Abstract
In Agrobacterium-mediated genetic transformation of plant cells, the bacterium exports a well defined transferred DNA (T-DNA) fragment and a series of virulence proteins into the host cell. Following its nuclear import, the single-stranded T-DNA is stripped of its escorting proteins, most likely converts to a double-stranded (ds) form, and integrates into the host genome. Little is known about the precise mechanism of T-DNA integration in plants, and no plant proteins specifically associated to T-DNA have been identified. Here we report the direct involvement of KU80, a protein that binds dsT-DNA intermediates. We show that ku80-mutant Arabidopsis plants are defective in T-DNA integration in somatic cells, whereas KU80-overexpressing plants exhibit increased susceptibility to Agrobacterium infection and increased resistance to DNA-damaging agents. The direct interaction between dsT-DNA molecules and KU80 in planta was confirmed by immunoprecipitation of KU80 dsT-DNA complexes from Agrobacterium-infected plants. Transformation of KU80-overexpressing plants with two separate T-DNA molecules resulted in an increased rate of extrachromosomal T-DNA to T-DNA recombination, indicating that KU80 bridges between dsT-DNAs and double-strand breaks. This last result further supports the notion that integration of T-DNA molecules occurs through ds intermediates and requires active participation of the host's nonhomologous end-joining repair machinery.
Keywords: Agrobacterium, DNA repair, double-strand breaks
Agrobacterium is the only known organism capable of transkingdom DNA transfer (1) to a wide number of species from different genera. These include plants (reviewed in refs. 2 and 3), yeast (4), fungi (5, 6), and even human cells (7). The genetic transformation process begins with the transfer of a single-stranded (ss) copy (T-strand) of the transferred DNA (T-DNA) segment located on the bacterial tumor-inducing plasmid and several virulence (Vir) proteins into the host cell and ends with integration of the T-DNA into the host cell genome (2, 8). T-DNA travels into the host cell as a protein-DNA complex (T-complex), with a single VirD2 attached to the 5′ end of the T-strand. Inside the host cell, the remainder of the T-strand is thought to be packed with numerous molecules of VirE2, a ssDNA-binding protein (reviewed in refs. 8 and 9). The T-complex is proposed to be imported into the host cell nucleus by interaction of the VirE2 and VirD2 proteins with the bacterial protein VirE3 (10) and the host factors VIP1 (11) and importin α (12). Once inside the nucleus, the T-complex is stripped of its escorting proteins, VirE2 and VIP1, by targeted proteolysis (13) and converts into a double-stranded (ds) intermediate, before its ultimate integration into the host genome, most likely as a ds molecule (14, 15).
It is currently accepted that Agrobacterium Vir proteins do not possess any known DNA-repair activity, and T-DNA integration is therefore most likely effected by host proteins (16). Under this scenario, the host cell DNA-repair machinery recognizes dsT-DNA molecules as genomic ds breaks (DSBs) and mediates their integration by a DNA-repair mechanism. Indeed, the roles of various DNA-repair proteins have been studied in yeast-based systems, demonstrating that key DNA-repair proteins are essential for T-DNA integration into the yeast cell genome by either homologous or nonhomologous (illegitimate) recombination (HR and NHR, respectively) (17, 18). Because in plant cells, T-DNA integration occurs mainly through NHR, it is most likely that nonhomologous end-joining (NHEJ) proteins will be required for the process of T-DNA integration. Interestingly AtLIG4, a NHEJ plant protein found to be important for T-DNA integration in yeast cells, is not required for T-DNA integration in plants (19). Moreover, the involvement of AtKU80, a key NHEJ protein, in T-DNA integration is inconclusive, because it has been reported to be both required (20) and dispensable (21) for T-DNA integration. Note that the latter contradictory results may simply have arisen from the nature of the flower-dip transformation method, which is performed under relatively uncontrolled conditions. Thus, the question of which host factors control T-DNA's integration in somatic cells remains open. Here we show that ku80-mutant Arabidopsis plants are defective in T-DNA integration in somatic cells, whereas KU80-overexpressing plants exhibit increased susceptibility to Agrobacterium infection and increased resistance to DNA-damaging agents. We also demonstrate a direct link between dsT-DNA molecules and KU80 in planta and show that overexpression of KU80 in transgenic plants increases the rate of extrachromosomal T-DNA to T-DNA recombination, thus indicating a function for KU80 in bridging between dsT-DNAs and DSBs.
Materials and Methods
Plant Materials, Tumorigenesis, and Transient T-DNA Expression Assays. Arabidopsis plants, ecotype Ws, were grown and maintained under standard growth conditions and were characterized for stable and transient Agrobacterium-mediated genetic transformation as described (22). Briefly, for tumor-formation assay, root segments from 10- to 14-day-old in vitro-grown Arabidopsis seedlings were infected with Agrobacterium tumefaciens A208, cultivated for 48 h at 25°C on hormone-free Murashige and Skoog (MS) medium and then washed and cultured for 4-5 weeks in hormone-free MS medium supplemented with timentin to kill the bacteria. Tumors were then counted and their morphology documented. For transient expression, root segments were infected with Agrobacterium tumefaciens EHA105 harboring a β-glucuronidase (GUS)-expressing binary vector and cultivated on hormone-free medium for 48 h, transferred to fresh medium for an additional 2 days, and stained with 5-bromo-4-chloro-3-indolyl β-d-glucuronide (X-Gluc). For quantification of GUS activity, transiently GUS-expressing root segments were ground and assayed as described (23). For regeneration assay, root segments were cultured on callus-induction medium (22), and regeneration was scored after 5-6 weeks of culture. For transgenic callus formation assay, A. tumefaciens EHA105 harboring a bar-expressing binary vector was used to infect root segments as described, and the roots were then cultured on callus-induction medium for 4-5 weeks in the presence of herbicide (for selection of transgenic callus) and timentin (to kill the bacteria). For complementation of the ku80 mutant (24), a genomic fragment containing the native KU80 gene and its regulatory sequences was PCR-amplified, cloned into a hygromycin-resistance-coding T-DNA, sequence-verified, and transformed into a ku80 mutant by using a flower-dip method (25). For overexpression, the KU80-coding sequence was cloned under the control of the cauliflower mosaic virus (CaMV) 35S promoter and transformed into wild-type Arabidopsis plants using the flower-dip method (25).
T-DNA Immunoprecipitation. The KU80-coding sequence was first fused to a HIS tag at its C terminus by PCR amplification of the KU80 ORF and cloning into the BamHI-SalI sites of pET28c(+). This plasmid was used to PCR amplify the HIS::KU80 fusion and clone it as an NcoI-SalI fragment into pSAT4-MCS (26), resulting in pSAT4-HIS-KU80 in which the expression of the HIS::KU80 fusion is under the control of the cauliflower mosaic virus (CaMV) 35S promoter. The HIS::KU80 expression cassette was then transferred as an I-SceI fragment from pSAT4-HIS-KU80 into pPZP-RCS2-hpt (26, 27), resulting in the Agrobacterium binary plasmid pRCS2-HIS-KU80. Root segments from 10- to 14-day-old in vitro-grown Arabidopsis seedlings were then infected with a mixture of two A. tumefaciens strains carrying a HIS-KU80-overexpressing cassette, a transient source for His-tagged KU80 protein, and a GUS-expressing cassette as the target T-DNA and cultivated on hormone-free medium. After 48 h, the tissues were vigorously washed of bacteria, transferred to fresh medium for an additional 2 days, fixed with formaldehyde and used for coimmunoprecipitation assay as described (28), by using T-DNA-specific primers (G-F, 5′-GGATTGATGTGATATCTCCACTG and G-R, 5′-CTTTTTCCAGTACCTTCTCTGCC) to amplify the immunoprecipitated DNA molecules. Primers specific to VirE2 (E2-Q-F, 5′-CACCGGGATGTCCGAAATTGATAT and E2-Q-R, 5′-GGCTCCAGGTTATGCAATTGTGC), and the binary plasmid's backbone (PZ-Q-F, 5′-CCGAGTTCGAGCGTTCCCTAATC and PZ-Q-R, 5′-CTGAAAGTTGACCCGCTTCATGG) were used to confirm the absence of contaminating bacteria and binary plasmids, respectively.
T-DNA Recombination Assay. The GUSA-intron-coding sequence was split after the first 112 nucleotides of its 173-bp intron by PCR amplification of each part individually. The C terminus of the uidA gene and its adjacent partial intron and the N terminus of uidA and its adjunct partial intron were cloned as NcoI-BamHI fragments into the same sites of pRTL2-GUS, replacing the GUS gene and resulting in pRTL2-GUint and pRTL2-IntS, respectively. Next, the CaMV 35S promoter and its adjacent GUint fragment, and the IntS fragment and its adjacent CaMV 35S polyA signal, non-GUS-expressing cassettes, were transferred separately as HindIII fragments into the Agrobacterium binary plasmid pPZP-RCS1 (26), producing p35Spro-GUint and pIntS-35Ster. The binary plasmids were mobilized separately to Agrobacterium cells and a freshly prepared culture of the two Agrobacterium strains in a 1:1 mix was used for Arabidopsis root transformation. Recombination events were monitored by 5-bromo-4-chloro-3-indolyl β-d-glucuronide (X-Gluc) staining of the infected roots.
Quantitative Real-Time PCR. Wild-type and KU80-overexpressing Arabidopsis root segments were infected with Agrobacterium harboring the hpt (encoding hygromycin resistance) gene-containing binary plasmid as described, and were placed on callus-induction medium (22) without selection. DNA was extracted from same-weight amounts of callus collected from individual transformation events and was used for quantitative real-time PCR analysis using iQ SYBR Green Supermix (Bio-Rad) on an ABI Prism 7700 sequence detection system (Applied Biosystems). A total of seven KU80-overexpressing callus clusters were analyzed and the amount of T-DNA (amplified by HG-Q-F 5′-GGGCGTGGATATGTCCTGCGGGTAAATAG and HG-Q-R 5′-TGAATTCCCCAATGTCAAGCACTTCCGGAA) for each transformant was calculated relative to the amount of T-DNA from an average of five wild-type callus clusters, using actin (amplified by AC-Q-F 5′-CTCCTGCTATGTATGTTGCCATTCAAGCTGTTC and AC-Q-R 5′-GCGTAACCCTCGTAGATTGGTACCGTGT) for quantitative measurements. Samples were also analyzed for the presence of contaminating bacteria (by amplification of the VirE2 gene, using E2-Q-F and E2-Q-R) or binary plasmids (by amplification of the binary plasmid's backbone by using PZ-Q-F and PZ-Q-R). For positive controls, bacterial or plasmid amplification was analyzed in the presence of plant DNA by adding bacterial or plasmid DNA to wild-type callus clusters. Relative DNA amounts were calculated according to the ABI Prism 7700 sequence detection system (Applied Biosystems) instruction manual. Briefly, the ΔCt value (the difference between threshold t for each sample and its control) was calculated for each sample and the ΔΔCt value for each sample was calculated by subtracting its ΔCt value from its control. Finally, the calculated ratio between the amount of DNA in the sample and its control was expressed by 2-ΔΔCt.
Results
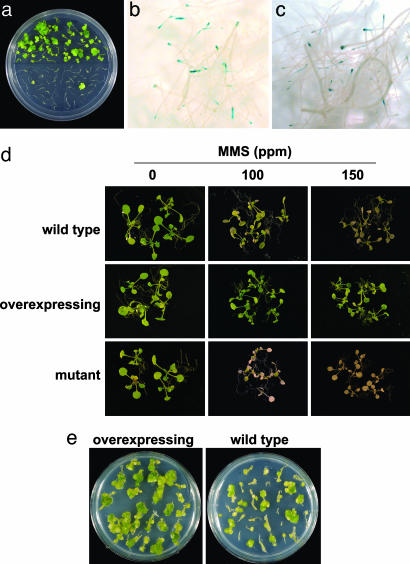
The Arabidopsis ku80 Mutant Is Defective in T-DNA Integration but Not Transient T-DNA Expression. Because recognition and repair of DNA breaks in plants are manifested by a set of NHEJ proteins, we reasoned that KU80 (29), a key protein in the NHEJ complex (30), may function during T-DNA integration. In accordance with our scenario, dsT-DNA intermediates would be recognized by KU80 as simple DSBs and would be directed to integration by a NHEJ mechanism. Thus, one would expect disruption of KU80 in plant cells to affect the T-DNA integration process. Indeed, Fig. 1 and Table 1 show that Agrobacterium-mediated genetic transformation of somatic cells is blocked in an Arabidopsis ku80 mutant. After infection with wild-type Agrobacterium, significantly reduced tumor formation was observed in the Arabidopsis ku80 mutant (Fig. 1a, bottom) as compared with wild-type plants (Fig. 1a, top). No differences, however, were observed between wild-type (Fig. 1b) and ku80-mutant (Fig. 1c) plants in transient GUS expression after infection with a disarmed Agrobacterium strain that carries the uidA-intron gene. A quantitative 4-methylumbelliferyl β-d-galactoside (MUG) assay confirmed the similar uidA-intron expression levels in the ku80-mutant and wild-type plants (Table 1), thus indicating that ku80-mutant plants show similar nuclear import, conversion of the T-DNA molecule to ds form, and transient expression, as the wild-type plants. A second assay was performed to further investigate the specific role of ku80 in T-DNA integration. After infection with disarmed Agrobacterium carrying a T-DNA encoding an herbicide-resistance gene, significantly reduced transgenic-callus formation was observed in the Arabidopsis ku80 mutant relative to the wild-type plants (Table 2). That no significant differences were observed in callus regeneration among noninfected wild-type and ku80-mutant plants after cultivation of root segments on nonselective callus-regeneration medium (Table 2) further supports a specific link between KU80 and the integration process. The susceptibility of the ku80 mutant to Agrobacterium-mediated transformation by flower dip (21), a phenomenon typical to many other mutants that are resistant to Agrobacterium transformation (rat) (31), enables genetic complementation of the ku80 mutant with its native gene and restoration of the complemented mutant's susceptibility to infection by a tumorigenic Agrobacterium strain (Table 1). This demonstrates that the ku80 phenotype most likely results from disruption of the native ku80 function by the mutation in the gene.
Fig. 1.
ku80 phenotype: effect on DNA repair and transient and stable genetic transformation by Agrobacterium. ku80-mutant plants are blocked at the T-DNA integration but not the transient-expression step of the Agrobacterium-mediated genetic transformation process. (a) Stable transformation of wild-type (top) and ku80-mutant (bottom) Arabidopsis plants. Root explants were infected with a tumorigenic Agrobacterium strain, and galls were scored 4-5 weeks postinoculation, as described (32). (b and c) Transient GUS expression of wild-type (b) and ku80-mutant (c) Arabidopsis plants. Root explants were infected with a disarmed Agrobacterium strain carrying the uidA-intron gene and were stained 4-5 days postinoculation. KU80-overexpressing plants are resistant to DNA damage and show high susceptibility to Agrobacterium infection. (d) Wild-type, ku80-mutant, and KU80-overexpressing plants were germinated on MS agar, and 1-week-old plants were transferred to MS agar with or without methyl methane sulfonate (MMS), grown for an additional 5 days, and photographed. (e) Stable transformation of wild-type and KU80-overexpressing plants. Root explants were infected with a tumorigenic Agrobacterium strain and galls were scored 4-5 weeks postinoculation.
Table 1. Quantitative analysis of ku80-mutant, complemented ku80-mutant, wild-type, and KU80-overexpressing plants.
| Plant | Roots producing galls,* % | Tumor morphology† | GUS expression,‡ % of control |
|---|---|---|---|
| High-concentration inoculum§ | |||
| Wild type | 92 ± 3 | Large and green | 100 |
| ku80 mutant | 7 ± 1 | Very small, yellow | 95 ± 2 |
| Rescued ku80 mutant | 89 ± 4 | Large and green | 96 ± 3 |
| Low-concentration inoculum | |||
| KU80 overexpression | 88 ± 3 | Very large and green | 52 ± 4 |
| Wild type | 42 ± 5 | Large and green | 50 ± 3 |
Numbers represent the percentages of root segments producing galls 5–6 weeks postinoculation (n = 10 plates)
Characterization of tumor morphology, as described (22)
GUS expression levels were calculated as percentages of the control (wild-type plants infected with a high concentration of Agrobacterium inoculum) (n = 10 plates)
High-concentration Agrobacterium inoculum was used for comparison between the wild type, ku80 mutant, and rescued ku80 mutant, whereas low-concentration Agrobacterium inoculum was used for comparison between wild-type and KU80-overexpressing plants
Table 2. Callus formation from ku80-mutant, wild-type, and KU80-overexpressing plants.
| Basta-resistant calli†
|
|||
|---|---|---|---|
| Plant | Nontransformed calli* | High-concentration inoculum‡ | Low-concentration inoculum‡ |
| Wild type | 89 ± 7§ | 76 ± 5 | 34 ± 7 |
| ku80 mutant | 84 ± 2 | 4 ± 1 | ND |
| KU80 overexpression | 89 ± 4 | ND | 68 ± 3 |
Nontransformed callus was induced on nonselective callus-inducing medium, without inoculation with Agrobacterium
Transgenic callus was induced on herbicide containing callus-inducing medium, after inoculation with Agrobacterium
High-concentration Agrobacterium inoculum was used for comparison between the wild-type and ku80 mutant, whereas low-concentration Agrobacterium inoculum was used for comparison between wild-type and KU80-overexpressing plants
Numbers represent the percentages of root segments producing calli after 4–5 weeks of culture (n = 6 plates)
KU80-Overexpressing Transgenic Arabidopsis Plants Are Resistant to DNA Damage and Are More Susceptible to Agrobacterium Infection. To further determine the molecular role played by KU80 in the transformation process, we produced transgenic Arabidopsis plants that overexpress the KU80-coding sequence under a 35S promoter. First, we evaluated the in vivo effect of KU80 overexpression on DNA repair by comparing the sensitivity of wild-type, KU80-overexpressing, and ku80-mutant plants to methyl methane sulfonate (MMS) (a DNA-damaging agent). Fig. 1d shows that, whereas the ku80 mutant was hypersensitive, relative to controls, to 100-ppm MMS, KU80-overexpressing plants exhibited enhanced resistance to MMS, even at 150 ppm. Similarly, KU80-overexpressing plants exhibited enhanced resistance to bleomycin (another DNA-damaging agent). Thus, overexpression of KU80 in Arabidopsis plants can specifically enhance the plant's cellular resistance to DNA damage. Next, we performed both tumorigenesis and transient gene-expression assays (22) on wild-type and KU80-overexpressing plants. Because the Arabidopsis ecotype Ws is highly susceptible to Agrobacterium transformation, we altered the transformation assays as described (32) by using a 1,000-fold lower concentration of Agrobacterium inoculum to differentiate between wild-type and KU80-overexpressing plants. Fig. 1e shows that even at a low Agrobacterium concentration, infected roots from wild-type plants produced galls. However, the number of gall-producing roots was 2-fold higher with the KU80-overexpressing plants, and the average size of the transgenic galls was much larger on these roots, as compared with the wild type (Fig. 1d and Table 1). No differences in GUS expression level were observed between wild-type and KU80-overexpressing Arabidopsis plants (Table 1) after infection with the uidA-intron-carrying T-DNA. KU80-overexpressing plants were also more susceptible to infection with disarmed Agrobacterium carrying a T-DNA encoding a herbicide-resistance gene but exhibited a callus-regeneration response on nonselective media that was similar to that of wild-type plants (Table 2). Furthermore, the regenerated calli were similar in size and shape in wild-type and KU80-overexpressing plants, under both control and selective conditions. These results indicate that overexpression of KU80 does not affect transient gene expression from the T-DNA but increases the extent of T-DNA integration into the host genome, and that KU80 does not nonspecifically affect the plant's regeneration potential.
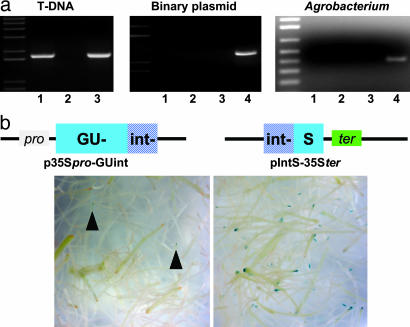
KU80 Binds to and Recombines dsT-DNA Molecules in Planta. Detection and repair of DNA damage in living cells begin with the recognition of DNA lesions by a KU protein complex. In addition, one line of experimental evidence suggests that dsT-DNA molecules, which are rapidly converted from T-strands to their ds form early in the infection process (33), are essential intermediates of T-DNA integration (reviewed in ref. 16). To test whether KU80 physically recognizes and interacts with dsT-DNA molecules in planta, we immunoprecipitated T-DNA molecules with KU80 from Agrobacterium-infected Arabidopsis cells, which were transiently expressing His-tagged KU80 (Fig. 2a). No PCR amplification of bacterial genes or the binary plasmid's backbone was observed in the various immunoprecipitation extracts (Fig. 2a), indicating the specific immunoprecipitation of dsT-DNA molecules by His-tagged KU80. Because KU80 binds to ds- but not ssDNA molecules during DSB repair (34), our results clearly indicate that dsT-DNA molecules are recognized as DSBs by the NHEJ repair machinery. It follows that KU80 should also be able to recognize the presence of two or even more T-DNA molecules and to facilitate their extrachromosomal recombination to each other. To explore this possibility, we designed an assay that would enable us to monitor the extent of recombination events between two specific T-DNA molecules in planta. In this assay, we split a uidA-intron-expressing cassette into two parts and cloned each part on a different Agrobacterium binary plasmid (Fig. 2b). In the 35Spro-GUint binary plasmid, the C terminus of the uidA gene and part of the 173-bp intron were cloned under the control of the tandem 35S promoter, whereas in pIntS-35Ster, the N terminus of the uidA gene and part of the 173-bp intron were cloned downstream of the 35S polyA signal. Although functional expression of the uidA-reporter gene could not be achieved by transfection of plant cells with either plasmid alone (data not shown), reconstruction of the uidA-intron expression cassette by directional recombination of two T-DNA molecules in plant cells was expected to result in functional GUS expression. After infection with a mix of the two Agrobacterium strains, functional GUS expression was indeed observed in wild-type Arabidopsis roots (Fig. 2b). However, events of T-DNA to T-DNA recombination leading to GUS expression in wild-type plants were rare (an average of four events per 150 root segments) as compared with KU80-overexpressing plants (an average of 40 events per 150 root segments) thus indicating the crucial role of KU80 in recognizing T-DNA molecules in planta.
Fig. 2.
The KU80 protein binds to and recombines dsT-DNA molecules in planta. (a) PCR analysis of T-DNA molecules immunoprecipitated and PCR-amplified as described (28) with antibodies against a His-tagged KU80 protein and using primers specific to T-DNA (Left), binary plasmid (center) and Agrobacterium VirE2 (Right). Lanes 1 (a and b) Precipitated extract treated with anti-His antibodies; lanes 2 (a and b) mock-precipitated extract to which no antibody was added; lanes 3 (a and b) untreated total DNA extract; lane 4 (Center) amplification of plasmid DNA and lane 4 (Right) amplification of bacterial DNA. (b) T-DNA to T-DNA recombination assay. Wild-type (Left) and KU80-overexpressing (Right) root segments were cotransformed with two binary plasmids, each carrying a partial GUS expression cassette (i.e., promoter-GUSint and intGUS-terminator). T-DNA to T-DNA recombination and expression of the GUS reporter gene were monitored by 5-bromo-4-chloro-3-indolyl β-d-glucuronide (X-Gluc) staining. Arrows (Left) indicate the presence of GUS-stained tissue in wild-type root segments.
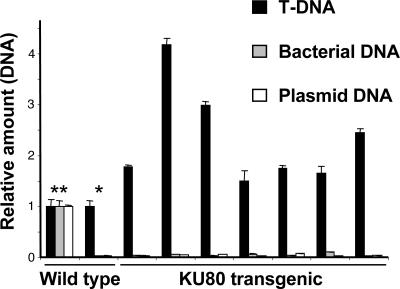
KU80 Increases the Rate of T-DNA Integration. The efficiency of KU80 binding to dsT-DNA molecules in transgenic plants is expected to affect the overall efficiency of T-DNA integration into the plant genome, even without applying selection pressure. To study this question, we used real-time PCR to quantify the rate of T-DNA integration after infection of transgenic and wild-type Arabidopsis roots and found the number of T-DNA integration events in KU80-transgenic plants to be 1.5- to 4.1-fold higher than in wild type (Fig. 3). PCR amplification of bacterial genes and binary plasmid backbone could be observed in samples where bacterial or plasmid DNA had been intentionally added but not in wild-type or KU80-transgenic clusters, indicating the specific amplification of integrated T-DNA molecules (Fig. 3).
Fig. 3.
Quantitative real-time PCR analysis of T-DNA integration in wild-type and KU80-transgenic plants. The amounts of integrating T-DNA molecules (black), bacterial DNA (gray), and binary plasmid DNA (white) were normalized against actin, serving as an internal control. The amounts of T-DNA molecules from individual KU80-overexpressing calli are presented in relative amounts in comparison to an average T-DNA amount from five individual wild-type calli. The amounts of bacterial and plasmid DNA molecules amplified from wild-type and KU80-overexpressing calli (*) are presented in relative amounts in comparison with control experiments, where bacterial and plasmid DNA was added to wild-type calli (**). Each reaction was repeated three times.
Discussion
Agrobacterium is a unique organism capable of genetically transforming many eukaryotic cells. To achieve its goal of transferring its T-DNA molecule and integrating it into the host cell genome, Agrobacterium relies not only on a set of its virulence proteins but also on their cellular partners and on various basic cellular processes (reviewed in refs. 2 and 8). For example, dynein-motor-assisted movement on microtubules has been proposed to be the driving force behind T-DNA transport through the host cell cytoplasm (35), and the interactions of the VirE2 and VirD2 proteins with the host factors VIP1 (11) and importin α (12) have been found to be important for T-complex import into the host cell nucleus (36). Inside the nucleus, T-DNA interactions with several host proteins have been suggested to facilitate its movement to points of integration (37, 38) and its dissociation from its escorting proteins (13). The last steps of the transformation process, i.e., conversion of the T-DNA to a ds molecule and its integration into the host cell genome, are most likely effected exclusively by host proteins and the host cell DNA-repair machinery (16). Indeed, genetic analysis has revealed the importance of various DNA-repair proteins for T-DNA integration in yeast cells (17, 18), and the plant's DNA-packaging protein H2A has been found essential for T-DNA integration in plants (32). Yet the key DNA-repair proteins and the molecular mechanism for T-DNA integration in plants has remained mostly unknown. Our results provide molecular evidence for the function of the plant protein KU80, a key protein in the NHEJ pathway, in T-DNA integration, suggesting that KU80 serves as the first point of contact between the T-DNA and the DNA-repair machinery. Many Arabidopsis mutants have been reported to be blocked in the last steps of the transformation process (22). However, although some of them may be blocked in the integration step itself (i.e., ref. 32), others may be deficient in various cellular processes leading to the formation of gall tissue, such as T-DNA expression, dedifferentiation of the transformed cells and redifferentiation into gall tissue. That an Arabidopsis ku80 mutant was blocked in one of the last steps of the genetic transformation process and that T-DNA nuclear import, conversion to the ds form and transient expression were not affected by the lack of KU80 protein support the notion that ku80 is not defective in T-DNA expression but most probably in T-DNA integration. Furthermore, that callus regeneration from ku80-mutant and KU80-overexpressing plants was similar to that from wild-type plants supports the notion that ku80 is not affected in dedifferentiation or redifferentiation of transformed or wild-type cells. KU80 overexpression resulted in larger tumors, but not callus clusters, after infection with wild-type or disarmed Agrobacterium, respectively. The herbicide-resistant callus clusters obtained from wild-type and KU80-overexpressing plants were similar in size, and the number of T-DNA integration events was higher in KU80-overexpressing plants. This suggests that the increased tumor size can be attributed to multiple integration events in the transformed tissues. The latter would lead to a larger number of oncogenic genes, and their expression would most likely result in higher cell division and proliferation rate. Interestingly, similar to many other rat mutants (31), the ku80 mutant can be transformed by Agrobacterium-mediated transformation via flower dip (20, 21). Because of the nature of the flower-dip transformation method, which is performed under relatively uncontrolled conditions, it is rather difficult to use it to clearly define the susceptibility or resistance of mutant lines; it can, however, serve to determine whether they are transformable or not. Indeed, controversy over the role of KU80 in flower-dip transformation still exists (20, 21), and additional data are required to clearly determine whether KU80 is essential during ovule transformation as it is in somatic cells. Nevertheless, it is likely that different and perhaps supplemental pathways govern the repair of DNA breaks in somatic and germ-line tissues; more data are required to accurately compare the mechanism of integration in these different tissues.
Previous studies have shown that T-DNA molecules, arriving from a single or two different agrobacteria, can integrate into the same location on the plant chromosome (39-42). Sequence analysis of some insertions has revealed precise fusion between two right-border ends, an event that can occur only through integration of dsT-DNA intermediates. Furthermore, one line of experimental evidence suggests that dsT-DNA molecules, which are rapidly converted from T-strands to their ds form early in the infection process (33), are dominant intermediates of T-DNA integration (14-16, 43) and do not necessarily represent a minor pathway of the integration process. These studies have also shown that dsT-DNA molecules can be directed to DSBs (14, 15, 43). Detection and repair of DNA damage in living cells begin with the recognition of DNA lesions by a protein sensor complex and are followed by its repair by DNA-repair proteins (30). More specifically, a simple DNA DSB is first recognized by a KU protein complex and then directed for repair by the XRCC4-LIG4 complex (44). Immunoprecipitation of dsT-DNA molecules with KU80 from infected plants and the high number of T-DNA to T-DNA recombination events in KU80-overexpressing plants clearly suggest that KU80 physically recognizes and interacts with dsT-DNA molecules, recognizing them as DSBs, and leads them to repair by the NHEJ repair machinery. This therefore suggests a simple DNA-repair mechanism for T-DNA integration into the genome.
Interestingly, the efficiency of T-DNA integration in KU80-transgenic plants was lower than expected when comparing the efficiency of extrachromosomal T-DNA to T-DNA recombination in KU80-transgenic and wild-type plants. Because naturally occurring DSBs may act as molecular “baits” for the incoming T-DNA molecules (14), and because their extent is most likely similar in wild-type and transgenic plants, it would be rational to assume that KU80 overexpression in transgenic plants leads to more efficient DNA-repair activity and to an overall reduction in DSB sites accessible for T-DNA integration. Thus, even a 1.5-fold increase in T-DNA integration rate in KU80-transgenic plants is, in fact, significant and further supports the crucial and direct participation of KU80 in the integration process.
Our data provide a mechanistic basis for the role of NHEJ in the integration process and suggest that dsT-DNA molecules are recognized as DSBs by the host's NHEJ machinery. We identified KU80 as a key protein in the T-DNA integration process and suggested that it recognizes and binds to dsT-DNA intermediates. Other DNA-repair proteins, such as KU70, RAD50, and MRE11, have been found to be important for T-DNA integration in yeast cells by NHEJ (17, 18), and it is crucial to study the function of their plant orthologues to further understand the mechanism of T-DNA integration in plant species.
Acknowledgments
We thank Dr. S. Gelvin for the stimulating discussion and the critical reading of the manuscript. This work was supported by a grant from the Human Frontiers Science Program (HFSP) and the U.S.-Israel Binational Agricultural Research and Development Fund (BARD) (to T.T.).
Author contributions: A.V., V.C., and T.T. designed research; J.L. and M.V. performed research; C.W. contributed new reagents/analytic tools; J.L. and T.T. analyzed data; and T.T. wrote the paper.
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: T-DNA, transferred DNA: ss, single stranded; ds, double stranded; T-strand, ss copy of T-DNA; Vir, virulence; DSB, double-strand break; NHEJ, nonhomologous end-joining; MS, Murashige and Skoog; GUS, β-glucuronidase.
References
- 1.Stachel, S. E. & Zambryski, P. C. (1989) Nature 340, 190-191. [DOI] [PubMed] [Google Scholar]
- 2.Gelvin, S. B. (2003) Microbiol. Mol. Biol. Rev. 67, 16-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gelvin, S. B. (2000) Annu. Rev. Plant Physiol. Plant Mol. Biol. 51, 223-256. [DOI] [PubMed] [Google Scholar]
- 4.Bundock, P., den Dulk-Ras, A., Beijersbergen, A. & Hooykaas, P. J. J. (1995) EMBO J. 14, 3206-3214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Groot, M. J., Bundock, P., Hooykaas, P. J. J. & Beijersbergen, A. G. (1998) Nat. Biotechnol. 16, 839-842. [DOI] [PubMed] [Google Scholar]
- 6.Gouka, R. J., Gerk, C., Hooykaas, P. J., Bundock, P., Musters, W., Verrips, C. T. & de Groot, M. J. (1999) Nat. Biotechnol. 17, 598-601. [DOI] [PubMed] [Google Scholar]
- 7.Kunik, T., Tzfira, T., Kapulnik, Y., Gafni, Y., Dingwall, C. & Citovsky, V. (2001) Proc. Natl. Acad. Sci. USA 98, 1871-1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tzfira, T. & Citovsky, V. (2002) Trends Cell Biol. 12, 121-129. [DOI] [PubMed] [Google Scholar]
- 9.Zupan, J., Muth, T. R., Draper, O. & Zambryski, P. C. (2000) Plant J. 23, 11-28. [DOI] [PubMed] [Google Scholar]
- 10.Lacroix, B., Vaidya, M., Tzfira, T. & Citovsky, V. (2005) EMBO J. 24, 428-437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tzfira, T., Vaidya, M. & Citovsky, V. (2001) EMBO J. 20, 3596-3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ballas, N. & Citovsky, V. (1997) Proc. Natl. Acad. Sci. USA 94, 10723-10728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tzfira, T., Vaidya, M. & Citovsky, V. (2004) Nature 431, 87-92. [DOI] [PubMed] [Google Scholar]
- 14.Tzfira, T., Frankmen, L., Vaidya, M. & Citovsky, V. (2003) Plant Physiol. 133, 1011-1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chilton, M.-D. M. & Que, Q. (2003) Plant Physiol. 133, 956-965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tzfira, T., Li, J., Lacroix, B. & Citovsky, V. (2004) Trends Genet. 20, 375-383. [DOI] [PubMed] [Google Scholar]
- 17.van Attikum, H., Bundock, P. & Hooykaas, P. J. J. (2001) EMBO J. 20, 6550-6558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van Attikum, H. & Hooykaas, P. J. J. (2003) Nucleic Acids Res. 31, 826-832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van Attikum, H., Bundock, P., Overmeer, R. M., Lee, L. Y., Gelvin, S. B. & Hooykaas, P. J. (2003) Nucleic Acids Res. 31, 4247-4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Friesner, J. & Britt, A. B. (2003) Plant J. 34, 427-440. [DOI] [PubMed] [Google Scholar]
- 21.Gallego, M. E., Bleuyard, J. Y., Daoudal-Cotterell, S., Jallut, N. & White, C. I. (2003) Plant J. 35, 557-565. [DOI] [PubMed] [Google Scholar]
- 22.Zhu, Y., Nam, J., Humara, J. M., Mysore, K., Lee, L. Y., Cao, H., Valentine, L., Li, J., Kaiser, A., Kopecky, A., et al. (2003) Plant Physiol. 132, 494-505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nam, J., Mysore, K. S., Zheng, C., Knue, M. K., Matthysse, A. G. & Gelvin, S. B. (1999) Mol. Gen. Genet. 261, 429-438. [DOI] [PubMed] [Google Scholar]
- 24.Gallego, M. E., Jalut, N. & White, C. I. (2003) Plant Cell 15, 782-789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ye, G. N., Stone, D., Pang, S. Z., Creely, W., Gonzalez, K. & Hinchee, M. (1999) Plant J. 19, 249-257. [DOI] [PubMed] [Google Scholar]
- 26.Tzfira, T., Tian, G.-W., Lacroix, B., Vyas, S., Li, J., Leitner-Dagan, Y., Krichevsky, A., Taylor, T., Vainstein, A. & Citovsky, V. (2005) Plant Mol. Biol. 57, 503-516. [DOI] [PubMed] [Google Scholar]
- 27.Chung, S. M., Frankman, E. L. & Tzfira, T. (2005) Trends Plant Sci. 10, 357-361. [DOI] [PubMed] [Google Scholar]
- 28.Gendrel, A. V., Lippman, Z., Yordan, C., Colot, V. & Martienssen, R. A. (2002) Science 297, 1871-1873. [DOI] [PubMed] [Google Scholar]
- 29.West, C. E., Waterworth, W. M., Story, G. W., Sunderland, P. A., Jiang, Q. & Bray, C. M. (2002) Plant J. 31, 517-528. [DOI] [PubMed] [Google Scholar]
- 30.Carr, A. M. (2003) Science 300, 1512-1513. [DOI] [PubMed] [Google Scholar]
- 31.Mysore, K. S., Kumar, C. T. & Gelvin, S. B. (2000) Plant J. 21, 9-16. [DOI] [PubMed] [Google Scholar]
- 32.Mysore, K. S., Nam, J. & Gelvin, S. B. (2000) Proc. Natl. Acad. Sci. USA 97, 948-953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Narasimhulu, S. B., Deng, X.-B., Sarria, R. & Gelvin, S. B. (1996) Plant Cell 8, 873-886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tamura, K., Adachi, Y., Chiba, K., Oguchi, K. & Takahashi, H. (2002) Plant J. 29, 771-781. [DOI] [PubMed] [Google Scholar]
- 35.Salman, H., Abu-Arish, A., Oliel, S., Loyter, A., Klafter, J., Granel, R. & Elbaum, M. (2005) Biophys. J. 89, 2134-2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tzfira, T., Lacroix, B. & Citovsky, V. (2005) in Nuclear Import and Export in Plants and Animals, eds. Tzfira, T. & Citovsky, V. (Landes Bioscience/Kluwer/Plenum, New York), pp. 83-99.
- 37.Li, J., Krichevsky, A., Vaidya, M., Tzfira, T. & Citovsky, V. (2005) Proc. Natl. Acad. Sci. USA 102, 5733-5738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bako, L., Umeda, M., Tiburcio, A. F., Schell, J. & Koncz, C. (2003) Proc. Natl. Acad. Sci. USA 100, 10108-10113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.De Block, M. & Debrouwer, D. (1991) Theor. Appl. Genet. 82, 257-263. [DOI] [PubMed] [Google Scholar]
- 40.Krizkova, L. & Hrouda, M. (1998) Plant J. 16, 673-680. [DOI] [PubMed] [Google Scholar]
- 41.De Buck, S., Jacobs, A., Van Montagu, M. & Depicker, A. (1999) Plant J. 20, 295-304. [DOI] [PubMed] [Google Scholar]
- 42.De Neve, M., De Buck, S., Jacobs, A., Van Montagu, M. & Depicker, A. (1997) Plant J. 11, 15-29. [DOI] [PubMed] [Google Scholar]
- 43.Salomon, S. & Puchta, H. (1998) EMBO J. 17, 6086-6095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Britt, A. B. & May, G. D. (2003) Trends Plant Sci. 8, 90-95. [DOI] [PubMed] [Google Scholar]