y, Soyano T, Machida C, Machida Y. The AtNACK1/HINKEL and STUD/TETRASPORE/AtNACK2 genes, which encode functionally redundant kinesins, are essential for cytokinesis in Arabidopsis. Genes Cells. 2004;9:1199–1211. doi: 10.1111/j.1365-2443.2004.00798.x. [DOI] [PubMed] [Google Scholar]
- Hays JL, Watowich SJ. Oligomerization-dependent changes in the thermodynamic properties of the TPR-MET receptor tyrosine kinase. Biochemistry. 2004;43:10570–10578. doi: 10.1021/bi0363275. [DOI] [PubMed] [Google Scholar]
- Salcini AE, Chen H, Iannolo G, De Camilli P, Di Fiore PP. Epidermal growth factor pathway substrate 15, Esp15. Int J Biochem Cell Biol. 1999;31:805–809. doi: 10.1016/S1357-2725(99)00042-4. [DOI] [PubMed] [Google Scholar]
- Weiner JA, Chun J. Png-1, a nervous system-specific zinc finger gene, identifies regions containing postmitotic neurons during mammalian embryonic development. J Comp Neurol. 1997;381:130–142. doi: 10.1002/(SICI)1096-9861(19970505)381:2<130::AID-CNE2>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- Samuels-Lev Y, O'Connor DJ, Bergamaschi D, Trigante G, Hsieh JK, Zhong S, Campargue I, Naumovski L, Crook T, Lu X. ASPP proteins specifically stimulate the apoptotic function of p53. Mol Cell. 2001;8:781–794. doi: 10.1016/S1097-2765(01)00367-7. [DOI] [PubMed] [Google Scholar]
- Munton RP, Vizi S, Mansuy IM. The role of protein phosphatase-1 in the modulation of synaptic and structural plasticity. FEBS Lett. 2004;567:121–128. doi: 10.1016/j.febslet.2004.03.121. [DOI] [PubMed] [Google Scholar]
- Yang J, Kim O, Wu J, Qiu Y. Interaction between tyrosine kinase Etk and a RUN domain- and FYVE domain-containing protein RUFY1. J Biol Chem. 2002;277:30219–30226. doi: 10.1074/jbc.M111933200. [DOI] [PubMed] [Google Scholar]
- Zhong R, Burk DH, Morrison H, III, Ye ZH. A kinesin-like protein is essential for oriented deposition of cellulose microfibrils and cell wall strength. Plant Cell. 2002;14:3101–3117. doi: 10.1105/tpc.005801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YRJ, Giang HM, Liu B. A novel plant kinesin-related protein specifically associates with the phragmoplast organelles. Plant Cell. 2001;13:2427–2439. doi: 10.1105/tpc.13.11.2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan R, Lee YRJ, Liu B. Localization of two homologous Arabidopsis kinesin-related proteins in the phragmoplast. Planta. 2004;220:156–164. doi: 10.1007/s00425-004-1324-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Prokaryotic coiled-coil proteins Tables S1-S14: Protein details of all long coiled-coil proteins predicted in the prokaryotic genomes analyzed in this study. Open file with Acrobat Reader.
Eukaryotic clusters of interest Figures S1-S6: Phylogenetic trees based on ClustalW alignments of the sequences, displayed using TreeView v.1.6.6. Open file with Acrobat Reader.
Sequence details for Figures S1-S6, supplement to additional file 2. Table S15: Protein information and prediction data for sequences contained in Figures S1-S6. AGI locus numbers from TAIR are used as sequence IDs for Arabidopsis, TIGR sequence IDs are used for rice and Synechocystis. All other sequence IDs correspond to the EBI identifiers in the downloaded FASTA files. Max. Coil Length, longest coiled-coil domain in the protein sequence; Coil Coverage, percent of sequence predicted to be in a coiled-coil. Open file with Microsoft Excel.
Yeast clusters Table S18: Protein information and prediction data for sequences in yeast clusters with two species (Saccharomyces cerevisiae and Schizosaccharomyces pombe) represented. Sequence IDs correspond to the EBI identifiers in the downloaded FASTA files. Max. Coil Length, longest coiled-coil domain in the protein sequence; Coil Coverage, percent of sequence predicted to be in a coiled-coil. Open file with Microsoft Excel.
Small mammalian clusters; supplement to Table 9. Table S16: Protein information and prediction data for sequences in mammalian clusters with two species (mouse, human) represented and less than 10 sequences per cluster. Sequence IDs correspond to the EBI identifiers in the downloaded FASTA files. Max. Coil Length, longest coiled-coil domain in the protein sequence; Coil Coverage, percent of sequence predicted to be in a coiled-coil. Open file with Microsoft Excel.
Small plant clusters; supplement to Table 10. Table S17: Protein information and prediction data for sequences in plant clusters with two species (Arabidopsis, rice) represents and less than 10 sequences per cluster. AGI locus numbers from TAIR or NCBI RefSeq numbers are used as sequence IDs for Arabidopsis, TIGR sequence IDs are used for rice. Max. Coil Length, longest coiled-coil domain in the protein sequence; Coil Coverage, percent of sequence predicted to be in a coiled-coil. Open file with Microsoft Excel.