Abstract
Global expression profiles of a consecutive series of 121 childhood acute leukemias (87 B lineage acute lymphoblastic leukemias, 11 T cell acute lymphoblastic leukemias, and 23 acute myeloid leukemias), six normal bone marrows, and 10 normal hematopoietic subpopulations of different lineages and maturations were ascertained by using 27K cDNA microarrays. Unsupervised analyses revealed segregation according to lineages and primary genetic changes, i.e., TCF3(E2A)/PBX1, IGH@/MYC, ETV6(TEL)/RUNX1(AML1), 11q23/MLL, and hyperdiploidy (>50 chromosomes). Supervised discriminatory analyses were used to identify differentially expressed genes correlating with lineage and primary genetic change. The gene-expression profiles of normal hematopoietic cells were also studied. By using principal component analyses (PCA), a differentiation axis was exposed, reflecting lineages and maturation stages of normal hematopoietic cells. By applying the three principal components obtained from PCA of the normal cells on the leukemic samples, similarities between malignant and normal cell lineages and maturations were investigated. Apart from showing that leukemias segregate according to lineage and genetic subtype, we provide an extensive study of the genes correlating with primary genetic changes. We also investigated the expression pattern of these genes in normal hematopoietic cells of different lineages and maturations, identifying genes preferentially expressed by the leukemic cells, suggesting an ectopic activation of a large number of genes, likely to reflect regulatory networks of pathogenetic importance that also may provide attractive targets for future directed therapies.
Acute leukemia is the most common malignancy in childhood, with an incidence of 4 and 0.7 cases of acute lymphoblastic leukemia (ALL) and acute myeloid leukemia (AML), respectively, per 100,000 children per year (1, 2). The outcome for children with ALL and AML has improved significantly over the last two decades, with the overall event-free survival approaching 80% and 50%, respectively (1, 3). The advances in the treatment of ALL and AML have, to a large extent, been achieved through individualizing therapy according to different prognostic factors (4, 5). For example, several acquired genetic changes have been shown to be strongly associated with outcome, such as high hyperdiploidy (>50 chromosomes), t(1;19)(q23;p13) (TCF3/PBX1), t(8;14)(q24;q32) (IGH@/MYC), t(9;22)(q34;q11) (BCR/ABL1), 11q23/MLL rearrangements, and t(12,21)(p13;q22) (ETV6/RUNX1) in ALL (6) and t(8;21)(q22;q22) (RUNX1/CBFA2T1), 11q23/MLL rearrangements, t(15;17)(q22;q12) (PML/RARA), and inv (16)(p13q22) (CBFB/MYH11) in AML (7).
With the recent introduction of microarrays, it has become increasingly clear that leukemias with specific chromosomal alterations display altered and distinct gene-expression profiles (8–13). However, it has proved more difficult to extract biologically important information from the vast amount of data generated in such analyses. Apart from a substantial technical and biological noise, critical regulatory oncogenic pathways are likely to be hidden within dominant gene-expression signatures associated with normal hematopoietic differentiation.
So far, only a few large-scale gene-expression analyses of childhood leukemia have been reported, and none has included gene profiling data on normal hematopoietic cells of different maturation levels (8, 9, 14). We have performed an extensive gene-expression analysis of childhood ALL and AML cases, normal bone marrows (NBMs), and purified normal hematopoietic subpopulations of different lineages and maturation stages. Using unsupervised analyses, we confirm and further extend previous findings showing that pediatric leukemias segregate based mainly on their lineage and primary genetic aberrations (8, 9, 14). Furthermore, we have identified genes that correlate with characteristic primary genetic changes and studied their expression patterns in different normal hematopoietic subpopulations.
Materials and Methods
Patient Material and Normal Cells. Bone marrow (BM) (n = 108) or peripheral blood (PB) (n = 13) samples from 121 children with ALL (87 B lineage and 11 T cell) or AML (n = 23) were obtained at the time of diagnosis. The leukemias were diagnosed and treated at Lund University (n = 89) or Linköping University (n = 32) Hospitals, under the same protocols (1, 3), representing ≈70% of all childhood leukemias diagnosed at these two hospitals during the study period (1997–2004). For inclusion of ALL cases, the blast frequencies in BM and PB had to exceed 60% and 25%, respectively. For the AML cases, no limit on the number of blasts was applied. The study was reviewed and approved by the Research Ethics committees of Lund and Linköping Universities.
As part of routine diagnostic procedures, all cases were analyzed cytogenetically and molecularly at the Department of Clinical Genetics (Lund), as described in ref. 12, and remaining cells were frozen in TRIzol (Invitrogen). The genetic features are summarized in Table 1, which is published as supporting information on the PNAS web site. For the samples from Linköping, which were sent by regular mail, there was, in contrast to the samples from Lund, an ≈24-hour delay before freezing in TRIzol.
NBMs from six healthy adult donors were obtained from the Department of Hematology at Lund Hospital. In addition, 10 selected normal subpopulations of different hematopoietic lineages and maturations, obtained from healthy adult donors, were included (see Supporting Methods and Table 2, which are published as supporting information on the PNAS web site). To obtain a sufficient amount of material for labeling, RNA from three to four purified subpopulations was pooled. For each purified cell population, two independent samples were hybridized.
Gene-Expression Profiling. cDNA microarray slides were obtained from the Swegene DNA Microarray Resource Center at Lund University (http://swegene.onk.lu.se). All samples were hybridized to 27K slides containing 25,648 clones representing 13,737 UniGene clusters and 11,592 LocusLink entries, according to UniGene build 180.
RNA extraction, amplification, labeling, hybridization, scanning, posthybridization washing, and feature analysis were performed as described in ref. 12. The quality of total and amplified RNA was assessed by using an Agilent 2100 BioAnalyzer (Agilent Technologies, Palo Alto, CA). As a reference, amplified RNA from the Universal Human Reference (Stratagene) was used.
Microarray-Data Analyses. The data matrix was up-loaded to the BioArray software environment (base) (15) and analyzed as described in ref. 12. In short, normalization was performed by using the Lowess method (16). For multiple reporters, measurements were merged, and the average expression value was used. To correct for bad-quality spots, an error-model correction was used (12). After this correction, a variation (SD ≥ 0.3) and presence filter (95%) was applied. To correct for an initially observed deviation of the gene-expression pattern with regard to sample referral site (Lund vs. Linköping University Hospitals; see Fig. 4, which is published as supporting information on the PNAS web site), mean-centering of the data with respect to hospital was performed before further analysis.
Hierarchical clustering analyses (HCAs) were performed in tmev (17), and principal component analyses (PCAs) were applied by using software developed at the Department of Mathematical Sciences at Lund University. By calculating the three principal components containing the largest variation for the normal hematopoietic cells and applying these components on the leukemic samples (represented by their centroids), the similarities between the malignant and normal cells were studied, and the square norm distance was used to determine the distance (see Supporting Methods).
Differentially expressed genes were identified by using the Golub score (18). A random permutation test was performed by using 5,000 sample label permutations. The P value for a score was calculated as the average number of reporters exceeding the score in a permutation test divided by the total number of reporters in the gene list. Genes with P ≤ 0.001 were considered significant. The differentially expressed genes were subjected to the gene ontology (GO) program ease (19). The ease tool was also used for basic pathway analysis and to investigate whether the deregulated genes were located at certain chromosomal regions, and a score <0.05 was considered significant. Primary data are available upon publication at ArrayExpress (www.ebi.ac.uk/arrayexpress).
Supervised Classification. The k-nearest-neighbors algorithm was used for supervised classification, where the class of a test sample is decided by the majority class among its k nearest neighbors. A cross-validation procedure was used to select the number of neighbors and genes used for classification, and the classifier approach was evaluated in a leave-one-out cross-testing procedure (for details, see Supporting Methods).
Molecular Analyses and Verification of Gene-Expression Data. FISH analyses were performed, as described in ref. 20, on cases in which the gene-expression patterns suggested the presence of a specific abnormality not detected by cytogenetics. To detect chromosomes commonly gained in hyperdiploid leukemias, probe cocktails containing probes derived from chromosomes X, 4, 6, 8, 10, 14, 17, 18, and 21 were used, as described in ref. 20. In addition, FISH probes (Vysis, Downers Grove, IL) and/or RT-PCR analyses for the ETV6/RUNX1 fusion gene were performed on selected cases.
To confirm the microarray results, real-time PCR analyses were performed on seven genes (see Tables 3–9, which are published as supporting information on the PNAS web site) by using standard protocols on an ABI Prism 7000 analyzer (Applied Biosystems). As a control, 18S was used. Primers were ordered from Applied Biosystems as assay-on-demand primers.
Comparison with External Data Sets. To verify our extracted lists of differentially expressed genes, each gene list was compared to two publicly available data sets (8, 9) by using the Fisher's inverse χ2 algorithm (21, 22). This algorithm is a metaanalytical technique that combines P values obtained separately in different data sets into a joint P value. The Fisher's algorithm yields low P values for genes that are differentially expressed in both data sets, whereas intermediate P values will be obtained for genes that are only differentially expressed in one of the data sets.
Results
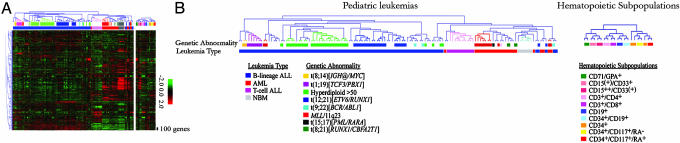
Unsupervised Analysis of Childhood Leukemias and Normal Cells. Unsupervised HCA resulted in near-perfect segregation according to lineage and primary genetic change (Fig. 1 A and B; and see Fig. 5, which is published as supporting information on the PNAS web site, for an enlarged version). A unique expression pattern, distinct from the ones in acute leukemias, was identified in NBMs. By using PCA, the NBMs did not segregate with the AMLs and the ALLs (see Fig. 6 A–C, which is published as supporting information on the PNAS web site).
Fig. 1.
Unsupervised analyses of the childhood acute leukemias. (A) HCA using Pearson correlation and average linkage on the 4,131 reporters [95% presence and a variation filter (standard deviation of 0.6)]. The majority of the leukemias segregate according to lineage and genetic rearrangement. To the right, the same gene order applied on the normal hematopoietic cells. (B) Enhanced dendogram of the leukemias and the normal hematopoietic subpopulations with the gene order maintained as in A. Samples are colored as in A. For an enlargement of this figure, see Fig. 5, in which the genetic status of the leukemias are indicated in detail.
HCA revealed that 64 (85%) of the 75 acute leukemias with primary genetic aberrations (Table 1), segregated according to their genetic change (Fig. 1B). The MLL-positive cases clustered according to lineage, consistent with more recent reports (8, 12, 13, 23). Seven ALL cases harbored a normal karyotype (NK), with few clustering together. Eleven of the cases with characteristic changes did not cluster with the cases harboring the same abnormality: four hyperdiploid cases, one P190 BCR/ABL1, one RUNX1/CBFA2T1, two ETV6/RUNX1, and three 11q23/MLL. By using PCA, however, two of the hyperdiploid cases and the three MLL-positive cases clustered with the respective primary abnormality (see Fig. 6 D–F). The remaining six cases clustered close to the NBMs.
Eight cases, negative for the above-mentioned genetic changes or with cytogenetic failure, nevertheless clustered with defined genetic subtypes. In three cases, we could confirm the subtype suggested by the microarray analysis. Among these three, two had a NK but were shown by FISH to be hyperdiploid, indicating selection of normal cells during culturing before chromosomal analysis. In one case with cytogenetic failure, an ETV6/RUNX1 fusion gene was detected by using RT-PCR. In five cases, however, we could not demonstrate the presence of the primary genetic change suggested by the expression profile. These cases included two samples with an expression pattern suggestive of hyperdiploidy and three with a profile suggestive of an ETV6/RUNX1 fusion. Interestingly, two of the latter cases were leukemias in children with a constitutional trisomy 21 [Down syndrome (DS)]. In one of the two cases, the cytogenetic analysis failed because of no analyzable metaphases, whereas the other case displayed an unbalanced translocation between chromosomes 3 and 7 in addition to the constitutional trisomy 21. The third DS-positive ALL harbored the BCR/ABL1 fusion gene and clustered separately from the other two DS–ALLs. To investigate whether the gene-expression similarities between the two DS-positive ALLs and ALLs with ETV6/RUNX1 reflected expression of genes located on chromosome 21, genes on this chromosome were removed before HCA, resulting in identical clustering, indicating that ETV6/RUNX1-positive ALLs and ALL in DS are similar at the global gene-expression level, independent of chromosome-21-encoded genes.
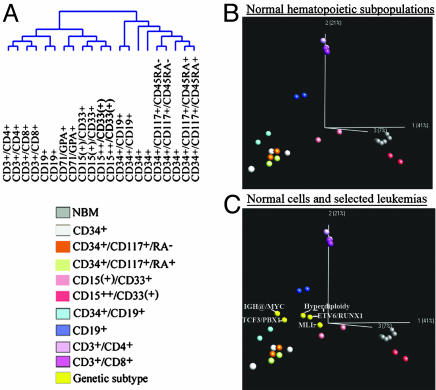
HCA was used to analyze the normal hematopoietic cells and showed that samples segregated according to lineages and maturations (Fig. 2A). The dendogram was divided in two clusters, the CD34+ cells constituting one and the CD34- cells the other. The CD34- cells were further subdivided in two clusters, one lymphoid and one myeloid. Interestingly, PCA revealed that the segregation of normal cells recapitulated their lineages and maturation stages (Fig. 2B). By applying the three principal components obtained by PCA of normal cells on leukemic samples, the relation of lineages and maturation stages of malignant and normal cells were efficiently vizualized (Fig. 2C). The square norm distance was used to calculate the distance between normal and malignant cells (see Table 10, which is published as supporting information on the PNAS web site). This analysis showed that the t(1;19)-positive ALLs clustered in the proximity of the CD34+ cells and closest to early pre-B cells. Intriguingly, the mature IGH@/MYC-positive cases also clustered closest to the early pre-B cells, despite the fact that this ALL subtype is considered to be a mature type of leukemia. The t(12;21)-positive and the hyperdiploid ALLs, on the other hand, clustered closer to the pre-B cells. The MLL-positive cases clustered close to CD15(+)/CD33+ cells, with some spreading toward the lymphoid compartment.
Fig. 2.
Hierarchical dendogram and PCA of the normal cells and visualization of the genetic subtypes in the principal components obtained from PCA of the normal cells. (A) Hierarchical dendogram using Pearson correlation and average linkage of the normal cells, showing that samples cluster according to lineage and that the CD34+ samples cosegregate and cluster separated from the mature cells. (B) PCA of normal cell populations, showing that the cells segregate according to their lineages and maturations. In all PCA analyses, the CD71/GPA+ cells were removed to better appreciate the structure of the data. (C) The pediatric ALLs, each subtype represented by their centroids (yellow), were projected onto the principal components determined by the normal cells. All PCAs can be viewed in Movies 1–10, which are published as supporting information on the PNAS web site.
Identification of Differentially Expressed Genes and Their Expression in Normal Hematopoietic Cells. Analyses of NBMs and purified hematopoietic cells versus B lineage ALL, T cell ALL, and AML revealed 2,750, 1,581, and 705 genes, respectively, to be differentially expressed (P ≤ 0.001) (see Figs. 7 and 8 A and B and Tables 11–14, which are published as supporting information on the PNAS web site). Differentially expressed genes for the primary genetic changes were identified by comparing each abnormality with samples of the same lineage (Table 11; and see Tables 15–23, which are published as supporting information on the PNAS web site). In ALLs, this analysis identified the following number of genes in the various genetic subgroups: 510 in TCF3/PBX1, 357 in IGH@/MYC, 22 in P190 BCR/ABL1, 683 in ETV6/RUNX1, 710 in hyperdiploidy >50 chromosomes, and 35 in cases with an NK (P ≤ 0.001). The number of genes in cases with P190 BCR/ABL1 or a NK was not higher than expected by chance. To extract genes associated with 11q23/MLL rearrangements, three different comparisons were made: (i) B lineage MLL versus the remaining B lineage ALLs, resulting in 89 genes, (ii) MLL-positive AMLs versus the remaining AMLs (147 genes), and (iii) MLL-positive acute leukemias versus the remaining leukemias, regardless of lineage (104 genes).
The expression patterns of the top-200-ranked genes for each genetic subtype were subsequently investigated in normal hematopoietic cells. For the MLL-positive cases, all differentially expressed genes (104 genes) were analyzed. This analysis provided a unique opportunity to identify leukemia-associated genes, i.e., genes preferentially expressed by the leukemic cells, to detect maturation-associated genes and aberrantly expressed genes, i.e., genes expressed by the leukemic cells and in normal cells of a different lineage (see below).
Biological Features of the Differentially Expressed Genes. Pathway and GO analyses of the differentially expressed genes from the comparison of leukemias of different lineage and normal cells (NBMs and purified cell populations) revealed similarities between the categories enriched. Both B lineage and T cell ALLs showed up-regulation of RNA-binding genes and enrichment of genes involved in transcription and in nucleic acid metabolism. In contrast, the AMLs displayed a high expression of genes implicated in metabolism of carbohydrates and complex lipids. Interestingly, all three subtypes (B lineage ALL, T cell ALL, and AML) showed down-regulation of cell adhesion and immune-response genes.
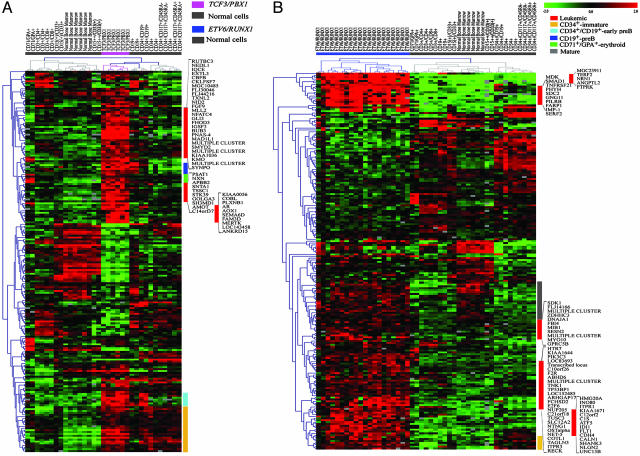
In ALLs with TCF3/PBX1, in contrast to the other genetic subtypes, analyses of the up-regulated genes revealed enrichment of genes involved in cell proliferation and in the cell cycle, e.g., the mitotic checkpoint genes CHEK1, MAD1L1, and MAD2L2. Interestingly, a significant correlation (ease score = 1.4 × 10-4) with the chromosomal location was seen, with 31 of the 309 up-regulated genes mapping to chromosome arm 1q. When studying the expression pattern of the top 200 TCF3/PBX1-ranked genes in the normal hematopoietic cells, 47 were found to be preferentially expressed in TCF3/PBX1-positive leukemias (Fig. 3A; and see Table 24, which is published as supporting information on the PNAS web site). GO analysis showed that these genes encoded proteins with transcription-factor activity or were involved in cell proliferation. In the t(1;19)-positive ALLs, we also identified genes that were expressed in normal hematopoietic cells of a different lineage. For example, PBX1, the top-ranked gene, was highly expressed in CD71/GPA+ (erythroid) cells. Furthermore, t(1;19)-positive ALLs overexpressed ≈30 genes that were also expressed in normal CD34+ populations and 13 genes in early pre-B and/or pre-B cells, likely reflecting signatures associated with maturation (Fig. 3A).
Fig. 3.
The expression of the top-200 ALL-associated genes in normal hematopoietic cells. (A) The top-200 TCF3/PBX1-associated genes applied on normal cells. (B) Top-200 ETV6/RUNX1-associated genes applied on normal cells.
Although only two B lineage ALLs harbored IGH@/MYC, it is noteworthy that both displayed a distinct signature with elevated expression of MYC and deregulated expression of a number of known MYC target genes, e.g., BCL2 and HMG1A (24, 25). In addition, several genes known to be up-regulated upon MYC expression, e.g., TRAP1, NCL, and EIF5A, were found (26). The top-ranked gene, BMP7, a member of the TGFB superfamily, has previously been identified by us to be elevated in cell lines harboring IGH@/MYC (12). Studying the expression pattern of the top-200 IGH@/MYC-ranked genes in normal hematopoietic cells revealed 53 that were expressed also by CD34+ cells (see Fig. 9A and Table 25, which are published as supporting information on the PNAS web site). In addition, 54 genes were preferentially expressed in the IGH@/MYC-positive cases. GO analysis revealed that many were RNA-binding genes, i.e., EIF4B, HNRPAB, and HNRPC, all involved in regulating translation.
In ALLs with ETV6/RUNX1, pathway analysis of the up-regulated genes showed enrichment of genes involved in the phosphatidylinositol signaling system. In this pathway, a number of protein tyrosine phosphatases (PTPN18, PTPRF, PTPRK, and PTPRM) were also identified. The latter three have been implicated in β-catenin signaling. Pathway analysis of the down-regulated genes revealed enrichment of genes involved in nucleotide metabolism. The expression pattern of the top 200 ETV6/RUNX1-ranked genes in normal cells displayed 67 that were preferentially expressed in ETV6/RUNX1-positive cases (Fig. 3B; and see Table 26, which is published as supporting information on the PNAS web site). GO analysis revealed that these genes were involved in transportation or implicated in cell growth and maintenance. Several of the differentially expressed genes were expressed by the normal CD34+ cells, and we also found genes expressed by NBMs or by myeloid CD15++/CD33(+) cells (Fig. 3B).
A striking feature of the hyperdiploid ALLs was the significant correlation between the chromosomal location of the up-regulated genes and the presence of trisomies/tetrasomies involving chromosomes X, 4, 6, 10, 14, 17, 18, and 21, likely reflecting a gene-dosage effect. When investigating the level of expression of the differentially expressed genes over the chromosomes, we noted that some displayed a higher expression level than would be expected by gain of a single chromosome. In addition, some genes located on the trisomic/tetrasomic chromosomes showed a marked decrease in their expression levels (data not shown). Studying the expression of the top-200 hyperdiploidy-associated genes in normal hematopoietic cells revealed 47 that displayed a preferential expression in hyperdiploid leukemias. Notably, approximately half were located on the X chromosome. Genes were also found that were expressed by CD71/GPA+ or by CD34+ cells (Fig. 9B; and see Table 27, which is published as supporting information on the PNAS web site).
To identify genes associated with MLL rearrangements, three different approaches were used. The first approach compared MLL-positive B lineage ALLs with the remaining B lineage ALLs, revealing that HOXA10 and the HOX-cofactor MEIS1 were up-regulated. Among the genes with a low expression, GO analysis revealed enrichment for genes with transcription-factor activity. In a second approach, we investigated the expression of genes in MLL-positive AMLs with the remaining AMLs, again identifying HOXA10, MEIS1, and PBX3. GO analysis revealed enrichment of genes involved in cell communication and adhesion. Several antiapoptotic genes were down-regulated, e.g., TNFRSF21, as were the tumor-suppressor genes BRCA1 and DLC1. In the third approach, we compared all MLL-positive leukemias with all remaining leukemias. Before this comparison, the leukemias were mean-centered with respect to lineage to remove the strong expression profile associated with this parameter. Again, enrichment for genes involved in cell adhesion was found. Moreover, HOXA10, HOXA4, MEIS1, and PBX3 were identified, suggesting that MLL-positive acute leukemias share a transcriptional program. This finding led us to investigate selectively the expression of genes encoding transcription factors. HCA using only this restricted set of genes revealed that leukemias with MLL rearrangements mainly clustered according to genetic change and not lineage (data not shown). Thus, MLL-positive leukemias express a common set of transcription factors, irrespective of lineage. Investigating the expression of MLL-associated genes in normal hematopoietic cells identified 26 with a higher expression in MLL-positive cases (Fig. 9C; and see Table 28, which is published as supporting information on the PNAS web site). GO analysis revealed that these genes were transcription factors or implicated in cell adhesion. Among the few genes that displayed low expression in CD34+ cells, HOXA10 and MEIS1 were identified. A small gene cluster was expressed both by MLL-positive cases and mature T cells, and another cluster was elevated in CD15++/CD33(+) and NBMs, likely reflecting the mixed lineage phenotype of acute leukemias with MLL abnormalities (Fig. 9C). A summary of the GO and pathway analyses performed as described in this article can be viewed in Tables 29–32, which are published as supporting information on the PNAS web site.
We verified our gene lists by comparing them with genes differentially expressed in the largest pediatric ALL and AML data sets published to date by Ross et al. (8, 9). By calculating a combined P value, we showed that, with a P ≤ 0.001, 77–86% of the genes identified in this study also were differentially expressed in the independent public data sets.
Supervised Classification. A classifier was built by using k nearest neighbors to predict genetic subtype among the B lineage ALLs (TCF3/PBX1, ETV6/RUNX1, and hyperdiploidy >50 chromosomes). The overall classification accuracy was 98.2%, with only one ETV6/RUNX1-positive case being misclassified (see Table 33, which is published as supporting information on the PNAS web site). The combined gene lists used for prediction can be viewed in Table 34, which is published as supporting information on the PNAS web site.
Discussion
In the present study, we have investigated the gene-expression profiles of 121 pediatric leukemias of different lineages (B lineage ALL, AML, and T cell ALL), collected consecutively during an 8-year period and analyzed at a single center. Using unsupervised methods, we confirm and further extend previous reports showing that childhood leukemias display a unique expression pattern with regard to leukemic and genetic subtype (8, 9, 14, 27, 28). The differentially expressed genes identified herein were verified by using the ALL and AML data sets published by Ross et al. (8, 9), providing an independent validation of our gene lists and showing a good overlap (77–86%) between the differentially expressed genes. On the other hand, about 14–23% of the genes presented herein have not been previously identified.
We identified a large number of genes correlating with leukemic subtype. Intriguingly, similar ontology and pathway categories were differentially expressed. For example, both B lineage and T cell ALLs showed an up-regulation of RNA-binding genes and of genes involved in transcription. An altered activity of RNA-binding genes has previously been associated with neoplastic transformation (29). In addition, all three subtypes showed down-regulation of genes involved in cell adhesion, indicating a common defect among all leukemic cases, regardless of lineage.
We also identified several genes correlating with primary genetic changes, providing biological insights into the different genetic subtypes of childhood leukemias. For example, in t(1;19)-positive ALLs, but not in the other genetic subtypes, an enrichment of cell-cycle- and cell-proliferation-associated genes was seen. When t(1;19) was first reported, patients with such ALLs were considered to have high-risk leukemia with leukocytosis and an increased risk of central nervous system involvement and relapse (30). Although the prognosis of patients with TCF3/PBX1-positive ALLs has improved by the use of more intensive therapies (31), it is likely that the finding of an increased expression of genes involved in cell proliferation reflects an aggressive phenotype. Furthermore, a correlation of the up-regulated genes to chromosome arm 1q was seen; whether this translates into the better prognosis suggested for ALLs with the unbalanced variant (31) remains to be elucidated.
In leukemias with the ETV6/RUNX1 fusion we found an up-regulation of genes involved in the phosphatidylinositol signaling system, mainly Ca2+ regulators, and, within this system, a number of protein tyrosine phosphatases (PTPs), not previously reported in this ALL type. The PTPs detected are involved in β-catenin signaling, maintaining its dephosphorylated state and, hence, the binding to cadherin (32). Recent data indicate that deregulation of β-catenin signaling may affect leukemic cell adhesion, proliferation, and survival (33). We also confirm the deregulated expression of genes involved in nucleotide metabolism in ETV6/RUNX1-positive leukemias (34).
Investigation of the chromosomal location of the differentially expressed genes in hyperdiploid malignancies confirmed that there is a correlation between highly expressed genes and chromosomes commonly gained in this leukemia type (28). Although many differentially expressed genes were identified, their pathogenetic importance remains unknown. When the expression of the identified genes was investigated over the chromosomes, genes were found that displayed either a substantially higher or, alternatively, lower expression than the average ratio (A.A. and T.F., unpublished data), suggesting that mechanisms other than a general chromosome-wide gene-dosage effect caused by trisomies/tetrasomies may be of pathogenetic importance in hyperdiploid leukemias.
MLL rearrangements result in acute leukemias of different lineages, and we identified and confirmed the presence of a gene signature able to define such leukemias, irrespective of lineage (8, 12, 35). This common gene-expression profile was first hidden among the dominant signatures associated with lineage, but after mean-centering of the data with respect to lineage, unsupervised analyses revealed segregation according to genetic change. Among the genes, the previously identified HOXA10, HOXA4, and the HOX-cofactors MEIS1 and PBX3 were found (8, 12, 23). HOXA10 can bind both MEIS and PBX1 in myeloid cells (36), but whether PBX3, identified as a top-ranked gene in this study, is capable of substituting for PBX1 in this complex remains to be elucidated.
Although malignant cells have been extensively studied by using microarrays, there is a lack of such studies on normal cells and, in particular, studies where malignant gene signatures have been investigated in subpopulations of normal hematopoietic cells. In this study, the expression of the top-200-ranked genes from each gene list was studied in purified normal hematopoietic cells. The salient result of this analysis was that several genes were preferentially and highly expressed by the leukemic cells, i.e., not in the various normal subpopulations, suggesting that leukemic cells display a deregulated activation of transcriptional programs not active in normal cells. Several genes also showed a lower expression in the leukemic cells, but those signatures were more heterogeneous and not as distinct as the genes showing an up-regulated expression. Furthermore, clusters of genes, expressed in selected normal subpopulations, were found. For example, in TCF3/PBX1-positive ALLs, a large gene signature, also expressed in CD34+ cells, was identified, an unexpected finding, because most t(1;19)-positive ALLs are CD34-negative (37). This finding could reflect the cellular origin of t(1;19)-positive leukemias or, alternatively, represent an aberrant or maintained expression of genes expressed in immature hematopoietic cells. Also IGH@MYC-positive ALLs displayed expression of an immature signature, and, despite the fact that this subtype is a mature B cell ALL, it expressed few genes also expressed in normal B cells.
Some genetic ALL subtypes expressed genes that were also highly expressed in normal cells of different lineages. For example PBX1, one of the top-ranked genes in cases with TCF3/PBX1, was highly expressed in erythroid progenitors. Also the ETV6/RUNX1-positive leukemias expressed genes that were up-regulated in normal myeloid cells. ALLs with this fusion often coexpress myeloid markers on their cell surface (38). Our findings indicate that not only single cell-surface markers display aberrant expression but that a large set of myeloid-associated genes is deregulated in this subtype.
PCA of the normal hematopoietic cells recapitulated their lineages and maturation stages. By projecting the primary leukemias onto the principal components determined only by the normal cells, the degree of maturation of childhood leukemias with characteristic genetic changes could be visualized. For example, the t(1;19)-positive ALLs clustered close to the CD34+/CD19+ cells, indicating an arrest at an early differentiation stage, despite their common lack of the CD34 marker. The t(12;21)-positive cases, on the other hand, clustered closer to the pre-B cells. Indeed, it has recently been shown that this fusion gene arises in a committed B cell progenitor (39).
A classifier using k nearest neighbors was built that predicted genetic subtype among B lineage ALLs with a high accuracy (98.2%), verifying previous reports that genetic subtype can be predicted with a high certainty (9, 13). Only one ETV6/RUNX1-positive case was misclassified as a hyperdiploid case, and this sample also clustered close to the NBM cells in the PCA and HCA, indicating contamination with normal cells.
We also identified five leukemic cases that clustered with previously defined genetic subtypes but that did not harbor the corresponding chromosomal or molecular changes. Two of these were DS–ALLs, which cosegregated with the ETV6/RUNX1-positive cases. We investigated whether the similarity in expression profile was a reflection of genes confined to chromosome 21, but no such association was found. The reasons behind this cosegregation are presently unknown, but it is likely a result of alternative mutational events occurring in DS B lineage ALL.
In conclusion, this microarray study compares the gene-expression profiles of leukemic cells to normal hematopoietic cells and investigates the gene-expression signatures of normal subpopulations of different lineages and maturations. Apart from providing important biological insights into the different genetic subtypes of childhood leukemias, this comparison revealed that several genes were preferentially expressed by the leukemic cells, suggesting an ectopic activation of a large number of genes that may not only reflect pathogenetically important regulatory pathways, but also suggest attractive targets for future directed therapies.
Supplementary Material
Acknowledgments
This work was supported by grants from the Swedish Cancer Society, the Swedish Children's Cancer Foundation, the Medical Faculty of Lund University, and the Inga-Britt and Arne Lundberg Foundation.
Author contributions: A.A. and T.F. designed research; A.A., T.O., D.L., B.N., C.R., P.E., C.L., J.R., M.F., H.M., J.H., M.B., F.M., M.H., B.J., and T.F. performed research; A.A. and C.R. analyzed data; and A.A. and T.F. wrote the paper.
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: ALL, acute lymphoblastic leukemia; AML, acute myeloid leukemia; BM, bone marrow; DS, Down syndrome; GO, gene ontology; HCA, hierarchical clustering analysis; NBM, normal bone marrow; NK, normal karyotype; PCA, principal component analysis.
References
- 1.Gustafsson, G., Schmiegelow, K., Forestier, E., Clausen, N., Glomstein, A., Jonmundsson, G., Mellander, L., Makipernaa, A., Nygaard, R. & Saarinen-Pihkala, U. M. (2000) Leukemia 14, 2267-2275. [DOI] [PubMed] [Google Scholar]
- 2.Forestier, E., Heim, S., Blennow, E., Borgstrom, G., Holmgren, G., Heinonen, K., Johannsson, J., Kerndrup, G., Andersen, M. K., Lundin, C., et al. (2003) Br. J. Haematol. 121, 566-577. [DOI] [PubMed] [Google Scholar]
- 3.Lie, S. O., Abrahamsson, J., Clausen, N., Forestier, E., Hasle, H., Hovi, L., Jonmundsson, G., Mellander, L. & Gustafsson, G. (2003) Br. J. Haematol. 122, 217-225. [DOI] [PubMed] [Google Scholar]
- 4.Silverman, L. B. & Sallan, S. E. (2003) Curr. Opin. Hematol. 10, 290-296. [DOI] [PubMed] [Google Scholar]
- 5.Chang, M., Raimondi, S. C., Ravindranath, Y., Carroll, A. J., Camitta, B., Gresik, M. V., Steuber, C. P. & Weinstein, H. (2000) Leukemia 14, 1201-1207. [DOI] [PubMed] [Google Scholar]
- 6.Johansson, B., Mertens, F. & Mitelman, F. (2004) Ann. Med. 36, 492-503. [DOI] [PubMed] [Google Scholar]
- 7.Grimwade, D. (2001) Best Pract. Res. Clin. Haematol. 14, 497-529. [DOI] [PubMed] [Google Scholar]
- 8.Ross, M. E., Mahfouz, R., Onciu, M., Liu, H. C., Zhou, X., Song, G., Shurtleff, S. A., Pounds, S., Cheng, C., Ma, J., et al. (2004) Blood 104, 3679-3687. [DOI] [PubMed] [Google Scholar]
- 9.Ross, M. E., Zhou, X., Song, G., Shurtleff, S. A., Girtman, K., Williams, W. K., Liu, H. C., Mahfouz, R., Raimondi, S. C., Lenny, N., et al. (2003) Blood 102, 2951-2959. [DOI] [PubMed] [Google Scholar]
- 10.Valk, P. J., Verhaak, R. G., Beijen, M. A., Erpelinck, C. A., Barjesteh van Waalwijk van Doorn-Khosrovani, S., Boer, J. M., Beverloo, H. B., Moorhouse, M. J., van der Spek, P. J., Lowenberg, B., et al. (2004) N. Engl. J. Med. 350, 1617-1628. [DOI] [PubMed] [Google Scholar]
- 11.Bullinger, L., Dohner, K., Bair, E., Frohling, S., Schlenk, R. F., Tibshirani, R., Dohner, H. & Pollack, J. R. (2004) N. Engl. J. Med. 350, 1605-1616. [DOI] [PubMed] [Google Scholar]
- 12.Andersson, A., Eden, P., Lindgren, D., Nilsson, J., Lassen, C., Heldrup, J., Fontes, M., Borg, A., Mitelman, F., Johansson, B., et al. (2005) Leukemia 19, 1042-1050. [DOI] [PubMed] [Google Scholar]
- 13.Haferlach, T., Kohlmann, A., Schnittger, S., Dugas, M., Hiddemann, W., Kern, W. & Schoch, C. (2005) Blood 106, 1189-1198. [DOI] [PubMed] [Google Scholar]
- 14.Yagi, T., Morimoto, A., Eguchi, M., Hibi, S., Sako, M., Ishii, E., Mizutani, S., Imashuku, S., Ohki, M. & Ichikawa, H. (2003) Blood 102, 1849-1856. [DOI] [PubMed] [Google Scholar]
- 15.Saal, L. H., Troein, C., Vallon-Christersson, J., Gruvberger, S., Borg, A. & Peterson, C. (2002) Genome Biol. 3, SOFTWARE0003. [DOI] [PMC free article] [PubMed]
- 16.Yang, Y. H., Dudoit, S., Luu, P., Lin, D. M., Peng, V., Ngai, J. & Speed, T. P. (2002) Nucleic Acids Res. 30, e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saeed, A. I., Sharov, V., White, J., Li, J., Liang, W., Bhagabati, N., Braisted, J., Klapa, M., Currier, T., Thiagarajan, M., et al. (2003) BioTechniques 34, 374-378. [DOI] [PubMed] [Google Scholar]
- 18.Golub, T. R., Slonim, D. K., Tamayo, P., Huard, C., Gaasenbeek, M., Mesirov, J. P., Coller, H., Loh, M. L., Downing, J. R., Caligiuri, M. A., et al. (1999) Science 286, 531-537. [DOI] [PubMed] [Google Scholar]
- 19.Hosack, D. A., Dennis, G., Jr., Sherman, B. T., Lane, H. C. & Lempicki, R. A. (2003) Genome Biol. 4, R70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Paulsson, K., Panagopoulos, I., Knuutila, S., Jee, K. J., Garwicz, S., Fioretos, T., Mitelman, F. & Johansson, B. (2003) Blood 102, 3010-3015. [DOI] [PubMed] [Google Scholar]
- 21.Fisher, R. (1925) Stastistical Methods for Research Workers (Oliver and Boyd, Harlow, Essex, U.K.).
- 22.Rhodes, D. R., Barrette, T. R., Rubin, M. A., Ghosh, D. & Chinnaiyan, A. M. (2002) Cancer Res. 62, 4427-4433. [PubMed] [Google Scholar]
- 23.Kohlmann, A., Schoch, C., Dugas, M., Schnittger, S., Hiddemann, W., Kern, W. & Haferlach, T. (2005) Leukemia. [DOI] [PubMed]
- 24.Wood, L. J., Mukherjee, M., Dolde, C. E., Xu, Y., Maher, J. F., Bunton, T. E., Williams, J. B. & Resar, L. M. (2000) Mol. Cell. Biol. 20, 5490-5502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eischen, C. M., Packham, G., Nip, J., Fee, B. E., Hiebert, S. W., Zambetti, G. P. & Cleveland, J. L. (2001) Oncogene 20, 6983-6993. [DOI] [PubMed] [Google Scholar]
- 26.Coller, H. A., Grandori, C., Tamayo, P., Colbert, T., Lander, E. S., Eisenman, R. N. & Golub, T. R. (2000) Proc. Natl. Acad. Sci. USA 97, 3260-3265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tsutsumi, S., Taketani, T., Nishimura, K., Ge, X., Taki, T., Sugita, K., Ishii, E., Hanada, R., Ohki, M., Aburatani, H., et al. (2003) Cancer Res. 63, 4882-4887. [PubMed] [Google Scholar]
- 28.Yeoh, E. J., Ross, M. E., Shurtleff, S. A., Williams, W. K., Patel, D., Mahfouz, R., Behm, F. G., Raimondi, S. C., Relling, M. V., Patel, A., et al. (2002) Cancer Cell 1, 133-143. [DOI] [PubMed] [Google Scholar]
- 29.Perrotti, D. & Calabretta, B. (2002) Oncogene 21, 8577-8583. [DOI] [PubMed] [Google Scholar]
- 30.Crist, W. M., Carroll, A. J., Shuster, J. J., Behm, F. G., Whitehead, M., Vietti, T. J., Look, A. T., Mahoney, D., Ragab, A., Pullen, D. J., et al. (1990) Blood 76, 117-122. [PubMed] [Google Scholar]
- 31.Uckun, F. M., Sensel, M. G., Sather, H. N., Gaynon, P. S., Arthur, D. C., Lange, B. J., Steinherz, P. G., Kraft, P., Hutchinson, R., Nachman, J. B., et al. (1998) J. Clin. Oncol. 16, 527-535. [DOI] [PubMed] [Google Scholar]
- 32.Stoker, A. W. (2005) J. Endocrinol. 185, 19-33. [DOI] [PubMed] [Google Scholar]
- 33.Chung, E. J., Hwang, S. G., Nguyen, P., Lee, S., Kim, J. S., Kim, J. W., Henkart, P. A., Bottaro, D. P., Soon, L., Bonvini, P., et al. (2002) Blood 100, 982-990. [DOI] [PubMed] [Google Scholar]
- 34.Zaza, G., Yang, W., Kager, L., Cheok, M., Downing, J., Pui, C. H., Cheng, C., Relling, M. V. & Evans, W. E. (2004) Blood 104, 1435-1441. [DOI] [PubMed] [Google Scholar]
- 35.Armstrong, S. A., Staunton, J. E., Silverman, L. B., Pieters, R., den Boer, M. L., Minden, M. D., Sallan, S. E., Lander, E. S., Golub, T. R. & Korsmeyer, S. J. (2002) Nat. Genet. 30, 41-47. [DOI] [PubMed] [Google Scholar]
- 36.Bromleigh, V. C. & Freedman, L. P. (2000) Genes Dev. 14, 2581-2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Borowitz, M. J., Hunger, S. P., Carroll, A. J., Shuster, J. J., Pullen, D. J., Steuber, C. P. & Cleary, M. L. (1993) Blood 82, 1086-1091. [PubMed] [Google Scholar]
- 38.Borowitz, M. J., Rubnitz, J., Nash, M., Pullen, D. J. & Camitta, B. (1998) Leukemia 12, 1764-1770. [DOI] [PubMed] [Google Scholar]
- 39.Castor, A., Nilsson, L., Astrand-Grundstrom, I., Buitenhuis, M., Ramirez, C., Anderson, K., Strombeck, B., Garwicz, S., Bekassy, A. N., Schmiegelow, K., et al. (2005) Nat. Med. 11, 630-637. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.