Abstract
The C-class MADS box gene AGAMOUS (AG) plays crucial roles in Arabidopsis thaliana development by regulating the organ identity of stamens and carpels, the repression of A-class genes, and floral meristem determinacy. To examine the conservation and diversification of C-class gene function in monocots, we analyzed two C-class genes in rice (Oryza sativa), OSMADS3 and OSMADS58, which may have arisen by gene duplication before divergence of rice and maize (Zea mays). A knockout line of OSMADS3, in which the gene is disrupted by T-DNA insertion, shows homeotic transformation of stamens into lodicules and ectopic development of lodicules in the second whorl near the palea where lodicules do not form in the wild type but carpels develop almost normally. By contrast, RNA-silenced lines of OSMADS58 develop astonishing flowers that reiterate a set of floral organs, including lodicules, stamens, and carpel-like organs, suggesting that determinacy of the floral meristem is severely affected. These results suggest that the two C-class genes have been partially subfunctionalized during rice evolution (i.e., the functions regulated by AG have been partially partitioned into two paralogous genes, OSMADS3 and OSMADS58, which were produced by a recent gene duplication event in plant evolution).
INTRODUCTION
Gene duplication is closely associated with the evolution of genes with new functions (Force et al., 1999; Hughes, 1999). After gene duplication, for example, one of the genes may be released from functional constraints, enabling it to accumulate mutations and to acquire a new function (neofunctionalization). Alternatively, several functions controlled by an ancestral gene may be partitioned into two genes produced by gene duplication (subfunctionalization). Repetitious gene duplication produces many genes from a single ancestral gene and gives rise to the diversification of gene function into multigene families. In some cases, a gene may lose its function and accumulate mutations as a pseudogene (nonfunctionalization).
The study of floral homeotic mutants in model eudicots, such as Arabidopsis thaliana and Antirrhinum majus, has established the ABC model, which explains the genetic mechanism underlying floral organ specification. The ABC model proposes that three classes of genes, termed A, B, and C, are active in two adjacent whorls, and combinatorial activities of these genes specify four types of floral organs (Bowman et al., 1991; Coen and Meyerowitz, 1991; reviewed in Lohmann and Weigel, 2002; Jack 2004). A genes alone specify sepals, A and B genes together specify petals, B and C genes together specify stamens, and C genes alone specify carpels. Most of the ABC genes belong to the MADS box gene family and encode MIKC-type MADS domain proteins that function as transcription factors (reviewed in Riechmann and Meyerowitz, 1997; Alvarez-Buylla et al., 2000). The Arabidopsis class E genes, such as SEPALLATA3 (SEP3), which forms a multiprotein complex with the B- and C-class MADS domain proteins, also belong to the MADS box family (Pelaz et al., 2000; Honma and Goto, 2001). Thus, the functional diversification of MADS box genes has contributed greatly to the precise and complex genetic regulation of flower development in Arabidopsis; in addition, the diversification of MADS box genes during evolution has been proposed to be a major driving force for floral diversity in land plant architecture (Theissen et al., 2000; Kramer et al., 2004; Irish and Litt, 2005).
The grasses, Poaceae, constitute a large family containing ∼10,000 species of monocotyledonous plants that diverged from their closest relatives, monocots in the families Joinvilleaceae and Ecdeiocoleaceae, around 55 to 70 million years ago (Kellogg, 2001). The grass inflorescences comprise characteristic structural units called spikelets that consist of florets (Schmidt and Ambrose, 1998; McSteen et al., 2000; Bommert et al., 2005). The florets do not have obvious petals and sepals but instead include lodicules and leaf-like organs called palea and lemma. It is of great interest to elucidate the genetic mechanisms that regulate developmental processes in grass flowers as they are highly diversified from eudicot flowers. Several lines of genetic evidence indicate that the functions of B-class genes are well conserved across grass species and eudicots (Ambrose et al., 2000; Nagasawa et al., 2003). Molecular genetic studies of the rice (Oryza sativa) gene SUPERWOMAN1 and the maize (Zea mays) gene Silky1 have revealed that these APETALA3 (AP3)-like genes specify lodicules (corresponding to petals) and stamens in both grasses, similar to the function of AP3 in Arabidopsis (Jack et al., 1992). In addition, the gene products of the maize B-class genes Silky1 and Zmm16 (PISTILLATA ortholog) work as heterodimers, as do Arabidopsis B-class proteins (Whipple et al., 2004).
In Arabidopsis, the C-class gene AGAMOUS (AG) plays a critical role in formation of the reproductive organs, stamens, and carpels (Yanofsky et al., 1990; Bowman et al., 1991, 1999). In loss-of-function mutants of AG, the stamens are homeotically transformed into petals, and the central carpels are replaced by another ag flower. This ag flower consists entirely of sepals and petals and continues to produce organs indeterminately. Thus, AG has functions that regulate stamen and carpel identity, that repress A-class gene activity in the inner two whorls, and that establish floral meristem determinacy. Furthermore, a negative autoregulatory mechanism involving AG and WUSCHEL terminates stem cell maintenance (Lenhard et al., 2001; Lohmann et al., 2001), and the function of AG in floral meristem determinacy can be genetically separated from its function in organ identity (Mizukami and Ma, 1995; Sieburth et al., 1995). SHATTER PROOF1 (SHP1) and SHP2, which belong to the other clade of C-class genes, are partly involved in carpel development and are required for differentiation of the dehiscence zone (Liljegren et al., 2000; Pinyopich et al., 2003). In Antirrhinum, PLENA (PLE), which is closely related to the SHP genes, has a function similar to that of AG. The other C-class gene FARINELLI, an ortholog of AG, is responsible for the development of male reproductive organs and is partly redundant to PLE (Bradley et al., 1993; Davies et al., 1999). Thus, C-class genes are diversified and involved in various aspects of flower development in eudicots.
The function of grass C-class genes is not well understood. In grasses, the function of C-class genes may have diversified and become partially redundant as a result of gene duplication (Mena et al., 1996). In maize, a loss-of-function mutation in a C-class gene, ZAG1, does not result in the loss of carpel identity, although floral meristem determinacy is partially lost. It has been proposed that the near-normal specification of carpels in the zag1 mutant is due to the activity of the other C-class gene, ZMM2, which is functionally redundant to ZAG1. Alternatively, the two C-class genes might have diversified with separate functions: ZMM2 for organ identity and ZAG1 for floral meristem determinacy. In spite of this attractive hypothesis, no further genetic analysis has been reported so far. A similar situation is also seen in rice; namely, antisense suppression of the C-class gene OSMADS3 produces flowers with transformed stamens and abnormal carpels, but it does not give rise to a clear homeotic change in the carpels (Kang et al., 1998). We have previously shown that the YABBY gene DROOPING LEAF (DL) has a crucial function in carpel specification in rice (Yamaguchi et al., 2004). In loss-of-function mutants of DL, carpels are completely transformed into stamens. DL is expressed specifically in the presumptive region where carpels subsequently develop and in the developing carpel primordia. Thus, the functions of C-class genes and their genetic relationship with DL are particularly interesting.
In this article, we describe the functions of two rice C-class genes, OSMADS3 and OSMADS58 and their interactions with genes involved in flower development, including DL. Loss-of-function mutants of OSMADS3 show severe defects in stamen identity and lodicule number. By contrast, downregulation of OSMADS58 expression by RNA silencing causes extremely severe defects in floral meristem determinacy and partial defects in carpel morphogenesis. Our studies show that OSMADS3 and OSMADS58 have been subfunctionalized such that they play more predominant roles in distinct whorls. In addition, our results also suggest that C-class genes are involved in the asymmetric distribution of lodicules in the rice flower.
RESULTS
Identification of a New Member of the C-Class Gene Family and Phylogenetic Analysis
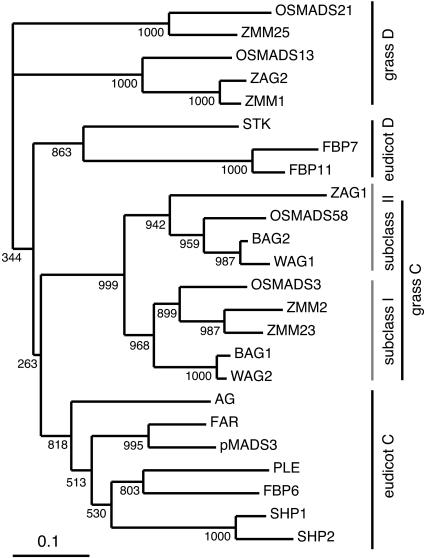
We identified a new member of the rice MADS box gene family, OSMADS58, which belongs to the AG subfamily in addition to the OSMADS3, OSMADS13, and OSMADS21 genes that have been reported so far (Kang et al., 1998; Lopez-Dee et al., 1999; Lee et al., 2003). OSMADS58 is most closely related to OSMADS3 among the rice MADS box genes. OSMADS3 and OSMADS58 share 96 and 67% sequence identity in the MADS domain and across the whole protein, respectively (see Supplemental Figure 1 online). OSMADS3 and OSMADS58 share similar genomic organization: they contain 10 exons and 9 introns at the same positions and have a large intron after the second exon, which partly contains the MADS box. These structural features suggest that the two genes arose by a duplication event relatively recently in evolution.
We therefore analyzed the phylogenetic relationship of MADS domain proteins belonging to the AG subfamily in several species of eudicots and monocots (Figure 1; see Supplemental Figure 2 online). The resulting tree indicates that OSMADS3 and OSMADS58 are classified into the C-lineage of the AG subfamily, whereas OSMADS13 and OSMADS21 are classified into the D-lineage, as shown previously (Kramer et al., 2004). Of the four species of the grass family examined, rice, wheat (Triticum aestivum), and barley (Hordeum vulgare) have two genes belonging to the C-lineage, whereas maize has three genes belonging to this lineage, and the genes are classified into two subgroups: subgroup I, including OSMADS3, ZMM2, ZMM23, WAG2, and BAG1; and subgroup II, including OSMADS58, ZAG1, WAG1, and BAG2. These results suggest that these genes arose by gene duplication before the divergence of the grass species. The presence of two maize genes in subgroup I may reflect the duplication of the maize genome after its divergence from other species (Gaut and Doebley, 1997).
Figure 1.
Phylogenetic Analysis of MADS Domain Proteins in the AG Subfamily.
Analysis was done by the neighbor-joining method, OSMADS21 and ZMM25 were used as outgroups, and the local bootstrap values after 1000 replicates are indicated near the branching points.
Comparison of OSMADS3 and OSMADS58 Expression Patterns
To determine the functions of the two C-class genes in rice, we first analyzed the spatial and temporal expression patterns of OSMADS3 and OSMADS58 during flower development in wild-type plants (Figures 2A to 2E and 2G to 2K).
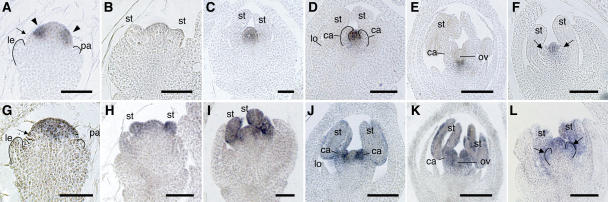
Figure 2.
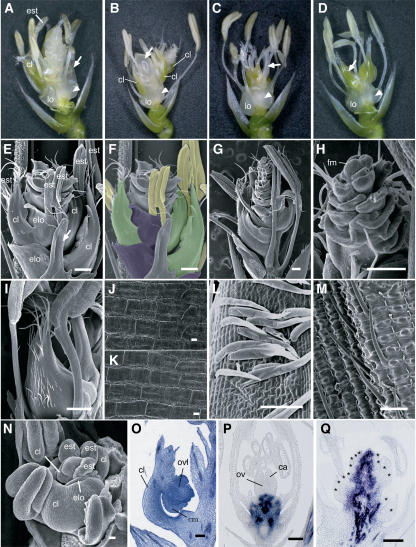
In Situ Localization of OSMADS3 and OSMADS58 Transcripts in Wild-Type and dl-sup1 Flowers.
(A) to (E) In situ localization of OSMADS3 transcripts in wild-type flowers. The stages of flower development proceed from (A) to (E).
(F) In situ localization of OSMADS3 transcripts in the dl-sup1 flower. The stage of flower development corresponds to the wild-type flower in (D).
(G) to (K) In situ localization of OSMADS58 transcripts in wild-type flowers. The stages of flower development proceed from (G) to (K).
(L) In situ localization of OSMADS58 transcripts in the dl-sup1 flower. The stage of flower development corresponds to the wild-type flower in (J).
Arrowheads in (A) indicate stamen anlagen where a high level of OSMADS3 transcripts accumulate. Arrows in (A) and (G) indicate the regions that later produce lodicule primordia and that do not accumulate OSMADS3 or OSMADS58 transcripts. Arrows in (F) and (L) indicate ectopic stamens in whorl 4. Lemma and palea primordia in (A) and (G), carpel primordia in (D), and ectopic stamen primordia in (L) are outlined. ca, carpel; le, lemma; lo, lodicule; ov, ovule; pa, palea; st, stamen. Bars = 20 μm.
Expression of OSMADS3 was first detected at the time when the lemma and palea primordia were just initiating (Figure 2A). At this stage, strong expression of OSMADS3 was observed mainly in whorl 3, where the stamen primordia subsequently initiate, and only a weak signal was detected in whorl 4 (Figure 2A). Soon after initiation of the stamen primordia, the OSMADS3 signal disappeared from the developing stamen primordia, but weak expression remained in whorl 4 (Figure 2B). Expression of OSMADS3 was highly elevated in whorl 4 just before carpel development initiated (Figure 2C), but it disappeared from the carpel primordia after emergence of the carpel bulge (Figure 2D). At this stage, strong expression of OSMADS3 was observed in the floral meristem, which later produces the ovule primordia (Figure 2D), but it was downregulated once the ovule primordia initiated (Figure 2E). Expression of OSMADS58 was initially detected at the same time as the first expression of OSMADS3 (Figure 2G) and was distributed uniformly in whorl 3 and whorl 4. OSMADS58 was expressed in the primordia of stamens, carpels, and ovules from their inception, and its expression persisted during the development of these floral organs (Figures 2H to 2K).
In summary, OSMADS3 and OSMADS58 show similar spatial patterns of expression in that they are both expressed exclusively in whorl 3 and whorl 4, suggesting that they have functions similar to those of typical C-class genes, such as AG in Arabidopsis. However, the temporal expression patterns of the two genes differ from each other. OSMADS3 is expressed strongly only in the presumptive region from which the stamen, carpel, and ovule primordia subsequently differentiate, whereas OSMADS58 is continually expressed in these primordia from their inception to later developmental stages.
Notably, both OSMADS3 and OSMADS58 were expressed asymmetrically in the floral meristem along the palea–lemma axis (Figures 2A and 2G). Both genes were expressed in the cells just adjacent to the palea primordium but not in the cells adjacent to the lemma primordium. In rice, two lodicules develop inside the lemma, but not on the palea side. The region where both OSMADS3 and OSMADS58 are downregulated corresponds to the region where the lodicule primordia initiate (see Discussion).
Isolation of Insertion Mutants of OSMADS3 and Silencing of OSMADS58 by RNA Interference
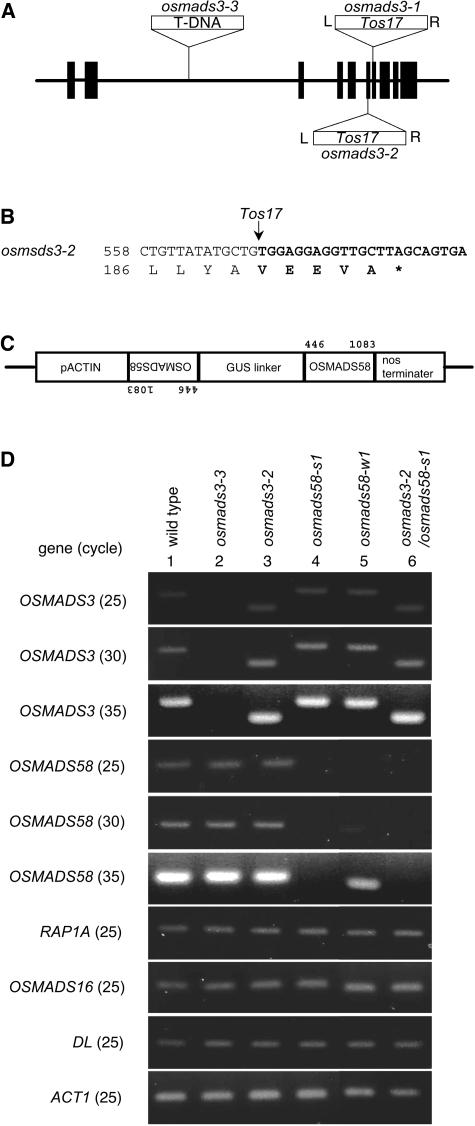
To investigate further the functions of the two C-class genes in rice, we used reverse genetic approaches. First, we tried to isolate insertional mutants from lines tagged with the retrotransposon Tos17 or with T-DNA (Hirochika et al., 1996; Hirochika, 2001; Jeong et al., 2002). We obtained two lines, osmads3-1 and osmads3-2, that had Tos17 insertions in OSMADS3 (Figure 3A). Although osmads3-1 showed no abnormality, osmads3-2, which lacked 86 amino acids in the C-terminal region (Figure 3B), showed a relatively weak phenotype. In osmads3-3, the T-DNA was inserted into the large second intron (Figure 3A), and this allele showed a severe phenotype. No OSMADS3 transcript was detected by RT-PCR analysis in RNA samples from osmads3-3 homozygotes (Figure 3D).
Figure 3.
Molecular Characterization of Insertional Mutants of OSMADS3 and RNAi Plants of OSMADS58.
(A) Genomic structure of OSMADS3 and position of the T-DNA or Tos17 insertion. Boxes indicate exons, and thick lines indicate introns.
(B) Insertion site of Tos17 in osmads3-2. Tos17 sequences are shown in bold. A possible chimeric translation product, terminating within the Tos17 element (asterisk), is shown beginning with amino acid 186.
(C) Schematic representation of the transgene construct, which can produce double-stranded RNA of the 3′-half the OSMADS58 sequence with a loop. The positions of the nucleotides of OSMADS58 used in this construct are shown.
(D) RT-PCR–based expression analysis of ABC MADS box genes and DL in young flowers of wild-type, osmads3 mutant, osmads58RNAi, and osmads3-2/osmads58RNAi lines. Rice ACT1 was analyzed as a control. The genes subjected to amplification are shown at the left. The number in parentheses indicates the PCR cycle number. Lines from which the RNA samples were isolated are shown at the top. To monitor OSMADS3 expression in osmads3-2, a gene-specific primer and a Tos17-specific primer were used, which resulted in a smaller fragment as compared with samples containing the wild-type allele.
Because we could not isolate any insertion lines for OSMADS58, we tried to repress OSMADS58 gene activity by RNA interference (RNAi) silencing (Figure 3C). Among 17 transgenic plants produced, 12 lines showed severe phenotypes and 5 lines showed weaker phenotypes. We further analyzed osmads58-s1 and osmads58-w1 as representative alleles for severe and weak phenotypes, respectively. In osmads58-s1, no OSMADS58 transcripts were detected even after 35 cycles of the RT-PCR reaction (Figure 3D). Normal amount of transcripts of the other MADS box genes were detected, even for OSMADS3, the gene most closely related to OSMADS58. These results indicate that RNAi silencing operates effectively and specifically on OSMADS58 in this line. In osmads58-w1, relatively few OSMADS58 transcripts were detected (Figure 3D). The effect of RNAi was also specific to OSMADS58 in this line.
Phenotypes of osmads3 Mutants
In the severe loss-of-function allele osmads3-3, almost all stamens in whorl 3 were homeotically transformed into lodicule-like organs (Figures 4B, 4E, 4F, and 8A, Table 1). Some stamens were completely transformed into lodicules that were indistinguishable from those in the second whorl. In other flowers, incomplete transformation was observed, that is, an organ of a mosaic of lodicules and stamens such that the anther-like organ was fused to the lodicule-like organ (Figure 4F). Analyses by scanning electron microscopy showed that the shape and arrangement of the epidermal cells of the transformed organs in osmads3-3 were almost identical to those of the lodicules in the wild type (Figures 4H and 4I). These observations demonstrate that OSMADS3 is necessary for stamen specification in rice.
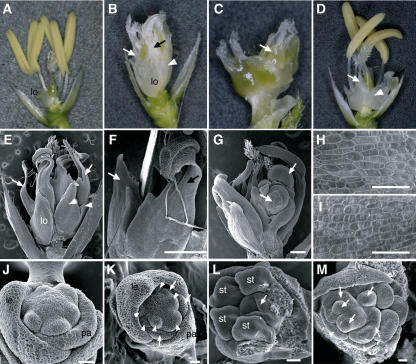
Figure 4.
Phenotypes of osmads3 Mutant Flowers.
(A) Wild-type flower.
(B) osmads3-3 flower showing the ectopic development of lodicules in whorl 2 and the homeotic transformation of stamens into lodicules in whorl 3.
(C) osmads3-3 flower showing the multiple carpels in whorl 4. For clarity, the organs in whorl 2 and whorl 3 have been removed. Arrow indicates ectopic carpels that formed interior to whorl 4 carpels.
(D) osmads3-2 flower.
(E) Scanning electron micrograph of the osmads3-3 flower.
(F) Scanning electron micrograph of transformed organs in whorl 3 of the osmads3-3 flower.
(G) Scanning electron micrograph of ectopic cell masses (arrows) emerging from osmads3-3 carpels.
(H) and (I) Scanning electron micrographs showing the epidermal morphology of a wild-type lodicule (H) and a transformed organ in whorl 3 of the osmads3-3 flower (I).
(J) and (L) Early development of the wild-type flower. For clarity, some stamens have been removed in (L).
(K) and (M) Early development of the osmads3-3 flower.
Arrowheads in (B), (D), and (E) indicate ectopic lodicules in whorl 2. White and black arrows in (B) and (D) to (F) indicate complete and incomplete transformation of stamens into lodicules in whorl 3, respectively. Arrows and arrowheads in (K) indicate ectopic lodicules in whorl 2 and in whorl 3, respectively. Arrows in (L) and (M) indicate carpel primordia. Arrowheads in (M) indicate transformed organs in whorl 3. le, lemma; lo, lodicule; pa, palea; st, stamen. Bars = 500 μm in (E) to (G) and 20 μm in (H) to (M).
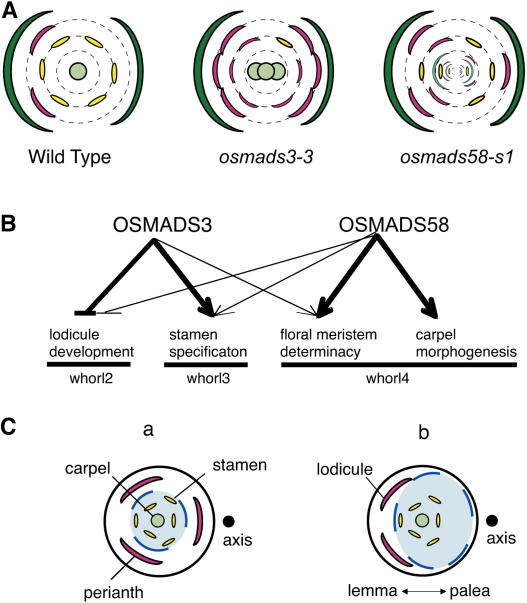
Figure 8.
Schematic Representation of the Function of OSMADS3 and OSMADS58.
(A) Typical flower phenotypes of the wild type, osmads3, and osmads58. Green, lemma (left) and palea (right); pink, lodicules; yellow, stamens; pale green, carpels or carpel-like organs.
(B) Functional diversification of OSMADS3 and OSMADS58. Thin arrows indicate a weak contribution of gene function.
(C) The asymmetric arrangement of lodicules and the expression domain of C-class genes. (a) A putative ancestral flower of rice. (b) A present-day rice flower. Blue colors indicate the expression domains of C-class genes.
Table 1.
Summary of Phenotypes
| Genotype | Second Whorlab | Third Whorla | Fourth Whorlc |
|---|---|---|---|
| Wild type | Lodicules | Stamens | Carpels |
| (2.0) [2.0] | (6.0) | (100%) | |
| osmads3-3 | Increased number of lodicules | Severe transformation of stamens into lodicules | Increased number of carpels |
| (4.4) [9.7] | (5.3) | (84%) | |
| osmads3-2 | Increased number of lodicules | Mild transformation of stamens into lodicules | Increased number of carpels |
| (3.8) [6.1] | (2.3) | (6%) | |
| osmads58-s1 | Increased number of lodicules | Weak transformation of stamens into lodicules | Indeterminate organ development |
| (3.2) [5.1] | (1.8) | (100%) | |
| osmads58-w1 | Increased number of lodicules | Weak transformation of stamens into lodicules | Increased number of carpels, indeterminate organ development |
| (2.7) [3.7] | (1.0) | (76%) (16%) | |
| osmads3-2/osmads58-s1 | Increased number of lodicules | Severe transformation of stamens into lodicules | Indeterminate organ development |
| (4.2) [8.4] | (4.2) | (100%) |
The numbers in parentheses indicate the average number of organs of that type found in that genotype.
The numbers in brackets indicate the total number of the lodicule/lodicule-like organs in second and third whorls on average.
The numbers in parentheses indicate the percentage of flowers of that type found in that genotype.
An increase in the carpel number in whorl 4 was observed in osmads3-3 (Figures 4C and 8A, Table 1). These carpels had developed from the same whorl and were fused at the base. The number of stigma also increased approximately fourfold in this line as compared with the wild type. The morphology of the carpels was almost normal, although undifferentiated cell masses sometimes emerged out of the carpels (Figure 4G). Occasionally, new carpels developed within the carpels of the fourth whorl (Figure 4C), suggesting that the number of floral whorls had slightly increased. Taken together, these phenotypes suggest that floral meristem determinacy is partially lost in osmads3-3.
Unexpectedly, loss of OSMADS3 function affected organ development in whorl 2. In the rice wild-type flower, two lodicules formed only on the lemma side (Figures 4A and 8A); in the osmads3-3 flower, however, ectopic lodicules developed on the palea side in whorl 2 (Figures 4B, 4E, and 8A). On average, 4.4 lodicules developed in whorl 2, but in an extreme case, six lodicules developed in this whorl (Table 1). These lodicules were often fused at the base and arranged in whorled phyllotaxy. This result suggests that in the wild type, OSMADS3 controls the number of lodicules in whorl 2 by repressing lodicule development, especially near the palea.
In the intermediate allele of osmads3-2, the homeotic transformation of stamens in whorl 3 and the increase in lodicule number in whorl 2 were also observed, although the extent of the abnormality was smaller than in osmads3-3 (Figure 4D, Table 1). The defects in whorl 4 were very weak, and an increase in carpel number was observed in only a few flowers (Table 1).
We examined the developmental defects in the osmads3-3 flower at early stages by scanning electron microscopy. Although osmads3-3 flower development proceeded normally until emergence of the lemma and palea primordia, abnormalities in osmads3-3 were detected at the stage when the stamen primordia initiated. Six stamen primordia of the wild-type flower were small and spherical in shape and were deposited at equal intervals (Figure 4J). By contrast, some of the third whorl organ primordia of the osmads3-3 flower were irregular in shape, and the positioning of the primordia was distorted, probably reflecting a homeotic change in the stamen (Figure 4K). Ectopic lodicule primordia at the palea side were detected at this stage (Figure 4K). After the multiple carpel primordia developed in whorl 4, the floral meristem was eventually consumed by these organs, suggesting that the defect in floral meristem determinacy is incomplete in osmads3-3 (Figure 4M).
Phenotypes of osmads58 RNAi Plants
The most prominent phenotype in the severely silenced line of osmads58 was the indeterminate development of floral organs (Figure 8A). The osmads58-s1 line produced florets that reiterated a set of floral organs consisting of lodicules, stamens/ectopic lodicules, and carpel-like organs (Figures 5A, 5B, and 5E to 5G; characteristics of the floral organs in each whorl are described below in detail). Observations of the early developmental pattern clearly showed a repetition of whorls forming the primordia of lodicules, stamens, and carpel-like organs (Figure 5N). This repetition produced >50 floral organs in the flower of osmads58-s1, although the organ number in each whorl gradually reduced toward the inner whorls. Even in an almost-mature flower that had mature anthers with long filaments, the flower meristem remained at the top of the flower, suggesting that floral meristem determinacy is severely lost in osmads58-s1 (Figures 5G and 5H). To confirm this, we examined the expression pattern of the class I KNOX gene, OSH1, which is a molecular marker of meristem-indeterminate cells in rice (Sato et al., 1996). In the wild-type flower, OSH1 expression was detected in the floral meristem at the early stages (Yamaguchi et al., 2004), but it completely disappeared when carpels developed (Figure 5P). By contrast, expression of OSH1 in the osmads58-s1 flower was maintained in the central region even after many floral organs had been produced (Figure 5Q). These results strongly indicate that determinacy of the floral meristem is completely disrupted in osmads58-s1.
Figure 5.
Phenotypes of osmads58RNAi Flowers.
(A) osmads58-s1 flower. Arrow indicates a whorl 3 stamen that has been partially transformed into a lodicule.
(B) osmads58-s1 flower. Arrow indicates an ectopic cell mass emerging from a carpel-like organ.
(C) osmads58-w1 flower. Arrow indicates an ectopic stamen produced interior to the multiple carpels in whorl 4.
(D) osmads58-w1 flower. Arrow indicates a carpel-like organ, similar to that formed in osmads58-s1. Arrowheads in (A) to (D) indicate ectopic lodicules in whorl 2.
(E) and (F) Scanning electron micrographs of the osmads58-s1 flower showing the reiterative development of lodicules, stamens, and carpel-like organs. Some organs in the outer whorls have been removed to show indeterminate organ development. Arrow in (E) indicates a partially transformed stamen into the lodicule. In (F), each floral organ is indicated by a different color: purple, lodicule; yellow, stamen; green, carpel-like organ.
(G) Scanning electron micrograph of the osmads58-s1 flower at anthesis. Some organs in the outer whorls have been removed to show indeterminate organ development.
(H) Close-up of (G). The floral meristem (fm), which still produces organs, can be observed.
(I) Scanning electron micrograph of a carpel-like organ in osmads58-s1.
(J) and (K) Scanning electron micrographs showing the epidermal morphology of a wild-type carpel (J) and the carpel-like organ of the osmads58-s1 flower (K).
(L) Scanning electron micrograph of trichomes in the carpel-like organs of the osmads58-s1 flower.
(M) Scanning electron micrograph of bristles in wild-type lemma.
(N) Scanning electron micrograph of an early stage of the osmads58-s1 flower.
(O) Longitudinal section of the carpel-like organ showing an ovule-like structure (ovl) in the osmads58-s1 flower.
(P) and (Q) In situ localization of OSH1 transcripts in late stage of the wild-type (P) and osmads58-s1 (Q) flowers. Asterisks in (Q) indicate floral organ primordia.
ca, carpel; cl, carpel-like organ; cm, cell mass; elo, ectopic lodicule; est, ectopic stamen developing reiteratively; lo, original lodicule; ov, ovule. Bars = 500 μm in (E) to (I) and 20 μm in (J) to (Q).
Although the number of lodicules in whorl 2 increased in osmads58-s1, as it did in osmads3, the defect was less severe in osmads58-s1 (Figures 5A, 5B, and 5E to 5G, Table 1). The homeotic transformation of stamens into lodicules in whorl 3 was also observed in osmads58-s1. The number of floral organs showing this transformation, however, was very low, and normal stamens developed in whorl 3 although the number of these was reduced (Figures 5A, 5B, and 5E to 5G, Table 1). These results indicate that, like OSMADS3, OSMADS58 plays a partial role in repressing in extra lodicule development in whorl 2 and in controlling stamen specification, but the contribution of OSMADS58 to these developmental functions seems to be weaker than that of OSMADS3.
Carpel morphology was strongly affected in osmads58-s1. In the wild type, carpel primordia initiate from one side and develop toward the opposite side; the carpels then finally fuse, resulting in the formation of a bottle-like structure with two stigmas at the top (Figure 4A; Yamaguchi et al., 2004). By contrast, the carpels developed into thick cup-like structures without fusing along their margins in osmads58-s1 (Figure 5I). These structures were pale green, like wild-type pistils, but they did not differentiate stigmatic tissues at the top, and on the epidermis, they produced many trichomes, which are not observed in wild-type carpels. Because wild-type palea and lemma are green and form trichomes, we examined the carpel-like structures of osmads58-s1 in detail. Scanning electron microscopy analyses showed that the shape and the arrangement of epidermal cells of the carpel-like organs of osmads58-s1 were almost identical to those of the wild-type carpel wall (Figures 5J and 5K). The trichomes on these organs were hairy (Figure 5L) and were distinct from bristles on the palea and lemma (Figure 5M). The surface morphology of the carpel-like organs also differed from those of wild-type palea and lemma (Figure 5M). In addition, longitudinal sections revealed that ovule-like organs were produced on the adaxial side of these organs (Figure 5O). Although these ovule-like organs were enclosed by outer and inner integuments similar to wild-type ovules, no embryo sac had differentiated inside them, and in some flowers, an ectopic cell mass formed around the integuments (Figure 5O). The cell mass occasionally emerged from the carpel-like organs (Figure 5B). Thus, the abnormal organs formed inside the stamens/ectopic lodicules in osmads58-s1 had carpel identities to a certain degree but their morphology was altered. These observations indicate that OSMADS58 may be required for the morphogenesis of carpels in rice.
In the weak line osmads58-w1, the defects in floral meristem determinacy were less severe, and multiple carpels developed in whorl 4. Interior to these carpels, a few lodicules or stamens often developed (Figure 5C). Thus, the number of floral whorls was slightly increased, but the floral meristem was not present in mature flowers. In many cases, the carpel morphology was normal, but in a few, abnormal carpel-like organs similar to those in osmads58-s1 developed (Figure 5D).
In summary, OSMADS58 plays a crucial role in regulating floral meristem determinacy. In addition, OSMADS58 is responsible for the normal morphogenesis of carpels and is weakly required both for stamen identity and for repressing the differentiation of ectopic lodicules.
Expression Analyses
To identify genetic interactions between C-class genes and other floral organ identity genes in rice, in situ hybridization analysis was performed.
In osmads3-3, expression of the rice B-class gene SPW1 was restricted to whorl 2 and whorl 3, as it is in the wild type (Figures 6A and 6B). In addition, DL was expressed specifically in the multiple carpels of osmads3-3 (Figure 6C). OSMADS58 was initially expressed in whorl 3 and whorl 4 of osmad3-3, as in the wild type, suggesting that OSMADS3 is not required for the initial expression of OSMADS58 (Figure 6D). At later developmental stages, the expression of OSMADS58 became weaker in the transformed organs in whorl 3, whereas it remained strong in the multiple carpel primordia (Figures 6E and 6F). The reduction in OSMADS58 expression in whorl 3 may be a secondary effect arising from the homeotic transformation of stamens into lodicules.
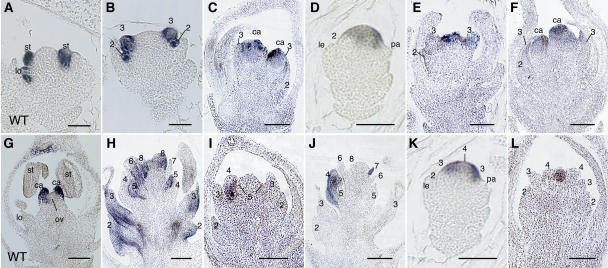
Figure 6.
In Situ Localization of Organ Identity Genes in Wild-Type, osmads3-3, and osmads58-s1 Flowers.
(A) and (G) In situ localization of SPW1 (A) and DL (G) transcripts in the wild-type flower.
(B) In situ localization of SPW1 transcripts in the osmads3-3 flower.
(C) In situ localization of DL transcripts in the osmads3-3 flower.
(D) to (F) In situ localization of OSMADS58 transcripts in the osmads3-3 flower. Flower development proceeds from (D) to (F).
(H) In situ localization of SPW1 transcripts in the osmads58-s1 flower.
(I) and (J) In situ localization of DL transcripts in the osmads58-s1 flower. Flower development proceeds from (I) to (J).
(K) and (L) In situ localization of OSMADS3 transcripts in the osmads58-s1 flower. Flower development proceeds from (K) to (L).
Whorls are numbered. ca, carpel; le, lemma; lo, lodicule; ov, ovule; pa, palea; st, stamen. Bars = 20 μm.
In osmads58-s1, expression of SPW1 was observed in the lodicules in whorl 2 and in the stamens/ectopic lodicules in whorl 3, but it was excluded from the carpel-like organs in whorl 4 and whorl 7 (Figure 6H). SPW1 expression was also observed in the organs corresponding to the lodicules and stamens in the inner flowers, but it was not detected in the carpel-like organs (Figure 6H). By contrast, DL was expressed specifically in the carpel-like organs of whorl 4 and that of the inner flowers (whorl 7) (Figures 6I and 6J). The signals of SPW1 and DL were complementary to each other; that is, SPW1 expression was excluded from the whorl (organs) where DL was expressed and vice versa. As above, these results demonstrate that flowers consisting of lodicules, stamens/ectopic lodicules, and a carpel-like organ develop reiteratively in osmads58-s1. In addition, the expression of DL in the carpel-like organs suggests that these organs retain carpel identity. The expression pattern of OSMADS3 was not altered in osmads58-s1. At an early developmental stage, strong expression in whorl 3 and relatively weak expression in whorl 4 were observed (Figure 6K). Later on, OSMADS3 continued to be expressed in the floral meristem (Figure 6L). This observation suggests that OSMADS3 is expressed independently of OSMADS58.
Next, we analyzed OSMADS3 and OSMADS58 expression in the severe dl mutant (dl-sup1), in which carpels are homeotically transformed into stamens (Nagasawa et al., 2003; Yamaguchi et al., 2004). At an early developmental stage, OSMADS3 and OSMADS58 were expressed in whorl 3 and whorl 4 of dl-sup1 as they were in the wild type (see Supplemental Figure 3 online). At the stage when ectopic stamens were produced in whorl 4 of dl-sup1, OSMADS3 was expressed in the floral meristem (Figure 2F), whereas OSMADS58 was expressed in stamens and ectopic stamens in whorl 3 and whorl 4 and in the floral meristem (Figure 2L). Thus, the expression domains of both OSMADS3 and OSMADS58 in dl-sup1 were similar to their domains in the wild type, indicating that DL does not act upstream of C-class genes.
Genetic Interactions between OSMADS3 and OSMADS58
To identify genetic interactions between the two C-function genes in rice, an RNAi construct targeting OSMADS58 was introduced into homozygotes of the intermediate allele osmads3-2, and four transgenic lines were isolated. Because all lines showed similar morphology, below we describe osmads3-2/osmads58-s1 as a representative line. In this line, expression of OSMADS58 was repressed specifically and strictly (Figure 3D).
In whorl 2, the number of ectopic lodicules was higher in osmads3-2/osmads58-s1 than in the osmads3-2 or osmads58-s1 single mutant (Figure 7A, Table 1). Similarly, the number of ectopic lodicules was higher in whorl 3 of osmads3-2/osmads58-s1 (Figure 7A, Table 1). These results suggest that OSMADS3 and OSMADS58 have overlapping roles in repressing extra lodicule development in whorl 2 and in specifying stamen identity in whorl 3.
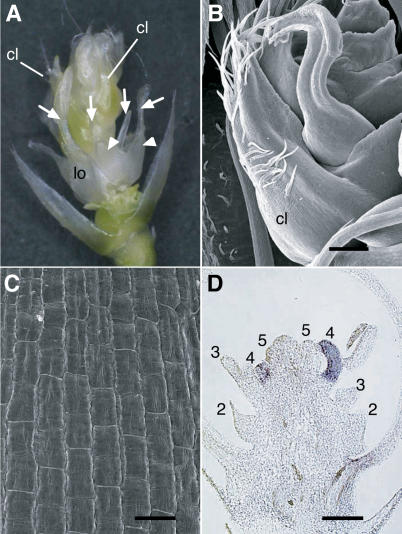
Figure 7.
Phenotypes of the osmads3-2/osmads58-s1 Flower.
(A) osmads3-2/osmads58-s1 flower. Arrowheads indicate ectopic lodicules in whorl 2. Arrows indicate transformed organs in whorl 3.
(B) Scanning electron micrograph of a carpel-like organ developing in the osmads3-2/osmads58-s1 flower.
(C) Scanning electron micrograph of epidermal morphology of a carpel-like organ in the osmads3-2/osmads58-s1 flower.
(D) In situ localization of DL transcripts in the osmads3-2/osmads58-s1 flower. Whorls are numbered.
cl, carpel-like organ; lo, original lodicule in whorl 2. Bars = 20 μm in (B) and (D) and 500 μm in (C).
Although abnormal carpel morphology was observed in osmads3-2/osmads58-s1 (Figure 7A), the overall morphology and epidermal characteristics of the carpel-like organs in osmads3-2/osmads58-s1 were indistinguishable from those in osmads58-s1 (Figures 7B and 7C). In addition, DL was expressed specifically in carpel-like organs, indicating that these organs retain carpel identity (Figure 7D). Thus, no additive defect in carpel morphology was detected in osmads3-2/osmads58-s1, suggesting that OSMADS3 is not involved in carpel morphogenesis.
The defect in floral meristem determinacy in osmads3-2/osmads58-s1 was also very similar to that in osmads58-s1, and almost the same numbers of organs developed (Table 1). This result strongly supports the idea that OSMADS58 plays a much more crucial role in regulating floral meristem determinacy than OSMADS3.
DISCUSSION
OSMADS58 Is Crucial for Regulating Floral Meristem Determinacy
RNA silencing of OSMADS58 caused a complete loss of floral meristem determinacy, resulting in the indeterminate development of flowers consisting of lodicules, stamens, and a carpel-like organ. In addition, OSH1 was still expressed in the mature flowers, suggesting that the floral meristem was not consumed by floral organs in the osamds58-silenced lines. By contrast, floral meristem determinacy was less affected by the loss of function of OSMADS3 because the repetitious flowers did not develop and the floral meristem was consumed by the carpels. In osmads3-2/osmads58-s1, the defect in the floral meristem was very similar to that observed in the osmads58-s1 single line. Taken together, these results indicate that OSMADS58 plays an essential role in regulating determinacy of the floral meristem in rice, whereas OSMADS3 weakly contributes to this function (Figure 8B).
In a previous study, we demonstrated that DL also regulates floral meristem determinacy because ectopic stamens develop repeatedly in whorl 4 and OSH1 is expressed in the floral meristem in the dl-sup1 flower (Yamaguchi et al., 2004). We found, however, that the defect in floral meristem determinacy in the osmads58-s1 flower is stronger than that in the dl-sup1 flower, suggesting that DL functions only partially in this regulation. Both OSMADS3 and OSMADS58 were expressed normally in dl-sup1, which suggests that DL does not regulate floral meristem determinacy by controlling C-class gene expression.
In maize, a loss-of-function mutation in ZAG1, which is classified in the same subgroup as OSMADS58, results in a partial defect in the floral meristem (Mena et al., 1996), suggesting that the genes in this clade have a similar function in regulating floral meristem determinacy in grass species. In Arabidopsis, the severe ag mutant repeatedly develops a set of floral organs (sepal-petal-petal) (Yanofsky et al., 1990; Bowman et al., 1991). This repetitious formation of a set of floral organs suggests that there is functional conservation between AG and OSMADS58 with respect to regulation of floral meristem determinacy. The osmads58-s1 flower, however, differs from the ag flower in that it contains carpels instead of sepals. In rice flower development, the floral meristem remains after the carpel primordia initiate (Suzaki et al., 2004; Yamaguchi et al., 2004), whereas in Arabidopsis flower development, the meristem is consumed by the carpel primordia. Therefore, this delayed consumption of the floral meristem in rice may be linked to the different type of repetitious flower formed in osmads58-s1, which involves carpels, as compared with the Arabidopsis ag mutant. In addition, the continued expression of DL also may be involved in carpel development in the osmads58-s1 flower. The lack of palea and lemma in the repetitious flowers of osmads58-s1 suggests that the region that is responsible for the development of lodicules, stamens, and carpels within the rice floral meristem corresponds to the floral meristem of Arabidopsis. Alternatively, DL may prevent the development of palea and lemma in osmads58-s1.
OSMADS58 Is Required for Normal Carpel Morphogenesis
We found that the severe silencing line of OSMADS58 develops abnormal carpels that are cup-shaped rather than bottle-like as wild-type carpels that lack stigmas and that differentiate trichomes. By contrast, carpel morphology was almost normal in osmads3-3, and no additive effect was observed in the carpel-like organs of osmads3-2/osmads58-s1 as compared with osmads58-s1. Taken together, these results strongly suggest that OSMADS58 is necessary for normal carpel morphogenesis, but OSMADS3 is not involved in this process (Figure 8B). DL was expressed specifically in the carpel-like organs in osmads58-s1 and osmads3-2/osmads58-s1, suggesting that DL is responsible for carpel-like organ development in osmads58-s1 and osmads3-2/osmads58-s1. This result also suggests that DL does not require C-class genes to be expressed; however, there remains the possibility that residual OSMADS3 activity could promote DL expression in osmads3-2/osmads58-s1.
The homeotic transformation of stamens into lodicules in osmads3 and osmads58 indicates that A-class genes are ectopically active in whorl 3, which suggests that C-class genes repress A-class gene activity in the inner whorls in the wild type. It is also plausible that ectopic A-class gene activity is involved in the abnormal morphogenesis of carpels in osmads58-s1. In line with this view, OSMADS3 seems not to repress A-class genes in whorl 4 because near-normal carpels were formed in the knockout line. This hypothesis could be verified by analyzing the expression patterns of A-class genes in osmads3 and osmads58. Alternatively, it is also possible that OSMADS58 represses some genes that cause the abnormal carpel morphology in osmads58-s1 via pathways that are independent of A-class genes.
OSMADS3 and OSMADS58 Regulate Stamen Identity but OSMADS3 Plays a More Crucial Role
Loss-of-function mutations of OSMADS3 or silencing of OSMADS58 caused the homeotic transformation of stamens into lodicules, suggesting that OSMADS3 and OSMADS58 are necessary for stamen specification in rice similar to typical C-class genes in eudicots (Yanofsky et al., 1990; Bowman et al., 1991; Coen and Meyerowitz, 1991). Our results are highly consistent with those of a previous study indicating that overexpression of OSMADS3 causes the homeotic transformation of lodicules into stamens (Kyozuka and Shimamoto, 2002).
Although the homeotic transformation of stamens was observed in both OSMADS3 and OSMADS58 loss-of-function lines, the contribution of the two genes to the function of the stamen is varied. Most of the stamens were homeotically transformed into lodicules in osmads3-3, whereas only a small number of stamens showed homeotic transformation even in the severe line osmads58-s1. In addition, the number of stamens transformed into lodicules was higher in the weaker allele osmads3-2 than in the severe allele osmads58-s1. These results, therefore, strongly indicate that OSMADS3 plays a more crucial role in stamen specification than OSMADS58 (Figure 8B). The additive effect on the transformation of stamens into lodicules in osmads3-2/osmads58-s1 may be due to genetic redundancy between OSMADS3 and OSMADS58. Antisense lines of OSMADS3 show weaker abnormalities in the flower than osmads3-3, suggesting that repression of this gene is not complete in the antisense lines (Kang et al., 1998).
We found that OSMADS3 is expressed only in the presumptive region (stamen anlagen) where the stamen later develop, whereas OSMADS58 is expressed in both the stamen anlagen and the developing stamen primordia. Thus, these findings suggest that early expression of OSMADS3 before stamen initiation is essential for specifying the stamen. Late expression of OSMADS58 in the developing stamen primordia may be required for maintaining stamen development, for example, for differentiation and/or tissue maturation, because the anthers in osmads58-s1 are white and shrunken, and few pollen are produced (T. Yamaguchi and H.-Y. Hirano, unpublished data).
Stamens are specified by the combinatorial activities of B- and C-class genes in eudicots. In rice, the loss of function of SPW1, a B-class gene, gives rise to the homeotic transformation of stamens into carpels (Nagasawa et al., 2003). Taking these findings together with our results, we conclude that the genetic mechanism that controls stamen specification is conserved in rice.
OSMADS3 and OSMADS58 Prevent Lodicule Development
Both the loss of function of OSMADS3 and the silencing of OSMADS58 led to ectopic lodicule formation. Ectopic lodicules were produced not only at the lemma side where lodicules are formed in the wild-type flower but also at the palea side where lodicule formation is not seen. Thus, OSMADS3 and OSMADS58 may together control the lodicule number by repressing ectopic lodicule formation. A greater number of ectopic lodicules, however, developed in osmads3-3 than in osmads58-s1, and an additive effect was observed in osmads3-2/osmads58-s1. Thus, OSMADS3 plays a more predominant role than OSMADS58 in controlling lodicule formation (Figure 8B).
The finding that OSMADS3 and OSMADS58 repress ectopic lodicule formation is unexpected considering the well-known general role of C-class genes. However, AG may be involved in a similar regulation in Arabidopsis. For example, strong ap2 mutations result in a loss of organs in whorl 2 where AG is ectopically expressed (Bowman et al., 1991; Drews et al., 1991), whereas organs develop in whorl 2 of the ap2 ag double mutant (Bowman et al., 1991). Similarly, high-level ectopic expression of AG in whorl 2 under the 35S or AP3 promoter leads to a loss of whorl 2 organs (Mizukami and Ma, 1992; Jack et al., 1997). These results suggest that AG, like C-class genes in rice, functions in repressing organ development in whorl 2 in Arabidopsis. Therefore, this function in preventing organ development in whorl 2 may be conserved between the C-class genes of rice and Arabidopsis, although the extent and type of effect on phenotype vary depending on the plant species.
The lodicules develop asymmetrically in the rice wild-type flower; that is, two lodicules are formed inside the lemma and no lodicules are produced inside the palea. We found that both OSMADS3 and OSMADS58 are expressed asymmetrically only on the palea side. Thus, the region where the two genes are downregulated corresponds to the region where the lodicule primordia later initiate. Many more lodicules developed on the palea side than on the lemma side when the activity of OSMADS3 or OSMADS58 was removed. Taken together, these results suggest that OSMADS3 and OSMADS58 expression just inside the palea is involved in repressing lodicule development also in the wild type and contributes to the formation of an asymmetric flower architecture.
Monocot flowers generally consist of trimerous floral organs, and Joinvilleaceae, the sister family of Poaceae (the grass family), has a trimerous perianth organ (six tapels), suggesting that flowers with three lodicules are an ancestral type in the grass family (Judd et al., 2002). Although many grass flowers have two lodicules, flowers with three lodicules are distributed in the basal grasses and in many species of Bambusoideae (Clifford, 1987; Grass Phylogeny Working Group, 2001). Bamusoideae is the subfamily closest to Ehrhartoideae, to which rice belongs, and has a higher frequency of species with three lodicules than other grass subfamilies. These morphological studies suggest that the rice flower may have been derived from an ancestral flower with three lodicules. It is therefore possible that the expansion of the expression domain of C-class genes toward the palea may be associated with a loss of lodicules near the palea, resulting in the asymmetric arrangement of lodicules in rice evolution (Figure 8B). The distribution of flowers with two or three lodicules is not correlated with phylogenetic relationships, indicating that a loss of lodicules may have occurred several times in grass evolution. It will be of great interest to determine the correlation between the asymmetric arrangement of lodicules and C-class gene expression in other species.
OSMADS3 and OSMADS58 Are Subfunctionalized with More Predominant Roles in Different Whorls
The C-class MADS box genes in eudicots have functions that specify stamens in whorl 3, repress A-class genes in whorl 3 and whorl 4, and control floral meristem determinacy in whorl 4. In addition to these well-known functions, the genes act in preventing organ development in whorl 2 as discussed above. OSMADS3 and OSMADS58 share these functions of C-class genes in rice. The relative contributions of each gene to individual function, however, differ depending on the floral whorl. In the outer two whorls, OSMADS3 plays a more predominant role in inhibiting lodicule development and in specifying stamen identity. By contrast, OSMADS58 contributes more to conferring floral meristem determinacy and to regulating carpel morphogenesis. These findings suggest that OSMADS3 and OSMADS58 have been subfunctionalized to have predominant functions in different whorls (Figure 8B).
Evolutionary analysis suggests that multiple C-class genes arose by gene duplication before the divergence of species in the grass family. The subfunctionalization of C-class genes may have been closely associated with this gene duplication event. In maize, a loss-of-function mutant of ZAG1 (zag1-mum1) shows a phenotype similar to that of osmads58-s1 (Mena et al., 1996). The floral meristem of zag1-mum1 has a defect in determinacy of the floral meristem, although the defect is less severe than that in osmads58-s1. Similar to the osmads58-s1 carpels, the zag1-mum1 carpels are not fused in the female flowers. By contrast, stamens are almost normal in the zag1-mum1 male flowers, suggesting that other C-class genes, such as ZMM2 and ZMM23 that are classified into subgroup I like OSMADS3, may be responsible for stamen specification. Thus, the subfunctionalization of C-class genes may have begun before the divergence of rice and maize; in addition, the fine-tuning of this subfunctionalization may have differed in both species because slight differences are observed between osmads58-s1 and zag1-mum1. It will be interesting to determine the phenotypes of loss-of-function mutants of ZMM2, ZMM23, or both genes.
An explanation for the whorl-dependent functional diversification of C-class genes in rice may lie in the difference in spatial expression patterns. For example, OSMADS3 expression is downregulated in whorl 4 before termination of the floral meristem, whereas OSMADS58 expression is maintained even after carpel initiation. However, the predominant function of OSMADS58 in whorl 4 cannot be explained only by its expression profile because it is expressed uniformly in both whorl 3 and whorl 4. Alternatively, differences in the specificity of protein–protein interactions may be associated with functional diversification. For example, OSMADS3 and OSMADS58 may interact specifically with factors that are preferentially expressed in whorl 3 and whorl 4, respectively. Possible candidates for protein partners might be the SEP-like MADS domain proteins (reviewed in Malcomber and Kellogg, 2005). In Arabidopsis, B- and C-class MADS domain proteins require SEP-like proteins to function, and these proteins form protein complexes (Pelaz et al., 2000; Honma and Goto, 2001). In some species, SEP-like proteins that have whorl-specific functions have been reported. For example, in Gerbera hybrida, downregulation of a SEP-like gene, GRCD1 (for Gerbera Regulator of Capitulum Development1), results in a homeotic transformation only in one whorl (i.e., staminodes are transformed into petals) (Kotilainen et al., 2000). In maize, the SEP-like gene ZMM14 is preferentially expressed in carpels (Cacharron et al., 1999). In rice, some SEP genes are expressed in inner whorls (reviewed in Malcomber and Kellogg, 2005). Identifying the protein partners of OSMADS3 and OSMADS58, together with their functional analyses, might elucidate the molecular nature of the whorl-dependent subfunctionalization of these genes in rice.
METHODS
Plant Materials
The rice strain used in this study was Oryza sativa spp japonica. Taichung 65 (T65) was used as the wild type for RT-PCR, histological observation, and in situ hybridization. The origin of dl-sup1 has been described previously (Nagasawa et al., 2003).
Identification of OSMADS58
OSMADS58 was identified by a database search as a genomic sequence with high similarity to OSMADS3 cDNA. The OSMADS58 sequence was determined from RT-PCR products that were amplified using the gene-specific primers and cDNA produced from RNA samples from developing rice flowers.
Isolation of Insertional Mutants of OSMADS3
osmads3-1 and osmads3-2 were isolated from the R2 population of plants (Nipponbare) that were putative mutant lines caused by insertion of the retrotransposon Tos17 (Hirochika et al., 1996). To screen for insertional mutants, DNA fragments were amplified by PCR using Tos17-specific primers and OSMADS3-specific primers from DNA pools obtained using the three-dimensional sampling method from 27,840 putative insertional lines (Hirochika, 2001; see Supplemental Table 1 online). The PCR products were separated on agarose gels and hybridized with a probe of the partial sequence of OSMADS3 cDNA. Positive products were gel purified and sequenced to verify the insertional lines isolated.
Isolation of a T-DNA–Tagged Line
The generation of T-DNA–tagged lines in japonica rice Hwayoung with pGA2715 has been reported previously (Jeong et al., 2002). The sequences flanking the T-DNA insertion sites were determined by inverse PCR analyses (Jeong et al., 2006). From the flanking sequence database, we isolated line 1A-19842, in which T-DNA was inserted into the 2nd intron of OSMADS3.
RNA Silencing of OSMADS58
To construct RNAi transgenic plants, the 3′-half of OSMADS58 (446 to 1083) was amplified by RT-PCR in both the sense and antisense directions. The sense and antisense RNAi fragments were amplified with primer pairs listed in Supplemental Table 1 online. The resulting PCR fragments were ligated between the Act1 promoter and the nopaline synthase terminator in the binary vector pACT-Nos (Sentoku et al., 2000) in a position sandwiching 700 bp of partial β-glucuronidase coding region. The pACT-Nos/RNAi OSMADS58 construct was verified by restriction mapping and sequencing, transferred into Agrobacterium tumefaciens strain EHA 105, and then transformed into rice as described (Hiei et al., 1994).
RT-PCR Analysis
Total RNA was isolated from a young inflorescence containing developing flowers using Trizol reagent (Life Technologies) according to the manufacturer's instructions. The first strand of cDNA was synthesized with Superscript II RT (Gibco BRL) from 2 μg of total RNA using oligo(dT)25 primers in 20 μL of the reaction mixture according to the manufacturer's instructions. We used 0.5 μL as a template for PCR amplification. PCR products were fractionated on agarose gels and stained by ethidium bromide. Two independent experiments for semiquantitative RT-PCR were performed with good agreement between the results.
In Situ Hybridization
Gene-specific regions at the 3′ end of OSMADS3 (676 to 1089) and OSMADS58 (464 to 1094) were amplified and cloned into a T-vector. The preparation of probes for DL, SPW1, and OSH1 has been described previously (Sato et al., 1996; Nagasawa et al., 2003; Yamaguchi et al., 2004). Probe synthesis, the preparation of sections, and in situ hybridization were performed as described previously (Yamaguchi et al., 2004).
Phylogenetic Analysis
The amino acid sequences were first aligned by ClustalW and readjusted manually in detail (see Supplemental Figure 2 online). The phylogenetic tree was constructed by the neighbor-joining method (Saitou and Nei, 1987). Programs used here were provided by the DNA database of Japan (http://www.ddbj.nig.ac.jp/search/clustalw-e.html).
Accession Numbers
Sequence data of the OSMADS58 cDNA has been deposited with DDBJ/EMBL/GenBank data libraries under accession number AB232157. The accession numbers of the sequence data used are as follows: AG (X53579), SHP1 (M55550), SHP2 (M55553), and STK (U20182) from Arabidopsis; PLE (S53900) and FAR (AJ239057) from Antirrhinum; FBP6 (X68675), pMADS3 (X72912), FBP7 (X81651), and FBP11 (X81852) from Petunia hybrida; OSMADS3 (L37528), OSMADS13 (AF151693), OSMADS21 (AP003379), and OSMADS58 (AB232157) from rice; ZAG1 (L18924), ZAG2 (L18925), ZMM1, (X81199), ZMM2 (X81200), ZMM23 (AJ430637), and ZMM25 (AJ430639) from maize; WAG1 (AB084577) and WAG2 (K. Murai, personal communication) from Triticum aestivum; BAG1 (AG486648) and BAG2 (AF486649) from Hordeum vulgare.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Table 1. Primers Used in This Work.
Supplemental Figure 1. Amino Acid Alignment of OSMADS3 and OSMADS58.
Supplemental Figure 2. Amino Acid Alignment of Proteins Encoded by the MADS Box Genes in the AG Subfamily.
Supplemental Figure 3. In Situ Localization of OSMADS3 and OSMADS58 in the Early Stages of dl Flowers.
Supplementary Material
Acknowledgments
We thank Y. Komeda for critically reading the manuscript. This research was supported by Grants-in-Aid for Scientific Research (15031208) from the Ministry of Education, Culture, Sports, Science, and Technology and by grants (Rice Genome Project, IP-1005) from the Ministry of Agriculture, Forestry, and Fisheries of Japan and the 21st Century Program (CG-1111) from Crop Functional Genomics Center, Republic of Korea.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Hiro-Yuki Hirano (hyhirano@biol.s.u-tokyo.ac.jp).
Online version contains Web-only data.
Open Access articles can viewed online without a subscription.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.105.037200.
References
- Alvarez-Buylla, E.R., Pelaz, S., Liljegren, S.J., Gold, S.E., Burgeff, C., Ditta, G.S., Ribas de Pouplana, L., Martinez-Castilla, L., and Yanofsky, M.F. (2000). An ancestral MADS-box gene duplication occurred before the divergence of plants and animals. Proc. Natl. Acad. Sci. USA 97 5328–5333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambrose, B.A., Lerner, D.R., Ciceri, P., Padilla, C.M., Yanofsky, M.F., and Schmidt, R.J. (2000). Molecular and genetic analyses of the silky1 gene reveal conservation in floral organ specification between eudicots and monocots. Mol. Cell 5 569–579. [DOI] [PubMed] [Google Scholar]
- Bommert, P., Satoh-Nagasawa, N., Jackson, D., and Hirano, H.-Y. (2005). Genetics and evolution of inflorescence and flower development in grasses. Plant Cell Physiol. 46 69–78. [DOI] [PubMed] [Google Scholar]
- Bowman, J.L., Baum, S.F., Eshed, Y., Putterill, J., and Alvarez, J. (1999). Molecular genetics of gynoecium development in Arabidopsis. Curr. Top. Dev. Biol. 45 155–205. [DOI] [PubMed] [Google Scholar]
- Bowman, J.L., Smyth, D.R., and Meyerowitz, E.M. (1991). Genetic interactions among floral homeotic genes of Arabidopsis. Development 112 1–20. [DOI] [PubMed] [Google Scholar]
- Bradley, D., Carpenter, R., Sommer, H., Hartley, N., and Coen, E. (1993). Complementary floral homeotic phenotypes result from opposite orientations of a transposon at the plena locus of Antirrhinum. Cell 72 85–95. [DOI] [PubMed] [Google Scholar]
- Cacharron, J., Saedler, H., and Theissen, G. (1999). Expression of MADS box genes ZMM8 and ZMM14 during inflorescence development of Zea mays discriminates between the upper and the lower floret of each spikelet. Dev. Genes Evol. 209 411–420. [DOI] [PubMed] [Google Scholar]
- Clifford, H.T. (1987). Spikelet and floral morphology. In Grass Systematics and Evolution, T.R. Soderstrom, K.W. Hilu, C.S. Campbell, and M.E. Barkworth, eds (Washington, DC: Smithsonian Institution Press), pp. 21–30.
- Coen, E.S., and Meyerowitz, E.M. (1991). The war of the whorls: Genetic interactions controlling flower development. Nature 353 31–37. [DOI] [PubMed] [Google Scholar]
- Davies, B., Motte, P., Keck, E., Saedler, H., Sommer, H., and Schwarz-Sommer, Z. (1999). PLENA and FARINELLI: Redundancy and regulatory interactions between two Antirrhinum MADS-box factors controlling flower development. EMBO J. 18 4023–4034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drews, G.N., Bowman, J.L., and Meyerowitz, E.M. (1991). Negative regulation of the Arabidopsis homeotic gene AGAMOUS by the APETALA2 product. Cell 65 991–1002. [DOI] [PubMed] [Google Scholar]
- Force, A., Lynch, M., Pickett, F.B., Amores, A., Yan, Y.L., and Postlethwait, J. (1999). Preservation of duplicate genes by complementary, degenerative mutations. Genetics 151 1531–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaut, B.S., and Doebley, J.F. (1997). DNA sequence evidence for the segmental allotetraploid origin of maize. Proc. Natl. Acad. Sci. USA 94 6809–6814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grass Phylogeny Working Group (2001). Phylogeny and subfamilial classification of the grasses (Poaceae). Ann. Mo. Bot. Gard. 88 373–457. [Google Scholar]
- Hiei, Y., Ohta, S., Komari, T., and Kumashiro, T. (1994). Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J. 6 271–282. [DOI] [PubMed] [Google Scholar]
- Hirochika, H. (2001). Contribution of the Tos17 retrotransposon to rice functional genomics. Curr. Opin. Plant Biol. 4 118–122. [DOI] [PubMed] [Google Scholar]
- Hirochika, H., Sugimoto, K., Otsuki, Y., Tsugawa, H., and Kanda, M. (1996). Retrotransposons of rice involved in mutations induced by tissue culture. Proc. Natl. Acad. Sci. USA 93 7783–7788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honma, T., and Goto, K. (2001). Complexes of MADS-box proteins are sufficient to convert leaves into floral organs. Nature 409 525–529. [DOI] [PubMed] [Google Scholar]
- Hughes, A.L. (1999). Adaptive Evolution of Genes and Genomes. (New York: Oxford University Press).
- Irish, V.F., and Litt, A. (2005). Flower development and evolution: Gene duplication, diversification and redeployment. Curr. Opin. Genet. Dev. 15 454–460. [DOI] [PubMed] [Google Scholar]
- Jack, T. (2004). Molecular and genetic mechanisms of floral control. Plant Cell 16 (suppl.), S1–S17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack, T., Brockman, L.L., and Meyerowitz, E.M. (1992). The homeotic gene APETALA3 of Arabidopsis thaliana encodes a MADS box and is expressed in petals and stamens. Cell 68 683–697. [DOI] [PubMed] [Google Scholar]
- Jack, T., Sieburth, L., and Meyerowitz, E. (1997). Targeted misexpression of AGAMOUS in whorl 2 of Arabidopsis flowers. Plant J. 11 825–839. [DOI] [PubMed] [Google Scholar]
- Jeong, D.-H., An, S., Kang, H.-G., Moon, S., Han, J.-J., Park, S., Lee, H.S., An, K., and An, G. (2002). T-DNA insertional mutagenesis for activation tagging in rice. Plant Physiol. 130 1636–1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong, D.-H., et al. (2006). Generation of flanking sequence tag database for activation tagging lines in japonica rice. Plant J., in press. [DOI] [PubMed]
- Judd, W.S., Campbell, C.S., Kellogg, E.A., Stevens, P.F., and Donoghue, M.J. (2002). Plant Systematics: A Phylogenetic Approach. (Sunderland, MA: Sinauer Associates).
- Kang, H.-G., Jeon, J.-S., Lee, S., and An, G. (1998). Identification of class B and class C floral organ identity genes from rice plants. Plant Mol. Biol. 38 1021–1029. [DOI] [PubMed] [Google Scholar]
- Kellogg, E.A. (2001). Evolutionary history of the grasses. Plant Physiol. 125 1198–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotilainen, M., Elomaa, P., Uimari, A., Albert, V.A., Yu, D., and Teeri, T.H. (2000). GRCD1, an AGL2-like MADS box gene, participates in the C function during stamen development in Gerbera hybrida. Plant Cell 12 1893–1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer, E.M., Jaramillo, M.A., and Di Stilio, V.S. (2004). Patterns of gene duplication and functional evolution during the diversification of the AGAMOUS subfamily of MADS box genes in angiosperms. Genetics 166 1011–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyozuka, J., and Shimamoto, K. (2002). Ectopic expression of OsMADS3, a rice ortholog of AGAMOUS, caused a homeotic transformation of lodicules to stamens in transgenic rice plants. Plant Cell Physiol. 43 130–135. [DOI] [PubMed] [Google Scholar]
- Lee, S., Kim, J., Son, J.-S., Nam, J., Jeong, D.-H., Lee, K., Jang, S., Yoo, J., Lee, J., Lee, D.-Y., Kang, H.-G., and An, G. (2003). Systematic reverse genetic screening of T-DNA tagged genes in rice for functional genomic analyses: MADS-box genes as a test case. Plant Cell Physiol. 44 1403–1411. [DOI] [PubMed] [Google Scholar]
- Lenhard, M., Bohnert, A., Jurgens, G., and Laux, T. (2001). Termination of stem cell maintenance in Arabidopsis floral meristems by interactions between WUSCHEL and AGAMOUS. Cell 105 805–814. [DOI] [PubMed] [Google Scholar]
- Liljegren, S.J., Ditta, G.S., Eshed, Y., Savidge, B., Bowman, J.L., and Yanofsky, M.F. (2000). SHATTERPROOF MADS-box genes control seed dispersal in Arabidopsis. Nature 404 766–770. [DOI] [PubMed] [Google Scholar]
- Lohmann, J.U., Hong, R.L., Hobe, M., Busch, M.A., Parcy, F., Simon, R., and Weigel, D. (2001). A molecular link between stem cell regulation and floral patterning in Arabidopsis. Cell 105 793–803. [DOI] [PubMed] [Google Scholar]
- Lohmann, J.U., and Weigel, D. (2002). Building beauty: The genetic control of floral patterning. Dev. Cell 2 135–142. [DOI] [PubMed] [Google Scholar]
- Lopez-Dee, Z.P., Wittich, P., Enrico Pe, M., Rigola, D., Del Buono, I., Gorla, M.S., Kater, M.M., and Colombo, L. (1999). OsMADS13, a novel rice MADS-box gene expressed during ovule development. Dev. Genet. 25 237–244. [DOI] [PubMed] [Google Scholar]
- Malcomber, S.T., and Kellogg, E.A. (2005). SEPALLATA gene diversification: Brave new whorls. Trends Plant Sci. 10 427–435. [DOI] [PubMed] [Google Scholar]
- McSteen, P., Laudencia-Chingcuanco, D., and Colasanti, J. (2000). A floret by any other name: Control of meristem identity in maize. Trends Plant Sci. 5 61–66. [DOI] [PubMed] [Google Scholar]
- Mena, M., Ambrose, B.A., Meeley, R.B., Briggs, S.P., Yanofsky, M.F., and Schmidt, R.J. (1996). Diversification of C-function activity in maize flower development. Science 274 1537–1540. [DOI] [PubMed] [Google Scholar]
- Mizukami, Y., and Ma, H. (1992). Ectopic expression of the floral homeotic gene AGAMOUS in transgenic Arabidopsis plants alters floral organ identity. Cell 71 119–131. [DOI] [PubMed] [Google Scholar]
- Mizukami, Y., and Ma, H. (1995). Separation of AG function in floral meristem determinacy from that in reproductive organ identity by expressing antisense AG RNA. Plant Mol. Biol. 28 767–784. [DOI] [PubMed] [Google Scholar]
- Nagasawa, N., Miyoshi, M., Sano, Y., Satoh, H., Hirano, H.-Y., Sakai, H., and Nagato, Y. (2003). SUPERWOMAN 1 and DROOPING LEAF genes control floral organ identity in rice. Development 130 705–718. [DOI] [PubMed] [Google Scholar]
- Pelaz, S., Ditta, G.S., Baumann, E., Wisman, E., and Yanofsky, M.F. (2000). B and C floral organ identity functions require SEPALLATA MADS-box genes. Nature 405 200–203. [DOI] [PubMed] [Google Scholar]
- Pinyopich, A., Ditta, G.S., Savidge, B., Liljegren, S.J., Baumann, E., Wisman, E., and Yanofsky, M.F. (2003). Assessing the redundancy of MADS-box genes during carpel and ovule development. Nature 424 85–88. [DOI] [PubMed] [Google Scholar]
- Riechmann, J.L., and Meyerowitz, E.M. (1997). MADS domain proteins in plant development. Biol. Chem. 378 1079–1101. [PubMed] [Google Scholar]
- Saitou, N., and Nei, M. (1987). The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4 406–425. [DOI] [PubMed] [Google Scholar]
- Sato, Y., Hong, S.K., Tagiri, A., Kitano, H., Yamamoto, N., Nagato, Y., and Matsuoka, M. (1996). A rice homeobox gene, OSH1, is expressed before organ differentiation in a specific region during early embryogenesis. Proc. Natl. Acad. Sci. USA 93 8117–8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt, R.J., and Ambrose, B.A. (1998). The blooming of grass flower development. Curr. Opin. Plant Biol. 1 60–67. [DOI] [PubMed] [Google Scholar]
- Sentoku, N., Sato, Y., and Matsuoka, M. (2000). Overexpression of rice OSH genes induces ectopic shoots on leaf sheaths of transgenic rice plants. Dev. Biol. 220 358–364. [DOI] [PubMed] [Google Scholar]
- Sieburth, L.E., Running, M.P., and Meyerowitz, E.M. (1995). Genetic separation of third and fourth whorl functions of AGAMOUS. Plant Cell 7 1249–1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzaki, T., Sato, M., Ashikari, M., Miyoshi, M., Nagato, Y., and Hirano, H.-Y. (2004). The gene FLORAL ORGAN NUMBER1 regulates floral meristem size in rice and encodes a leucine-rich repeat receptor kinase orthologous to Arabidopsis CLAVATA1. Development 131 5649–5657. [DOI] [PubMed] [Google Scholar]
- Theissen, G., Becker, A., Di Rosa, A., Kanno, A., Kim, J.T., Munster, T., Winter, K.U., and Saedler, H. (2000). A short history of MADS-box genes in plants. Plant Mol. Biol. 42 115–149. [PubMed] [Google Scholar]
- Whipple, C.J., Ciceri, P., Padilla, C.M., Ambrose, B.A., Bandong, S.L., and Schmidt, R.J. (2004). Conservation of B-class floral homeotic gene function between maize and Arabidopsis. Development 131 6083–6091. [DOI] [PubMed] [Google Scholar]
- Yamaguchi, T., Nagasawa, N., Kawasaki, S., Matsuoka, M., Nagato, Y., and Hirano, H.-Y. (2004). The YABBY gene DROOPING LEAF regulates carpel specification and midrib development in Oryza sativa. Plant Cell 16 500–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanofsky, M.F., Ma, H., Bowman, J.L., Drews, G.N., Feldmann, K.A., and Meyerowitz, E.M. (1990). The protein encoded by the Arabidopsis homeotic gene agamous resembles transcription factors. Nature 346 35–39. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.