Sensing and responding to light is a central feature of plant biology that is critical for tuning plant metabolism to the light environment. To accomplish this vital task, plants use a number of different photosensory proteins that perceive different bands of the electromagnetic spectrum (Batschauer, 2003; Briggs and Spudich, 2005; Schäfer and Nagy, 2005). For sensing red light–depleted (shade) and red light–enriched (full sun) conditions, plants use phytochromes, large (∼120 kD) proteins that possess the covalently linked linear tetrapyrrole (bilin) chromophore phytochromobilin (Figure 1A). First detected in 1959 by investigators at the USDA Plant Industry Station in Beltsville, MD (Butler et al., 1959), phytochromes share the characteristic that red light (R) irradiation converts the R-absorbing Pr state into the metastable, far-red light (FR)-absorbing Pfr state (Figure 1B). This photoconversion is reversible, with Pfr returning to Pr either upon absorption of an FR photon or upon prolonged incubation in the dark via a thermal process known as dark reversion (Sineshchekov, 1995; Braslavsky et al., 1997). We now know that this reversible photochemistry reflects the unique environment of a bilin chromophore that is buried within a highly conserved N-terminal photosensory core region. Recently, Katrina Forest, together with Richard Vierstra and colleagues from the University of Wisconsin, captured a snapshot of the structure of the conserved photosensory core of a phytochrome, a result that has profound implications for the structural and functional basis of phytochrome signaling in plants (Wagner et al., 2005).
Figure 1.
Bilin Chromophores, Photoconversion, and Domain Architecture of Phytochromes.
(A) The bilin chromophores derived from phycocyanobilin (PCB), used by cyanobacterial and algal phytochromes (right), phytochromobilin (PΦB), used by plant phytochromes (right), and biliverdin (BV), used by the BphP subfamily (left), are shown along with their respective thioether linkages to the phytochrome apoproteins. Ring names (bold) and carbon numbering are indicated.
(B) The photoconversion between Pr and Pfr phytochromes is shown along with the primary photochemical intermediates. The conversions of lumi-R into Pr and lumi-F into Pfr are thermal processes that lead to changes in the protein moieties that initiate signal output.
(C) Conserved domains in the phytochrome primary sequence are shown (see text for description of PAS, GAF, and PHY domains). PAS and GAF domains are red and blue, respectively. The fragment of DrBphP whose structure has been reported (Wagner et al., 2005; PDB code 1ZTU) is shown at bottom with the positions of key chromophore-interacting residues shown in the primary sequence. HKRD, histidine kinase–related domain.
Plant phytochromes (Phy family) are members of a more widespread family of photosensors that are found in cyanobacteria (cyanobacterial phytochromes Cph1 and Cph2) as well as in purple and nonphotosynthetic bacteria (bacteriophytochromes; BphP) and even fungi (fungal phytochromes; Fph family) (Montgomery and Lagarias, 2002; Blumenstein et al., 2005; Froehlich et al., 2005; Karniol et al., 2005). Members of the extended family of phytochrome proteins share a common N-terminal photosensory core, with three blocks of homology sometimes referred to as P2, P3, and P4 (Montgomery and Lagarias, 2002) (Figure 1C). Phytochrome C-terminal domains mediate the transmission of photosensory signals perceived by the N-terminal region to signal transduction pathways within the cell. This region typically contains a histidine kinase–related domain that has been shown to confer ATP-dependent protein phosphotransferase activity in several cases (Yeh et al., 1997; Yeh and Lagarias, 1998; Bhoo et al., 2001; Hübschmann et al., 2001; Lamparter et al., 2001; Karniol and Vierstra, 2003; Giraud et al., 2005; Tasler et al., 2005). While bacterial and cyanobacterial phytochromes typically employ classical two-component phosphotransfer relays, the mechanism of plant phytochrome signaling appears considerably more complex, involving light-mediated nuclear translocation and regulation of transcription factor function (Chen et al., 2004; Nagatani, 2004; Huq and Quail, 2005; Schäfer and Nagy, 2005). The C-terminal regulatory domains of plant phytochromes have been shown to mediate homodimerization and light-modulated nuclear targeting, both of which are required for signal transmission (Matsushita et al., 2003; Chen et al., 2005). Signaling by plant phytochromes also involves protein–protein interactions with the N-terminal part of the protein (Ni et al., 1999; Oka et al., 2004), although it is not yet known whether these interactions are actually mediated via the conserved photosensory core (P2-P3-P4), via the N-terminal Ser/Thr-rich extension specific to plant phytochromes, or both. In more primitive plants and algae, atypical phytochromes have been described in which the C-terminal region has been replaced by fortuitous gene fusions with phototropins and other eukaryotic Ser/Thr kinases (Thümmler et al., 1992; Nozue et al., 1998; Suetsugu et al., 2005). Bacteriophytochromes that lack recognizable kinase output domains have also been reported (Giraud et al., 2002; Karniol et al., 2005). It thus appears that the primary mechanism of light perception, arguably shared by the conserved photosensory core of all phytochromes, has been evolutionarily co-opted to regulate output domains with different molecular architectures.
The distinct bilin chromophores of the various phytochrome classes are all derived from the oxidative metabolism of heme. Heme oxygenases convert heme to biliverdin IXα (BV), which is the direct precursor of the chromophores of bacteriophytochromes (Bhoo et al., 2001; Giraud et al., 2002, 2005; Lamparter et al., 2003, 2004; Tasler et al., 2005) and probably also those of fungal phytochromes (Blumenstein et al., 2005; Froehlich et al., 2005). In cyanobacteria and algae, BV is converted to phycocyanobilin (PCB) via a four-electron reduction mediated by ferredoxin-dependent bilin reductases of the PcyA subfamily (Frankenberg et al., 2001). Higher plants instead convert BV to phytochromobilin (PΦB) through the action of the homologous two-electron bilin reductase phytochromobilin synthase (HY2) (Kohchi et al., 2001). In all phytochromes, the bilin chromophore precursor is covalently attached to the protein via a thioether linkage between a Cys residue and the bilin A-ring (Figure 1A). Plant and cyanobacterial phytochromes utilize a conserved Cys residue in the P3 domain (Lagarias and Rapoport, 1980; Hübschmann et al., 2001), whereas bacteriophytochromes (and probably fungal phytochromes) instead utilize a different Cys in the P2 region (Lamparter et al., 2004; Karniol et al., 2005; Tasler et al., 2005) (Figure 1C). Unlike the phycobiliproteins, which utilize C-S lyases for covalent attachment of their bilin chromophores (Schluchter and Glazer, 1999), plant phytochromes do not require enzymes or cofactors to assist the covalent assembly reaction (Terry et al., 1993). The same is true for cyanobacterial, fungal, and bacteriophytochromes (Karniol et al., 2005). While recent work implicates the importance of the P2 domain for proper holoprotein assembly of a cyanobacterial phytochrome (Zhao et al., 2004), the more distantly related phytochromes of the Cph2 subfamily lack this domain altogether but are nevertheless able to support bilin attachment (Wu and Lagarias, 2000). This suggests that the P2 domain performs an accessory role in holoprotein assembly, possibly by stabilizing proper folding of the chromophore binding pocket in the P3 domain. Interestingly, introduction of a P3 Cys into bacteriophytochromes supports assembly with PCB (Davis et al., 1999; Lamparter et al., 2004; Quest and Gärtner, 2004), arguing for structural conservation of the chromophore binding pocket.
In contrast with the P2 and P3 domains, P4 domains of plant, cyanobacterial, and bacteriophytochromes are dispensable for covalent attachment of the bilin chromophore (Vierstra, 1993; Wu and Lagarias, 2000; Oka et al., 2004; Karniol et al., 2005). The P4 domain is however necessary both for efficient photochemistry and for the normal Pr spectrum, which are measures of bilin chromophore conformation (Falk, 1989). P4 therefore seems critical for optimizing the bilin chromophore environment to enable efficient photoconversion, perhaps by conferring rigidity to the bound bilin and thereby minimizing competing radiationless deexcitation pathways (Wu and Lagarias, 2000; Fischer and Lagarias, 2004). It was previously reported that P2 and P3 are related to domains known to be involved in ligand binding and signal transduction (Montgomery and Lagarias, 2002): P2 domains of some phytochrome protein sequences have been identified by domain database searches to be PAS domains (named for period clock, ARNT, and single-minded proteins; Ponting and Aravind, 1997), while P3 is identified as a GAF domain (named for vertebrate cGMP-specific phosphodiesterases, cyanobacterial adenylate cyclases, and the transcription activator FhlA; Aravind and Ponting, 1997). Interestingly, the three-dimensional structures of representative PAS and GAF domain proteins exhibit similarity between the two protein folds (discussed in Montgomery and Lagarias, 2002). The P4 domain of phytochromes, classified as a PHY domain by domain family database searches, also exhibits some homology to GAF domains. However, the similarity between P4 and GAF domains is much weaker than that between P3 and GAF domains. Since P4 domains of representative members of the Cph2 subfamily of phytochromes bridge the sequence gap between GAF domains and PHY domains of other phytochromes, the P4 PHY domain is likely to adopt a GAF fold.
STRUCTURE AND ASSEMBLY OF THE PHYTOCHROME PHOTOSENSORY CORE
Despite the long-standing interest in phytochromes and the application of a wide range of biophysical and spectroscopic techniques, determination of phytochrome structures and elucidation of the photochemical mechanism have remained difficult objectives. The interactions between the PAS, GAF, and PHY domains have remained opaque, and there is considerable variability among PAS–PAS and GAF–GAF interactions in published protein structures (Ho et al., 2000; Martinez et al., 2002, 2005; Erbel et al., 2003; Kurokawa et al., 2004; Yildiz et al., 2005). It has thus been difficult to predict how the domains in phytochrome might assemble, much less how light absorption results in signal transmission to the C-terminal portion of the protein. The structure of the chromophore in both Pr and Pfr states has also attracted considerable attention without a clear consensus; for example, the Pr chromophore has variously been proposed to adopt C5-Z,anti/C10-Z,syn/C15-Z,syn, C5-Z,anti/C10-Z,syn/C15-Z,anti, C5-Z,anti/C10-E,anti/C15-Z,syn, or C5-Z,syn/C10-Z,syn/C15-Z,anti conformations on the basis of vibrational spectroscopy and theoretical approaches (Andel et al., 1996, 2000; Kneip et al., 1999; Mroginski et al., 2004; Fischer et al., 2005; see http://www.iupac.org/publications/compendium/index.html for conformational nomenclature).
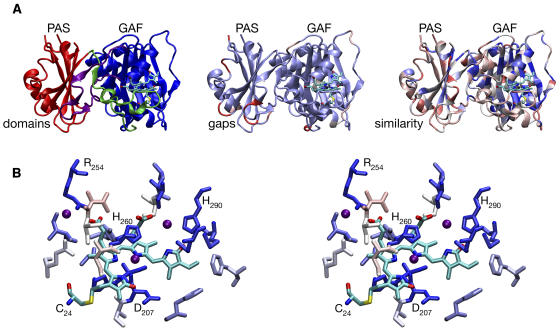
The phytochrome field is thus greatly advanced by the recent determination of the 2.5-Å crystal structure of the PAS and GAF domains of the bacteriophytochrome DrBphP holoprotein from Deinococcus radiodurans (Wagner et al., 2005). This breakthrough not only provides experimental confirmation of many suppositions, but it also yields surprises that shed exciting new light on how phytochromes work. As predicted, P2 and P3 adopt PAS and GAF folds, respectively (Figure 2A). The BV chromophore of DrBphP is deeply buried within the GAF domain with its A-ring vinyl side chain covalently linked to the conserved Cys24 (Figure 2B). Unexpectedly, Cys24 of DrBphP is apparently linked to the 32 carbon of the A-ring vinyl group of BV as opposed to the thioether linkage between the invariant GAF Cys and the 31 carbon of PΦB or PCB (Figure 1A) found in plant and cyanobacterial phytochromes (Lagarias and Rapoport, 1980; Hübschmann et al., 2001). The Met that substitutes for the nucleophilic GAF Cys found in plant and cyanobacterial phytochromes is located in close proximity to the 31 carbon of the A-ring vinyl group. Wagner et al. (2005) therefore argue that it is easy to envisage the molecular evolution of cyanobacterial (and plant) phytochromes from a bacteriophytochrome ancestor via the gain of the GAF Cys along with the appearance of bilin reductases capable of the conversion of BV to the more reduced bilins, PCB and PΦB. This result also confirms that the bilin binding pockets of cyanobacterial and plant phytochromes are similar to those of bacteriophytochromes. In addition, regions most conserved in the extended phytochrome family map to the DrBphP PAS and GAF cores, with sequence variability, gaps, and insertions falling within surface loops (Figure 2A). The photosensory region of plant phytochromes is therefore very likely to adopt a fold similar to that of DrBphP.
Figure 2.
Conservation of Structural Elements among Phytochromes.
(A) The DrBphP crystal structure (Wagner et al., 2005) is shown colored by domains (left), gaps (center), and similarity with other phytochromes (right). The BV chromophore and Cys24 (the site of covalent attachment in DrBphP) are shown colored by atom type (cyan, C; blue, N; red, O; yellow, S). Domains (left) are colored red (PAS domain), blue (GAF domain), green (N-terminal to the PAS domain, including part of the trefoil knot), and purple (lasso sequence inserted to the GAF domain and forming the remainder of the trefoil knot). Gaps (center) are colored with a continuous scale ranging from light blue (no gaps) to red (gaps ≥5 amino acids long) using an alignment of 122 members of the phytochrome family, including DrBphP. Similarity (right) is colored with a continuous scale ranging from dark blue (complete conservation) to red (variable) using a normalized BLOSUM62 substitution matrix (Henikoff and Henikoff, 1992) and the same sequence alignment.
(B) Stereo view of chromophore binding in DrBphP. Cys24 and the BV chromophore are colored by element as in (A). Other residues within 4 Å of chromophore are colored by similarity as in (A). Water molecules within 4 Å of the BV chromophore are shown as purple spheres. Figures 2 to 4 were prepared with VMD, Tachyon, STRIDE, and homolmapper (Frishman and Argos, 1995; Humphrey et al., 1996; Stone, 1998).
Astonishingly, the interface between the PAS and GAF domains forms a trefoil knot (Figure 3A) centered on the conserved Ile35, a residue that lies between Cys24 and the start of the PAS domain (Zhao et al., 2004; Wagner et al., 2005). The sequence between Cys24 and the start of the PAS domain is passed through a loop formed by a large insertion within the GAF domain (Wagner et al., 2005). This motif is thus a true knot with an approximate trefoil (three-lobed) symmetry (Figure 3B). The GAF insertion in the knot also contains the conserved Arg254, which interacts with the side chain carboxylate of the BV chromophore B-ring (Figure 3C). This knot creates a unique interface between the PAS and GAF domains, helping to anchor both Cys24 and a key chromophore-interacting residue. This is likely to result in a much more rigid structure than might have been expected for a more conventional domain–domain interaction. Wagner et al. (2005) propose that this extra rigidity facilitates the photoconversion process by reducing nonproductive deexcitation associated with protein vibrations and/or domain–domain motions.
Figure 3.
The Trefoil Knot Interface between the PAS and GAF Domains.
(A) A stereo view of the conserved knot core is shown colored as in Figure 2A (left). Amino acids 27 to 38 (green) are upstream of the PAS domain (red) and form the knot with amino acids 228 to 256 (the lasso, purple) that are inserted between β4 and α4 of the GAF domain (blue).
(B) A simplified illustration of the trefoil knot topology is shown with colors as in (A). The GAF domain is formed by sequences on either side of the lasso, and residues that were not experimentally resolved in the loop between the PAS and GAF domains (Wagner et al., 2005) are shown as a dashed line. This schematic emphasizes the trefoil topology of the knot.
(C) The interaction between the knot region and the chromophore is shown in stereo. The protein backbone is shown as a transparent trace colored as in (A), with residues 24 to 38 shown in green and Cys24, Arg254, and BV shown colored by atom type as in Figure 2.
Such a structure also poses unique problems for DrBphP protein folding: how can this knot form? The formation of such deep knots is not yet well characterized; indeed, such structures have only been recognized fairly recently (Taylor, 2000; Nureki et al., 2002). In the case of DrBphP, Wagner et al. (2005) propose that knot formation occurs cotranslationally. The PAS domain folds completely, leaving the Cys-containing N-terminal peptide as an unstructured coil. Knot formation is envisaged to proceed concomitant with translation of the large insert in the GAF domain; this insert lassos the central β-strand containing Ile35 prior to translation of the C-terminal portion of the GAF domain and folding of the central GAF β-sheet. Such a mechanism could also explain why phytochromes are intolerant of chimeric fusions N-terminal to the photosensory domain (J.C. Lagarias, unpublished data): placement of rapidly folding protein sequences proximal to the PAS domain could well interfere with proper formation of the knot and result in severe defects in folding.
The facile in vitro assembly of apoprotein with chromophore indicates that bilin binding is not necessary for folding of these two domains, as is implicit in the above proposal. In this regard, examination of chromophore assembly with Cph1 by stopped-flow spectrophotometry (Borucki et al., 2003) demonstrated that assembly involves three key steps: initial noncovalent binding of PCB to the Cph1 apoprotein in a cyclic, porphyrin-like conformation, formation of a more extended red-shifted PCB·apoCph1 intermediate, and covalent bond formation concomitant with a spectral blue shift. The red-shifted intermediate is thought to reflect chromophore protonation and adoption of the more extended configuration characteristic of Pr. A recent theoretical study of PCB indicates that the extended conformation is primarily responsible for enhanced red absorbance, with protonation driving the red shift (Göller et al., 2005).
The DrBphP crystal structure provides strong evidence for either His260 or Asp207 as potential proton donors for the BV chromophore. However, while recent NMR studies provide evidence that all four chromophore nitrogens in both Pr and Pfr chromophores of Cph1 are protonated (Strauss et al., 2005), such studies have not yet been extended to DrBphP. The assembly reaction and spectral properties of BV-utilizing phytochromes such as DrBphP have also not been reported. Since it is not possible to ascertain the protonation state of the BV prosthetic group of DrBphP from the structure of Wagner et al. (2005), the hypothesis that its chromophore may not be fully protonated remains a formal, albeit less appealing, possibility. In this regard, removal of the PHY domains of plant, cyanobacterial, and bacteriophytochromes is known to strongly alter the spectroscopic and photochemical properties of the truncated holoproteins (Vierstra, 1993; Wu and Lagarias, 2000; Karniol et al., 2005). Structural information for the PHY domain, as well as additional spectroscopic measurements, will aid in interpretation of this aspect of phytochrome structure, as would determination of the structures of PAS and GAF domains for other phytochromes. The structure reported for DrBphP will hopefully be the first of many.
LIGHT SENSING AND SIGNAL TRANSDUCTION: NEW INSIGHTS
The crystal structure of the chromophore binding domains of DrBphP provides a wealth of new information about the nature of the chromophore binding pocket and, by analogy, those of plant phytochromes. The paramount question of chromophore configuration in the Pr state is now conclusively answered: the chromophore adopts a C5-Z,syn/C10-Z,syn/C15-Z,anti conformation (Figure 1A). In addition, the DrBphP structure exhibits many interactions between the chromophore and conserved residues (Figure 2B). The entire structure of the chromophore is buried, surprisingly including the hydrophilic carboxylate side chains. The burial of the water-soluble carboxylates appears counterintuitive on first principles but for the observation that one carboxylate interacts with the highly conserved Arg254 that lies within the knot region (Figure 3C). Such a buried salt bridge is likely to be energetically favorable for the assembly process. It is particularly interesting that three ordered water molecules are found within 4 Å of the BV chromophore, one of which is in close contact with its conjugated ring system (Figure 2B). Since the exclusion of solvent water from the chromophore binding pocket is likely a key factor in ensuring efficient photochemistry, it will be interesting to assess how the position of these waters is affected by the presence of the PHY domain that is missing from the DrBphP structure (Figure 1C).
The chromophore ring system of DrBphP makes a number of contacts with conserved residues, including the universally conserved Asp207-Ile208-Pro209 motif. Strikingly, the D-ring is less tightly packed by protein residues than other parts of the chromophore, presumably making it easier for this ring to rotate during the photoconversion process. The hydrophobic contacts that the D-ring does make are primarily with conserved aromatic residues (Figure 4A); these may provide rigidity to modulate the photochemical reaction. One of the residues lining the D-ring cavity, Tyr176, is known to be critical for proper photochemistry, perhaps by gating the rotation of the chromophore D-ring (Fischer and Lagarias, 2004). The photochemical gating role for this conserved Tyr residue in the bacteriophytochromes was more recently questioned (Fischer et al., 2005).
Figure 4.
Stereo Views of Interactions between the BV Chromophore and Nearby Residues in the DrBphP Apoprotein.
(A) Residues within 4.5 Å of the chromophore D-ring are shown colored by atom type and as a single solvent-accessible surface prepared with MSMS (Sanner et al., 1996) colored by similarity as in Figure 2. Cys24 and the BV chromophore are shown in purple. The color highlighting of the protein residues ranges from dark blue (absolutely conserved) to light blue (strongly conserved). No red-shaded (variable) residues are located near the chromophore.
(B) Tight packing of His260, Ile208, and Tyr216 about C10 is shown with Cys24 and chromophore in purple. Amino acid residues are shown colored by atom type as in Figure 2A.
Primary photochemistry is generally accepted to involve a Z-to-E isomerization about the C15,16 double bond (Braslavsky et al., 1997). Consistent with this hypothesis, a recent theoretical study of PCB provided evidence for considerable double-bond character about the C15,16 bond in the ground state but not in the excited state (Göller et al., 2005). The DrBphP structure provides new insight into the process of photoconversion. Since the bilin A-ring is covalently attached to the protein matrix and deeply buried between two α-helices, rotation about the C4,5 double bond is unlikely. The B- and C-rings are tightly packed by His260, Asp207, Ile208, and Tyr216 (Figure 4B), making it difficult to envision rotation about C10. These structural constraints explain the observation that holophytochromes assembled with phycoerythrobilin, a bilin that is missing the C15,16 double bond, are intensely fluorescent, nonphotoconvertible biliproteins (Li et al., 1995; Murphy and Lagarias, 1997). In the DrBphP structure, the BV D-ring is twisted out of plane relative to the A-, B-, and C-rings. This torsional strain would reduce the energetic barrier for rotation of the D-ring by destabilizing the ground state, thereby facilitating photochemistry. It is also conceivable that photoconversion might proceed via a hula twist mechanism involving simultaneous rotation about two bonds rather than via a single-bond rotation mechanism, as has been described for other photoprotein systems (Liu and Hammond, 2000, 2001; Andresen et al., 2005). However, a hula twist about C5 would introduce a steric clash between the A- and B-rings, since both are tightly packed by protein residues and the A-ring is covalently anchored to the protein matrix. A hula twist about C10 also would be energetically disfavored due to close interactions between the BV chromophore and the side chains of the conserved His260, Ile208, and Tyr216 residues (Figure 4B). Moreover, assembly of the closely related bacteriophytochrome Agp1 from Agrobacterium tumefaciens with synthetic, sterically locked biliverdin derivatives (Inomata et al., 2005) strongly supports the conclusion that the Pr chromophore adopts a C15-Z,anti configuration, while the Pfr chromophore assumes a C15-E,anti configuration (Figure 5). For this reason, as well as the observed bathochromic shift of the primary Lumi-R intermediate (Sineshchekov, 1995), a hula twist about C15 also can be ruled out in the Pr-to-Pfr photoconversion pathway.
Figure 5.
Structures of Synthetically Locked Bilins and Spectra of Their Holophytochrome Adducts.
Wild-type BV (top panels) or two synthetic analogs locked in either the C15-Z,anti (middle panels) or C15-E,anti (bottom panels) configuration are shown assembled with the bacteriophytochrome Agp1. Absorption spectra of the resulting adducts are shown on the left, and structures of the bilin chromophores are shown on the right. Atoms determining the configuration about C15 are highlighted in blue. The wild-type BV adduct is shown in the Pr state (solid) and as a calculated pure Pfr spectrum (dashed line). Figures were prepared from raw data kindly provided by Tilman Lamparter and Katsuhiko Inomata (Inomata et al., 2005).
The DrBphP structural data also provide an exciting new template for experiments to explore the thermal pathways that follow the primary photochemical interconversion of Pr to Lumi-R. In this regard, the light-independent interconversion of Lumi-R to Pfr has been proposed to involve a further rotation about the C14,15 bond (Fodor et al., 1990; Andel et al., 1996; Kneip et al., 1999). This hypothesis requires that the Pfr chromophore would adopt a C15-E,syn conformation. Based on the DrBphP structure, however, this type of rotation would cause severe steric clashes with conserved Asp207, Tyr176, or Tyr263 (Wagner et al., 2005). Taken together with the locked bilin study (Inomata et al., 2005), the DrBphP crystal structure provides convincing evidence that the complete Pr-to-Pfr photoconversion involves a single light-dependent rotation about the C15,16 bond, with subsequent light-independent processes such as proton transfers and/or protein motions contributing to the overall conversion.
Bilin photoisomerization must be translated into changes in the biochemical activity of the C-terminal region to transduce the light absorption signal. Wagner et al. (2005) suggest that chromophore rearrangement may disrupt the interaction between the B-ring carboxylate of BV and conserved Arg254, which is part of the trefoil knot (Figure 3C). Such shifts could loosen the interaction between the PAS and GAF domains and permit the PAS domain to transduce the light signal, analogous to signal transduction by the PAS domains of the phototropins (Harper et al., 2003, 2004a, 2004b). Alternately, photochemistry could be transmitted through the GAF domain β-sheet to the backside helices, a mechanism suggested by other GAF domains (Martinez et al., 2002, 2005). Resolution of these and other possible mechanisms for signal transduction is an exciting area for future study.
IMPLICATIONS AND UNANSWERED QUESTIONS
Workers on plant phytochromes may well wonder how applicable the lessons from the DrBphP crystal structure will prove for their own research. For instance, will this information permit development of new phytochrome alleles whose expression in transgenic plants will enhance grain yield, inhibit shade avoidance, or confer other agronomically important traits to crop plants (Smith, 1994; Sawers et al., 2005)? To answer this question, one needs to consider the extent to which the photochemical and signaling mechanisms will be conserved between DrBphP and plant phytochromes. All phytochromes exhibit sequence conservation in the PAS and GAF regions whose structures have now been solved (Figure 2A); moreover, key residues in the knot between these two domains are also conserved (Wagner et al., 2005), indicating that the architecture of this photosensory core is very likely to be conserved in plant phytochromes.
The quantum yield for photoconversion for at least one bacteriophytochrome-BV adduct is known to be lower than that of the Cph1-PCB adduct (Lamparter et al., 2002). It is also known that at least some members of this subfamily have higher intrinsic fluorescence than Cph1 or plant phytochromes, suggesting that photoconversion may be generally less efficient for the bacteriophytochrome subfamily (Fischer et al., 2005; Giraud et al., 2005). Nevertheless, the sequence identity among all phytochromes is highest in the GAF domain, which supplies almost all of the protein residues contacting the chromophore (Figure 2). Moreover, the residues surrounding the bilin D-ring are very highly conserved between plant phytochromes that utilize PΦB as chromophore, cyanobacterial phytochromes that utilize PCB, and bacteriophytochromes that utilize BV (Figure 4A). It therefore seems likely that the key chromophore motions and protein conformational changes associated with the photochemical reaction pathway will be conserved among all members of the phytochrome family, perhaps even those bacteriophytochromes that adopt the Pfr state at thermal equilibrium (Karniol and Vierstra, 2003; Tasler et al., 2005) or others that interconvert between Pr and a blue-shifted Pnr (for pigment absorbing in the near red) state (Giraud et al., 2005).
The determination of the DrBphP crystal structure will permit much more detailed elucidation of the phytochrome photocycle as well as development of the next generation of structural hypotheses about phytochrome signaling in many organisms. This structure reveals several key surprises that could not have been reliably predicted, including the chromophore configuration and the trefoil knot. While not directly addressing the molecular mechanism of signal transmission to the C-terminal domains, these structural data will prove invaluable in attacking all aspects of phytochrome research. A single, static crystal structure may not answer all questions about the dynamic processes of photochemistry and signal transduction in phytochromes, but it at last provides a much firmer experimental foundation for asking the right questions.
Acknowledgments
We thank Katrina Forest, Richard Vierstra, Joseph Brunzelle, and Jeremiah Wagner for supplying the coordinates for the DrBphP structure prior to publication. We also thank Katsuhiko Inomata and Tilman Lamparter for providing the raw spectral data for Figure 5. J.C.L. is supported by grants from the National Institutes of Health (GM068552) and from the National Science Foundation Center for Biophotonics Science and Technology (PHY-0120999). We also acknowledge support from the National Institutes of Health Biomedical Supercomputing Iniative (MCB930017P) for bioinformatic analyses performed at the Pittsburgh Supercomputing Center.
References
- Andel, F., Lagarias, J.C., and Mathies, R.A. (1996). Resonance Raman analysis of chromophore structure in the lumi-R photoproduct of phytochrome. Biochemistry 35 15997–16008. [DOI] [PubMed] [Google Scholar]
- Andel, F., Murphy, J.T., Haas, J.A., McDowell, M.T., van der Hoef, I., Lugtenburg, J., Lagarias, J.C., and Mathies, R.A. (2000). Probing the photoreaction mechanism of phytochrome through analysis of resonance Raman vibrational spectra of recombinant analogues. Biochemistry 39 2667–2676. [DOI] [PubMed] [Google Scholar]
- Andresen, M., Wahl, M.C., Stiel, A.C., Grater, F., Schafer, L.V., Trowitzsch, S., Weber, G., Eggeling, C., Grubmuller, H., Hell, S.W., and Jakobs, S. (2005). Structure and mechanism of the reversible photoswitch of a fluorescent protein. Proc. Natl. Acad. Sci. USA 102 13070–13074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravind, L., and Ponting, C.P. (1997). The GAF domain: An evolutionary link between diverse phototransducing proteins. Trends Biochem. Sci. 22 458–459. [DOI] [PubMed] [Google Scholar]
- Batschauer, A. (2003). Photoreceptors and Light Signaling. (Cambridge, UK: Royal Society of Chemistry).
- Bhoo, S.H., Davis, S.J., Walker, J., Karniol, B., and Vierstra, R.D. (2001). Bacteriophytochromes are photochromic histidine kinases using a biliverdin chromophore. Nature 414 776–779. [DOI] [PubMed] [Google Scholar]
- Blumenstein, A., Vienken, K., Tasler, R., Purschwitz, J., Veith, D., Frankenberg-Dinkel, N., and Fischer, R. (2005). The Aspergillus nidulans phytochrome FphA represses sexual development in red light. Curr. Biol. 15 1833–1838. [DOI] [PubMed] [Google Scholar]
- Borucki, B., Otto, H., Rottwinkel, G., Hughes, J., Heyn, M.P., and Lamparter, T. (2003). Mechanism of Cph1 phytochrome assembly from stopped-flow kinetics and circular dichroism. Biochemistry 42 13684–13697. [DOI] [PubMed] [Google Scholar]
- Braslavsky, S.E., Gartner, W., and Schaffner, K. (1997). Phytochrome photoconversion. Plant Cell Environ. 20 700–706. [Google Scholar]
- Briggs, W.R., and Spudich, J.A. (2005). Handbook of Photosensory Receptors. (Weinheim, Germany: Wiley VCH).
- Butler, W.L., Norris, K.H., Seigelman, H.W., and Hendricks, S.B. (1959). Detection, assay, and preliminary purification of the pigment controlling photoresponsive development of plants. Proc. Natl. Acad. Sci. USA 45 1703–1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, M., Chory, J., and Fankhauser, C. (2004). Light signal transduction in higher plants. Annu. Rev. Genet. 38 87–117. [DOI] [PubMed] [Google Scholar]
- Chen, M., Tao, Y., Lim, J., Shaw, A., and Chory, J. (2005). Regulation of phytochrome B nuclear localization through light-dependent unmasking of nuclear-localization signals. Curr. Biol. 15 637–642. [DOI] [PubMed] [Google Scholar]
- Davis, S.J., Vener, A.V., and Vierstra, R.D. (1999). Bacteriophytochromes: Phytochrome-like photoreceptors from nonphotosynthetic eubacteria. Science 286 2517–2520. [DOI] [PubMed] [Google Scholar]
- Erbel, P.J., Card, P.B., Karakuzu, O., Bruick, R.K., and Gardner, K.H. (2003). Structural basis for PAS domain heterodimerization in the basic helix–loop–helix-PAS transcription factor hypoxia-inducible factor. Proc. Natl. Acad. Sci. USA 100 15504–15509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falk, H. (1989). The Chemistry of Linear Oligopyrroles and Bile Pigments. (Vienna, Austria: Springer-Verlag).
- Fischer, A.J., and Lagarias, J.C. (2004). Harnessing phytochrome's glowing potential. Proc. Natl. Acad. Sci. USA 101 17334–17339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer, A.J., Rockwell, N.C., Jang, A.Y., Ernst, L.A., Waggoner, A.S., Duan, Y., Lei, H., and Lagarias, J.C. (2005). Multiple roles of a conserved GAF domain tyrosine residue in cyanobacterial and plant phytochromes. Biochemistry 44 15203–15215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fodor, S.P.A., Lagarias, J.C., and Mathies, R.A. (1990). Resonance Raman analysis of the Pr and Pfr forms of phytochrome. Biochemistry 29 11141–11146. [DOI] [PubMed] [Google Scholar]
- Frankenberg, N., Mukougawa, K., Kohchi, T., and Lagarias, J.C. (2001). Functional genomic analysis of the HY2 family of ferredoxin-dependent bilin reductases from oxygenic photosynthetic organisms. Plant Cell 13 965–978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frishman, D., and Argos, P. (1995). Knowledge-based protein secondary structure assignment. Proteins 23 566–579. [DOI] [PubMed] [Google Scholar]
- Froehlich, A.C., Noh, B., Vierstra, R.D., Loros, J., and Dunlap, J.C. (2005). Genetic and molecular analysis of phytochromes from the filamentous fungus Neurospora crassa. Eukaryot. Cell, in press. [DOI] [PMC free article] [PubMed]
- Giraud, E., Fardoux, J., Fourier, N., Hannibal, L., Genty, B., Bouyer, P., Dreyfus, B., and Verméglio, A. (2002). Bacteriophytochrome controls photosystem synthesis in anoxygenic bacteria. Nature 417 202–205. [DOI] [PubMed] [Google Scholar]
- Giraud, E., Zappa, S., Vuillet, L., Adriano, J.M., Hannibal, L., Fardoux, J., Berthomieu, C., Bouyer, P., Pignol, D., and Verméglio, A. (2005). A new type of bacteriophytochrome acts in tandem with a classical bacteriophytochrome to control the antennae synthesis in Rhodopseudomonas palustris. J. Biol. Chem. 280 32389–32397. [DOI] [PubMed] [Google Scholar]
- Göller, A.H., Strehlow, D., and Hermann, G. (2005). The excited-state chemistry of phycocyanobilin: A semiempirical study. Chemphyschem 6 1259–1268. [DOI] [PubMed] [Google Scholar]
- Harper, S.M., Christie, J.M., and Gardner, K.H. (2004. a). Disruption of the LOV-J alpha helix interaction activates phototropin kinase activity. Biochemistry 43 16184–16192. [DOI] [PubMed] [Google Scholar]
- Harper, S.M., Neil, L.C., Day, I.J., Hore, P.J., and Gardner, K.H. (2004. b). Conformational changes in a photosensory LOV domain monitored by time-resolved NMR spectroscopy. J. Am. Chem. Soc. 126 3390–3391. [DOI] [PubMed] [Google Scholar]
- Harper, S.M., Neil, L.C., and Gardner, K.H. (2003). Structural basis of a phototropin light switch. Science 301 1541–1544. [DOI] [PubMed] [Google Scholar]
- Henikoff, S., and Henikoff, J.G. (1992). Amino acid substitution matrices from protein blocks. Proc. Natl. Acad. Sci. USA 89 10915–10919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho, Y.S.J., Burden, L.M., and Hurley, J.H. (2000). Structure of the GAF domain, a ubiquitous signaling motif and a new class of cyclic GMP receptor. EMBO J. 19 5288–5299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hübschmann, T., Börner, T., Hartmann, E., and Lamparter, T. (2001). Characterization of the Cph1 holo-phytochrome from Synechocystis sp PCC 6803. Eur. J. Biochem. 268 2055–2063. [DOI] [PubMed] [Google Scholar]
- Humphrey, W., Dalke, A., and Schulten, K. (1996). VMD: Visual molecular dynamics. J. Mol. Graph. 14, 33–38, 27–28. [DOI] [PubMed]
- Huq, E., and Quail, P.H. (2005). Phytochrome Signaling. In Handbook of Photosensory Receptors, W.R. Briggs and J.A. Spudich, eds (Weinheim, Germany: Wiley VCH), pp. 151–170.
- Inomata, K., Hammam, M.A., Kinoshita, H., Murata, Y., Khawn, H., Noack, S., Michael, N., and Lamparter, T. (2005). Sterically locked synthetic bilin derivatives and phytochrome Agp1 from Agrobacterium tumefaciens form photoinsensitive Pr- and Pfr-like adducts. J. Biol. Chem. 280 24491–24497. [DOI] [PubMed] [Google Scholar]
- Karniol, B., and Vierstra, R.D. (2003). The pair of bacteriophytochromes from Agrobacterium tumefaciens are histidine kinases with opposing photobiological properties. Proc. Natl. Acad. Sci. USA 100 2807–2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karniol, B., Wagner, J.R., Walker, J.M., and Vierstra, R.D. (2005). Phylogenetic analysis of the phytochrome superfamily reveals distinct microbial subfamilies of photoreceptors. Biochem. J. 392 103–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kneip, C., Hildebrandt, P., Schlamann, W., Braslavsky, S.E., Mark, F., and Schaffner, K. (1999). Protonation state and structural changes of the tetrapyrrole chromophore during the P-r -> P-fr phototransformation of phytochrome: A resonance Raman spectroscopic study. Biochemistry 38 15185–15192. [DOI] [PubMed] [Google Scholar]
- Kohchi, T., Mukougawa, K., Frankenberg, N., Masuda, M., Yokota, A., and Lagarias, J.C. (2001). The Arabidopsis HY2 gene encodes phytochromobilin synthase, a ferredoxin-dependent biliverdin reductase. Plant Cell 13 425–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurokawa, H., Lee, D.S., Watanabe, M., Sagami, I., Mikami, B., Raman, C.S., and Shimizu, T. (2004). A redox-controlled molecular switch revealed by the crystal structure of a bacterial heme PAS sensor. J. Biol. Chem. 279 20186–20193. [DOI] [PubMed] [Google Scholar]
- Lagarias, J.C., and Rapoport, H. (1980). Chromopeptides from phytochrome. The structure and linkage of the Pr form of the phytochrome chromophore. J. Am. Chem. Soc. 102 4821–4828. [Google Scholar]
- Lamparter, T., Carrascal, M., Michael, N., Martinez, E., Rottwinkel, G., and Abian, J. (2004). The biliverdin chromophore binds covalently to a conserved cysteine residue in the N-terminus of Agrobacterium phytochrome Agp1. Biochemistry 43 3659–3669. [DOI] [PubMed] [Google Scholar]
- Lamparter, T., Esteban, B., and Hughes, J. (2001). Phytochrome Cph1 from the cyanobacterium Synechocystis PCC6803. Purification, assembly, and quaternary structure. Eur. J. Biochem. 268 4720–4730. [DOI] [PubMed] [Google Scholar]
- Lamparter, T., Michael, N., Caspani, O., Miyata, T., Shirai, K., and Inomata, K. (2003). Biliverdin binds covalently to Agrobacterium phytochrome Agp1 via its ring A vinyl side chain. J. Biol. Chem. 278 33786–33792. [DOI] [PubMed] [Google Scholar]
- Lamparter, T., Michael, N., Mittmann, F., and Esteban, B. (2002). Phytochrome from Agrobacterium tumefaciens has unusual spectral properties and reveals an N-terminal chromophore attachment site. Proc. Natl. Acad. Sci. USA 99 11628–11633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, L., Murphy, J.T., and Lagarias, J.C. (1995). Continuous fluorescence assay of phytochrome assembly in vitro. Biochemistry 34 7923–7930. [DOI] [PubMed] [Google Scholar]
- Liu, R.S., and Hammond, G.S. (2000). The case of medium-dependent dual mechanisms for photoisomerization: One-bond-flip and hula-twist. Proc. Natl. Acad. Sci. USA 97 11153–11158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, R.S., and Hammond, G.S. (2001). Examples of hula-twist in photochemical cis-trans isomerization. Chemistry 7 4537–4544. [DOI] [PubMed] [Google Scholar]
- Martinez, S.E., Bruder, S., Schultz, A., Zheng, N., Schultz, J.E., Beavo, J.A., and Linder, J.U. (2005). Crystal structure of the tandem GAF domains from a cyanobacterial adenylyl cyclase: Modes of ligand binding and dimerization. Proc. Natl. Acad. Sci. USA 102 3082–3087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez, S.E., Wu, A.Y., Glavas, N.A., Tang, X.B., Turley, S., Hol, W.G.J., and Beavo, J.A. (2002). The two GAF domains in phosphodiesterase 2A have distinct roles in dimerization and in cGMP binding. Proc. Natl. Acad. Sci. USA 99 13260–13265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsushita, T., Mochizuki, N., and Nagatani, A. (2003). Dimers of the N-terminal domain of phytochrome B are functional in the nucleus. Nature 424 571–574. [DOI] [PubMed] [Google Scholar]
- Montgomery, B.L., and Lagarias, J.C. (2002). Phytochrome ancestry. Sensors of bilins and light. Trends Plant Sci. 7 357–366. [DOI] [PubMed] [Google Scholar]
- Mroginski, M.A., Murgida, D.H., von Stetten, D., Kneip, C., Mark, F., and Hildebrandt, P. (2004). Determination of the chromophore structures in the photoinduced reaction cycle of phytochrome. J. Am. Chem. Soc. 126 16734–16735. [DOI] [PubMed] [Google Scholar]
- Murphy, J.T., and Lagarias, J.C. (1997). The Phytofluors: A new class of fluorescent protein probes. Curr. Biol. 7 870–876. [DOI] [PubMed] [Google Scholar]
- Nagatani, A. (2004). Light-regulated nuclear localization of phytochromes. Curr. Opin. Plant Biol. 7 708–711. [DOI] [PubMed] [Google Scholar]
- Ni, M., Tepperman, J.M., and Quail, P.H. (1999). Binding of phytochrome B to its nuclear signalling partner PIF3 is reversibly induced by light. Nature 400 781–784. [DOI] [PubMed] [Google Scholar]
- Nozue, K., Kanegae, T., Imaizumi, T., Fukuda, S., Okamoto, H., Yeh, K.C., Lagarias, J.C., and Wada, M. (1998). A phytochrome from the fern Adiantum with features of the putative photoreceptor NPH1. Proc. Natl. Acad. Sci. USA 95 15826–15830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nureki, O., et al. (2002). An enzyme with a deep trefoil knot for the active-site architecture. Acta Crystallogr. D Biol. Crystallogr. 58 1129–1137. [DOI] [PubMed] [Google Scholar]
- Oka, Y., Matsushita, T., Mochizuki, N., Suzuki, T., Tokutomi, S., and Nagatani, A. (2004). Functional analysis of a 450-amino acid N-terminalfragment of phytochrome B in Arabidopsis. Plant Cell 16 2104–2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponting, C.P., and Aravind, L. (1997). PAS: A multifunctional domain family comes to light. Curr. Biol. 7 R674–R677. [DOI] [PubMed] [Google Scholar]
- Quest, B., and Gärtner, W. (2004). Chromophore selectivity in bacterial phytochromes: Dissecting the process of chromophore attachment. Eur. J. Biochem. 271 1117–1126. [DOI] [PubMed] [Google Scholar]
- Sanner, M.F., Olson, A.J., and Spehner, J.C. (1996). Reduced surface: An efficient way to compute molecular surfaces. Biopolymers 38 305–320. [DOI] [PubMed] [Google Scholar]
- Sawers, R.J., Sheehan, M.J., and Brutnell, T.P. (2005). Cereal phytochromes: Targets of selection, targets for manipulation? Trends Plant Sci. 10 138–143. [DOI] [PubMed] [Google Scholar]
- Schluchter, W.M., and Glazer, A.N. (1999). Biosynthesis of phycobiliproteins in cyanobacteria. In The Photosynthetic Prokaryotes, G.A. Peschek, W. Loffelhardt, and G. Schmetterer, eds (New York: Kluwer Academic/Plenum Press), pp. 83–95.
- Schäfer, E., and Nagy, F. (2005). Photomorphogenesis in Plants and Bacteria: Function and Signal Transduction Mechanisms, 3rd ed. (Dordrecht, The Netherlands: Springer-Verlag).
- Sineshchekov, V.A. (1995). Photobiophysics and photobiochemistry of the heterogeneous phytochrome system. Biochim. Biophys. Acta 1228 125–164. [Google Scholar]
- Smith, H. (1994). Phytochrome transgenics: Functional, ecological and biotechnological applications. Semin. Cell Biol. 5 315–325. [DOI] [PubMed] [Google Scholar]
- Stone, J. (1998). An Efficient Library for Parallel Ray Tracing and Animation. PhD dissertation (Rolla, MO: University of Missouri).
- Strauss, H.M., Hughes, J., and Schmieder, P. (2005). Heteronuclear solution-state NMR studies of the chromophore in cyanobacterial phytochrome Cph1. Biochemistry 44 8244–8250. [DOI] [PubMed] [Google Scholar]
- Suetsugu, N., Mittmann, F., Wagner, G., Hughes, J., and Wada, M. (2005). From the Cover: A chimeric photoreceptor gene, NEOCHROME, has arisen twice during plant evolution. Proc. Natl. Acad. Sci. USA 102 13705–13709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasler, R., Mioises, T., and Frankenberg-Dinkel, N. (2005). Biochemical and spectroscopic characterization of the bacterial phytochrome of Pseudomonas aeruginosa. J. Biol. Chem. 272 1927–1936. [DOI] [PubMed] [Google Scholar]
- Taylor, W.R. (2000). A deeply knotted protein structure and how it might fold. Nature 406 916–919. [DOI] [PubMed] [Google Scholar]
- Terry, M.J., Wahleithner, J.A., and Lagarias, J.C. (1993). Biosynthesis of the plant photoreceptor phytochrome. Arch. Biochem. Biophys. 306 1–15. [DOI] [PubMed] [Google Scholar]
- Thümmler, F., Dufner, M., Kreisl, P., and Dittrich, P. (1992). Molecular cloning of a novel phytochrome gene of the moss Ceratodon purpureus which encodes a putative light-regulated protein kinase. Plant Mol. Biol. 20 1003–1017. [DOI] [PubMed] [Google Scholar]
- Vierstra, R.D. (1993). Illuminating phytochrome functions. Plant Physiol. 103 679–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner, J.R., Brunzelle, J.S., Forest, K.T., and Vierstra, R.D. (2005). A light-sensing knot revealed by the structure of the chromophore binding domain of phytochrome. Nature 438 325–331. [DOI] [PubMed] [Google Scholar]
- Wu, S.H., and Lagarias, J.C. (2000). Defining the bilin lyase domain: Lessons from the extended phytochrome superfamily. Biochemistry 39 13487–13495. [DOI] [PubMed] [Google Scholar]
- Yeh, K.-C., and Lagarias, J.C. (1998). Eukaryotic phytochromes: Light-regulated serine/threonine protein kinases with histidine kinase ancestry. Proc. Natl. Acad. Sci. USA 95 13976–13981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh, K.-C., Wu, S.-H., Murphy, J.T., and Lagarias, J.C. (1997). A cyanobacterial phytochrome two-component light sensory system. Science 277 1505–1508. [DOI] [PubMed] [Google Scholar]
- Yildiz, O., Doi, M., Yujnovsky, I., Cardone, L., Berndt, A., Hennig, S., Schulze, S., Urbanke, C., Sassone-Corsi, P., and Wolf, E. (2005). Crystal structure and interactions of the PAS repeat region of the Drosophila clock protein PERIOD. Mol. Cell 17 69–82. [DOI] [PubMed] [Google Scholar]
- Zhao, K.H., Ran, Y., Li, M., Sun, Y.N., Zhou, M., Storf, M., Kupka, M., Böhm, S., Bubenzer, C., and Scheer, H. (2004). Photochromic biliproteins from the cyanobacterium Anabaena sp. PCC 7120: Lyase activities, chromophore exchange, and photochromism in phytochrome AphA. Biochemistry 43 11576–11588. [DOI] [PubMed] [Google Scholar]