Abstract
Rust fungi, obligate biotrophs that cause disease and yield losses in crops such as cereals and soybean (Glycine max), obtain nutrients from the host through haustoria, which are specialized structures that develop within host cells. Resistance of flax (Linum usitatissimum) to flax rust (Melampsora lini) involves the induction of a hypersensitive cell death response at haustoria formation sites, governed by gene-for-gene recognition between host resistance and pathogen avirulence genes. We identified genes encoding haustorially expressed secreted proteins (HESPs) by screening a flax rust haustorium-specific cDNA library. Among 429 unigenes, 21 HESPs were identified, one corresponding to the AvrL567 gene. Three other HESPs cosegregated with the independent AvrM, AvrP4, and AvrP123 loci. Expression of these genes in flax induced resistance gene–mediated cell death with the appropriate specificity, confirming their avirulence activity. AvrP4 and AvrP123 are Cys-rich proteins, and AvrP123 contains a Kazal Ser protease inhibitor signature, whereas AvrM contains no Cys residues. AvrP4 and AvrM induce cell death when expressed intracellularly, suggesting their translocation into plant cells during infection. However, secreted AvrM and AvrP4 also induce necrotic responses, with secreted AvrP4 more active than intracellular AvrP4, possibly as a result of enhanced formation of endoplasmic reticulum–dependent disulfide bonds. Addition of an endoplasmic reticulum retention signal inhibited AvrM-induced necrosis, suggesting that both AvrM and AvrP4 can reenter the plant cell after secretion in the absence of the pathogen.
INTRODUCTION
Rust fungi (order Uredinales) use one of the most specialized infection strategies of plant pathogens. These obligate biotrophs obtain their nutritional resources from the living cells of their host and are completely dependent on living plant tissue for their growth and propagation. Fungal hyphae grow in the intercellular space of the leaf but form a close association with host plant cells through haustoria. These specialized infection structures differentiate from haustorial mother cells and penetrate the plant cell wall. The plant cell membrane is invaginated and forms an extrahaustorial membrane, with a region between this and the fungal membrane termed the extrahaustorial matrix. Haustoria are thought to play the primary role in nutrient acquisition from the host (Hahn and Mendgen, 2001) and apparently also influence host metabolism and defense responses (Voegele and Mendgen, 2003) and induce the reorganization of the host cell cytoskeleton and nuclear DNA (Kobayashi et al., 1994; Heath, 1997). Thus, the haustorium–host cell interface appears to be a dynamic interaction involving extensive molecular trafficking and signaling processes. However, the nature of these exchanges remains unknown. Other obligate biotrophs, such as the oomycete downy mildew pathogens and the ascomycete powdery mildews, have independently evolved a similar infection process (Panstruga, 2003), although the powdery mildews propagate aerial hyphae on the leaf surface and form haustoria only in epidermal cells. Some hemibiotrophic fungi and oomycetes, such as Phytophthora, also form haustoria during their initial stage of infection, but they later enter a necrotrophic stage during which they induce host cell death (Perfect and Green, 2001).
Frequently, plant disease resistance relies on the recognition of pathogen avirulence (Avr) products by host Resistance (R) proteins, which induces a hypersensitive response (HR). Avr proteins are generally thought to be pathogenicity effectors with positive roles in establishing infection, which the plants' R protein surveillance mechanism has evolved to recognize as indicators of invasion. The nature and functions of these effector molecules differ between pathogen classes. For instance, most bacterial Avr proteins are delivered directly into the plant cytoplasm via the type III secretion system (Lahaye and Bonas, 2001). Although the functions of many of these effectors remain unknown, some are targeted to the host nucleus and may act as transcription factors (Lahaye and Bonas, 2001; Deslandes et al., 2003), whereas others are proteases that cleave specific cytoplasmic host proteins (Orth et al., 2000; Axtell et al., 2003; Shao et al., 2003; Coaker et al., 2005). These bacterial Avr proteins are generally recognized by intracellular R proteins containing nucleotide binding site (NBS) and leucine-rich repeat (LRR) domains (Luderer and Joosten, 2001). By contrast, Avr proteins from extracellular fungal pathogens are secreted into the apoplast. These fungal effectors, such as AVR2 (Luderer et al., 2002), AVR4 (Joosten et al., 1994), and AVR9 (van den Ackerveken et al., 1992) from Cladosporium fulvum, NIP1 from Rhynchosporium secalis (Rohe et al., 1995), and SIX1 from Fusarium oxysporum f sp lycopersici (Rep et al., 2004), are Cys-rich, and for most of these proteins the formation of disulfide bonds is critical for their structure and stability in the protease-rich apoplast (van den Hooven et al., 2001; van den Burg et al., 2003; van't Slot et al., 2003). The biochemical activities of some of these proteins are known. For instance, AVR4 is a chitin binding protein that protects fungal cell walls from degradation by plant chitinases (van den Burg et al., 2003), and AVR2 is an inhibitor of the extracellular Cys protease Rcr3 in tomato (Lycopersicon esculentum) (Rooney et al., 2005). R proteins recognizing the Cladosporium extracellular Avr products are membrane-bound receptors with extracellular LRR domains (Luderer and Joosten, 2001).
In contrast with these well-studied pathogen classes, there is a dearth of information concerning virulence and avirulence factors and pathogenicity mechanisms of rust and mildew pathogens, largely because of the difficulties of studying these obligate biotrophs. Identification of the first rust Avr proteins, the AvrL567 proteins of the flax rust Melampsora lini (Dodds et al., 2004), led to the realization that these biotrophic fungi, like bacterial pathogens, may deliver proteins directly into the plant cytoplasm. AvrL567 proteins are expressed in haustoria and contain predicted signal peptides, suggesting that they are secreted into the extrahaustorial matrix. However, expression of these proteins in the plant cytoplasm induces an HR dependent on the cytoplasmic NBS-LRR L5, L6, or L7 resistance proteins of flax (Linum usitatissimum). This finding suggests that there may be a specific translocation mechanism that delivers these proteins across the plant-derived extrahaustorial membrane and into the host cell cytoplasm during rust infection. However, the nature of this mechanism remains unresolved. Avr proteins recently identified from biotrophic and hemibiotrophic oomycetes, such as ATR13 and ATR1NdWsB from Hyaloperonospora parasitica (Allen et al., 2004; Rehmany et al., 2005), Avr1b from Phytophthora sojae (Shan et al., 2004), and Avr3a from Phytophthora infestans (Armstrong et al., 2005), also contain signal peptides. Like AvrL567, ATR13, ATR1NdWsB, and Avr3a trigger a HR response when localized inside the host cell, suggesting that they may be translocated during infection. A conserved motif in these oomycete proteins is similar to a motif required for the transport of proteins secreted by the malaria parasite (Plasmodium falciparum) proteins across the erythrocyte vacuolar membrane (Hiller et al., 2004; Marti et al., 2004), suggesting the possibility of a conserved translocation mechanism used by these highly diverged organisms.
To further investigate rust fungal pathogenicity mechanisms, we have attempted to isolate additional Avr genes from flax rust. Because the HR that occurs during rust resistance appears to be focused on the emerging haustoria (Kobayashi et al., 1994; Heath, 1997), we reasoned that rust Avr proteins should be expressed in these structures. In addition, the precedent of AvrL567 and the observation that all 19 of the cloned flax R genes encode predicted cytoplasmic NBS-LRR proteins (Lawrence et al., 1995; Anderson et al., 1997; Dodds et al., 2001a, 2001b) suggested that these would be secreted proteins. Therefore, we generated a haustorium-specific cDNA library and searched for ESTs encoding putative secreted proteins. Three of these cosegregated with Avr loci in an F2 mapping family, and transient expression assays showed that these genes function as avirulence determinants to induce R gene–dependent cell death in flax.
RESULTS
Isolation of Haustorially Expressed cDNAs Encoding Predicted Secreted Proteins
A haustorium-specific cDNA library was searched for ESTs encoding putative signal peptides. Haustorial cells were isolated from flax leaves infected with rust strain CH5 using affinity chromatography with a Sepharose–concanavalin A column (Hahn and Mendgen, 1992). The majority of isolated haustorial cells remained intact, and contaminating plant cells represented <1% of haustoria numbers. A cDNA library was constructed from haustoria RNA samples and consisted of 3.5 × 106 independent clones. Preliminary sequencing revealed a high number of clones containing rRNA genes, and subsequent clones were prescreened by colony blot hybridization using probes against a subset of flax rust rRNAs. A total of 822 clones were sequenced, of which 150 matched rRNA genes. The remaining 672 ESTs generated 429 unique sequences. BLAST (Altschul et al., 1997) searches against the public databases identified related sequences for 46% of the unique ESTs (E values < 1 × 10−4). Approximately 5% of sequences were significantly similar to known plant genes and were probably derived from contaminating flax cells. Four ESTs were homologous with genes identified from a Uromyces fabae (broad bean rust) haustorium-specific library: MAD1 (mannitol dehydrogenase; Voegele et al., 2005); BGL1 (β-glucosidase; Haerter and Voegele, 2004); HXT1 (hexose transporter; Voegele et al., 2001); and PIG28 (a putative cyclophilin; Hahn and Mendgen, 1997).
Among the initial sequences, three ESTs were found to encode the secreted flax rust AvrL567 proteins. To identify additional secreted proteins, we selected a subset of 167 unique ESTs based on the presence of a potential full-length open reading frame that included an initiating Met codon and lacked homology with proteins of known intracellular function (e.g., ribosomal proteins). To search for potential secretory signals, the predicted protein sequences of these cDNAs were analyzed with the SignalP 3.0 algorithm (Bendtsen et al., 2004; http://www.cbs.dtu.dk/services/SignalP/). This screen identified 20 sequences encoding proteins with predicted secretory signal peptides (Table 1). To support the SignalP results, the protein sequences for each of the haustorially expressed secreted proteins (HESPs) were also entered into other signal peptide–predicting software. iPSORT (Bannai et al., 2002) predicted 17 of 20 proteins to be secreted, and PSORTII (Nakai and Horton, 1999) predicted 19 of 20 with McGeoch's method. These HESPs encode small proteins (73 to 384 amino acids), most with no significant similarity to known protein sequences. Four HESPs were homologous with predicted fungal proteins of unknown function, and two contained motifs indicative of Kazal-type Ser protease inhibitors (C51 = AvrP123-A; Figure 1A) or trehalose phosphatases (C61). Eight HESPs encode Cys-rich proteins with even numbers of Cys residues that may be involved in disulfide bonds.
Table 1.
Flax Rust HESPs
| HESP No. | Redundancya | Protein Size (Amino Acids) | No. of Cys Residues | Top Hit with BLASTP Search (Matches Are to Predicted Proteins) | E Value | Protein Signatureb |
|---|---|---|---|---|---|---|
| 178 | 1 | 146 | 8 | Magnaporthe grisea (BAC65876) | 1E-05 | CFEM |
| 270 | 1 | 267+ | ||||
| 327 | 1 | 79 | ||||
| 376 | 1 | 142 | ||||
| 379 | 1 | 205 | Ustilago maydis (EAK82919) | 3E-10 | ||
| 417 | 1 | 126 | 4 | |||
| 570 | 1 | 222 | ||||
| 642 | 1 | 140 | 6 | |||
| 735 | 1 | 169 | ||||
| 767 | 1 | 85 | Cryptococcus neoformans var neoformans (EAL23156) | 4E-24 | ||
| 897 | 1 | 142 | 10 | |||
| 937 | 1 | 113 | ||||
| C29 | 5 | 343 | ||||
| C48 | 2 | 95 | 6 | |||
| C49 | 2 | 176 | 12 | |||
| C51 | 4 | 117 | 10 | KAZAL | ||
| C55 | 3 | 122 | ||||
| C61 | 5 | 327 | Yarrowia lipolytica (CAG81011) | 5E-13 | Trehalose PPase | |
| C63 | 3 | 197 | ||||
| C66 | 3 | 73 | 8 |
Number of clones in 822 sequences from a haustorium-specific cDNA library.
Protein signatures are as follows: CFEM, fungal-specific eight-Cys-containing domain of extracellular membrane proteins, found in some proteins with proposed roles in fungal pathogenesis; KAZAL, family of Ser protease inhibitors; Trehalose PPase, trehalose phosphatases catalyze the dephosphorylation of trehalose-6-phosphate to trehalose and orthophosphate.
Figure 1.
The Kazal-Like AvrP123-A Is Recognized by P1 and P2 Resistance Genes in Flax.
(A) Amino acid sequence of the predicted AvrP123 protein. The predicted signal peptide is underlined, and the consensus residues of the KAZAL family of Ser protease inhibitors are indicated by black arrowheads with the residue at the P1 active site indicated. The Cys residues and the GIAR motif identified by Multiple Expectation Maximization for Motif Elicitation (MEME) analysis are shaded in gray.
(B) Leaves of four near-isogenic flax lines containing P1 (B10×P1), P2 (B4×P2), P3 (B13×P3), or no P resistance gene (Bison) were infiltrated with A. tumefaciens cultures containing T-DNA plasmids encoding the full-length AvrP123-A. The photograph was taken 10 d after infiltration.
The AvrM, AvrP4, and AvrP123 Loci Encode HESPs
DNA gel blot hybridization confirmed that all of the HESPs were derived from the rust genome. Sixteen HESPs detected restriction fragment length polymorphisms that were mapped among a flax rust F2 family of 74 individuals (self-progeny of rust CH5) that segregate for 16 different avirulence specificities located at 10 loci (Lawrence et al., 1981a). Two HESPs, C29 and C48, detected restriction fragment length polymorphisms that cosegregated with Avr genes corresponding to the flax resistance genes M and P4, respectively. A third HESP, C51, detected several restriction fragment length polymorphisms that either cosegregated with or were closely linked to a complex Avr locus corresponding to the P1, P2, and P3 resistance genes in flax. These avirulence specificities are tightly linked but have been separated by recombination or mutation events in several individuals found in a large screening procedure (Lawrence et al., 1981b). The remaining 13 HESPs showed no genetic association with other known Avr loci segregating in this rust family. The HR is one of the primary manifestations of R–Avr recognition, and in planta expression of corresponding R and Avr genes has been shown to induce a HR-like cell death (Erickson et al., 1999; van der Hoorn et al., 2000; Allen et al., 2004; Dodds et al., 2004). Therefore, we generated T-DNA constructs in which the Avr gene candidates were under the control of the 35S promoter and used Agrobacterium tumefaciens infiltration to express these constructs in plants containing the corresponding resistance genes. Expression of C51 induced a HR-like necrotic response on near-isogenic flax plants containing the P1 and P2 resistance genes but not on those containing P3 or the recurrent backcross parent, Bison (Figure 1B). This finding indicates that this gene is indeed an active member of the AvrP1, P2, P3 complex locus, and we have designated it AvrP123-A. Hybridization of this probe to genomic DNA gel blots indicated that several additional homologs occur at this locus, but these have not been examined further in this study. In planta expression assays (see below) also showed that the C29 and C48 genes triggered R gene–dependent cell death with the appropriate specificity; therefore, we designated them AvrM and AvrP4. These two Avr loci are described in greater detail below.
The AvrM Locus Contains Multiple Homologs
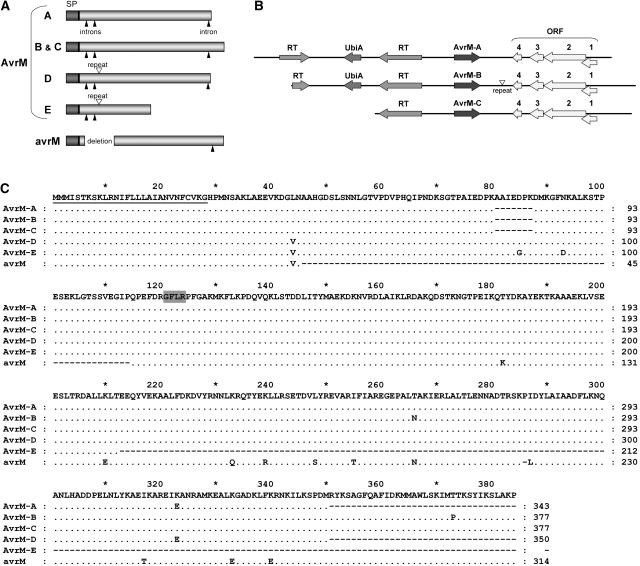
DNA gel blot analysis detected up to eight DNA fragments hybridizing to the AvrM probe in rust strain CH5, depending on the restriction enzyme used. Most of the fragments segregated with the avirulence allele, whereas a single fragment occurred at the virulence allele (data not shown). Sequence analysis of RT-PCR products amplified from RNA of leaves infected with an F2 rust strain homozygous for the virulence allele detected a single sequence variant, avrM. By contrast, four sequence variants (AvrM-A, -B, -C, and -D) were detected in RT-PCR products amplified from leaves infected with a homozygous avirulent F2 rust strain. Screening the haustorium cDNA library with the C29 probe identified one additional full-length sequence variant (AvrM-E) as well as two partial cDNA clones with further sequence differences that may represent additional AvrM variants. Thus, the AvrM avirulence allele contains at least five closely related sequences, whereas a single copy occurs at the virulence allele. The AvrM proteins show no significant sequence similarity (BLASTP; Altschul et al., 1997) to other known proteins or peptide sequence motifs.
The six predicted AvrM proteins contain several sequence differences and also vary in size (Figure 2). AvrM-B and -C both contain 377 amino acid residues, whereas AvrM-A has a 34–amino acid C-terminal truncation, resulting in a 343-residue protein. AvrM-D shares this truncation but also contains a 7–amino acid insertion (corresponding to a 21-bp perfect repeat in the coding sequence), resulting in a 350–amino acid protein. AvrM-E also contains this insertion but is truncated to 212 amino acids as a result of a nucleotide change that introduces a premature stop codon in the coding sequence. The avrM protein derived from the virulent rust parent contains 314 amino acid residues as a result of two in-frame deletions of 207 and 3 bp. In addition to these large structural differences, there are 14 polymorphic sites within the protein sequence. AvrM-B and -C share the greatest sequence similarity, with only 3 amino acid differences between them; the greatest variation is seen with avrM, which has two deletions and 10 unique amino acid differences (Figures 2A and 2C). The first 28 residues of all of these proteins are not polymorphic and are predicted to be a cleaved secretion signal peptide. The presence of upstream stop codons in the cDNA sequences indicates that the predicted open reading frames are full length.
Figure 2.
Characterization of the Flax Rust AvrM Locus.
(A) Scheme showing the variation at the AvrM locus. For each open reading frame, the signal peptide (SP) and intron positions in the corresponding genomic DNA (black arrowheads) are shown. The positions of the 207-bp deletion in avrM and the 22-bp repeat (white arrowhead) in AvrM-D and -E are indicated.
(B) Scheme of three avirulence genes at the AvrM locus. Three regions from the avirulence allele (16.3, 15.9, and 11.8 kb) contain the AvrM-A, AvrM-B, and AvrM-C genes, respectively. The positions of several common features surrounding the Avr open reading frames are shown. Upstream are two regions homologous with a reverse transcriptase (RT) and a third region related to UbiA prenyltransferase (UbiA). Downstream are four codirectional open reading frames (ORF; no similarity to database sequences) that may represent a single open reading frame separated by introns. The position of a 22-bp repeat that occurs downstream of AvrM-B is shown as a white arrowhead.
(C) Amino acid sequence alignment of the predicted AvrM proteins. Only those amino acids that differ from the consensus (top line) are shown, with identical residues indicated by dots and gaps indicated by dashes. The signal peptide is underlined, and the GFLR motif identified by MEME analysis is shaded gray.
An AvrM probe was used to isolate clones from a genomic DNA library prepared from the rust strain CH5. Of five sequenced clones, three contained AvrM-C (the largest sequenced region being 11.8 kb) and the remaining two encoded AvrM-A (16.3 kb sequenced) and AvrM-B (15.9 kb sequenced). Because these sequences do not overlap, the AvrM-A, -B, and -C genes must be separated by at least 10 to 15 kb on the chromosome. Three introns of 74, 70, and 98 bp are present within the coding region, although the first two intron sites are absent from avrM as a result of the large internal deletion within the genomic DNA (Figure 2A). Significant sequence identity (99.9%) in the DNA surrounding the AvrM genes suggests that they evolved from large duplication events (Figure 2B). Upstream of the Avr open reading frame are two regions with similarity to reverse transcriptase sequences and a predicted gene related to UbiA prenyltransferase genes. Downstream of the Avr genes are four codirectional open reading frames that may represent a single open reading frame separated by introns. No proteins with significant similarity to these sequences were detected in public databases.
Transient Expression of AvrM Genes in Flax Causes M Gene–Dependent Cell Death
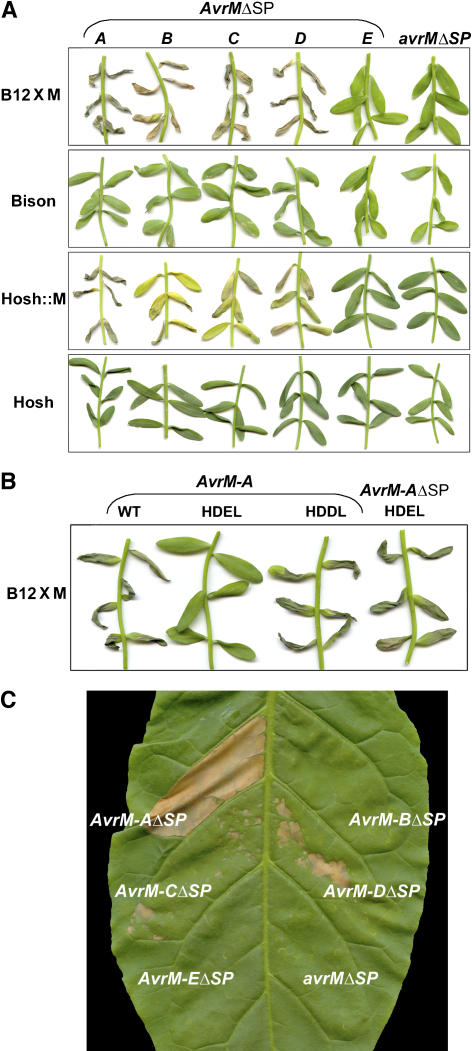
We used A. tumefaciens infiltration to transiently express the AvrM genes in flax plants with or without the M resistance gene. Two sets of T-DNA plasmids were constructed with cDNA clones of each of the AvrM genes, one containing the entire open reading frame and another that excluded the predicted signal peptide leader sequence but included an additional ATG start codon. All constructs contained the 35S promoter of Cauliflower mosaic virus to drive transcription and the nopaline synthase terminator of A. tumefaciens. A. tumefaciens strains containing AvrM constructs were infiltrated into the leaves of four flax genotypes: Bison (m/m) and its near-isogenic line B12×M containing the M gene, Hoshangabad (m/m), and a transgenic derivative transformed with the M gene (Hosh::M) (Anderson et al., 1997) (Figure 3). Transient expression of AvrM genes without the predicted signal peptides in the leaves of Bison and Hoshangabad gave no phenotype. However, expression of AvrM-A, -B, -C, or -D in leaves containing the M gene, either B12×M or Hosh::M, induced a necrotic response indicating that all four proteins are recognized by the M protein. Expression of avrM encoded by the homologous gene from the virulent parental rust strain did not induce necrosis in any flax genotype. Similarly, the truncated AvrM-E protein was not recognized by M. RNA gel blot analysis showed that the AvrM constructs were each expressed at similar levels in A. tumefaciens–infiltrated leaves, except for avrM, which showed a reduced level of expression (data not shown).
Figure 3.
Transient Expression of AvrM Genes in Flax Causes M Gene–Dependent Necrosis.
(A) Leaves of four different flax genotypes with or without the M resistance gene were infiltrated with A. tumefaciens cultures containing T-DNA plasmids encoding AvrM-A, -B, -C, -D, -E, or avrM. Photographs were taken 12 d after infiltration. All constructs excluded the predicted signal peptide sequence but included an additional ATG start codon (ΔSP). B12×M and Hosh::M are flax lines containing the M gene, whereas the Bison and Hoshangabad (Hosh) varieties do not.
(B) Leaves of flax variety B12×M, carrying the M resistance gene, were infiltrated with A. tumefaciens cultures containing T-DNA plasmids encoding the full-length wild-type AvrM-A protein, the full length AvrM-A with a C-terminal HDEL or HDDL extension, or the truncated AvrMΔSP with the HDEL extension. The photograph was taken 4 d after infiltration.
(C) Leaves of transgenic tobacco W38 carrying the flax rust resistance gene M were infiltrated with A. tumefaciens cultures containing T-DNA plasmids encoding AvrM-A, -B, -C, -D, -E, or avrM without the predicted signal peptide. The photograph was taken 4 d after infiltration.
The necrotic response seen in the infiltration experiments varied between the AvrM genes and between the different M gene flax lines (Figure 3). In B12×M, AvrM-A gave the quickest necrotic response, followed by AvrM-D, whereas the necrosis seen with AvrM-B and -C was considerably slower to develop (8 to 10 d compared with 3 to 4 d). When these genes were transiently expressed in Hosh::M, the overall response was reduced and delayed by 3 to 4 d, except for that of AvrM-A, which gave an equivalent and strong response in both flax lines. Induction of R gene–dependent necrosis by AvrM lacking the predicted signal peptides suggests that recognition occurs within the plant cell, which agrees with the predicted cytoplasmic location of the M resistance protein. However, expression of the full-length AvrM genes including the signal peptides also induced a necrotic response. In the case of AvrM-A and -D, this response was similar to that induced by the truncated genes, but for AvrM-B and -C, the response was slower to develop for the full-length versions and was characterized by chlorosis rather than necrosis (see Supplemental Figure 1 online).
To investigate whether the leader peptide functions as a secretion signal in planta to direct the AvrM proteins through the endoplasmic reticulum (ER) secretory pathway, we expressed this protein with the HDEL tetrapeptide ER retrieval signal (Denecke et al., 1992, 1993) fused to the C terminus. If the full-length protein is secreted through this pathway, retention in the ER would prevent accumulation of the protein in the apoplast and subsequent reentry into the plant cytoplasm. Addition of the HDEL motif inhibited the necrotic response induced by the full-length AvrM-A protein (Figure 3B). However, the chemically similar tetrapeptide HDDL, which does not function as an ER retrieval signal in plants (Denecke et al., 1992, 1993), did not affect AvrM recognition. Furthermore, recognition of the nonsecreted version of AvrM-A was not affected by the addition of HDEL (Figure 3B). This finding demonstrates that the presence of this motif does not interfere with AvrM recognition, and the inhibition of HR induction by the full length AvrM-A–HDEL protein is likely attributable to the retention of the protein in the ER. These results confirm that the predicted signal peptide is functional in plants and suggest that secreted AvrM protein reenters the plant cell from the apoplast.
The flax rust AvrL567 genes were found to induce a HR when coexpressed in tobacco (Nicotiana tabacum) with the flax resistance gene L6 (Dodds et al., 2004). To determine whether AvrM could also give a recognition interaction in an unrelated species, they were transiently expressed in transgenic W38 tobacco containing the M gene (W38::M). Expression of AvrM-A gave a very strong necrotic response, AvrM-C and -D gave much weaker responses, and no necrotic response was seen for AvrM-E and avrM (Figure 3C). Although no necrosis was seen for AvrM-B, these results generally reflected the recognition specificity seen in flax. No response was seen with any of the infiltrated AvrM genes in nontransgenic W38 tobacco (data not shown).
AvrP4 Encodes a Small Cys-Rich Secreted Protein
In contrast with the complex AvrM locus, DNA gel blot analysis with HESP C48 detected a single hybridizing fragment segregating with each allele of AvrP4. RT-PCR amplification of RNA from leaves infected with F2 rust strains homozygous for the avirulence (AvrP4) or virulence (avrP4) alleles identified a single sequence variant for each allele. Both sequences were represented in cDNAs isolated from the CH5 haustorium cDNA library. To identify further sequence variation at the AvrP4 locus, RT-PCR was performed on several independent rust strains. An additional sequence variant (AvrP4WA) was found in rust strain WA, which was isolated from the Australian native flax species Linum marginale and is avirulent on flax varieties containing the P4 resistance gene (Lawrence, 1989).
Each AvrP4 homolog contains a 285-bp open reading frame encoding a 95–amino acid Cys-rich protein with a predicted 28–amino acid cleavable secretion signal peptide (Figure 4). These predicted proteins show no significant sequence homology (BLASTP; Altschul et al., 1997) with other proteins and have no predicted motif structures. However, they do contain six Cys residues in the last 28 amino acids of the C terminus that are spaced according to the consensus of an inhibitor Cys knot structure (CX3-7CX4-6CX0-5CX1-4CX4-10C; Figure 4), which is commonly found in toxins and inhibitors of receptors or proteases (Pallaghy et al., 1994). Within the presumed 67 amino acid mature protein, there are only seven polymorphisms, all of which occur within the last 22 amino acids of the C terminus, a significantly (χ2; P < 2 × 10−4) nonrandom distribution. None of the six Cys residues represents a polymorphic site.
Figure 4.
Amino Acid Sequence Alignment of the Predicted AvrP4 Proteins.
Only those amino acids that differ from the consensus (top line) are shown, with identical residues indicated by dots. The signal peptide is underlined, and Cys residues and the GFSR motif identified by MEME analysis are shaded in gray.
An AvrP4 probe was hybridized with the genomic DNA library prepared from the F1 rust CH5 (Ayliffe et al., 2001), and five identical λ clones were isolated containing the AvrP4 sequence plus 2.4 kb of flanking DNA. Because the alternative allele was not recovered from the genomic DNA library, the corresponding sequence was amplified by long-distance PCR from an F2 rust strain homozygous for this allele. Comparison of the genomic and cDNA sequences revealed two introns of 92 and 86 bp within the 5′ untranslated leader of AvrP4. The avrP4 gene contains an additional 5′ splice site at the first intron, leading to two alternative transcripts with either 92 or 113 bp removed. Both transcripts were represented at similar levels in the cDNA library and among cloned RT-PCR products and contained an identical open reading frame. A 600-bp region that has been duplicated at the avrP4 allele begins 1028 bp upstream of the AvrP4 start codon.
Transient Expression of AvrP4 in Flax Causes P4-Dependent Cell Death
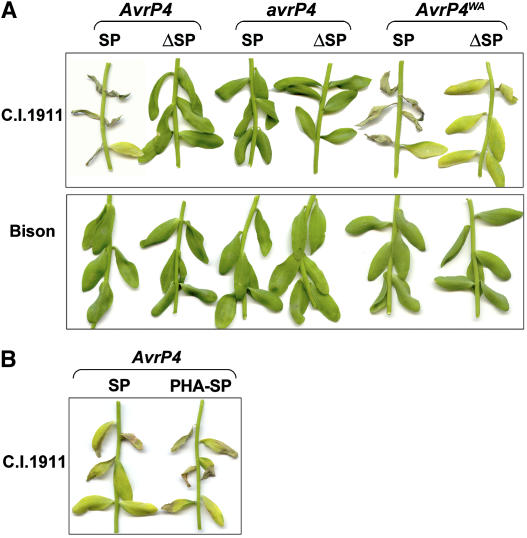
The AvrP4 genes were transiently expressed in the flax varieties Bison (which lacks P4 resistance) and C.I.1911 (which contains P4) by A. tumefaciens infiltration (Figure 5A). As described above, two sets of T-DNA plasmids were constructed with each gene, one including the entire open reading frame and the other with Met-26 as the first translated codon, which excluded the first 25 amino acids of the 28–amino acid predicted signal peptides. Expression of either AvrP4 or AvrP4WA, but not avrP4, induced necrosis in C.I.1911 plants but not in Bison, consistent with the predicted avirulence function of these proteins. However, differences were seen between the secreted and nonsecreted versions. For both AvrP4 and AvrP4WA, the full-length construct encoding a secreted protein induced a strong necrotic response. However, truncated AvrP4WA gave a delayed necrotic response relative to the full-length version, and the truncated AvrP4 gave no response. The response to the truncated AvrP4WA construct indicates that recognition of the encoded protein occurs inside the plant cell, as observed for AvrM and AvrL567 (Dodds et al., 2004). However, the stronger response observed for the full-length constructs suggests that the presence of the 28–amino acid N-terminal region enhances recognition of the encoded proteins. RNA gel blot analysis on infiltrated flax leaves confirmed that all constructs were expressed at similar levels (data not shown).
Figure 5.
Transient Expression of AvrP4 in Flax Causes P4 Gene–Dependent Necrosis.
(A) Leaves of the flax variety Bison or C.I.1911, which contains P4, were infiltrated with A. tumefaciens cultures containing T-DNA plasmids encoding AvrP4, AvrP4WA, or avrP4. Photographs were taken 10 d after infiltration. Constructs included either the complete transcript, including the signal peptide (SP), or excluded the first 25 amino acids of the 28–amino acid predicted signal peptide, with Met-26 as the first translated codon (ΔSP).
(B) Leaves of the flax variety C.I.1911 were infiltrated with A. tumefaciens cultures containing T-DNA plasmids encoding the native predicted signal peptide (AvrP4 SP) or a plant signal peptide (AvrP4 PHA-SP). The photograph was taken 10 d after infiltration.
As with AvrM, we tested whether the presence of an ER retention signal would inhibit recognition of the secreted AvrP4 protein. However, addition of either the HDEL (ER-retrievable) or HDDL (nonretrievable) motif to AvrP4 abolished recognition (data not shown), indicating that the presence of these additional amino acids interfered with recognition irrespective of ER retention. As an alternative means to determine whether the leader peptide functions as a secretion signal, we replaced the 28–amino acid predicted signal peptides of AvrP4 with the 44–amino acid leader sequence that directs the secretion of bean phytohemagglutinin (PHA) (Tague et al., 1990). Transient expression of AvrP4 PHA-SP induced necrosis in C.I.1911 plants but not in Bison (Figure 5B). One likely explanation for the requirement of secretion for full recognition is that the six Cys residues in this protein form disulfide bonds during passage through the oxidizing environment of the ER, which may be important for protein structure and/or stability. Consistent with this hypothesis, mutation of either Cys-84 or Cys-93 to Ala abolished the P4-dependent necrosis-inducing activity of AvrP4 (data not shown). These results suggest that secretion is required for full recognition of the AvrP4 protein expressed in planta, probably because it allows optimal folding and disulfide bond formation.
A yeast signal peptide screen was also used to investigate the predicted signal peptides of AvrM and AvrP4. The nucleotide sequence encoding the first 32 amino acids of both AvrM and AvrP4 was cloned into a yeast shuttle vector upstream of the reporter gene invertase, which lacks its own start codon and signal peptides. The AvrM signal peptide was able to direct secretion of the invertase protein, which was detected as growth on medium containing sucrose as the sole carbon source (see Supplemental Figure 2 online). Although the AvrP4-SP invertase fusion construct did not allow the growth of yeast on sucrose medium, we do not know whether this fusion protein was efficiently expressed, so this negative result is inconclusive and the weight of other evidence suggests that this is a functional signal sequence.
A Short Conserved Motif Is Present in Flax Rust Avr Proteins
More than 40 predicted oomycete-secreted proteins, including several identified Avr proteins, contain the sequence motif RxLRx5-21ddEER (Armstrong et al., 2005; Rehmany et al., 2005), which is related to a host translocation signal in secreted proteins of the malaria parasite (Hiller et al., 2004; Marti et al., 2004). However, the oomycete RxLR motif is absent from all 21 flax rust HESPs (including AvrL567). To search for other conserved motifs, these sequences were analyzed by MEME (Bailey and Elkan, 1994). This analysis identified a short motif in the N-terminal region after the signal peptides of AvrL567 (GYTR), AvrM (GFLR), and AvrP4 (GFSR) and in the C-terminal region of AvrP123 (GIAR). Similar sequences occur in the C-terminal regions of some of the other HESPs. To examine whether this motif may play a role in the recognition or uptake of these proteins in planta, we generated mutations of this motif in AvrM and AvrP4. All four amino acids were converted to Ala in AvrM and AvrMΔSP and also in AvrP4 and AvrP4WAΔSP. When transiently expressed in flax plants containing the corresponding R genes, these constructs all induced a necrotic response similar to the wild type (data not shown). Therefore, this motif is not important for either recognition of these proteins in the cytoplasm or uptake of the secreted proteins into the cytoplasm during in planta expression.
AvrM and AvrP4 Have Different Expression Profiles
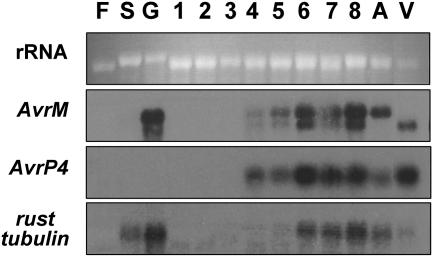
The expression profiles of AvrP4 and AvrM were examined by RNA gel blot hybridization (Figure 6). Transcripts for AvrP4 were not detected in RNA samples from rust spores or in vitro–germinated spores, but they were detected in RNA from flax leaves infected with rust strain CH5, indicating that it is induced during infection of the leaf. The increase in abundance from 4 to 8 d after infection probably reflects the increase in fungal biomass during this period. The expression profile of AvrM differed from that of AvrP4, with AvrM transcripts also found in RNA from in vitro–germinated rust spores but not from spore RNA. This finding implies that AvrM is induced during germination but before infection of the host has been established and is not dependent on any plant signal (Figure 6). As for AvrP4, the greatest amount of transcript was observed in the later stages of infection, when there is a greater fungal biomass. Conversely, the flax rust β-tubulin gene (Ayliffe et al., 2001) is expressed in both germinated and ungerminated spores as well as during infection.
Figure 6.
AvrM but Not AvrP4 Is Expressed in in Vitro–Germinated Spores.
RNA samples (5 μg) from uninfected flax leaves (F), CH5 rust spores (S), in vitro–germinated CH5 rust spores (G), rust CH5–infected flax leaves 1 to 8 d after infection (1 to 8), and rust strains homozygous for either the avirulence (A) or virulence (V) allele (isolated from infected leaves 7 d after inoculation) were separated on a 1.5% agarose gel, which was stained with ethidium bromide to detect rRNA loading (top). RNA from the gel was transferred to a nylon membrane and hybridized with probes for the AvrM, AvrP4, and β-tubulin genes of M. lini.
We also examined the expression of AvrP4 and AvrM genes in F2 rust individuals homozygous for either the avirulence or virulence alleles of these loci. Both AvrP4 and avrP4 transcripts were detected in RNA samples from rust-infected flax leaves, indicating that both alleles are expressed during infection. Interestingly, the level of expression for avrP4 appears to be greater than that for AvrP4. Consistent with this, of nine AvrP4-hybridizing clones isolated from the haustorium-specific cDNA library, seven corresponded to avrP4 and two corresponded to AvrP4. These clones represented ∼0.04% of the library. Higher expression of avrP4 transcripts was also seen in RNA gel blots of the flax leaves infiltrated with the in planta expression constructs for AvrP4 (data not shown), suggesting that this increased level of message may be an intrinsic feature of the avrP4 coding sequence. No differences in expression levels from the AvrM locus were observed between rust strains homozygous for either the avirulence or the virulence allele. This despite the fact that the avirulence allele contains at least five AvrM homologs compared with a single copy at the virulence allele. This observation is supported by the frequency of cDNA clones recovered from the haustorium-specific cDNA library, which included eight of avrM, two of AvrM-A, two of AvrM-D, and one each of AvrM-B and AvrM-E. Although AvrM-C was not identified among these 14 clones, it was recovered by RT-PCR and therefore is expressed. Genes at the AvrM locus represent ∼0.15% of clones in the haustorium-specific cDNA library.
Evidence for Diversifying Selection
Nucleotide variation at the AvrM and AvrP4 loci was analyzed for evidence of positive selection. Among the AvrM homologs, most of the amino acid polymorphisms occur between avrM and the five AvrM proteins. Comparing the nucleotide sequences of avrM and the other genes, we found a significant excess of nonsynonymous over synonymous changes (ratio = 2.1; t test, P < 0.002). A significant excess of nucleotide substitutions was also found in the coding regions (5 in 1134 bp) relative to the flanking DNA (12 in 9700 bp) when we compared the AvrM-A, -B, and -C genomic sequences (χ2 test, P < 0.011). These results suggest that selection has contributed to the accumulation of sequence differences between AvrM homologs. Although all but one of the nucleotide changes within the coding sequences of the three AvrP4 genes are nonsynonymous, this difference is not statistically significant. However, a significant excess of nucleotide substitutions was observed in the coding regions (6 in 285) relative to the flanking DNA (11 in 1502) when the AvrP4 and avrP4 genomic sequences were aligned (χ2, P < 0.025), suggesting that diversifying selection has acted upon this gene.
DISCUSSION
The AvrL567 genes from flax rust, which were previously isolated by map-based cloning, encode secreted proteins and are expressed in haustoria (Dodds et al., 2004). We reasoned that these two features may be characteristic of other Avr proteins in flax rust, because hypersensitive cell death in rust resistance occurs exclusively at sites of haustoria formation (Heath, 1997) and all of the cloned rust resistance genes from flax encode predicted cytoplasmic proteins. Therefore, we searched a haustorium-specific cDNA library for sequences encoding putative secreted proteins. Mapping the identified HESPs in a segregating rust family and in planta expression analysis led to the identification of three additional Avr genes, AvrM, AvrP4, and AvrP123. At the AvrM locus, at least five closely related genes segregate with the avirulence allele in CH5 (AvrM-A, B, C, D, and E), although it is possible that additional homologs may occur at this haplotype. Genomic sequences flanking three of these genes (A, B, and C) indicate that they are separated by at least 10 to 15 kb. Four of these genes caused M gene–dependent cell death when expressed transiently in flax leaves (Figure 3), whereas avrM, the single gene at the virulence allele, did not cause necrosis. At the AvrP4 locus in rust CH5, a single gene is present at each allele. The gene derived from the avirulence allele, AvrP4, induced cell death in P4 plants when transiently expressed in flax leaves, as did a homologous gene (AvrP4WA) from another rust strain avirulent on P4, whereas the virulence allele, avrP4, eludes this detection (Figure 5). AvrP123 is a complex locus in flax rust and encodes avirulence on three resistance genes, P1, P2, and P3. These specificities are tightly linked, but they have been separated by recombination and mutation events in a large-scale screen (Lawrence et al., 1981b). We identified one gene, AvrP123-A, that is recognized in planta by the P1 and P2 resistance genes (Figure 1). At least three homologs of this gene occur at this locus and may correspond to P3.
Intracellular Recognition of AvrM and AvrP4
When AvrM genes were expressed without the signal peptides, a strong M gene–dependent necrotic response was observed in flax and tobacco leaves (Figure 3), which implies that recognition of AvrM occurs within the plant cell, similar to AvrL567 (Dodds et al., 2004). This intracellular recognition is consistent with the predicted cytoplasmic location of the NBS-LRR class L and M resistance proteins (Lawrence et al., 1995; Anderson et al., 1997). Interestingly, although the L and M proteins are closely related homologs (75% amino acid identity, 84% similarity), the AvrL567 and AvrM proteins are completely unrelated. As with the L6–AvrL567 interaction (Dodds et al., 2004), the M–AvrM interaction resulted in HR induction in tobacco (Figure 3B). This finding indicates that host factors required for this recognition event and the downstream signaling pathway are sufficiently conserved in tobacco to allow a productive interaction. Expression of AvrP4WA without its signal peptides also induced P4 gene–dependent necrosis in flax (Figure 5A), implying that recognition of this protein also occurs inside the plant cell. Although the P4 resistance gene has not been cloned, the allelic P and P2 genes encode predicted cytoplasmic NBS-LRR proteins (Dodds et al., 2001b), which agrees with an intracellular Avr recognition site. We predict that during infection, AvrL567, AvrM, and AvrP4 proteins are secreted from the rust and translocated into the plant cell, where they are recognized by the corresponding cytoplasmic resistance proteins.
AvrP4 and AvrP123 Are Cys-Rich Proteins
AvrP4 encode a predicted 67–amino acid protein after cleavage of the signal peptide, with six Cys residues in the last 28 amino acids. These residues comply with the spacing consensus of inhibitor Cys knot structures (Figure 4), such as the C. fulvum AVR9 protein (van den Hooven et al., 2001), which form three disulfide bridges. All seven polymorphic residues that distinguish the mature AvrP4 proteins occur within this C-terminal region, which may represent a distinct structural domain of the AvrP4 protein. It is possible that this protein may be subject to processing to remove the non-Cys-rich N-terminal region, similar to the processing of Avr9 (van den Ackerveken et al., 1993). The SIX1 protein of F. oxysporum, which determines avirulence on the I-3 resistance gene in tomato, is also a Cys-rich protein that is subject to posttranslational processing. In this case, a 12-kD protein containing six Cys residues occurs in xylem sap of infected plants and corresponds to the central region of the predicted 30-kD precursor protein (Rep et al., 2004). Given these considerations, passage of AvrP4 through the oxidizing environment of the ER during secretion may be necessary for efficient folding and formation of disulfide bonds or posttranslational processing. Indeed, mutation of two of these residues, Cys-84 and Cys-93, caused a loss of recognition, which would be expected if these were involved in structurally important disulfide bonds. Similarly, altered Cys residues in some virulence alleles of the C. fulvum AVR4 protein lead to instability of these proteins as a result of the disruption of an internal disulfide bond (Joosten et al., 1994). This hypothesis would explain the requirement for the signal peptides for full recognition of AvrP4 and the stronger response induced by AvrP4WA when the signal peptide is retained (Figure 5A). In this situation, the weaker recognition of AvrP4WAΔSP may be attributable to the presence of a small amount of correctly folded AvrP4WA in the cytoplasm. The difference between the responses induced by the AvrP4ΔSP and AvrP4WAΔSP proteins may reflect either their relative efficiency of folding in the cytoplasm or intrinsic differences in recognition strength. Because the PHA signal peptide was able to substitute for the N-terminal 28 amino acids of AvrP4 to allow recognition (Figure 5B), it is likely that the requirement for this region relates directly to its role in secretion.
AvrP123-A is also a Cys-rich protein and contains a signature (CX7CX6YX3CX2-3C; Figure 1A) characteristic of the Kazal family of Ser protease inhibitors. Two extracellular Kazal-like Ser protease inhibitors from the oomycete P. infestans were found to inhibit the pathogenesis-related P69B protease in tomato (Tian et al., 2004, 2005). Interestingly, previous database searching (Tian et al., 2004) identified 35 predicted proteins with Kazal motifs from oomycetes but none from fungi. The oomycete Kazal proteins generally contain an Asp residue at the P1 position (Tian et al., 2004), which is important in determining protease interaction specificity, whereas the corresponding residue in AvrP123-A is a Cys. Thus, it is possible that AvrP123 proteins may be inhibitors of host proteases, as is also observed for the Avr2 protein from C. fulvum, which inhibits the Cys protease Rcr3 in tomato (Rooney et al., 2005).
The Role of Sequence Variation in AvrM and AvrP4 Recognition
Positive selection appears to have contributed to the accumulation of sequence differences within the coding sequences of the AvrP4 and AvrM genes. Genes at both loci show an excess of nucleotide differences in the coding region relative to the flanking genomic DNA, and we found a significant excess of nonsynonymous over synonymous substitutions that distinguish the virulence allele avrM from AvrM genes at the avirulence allele. In the case of avrM, lack of recognition may be attributable to the 10 unique polymorphic amino acids that are concentrated in the C-terminal region (Figure 2B) or to the 69-residue internal deletion in the N-terminal region. As high levels of transcript from this gene can be found in both avirulent and virulent homozygotes, escaping recognition does not appear to be caused by an absence or low rate of gene expression, although we do not know whether the mRNA is translated to a stable protein. Of the five AvrM genes from the avirulence allele, the only one to elude detection is AvrM-E, which encodes a truncated protein lacking 131 amino acids from its C terminus (Figures 2A and 2B). The absence of this region may prevent recognition or lead to protein instability. The presence of a 34–amino acid C-terminal extension in some AvrM proteins (Figures 2A and 2B) also has some effect on recognition, because AvrM-A and -D, which lack this region, gave a stronger response than AvrM-B and -C.
Differences in recognition between AvrP4 proteins apparently result from the amino acid differences in the C-terminal Cys-rich domain (Figure 4), suggesting that this region is important for recognition. Furthermore, AvrP4 recognition was abolished when additional amino acids (HDEL and HDDL) were added to the C terminus of this protein, further supporting the involvement of the C-terminal region in recognition. Again, the AvrP4 and avrP4 alleles are both expressed at the mRNA level, with avrP4 transcripts actually more abundant. Thus, escaping recognition does not appear to be attributable to an absence of gene expression, although, as for AvrM, we do not know whether the translated protein is stable and able to accumulate. As mentioned above, the generation of unstable AVR4 isoforms allows virulent strains to avoid Cf-4 recognition (Joosten et al., 1994, 1997), whereas, conversely, C. fulvum strains virulent on plants carrying Hcr9-4E can evade detection by producing a stable Avr4E protein with only two amino acid substitutions (Westerink et al., 2004). Evidence for selection underlying amino acid variation was also found between the AvrL567 proteins (Dodds et al., 2004) and the H. parasitica ATR13 and ATR1NdWsB proteins (Allen et al., 2004; Rehmany et al., 2005). In all cases, selection imposed by the presence of corresponding R genes in the host appears to be the driver for the accumulation of Avr variation, leading to escape from recognition.
Translocation of Avirulence Proteins into Host Cells
The identification of three flax rust avirulence loci that encode secreted proteins expressed in haustoria and recognized inside plant cells supports the hypothesis that, like bacterial pathogens, rust fungi secrete a suite of effector proteins into infected host cells. These proteins are likely to have important roles in pathogenesis-related processes. However, the mechanism of translocation from the extrahaustorial matrix across the plant cell membrane is not known. We consider there to be two main possibilities for this process. It may be mediated by a specialized translocation apparatus produced by the rust haustorium, or it may rely on intrinsic plant cross-membrane transport mechanisms. The AvrM-B and -C proteins induced a weaker necrotic response when secreted (data not shown), as did the AvrL567 proteins recognized by L6 (Dodds et al., 2004). Although this is consistent with the hypothesis that these proteins may reenter the cell after secretion in the absence of the rust pathogen, these observations could be explained by the accumulation of small amounts of these proteins in the cytoplasm as a result of aberrant processing of the signal peptide or inefficient secretion. However, the secreted versions of AvrM-A and -D induce a very strong necrotic response, which in the case of AvrM-A is equivalent to the nonsecreted version (data not shown), which is more difficult to explain on this basis. Furthermore, the addition of an ER retrieval signal to the full-length AvrM-A protein inhibited the necrotic response (Figure 3B), indicating that this protein is directed through the secretory pathway in planta and that the secreted version is able to enter the host cytoplasm from the apoplast in the absence of the pathogen. Addition of the HDEL motif to a secreted Avr9 protein also abolished Cf-9–Avr9-dependent necrosis (Wilson et al., 2005). Similarly, as discussed above, it is likely that the much stronger recognition of the secreted AvrP4 proteins compared with the nonsecreted versions is the result of the more efficient formation of disulfide bonds in the ER during secretion. Thus, the enhanced recognition would be conferred by correctly folded AvrP4 protein that has passed through the secretory pathway and then reentered the cell. The question remains whether this observation represents an experimental artifact that may be caused by artificially high levels of these proteins in the transient assay system, or do these rust proteins hijack a plant-derived translocation mechanism to gain entry to host cells?
Several avirulence products identified from oomycetes are also thought to be recognized after transport into the host cell. Like rusts, these pathogens grow intercellular hyphae and form haustoria that are surrounded by a plant-derived plasma membrane. The secreted protein ATR13 from H. parasitica triggers RPP13 resistance in Arabidopsis thaliana when transiently expressed in planta without its signal peptide (Allen et al., 2004). This intracellular recognition implies that ATR13 is transported into the plant cell during infection. A similar situation was observed with ATR1NdWsB also from H. parasitica (Rehmany et al., 2005) and Avr3a from P. infestans (Armstrong et al., 2005). In each case, the R protein responsible for recognition of the Avr elicitor is a cytoplasmic NBS-LRR protein (Botella et al., 1998; Allen et al., 2004; Armstrong et al., 2005). Therefore, the predicted localization of the R protein is consistent with the Avr recognition site. Similar to AvrM-A and -D, the presence or absence of the signal peptides for the ATR13 avirulence product from H. parasitica did not affect its activity in transient expression analysis in planta (Allen et al., 2004), whereas a reduced response was observed with the inclusion of the signal peptides for Avr3a and ATR1NdWsB (Armstrong et al., 2005; Rehmany et al., 2005). In another example, infiltration of recombinantly expressed Avr1b-1 protein into the apoplast of soybean (Glycine max) cultivars containing the Rps1b resistance gene resulted in necrosis, suggesting that this secreted Avr protein from P. sojae may enter the plant cell in the absence of the pathogen (Shan et al., 2004), although the site of recognition has not been confirmed.
The oomycete Avr proteins all contain a sequence motif (RxLRx5-21ddEER) (Armstrong et al., 2005; Rehmany et al., 2005) that is similar to the host-targeting signal (RxLxE/Q) that directs the transport of secreted malaria proteins across the parasitiphorous vacuolar membrane into the cytoplasm of erythrocytes (Hiller et al., 2004; Marti et al., 2004). This similarity suggests that the oomycete motif may also be involved in the translocation of proteins into the host plant cell. This motif is also present in at least 40 other predicted oomycete-secreted proteins and may define a set of translocated effector proteins in these pathogens; interestingly, this motif appears to be absent from secreted Cys-rich proteins (Rehmany et al., 2005). The oomycete RxLR motif is not present in the flax rust Avr proteins, suggesting that secreted rust effector proteins may use a different translocation mechanism. We did identify a short motif (Gx1x2R,where x1 is a hydrophobic or aromatic amino acid) in the N-terminal region of AvrL567, AvrM, and AvrP4 and in the C-terminal region of AvrP123, which may represent a motif common to rust avirulence proteins. Mutations of this motif did not affect the recognition of either secreted or nonsecreted versions of AvrM or AvrP4 expressed in planta. Although this indicates that this motif is not important for the uptake of the plant-produced secreted proteins, we cannot exclude a role in the translocation of rust-derived proteins during infection.
It is noteworthy that a relatively modest sequencing effort of only 672 ESTs yielded 21 HESPs that included four confirmed avirulence genes: AvrL567, AvrM, AvrP4, and AvrP123. It is possible that some of the other HESPs may also encode avirulence genes, because only 16 of the >30 known avirulence specificities segregate in the CH5-derived mapping family. Thus, this approach of screening haustorium ESTs for those encoding predicted signal peptides has proven to be a powerful method to isolate Avr genes from flax rust. It is likely that this approach will also be applicable to other rust species that cause severe disease on crops such as soybean and wheat (Triticum aestivum). It is likely that rust HESPs include proteins with important pathogenicity functions, and further study of these genes may provide insights into the parasitic interaction between rust fungi and their hosts.
METHODS
Rust and Plant Material
A flax rust (Melampsora lini) F2 family derived from selfing rust strain CH5, the F1 hybrid of a cross between parental strains C and H, was described by Lawrence et al. (1981a). The B12×M, B10×P1, B4×P2, and B13×P3 flax (Linum usitatissimum) lines contain the M, P1, P2, and P3 resistance genes backcrossed for 12, 10, 4, or 13 generations, respectively, into the variety Bison (Lawrence et al., 1981a). The flax line C.I.1911, which contains P4, was described by Zimmer and Comstock (1973). A flax line containing the M transgene in the variety Hoshangabad was described by Anderson et al. (1997), and the same M gene construct was transformed into the tobacco (Nicotiana tabacum) line W38 as described by Frost et al. (2004). Rust inoculations were performed as described by Lawrence et al. (1981a). Rust spores were germinated overnight on water, and genomic DNA was isolated from the resulting mycelial mat as described by Anderson et al. (1997).
Isolation of Flax Rust Haustorial Cells
Haustoria were isolated from flax leaves of the rust-susceptible variety Hoshangabad at 6 d after inoculation with rust strain CH5 by affinity chromatography essentially as described by Hahn and Mendgen (1992). A 100-μm pore size nylon mesh was used to remove the bulk of the plant cell material from the crude preparation, which was then passed through an 11-μm pore size nylon mesh to remove intact plant cells. An affinity column was prepared by covalently attaching concanavalin A (Pharmacia Biotech) to cyanogen bromide–activated Sepharaose 6MB (Pharmacia Biotech) as described in the manufacturer's protocol. Samples of purified haustoria were examined by immunofluorescence labeling with monoclonal antibody ML1 as described by Murdoch et al. (1998), except that samples were fixed to slides in 0.5% gelatin, 0.05% chromic potassium sulfate, and 0.02% NaN3 and the secondary antibody used was the goat anti-mouse coupled to the fluorophore Alexa 488 (Molecular Probes). Slides were examined on a Leica TCS SP2 confocal microscope under light or under excitation by an argon ion laser at a wavelength of 488 nm. Images were collected at 500 to 530 nm with pseudocolor green.
Haustorium-Specific cDNA Library and Sequence Analysis
Total RNA was isolated from frozen haustoria samples using the Qiagen Plant RNeasy kit, then treated with DNase and reisolated on the Qiagen Plant RNeasy column. A cDNA library was prepared from 1 μg of total RNA from haustoria using the SMART cDNA library kit (Clontech) according to the manufacturer's instructions. Library clones were colony-blotted and then probed with a subset of flax rust rRNA genes as described below, and positives were eliminated before sequencing. DNA sequence data were obtained from PCR cycle sequencing using the enzymatic dideoxy chain-termination method with an ABI PRISM BigDye primer cycle sequencing kit. BLAST searches were made at the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/BLAST/). Searches for protein signatures were performed by Motif Scan (http://myhits.isb-sib.ch/cgi-bin/motif_scan). The AvrM and AvrP4 coding sequences or flanking sequences were aligned, and nucleotide sequence distances were calculated across all sites or specifically for nonsynonymous or synonymous sites using the Jukes–Cantor algorithm of the Molecular Evolutionary Genetics Analysis software version 1.02 (http://evolgen.biol.metro-u.ac.jp/MEGA/). The significance of differences between average pairwise nucleotide distances was assessed by t test.
Colony and Plaque Lifts and Gel Blot Analysis
Plaques or colonies from the haustorium-specific cDNA library or plaques from a λ EMBL3 genomic DNA library prepared from rust strain CH5 (Ayliffe et al., 2001) were transferred to Hybond N+ nylon membranes (Amersham). Restriction enzyme–digested genomic DNA was separated on 1.0% agarose gels and transferred to Hybond N+ nylon membranes. RNA samples were separated on 1.5% agarose gels and transferred to Hybond N+. Prehybridization and hybridization with [32P]dCTP-labeled DNA probes were performed in 7% (w/v) SDS, 1% (w/v) BSA, 0.5 M sodium phosphate, pH 7.2, and 1 mM EDTA at 65°C, and washing was in 1× SSC (0.15 M NaCl and 0.015 M sodium citrate) and 0.1% SDS at 65°C.
Transcript Analysis by RT-PCR
Total RNA was reverse-transcribed using Superscript reverse transcriptase (Gibco BRL) with an oligo(dT25) primer and then amplified by Taq polymerase with the following thermal profile: 94°C for 2 min; 35 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 1 to 2 min; 72°C for 5 min. AvrM transcripts were amplified with the primers F1-C29 (5′-GGTACCTACCTTTTGAACC-3′) and R1-C29 (5′-CGCATCCAAGAATCACAATC-3′) or FA-C29 (5′-CCACGGAGATAGCTTGTC-3′) and Rstop-C29 (5′-GCTTTCAAGGTTTTGCTAATG-3′). AvrP4 transcripts were amplified with F3-C48 (5′-CATCAAAATCTAACCCGTAC-3′) and R1-C48 (5′-GTAGCATTGAGATCCATGG-3′). RT-PCR products were cloned into pGEM T-Easy, and independent clones were sequenced.
Transient in Planta Expression Assays
Gene expression constructs contained the AvrM, AvrP4, and AvrP123 coding sequences inserted between a 35S cauliflower mosaic virus promoter and a nopaline synthase terminator in the binary vector pTNotTReg (Anderson et al., 1997). The AvrM and AvrP123 expression constructs were prepared by inserting a cloned PCR product as an EcoRI fragment. Constructs encoding the truncated AvrM proteins, lacking the signal peptide, begin at nucleotide position 79 relative to the first ATG and have a GGA-to-ATG alteration at positions 82 to 84 to incorporate a start Met at amino acid 28, the last amino acid of the predicted signal peptide. Constructs encoding the full-length AvrM proteins contain the entire coding sequences as well as four nucleotides 5′ to the first ATG. The construct encoding the full-length AvrP123 protein contains the entire coding sequences as well as 87 nucleotides 5′ to the first ATG and 194 nucleotides of the 3′ untranslated region. The AvrP4 expression constructs were constructed by inserting a cloned PCR product as a BamHI-SacI fragment. Constructs encoding the truncated AvrP4 proteins begin at nucleotide position 49 relative to the first ATG, making the start of the open reading frame at Met-26, the third last amino acid of the 28–amino acid predicted signal peptide. Constructs encoding the full-length AvrP4 proteins contain the entire coding sequences as well as 22 nucleotides 5′ to the first ATG. Cys-to-Ala mutations were generated by PCR amplification using a 3′ primer to incorporate GCT at codon position 84 or 93. The constructs containing the PHA signal peptides have the AvrP4 gene sequence starting at nucleotide position 85 relative to the first ATG plus a BamHI fragment encoding the first 44 amino acids of the bean PHA (Tague et al., 1990). Between the PHA and AvrP4 sequences are the additional residues Gly-Ser, resulting from the BamHI cloning site. AvrM-A and AvrP4 constructs encoding an ER retrieval signal contain 12 additional bases (CATGATGAGCTC, encoding HDEL, or CATGATGACCTC, encoding HDDL) inserted immediately before the stop codon. Quadruple Ala mutants in the GFLR and GFSR motifs of AvrM-A and AvrP4 were generated by recombinant PCR amplification. All constructs were fully sequenced to confirm their integrity. Agrobacterium tumefaciens cultures of strain GV3101 containing the binary vector expression constructs were prepared at an OD600 of 1.0 in liquid Murashige and Skoog medium containing 200 μM acetosyringone and infiltrated into flax or tobacco leaves by syringe.
Yeast Signal Peptide Screen
The nucleotide sequences encoding the first 28 amino acids of AvrM or AvrP4 were cloned as an EcoRI–NotI fragment into the yeast shuttle vector pSMASH V2, which contains ura3 as a selectable marker (Goo et al., 1999). Constructs were transformed into the yeast strain DBYα2445, and cells were grown on yeast minimal medium with sucrose in place of glucose. The pSMASH V2 vector containing the invertase signal peptides or just a start Met were used as positive or negative controls, respectively. The 44–amino acid leader sequence of bean PHA was also used as a positive control.
Accession Numbers
Sequence data from this article can be found in the GenBank data library under accession numbers DQ279864 to DQ279890.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Transient Expression of the Full-Length AvrM Genes in Flax.
Supplemental Figure 2. Activity of AvrM and AvrP4 Signal Peptides in Yeast.
Supplementary Material
Acknowledgments
We thank Valerie Ryle, Patricia Atkinson, and Kim Newell for providing excellent technical assistance. The ML1 monoclonal antibody was kindly provided by Adrienne Hardham (Research School of Biological Science, Australian National University). This research was supported by grants from the Grains Research and Development Corporation. A.-M.C. was supported by an Australian Postgraduate Award funded by the Department of Education, Science, and Training and by a Commonwealth Scientific and Industrial Research Organization partial scholarship.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Peter N. Dodds (peter.dodds@csiro.au).
Online version contains Web-only data.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.105.035980.
References
- Allen, R.L., Bittner-Eddy, P.D., Grenville-Briggs, L.J., Meitz, J.C., Rehmany, A.P., Rose, L.E., and Beynon, J.L. (2004). Host-parasite coevolutionary conflict between Arabidopsis and downy mildew. Science 306 1957–1960. [DOI] [PubMed] [Google Scholar]
- Altschul, S.F., Madden, T.L., Schaffer, A.A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D.J. (1997). Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson, P.A., Lawrence, G.J., Morrish, B.C., Ayliffe, M.A., Finnegan, E.J., and Ellis, J.G. (1997). Inactivation of the flax rust resistance gene M associated with loss of a repeated unit within the leucine-rich repeat coding region. Plant Cell 9 641–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong, M.R., et al. (2005). An ancestral oomycete locus contains late blight avirulence gene Avr3a, encoding a protein that is recognised in the host cytoplasm. Proc. Natl. Acad. Sci. USA 102 7766–7771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axtell, M.J., Chisholm, S.T., Dahlbeck, D., and Staskawicz, B.J. (2003). Genetic and molecular evidence that the Pseudomonas syringae type III effector protein AvrRpt2 is a cysteine protease. Mol. Microbiol. 49 1537–1546. [DOI] [PubMed] [Google Scholar]
- Ayliffe, M.A., Dodds, P.N., and Lawrence, G.J. (2001). Characterisation of the beta-tubulin genes from Melampsora lini and comparison of fungal beta-tubulin genes. Mycol. Res. 105 818–826. [Google Scholar]
- Bailey, T.L., and Elkan, C. (1994). Fitting a mixture model by expectation maximization to discover motifs in biopolymers. In Proceedings of the Second International Conference on Intelligent Systems for Molecular Biology. (Menlo Park, CA: AAAI Press), pp. 28–36. [PubMed]
- Bannai, H., Tamada, Y., Maruyama, O., Nakai, K., and Miyano, S. (2002). Extensive feature detection of N-terminal protein sorting signals. Bioinformatics 18 298–305. [DOI] [PubMed] [Google Scholar]
- Bendtsen, J.D., Nielsen, H., von Heijne, G., and Brunak, S. (2004). Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340 783–795. [DOI] [PubMed] [Google Scholar]
- Botella, M.A., Parker, J.E., Frost, L.N., Bittner-Eddy, P.D., Beynon, J.L., Daniels, M.J., Holub, E.B., and Jones, J.D.G. (1998). Three genes of the Arabidopsis RPP1 complex resistance locus recognize distinct Peronospora parasitica avirulence determinants. Plant Cell 10 1847–1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coaker, G., Falick, A., and Staskawicz, B. (2005). Activation of a phytopathogeneic bacterial effector protein by a eukaryotic cyclophilin. Science 308 506–508. [DOI] [PubMed] [Google Scholar]
- Denecke, J., DeRyk, R., and Botterman, J. (1992). Plant and mammalian sorting signals for protein retention in the endoplasmic reticulum contain a conserved epitope. EMBO J. 11 2345–2355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denecke, J., Ek, B., Caaspers, M., Sinjorgo, K.M., and Palva, E.T. (1993). Analysis of sorting signals responsible for the accumulation of soluble reticuloplasmins in the plant endoplasmic reticulum. J. Exp. Bot. 44 213–221. [Google Scholar]
- Deslandes, L., Olivier, J., Peeters, N., Feng, D.X., Khounlotham, M., Boucher, C., Somssich, I., Genin, S., and Marco, Y. (2003). Physical interaction between RRS1-R, a protein conferring resistance to bacterial wilt, and PopP2, a type III effector targeted to the plant nucleus. Proc. Natl. Acad. Sci. USA 100 8024–8029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodds, P.N., Lawrence, G.J., Catanzariti, A., Ayliffe, M.A., and Ellis, J.G. (2004). The Melampsora lini AvrL567 avirulence genes are expressed in haustoria and their products are recognized inside plant cells. Plant Cell 16 755–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodds, P.N., Lawrence, G.J., and Ellis, J.G. (2001. a). Contrasting modes of evolution acting on the complex N locus for rust resistance in flax. Plant J. 27 439–453. [DOI] [PubMed] [Google Scholar]
- Dodds, P.N., Lawrence, G.J., Pryor, T., and Ellis, J.G. (2001. b). Six amino acid changes confined to the leucine-rich repeat β-strand/β-turn motif determine the difference between the P and P2 rust resistance specificities in flax. Plant Cell 13 163–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson, F.L., Holzberg, S., Calderon-Urrea, A., Handley, V., Axtell, M., Corr, C., and Baker, B. (1999). The helicase domain of the TMV replicase proteins induces the N-mediated defence response in tobacco. Plant J. 18 67–75. [DOI] [PubMed] [Google Scholar]
- Frost, D., Way, H., Howles, P., Luck, J., Manners, J., Hardham, A., Finnegan, J., and Ellis, J. (2004). Tobacco transgenic for the flax rust resistance gene L expresses allele-specific activation of defense responses. Mol. Plant Microbe Interact. 17 224–232. [DOI] [PubMed] [Google Scholar]
- Goo, J.H., Park, A.R., Park, W.J., and Park, O.K. (1999). Selection of Arabidopsis genes encoding secreted and plasma membrane proteins. Plant Mol. Biol. 41 415–423. [DOI] [PubMed] [Google Scholar]
- Haerter, A.C., and Voegele, R.T. (2004). A novel β-glucosidase in Uromyces fabae: Feast or fight? Curr. Genet. 45 96–103. [DOI] [PubMed] [Google Scholar]
- Hahn, M., and Mendgen, K. (1992). Isolation by ConA binding of haustoria from different rust fungi and comparison of their surface qualities. Protoplasma 170 95–103. [Google Scholar]
- Hahn, M., and Mendgen, K. (1997). Characterization of in planta-induced rust genes isolated from a haustorium-specific cDNA library. Mol. Plant Microbe Interact. 10 427–437. [DOI] [PubMed] [Google Scholar]
- Hahn, M., and Mendgen, K. (2001). Signal and nutrient exchange at biotrophic plant-fungus interfaces. Curr. Opin. Plant Biol. 4 322–327. [DOI] [PubMed] [Google Scholar]
- Heath, M.C. (1997). Signalling between pathogenic rust fungi and resistant or susceptible host plants. Ann. Bot. (Lond.) 80 713–720. [Google Scholar]
- Hiller, N.L., Bhattacharjee, S., van Ooij, C., Liolios, K., Harrison, T., Lopez-Estraño, C., and Haldar, K. (2004). A host-targeting signal in virulence proteins reveals a secretome in malarial infection. Science 306 1934–1937. [DOI] [PubMed] [Google Scholar]
- Joosten, M.H.A.J., Cozijnsen, T.J., Verberne, M.C., and de Wit, P.J.G.M. (1994). Host resistance to a fungal tomato pathogen lost by a single base-pair change in an avirulence gene. Nature 367 384–386. [DOI] [PubMed] [Google Scholar]
- Joosten, M.H.A.J., Vogelsang, R., Cozijnsen, T.J., Verberne, M.C., and de Wit, P.J.G.M. (1997). The biotrophic fungus Cladosporium fulvum circumvents Cf-4-mediated resistance by producing unstable AVR4 elicitors. Plant Cell 9 367–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi, I., Kobayashi, Y., and Hardham, A.R. (1994). Dynamic reorganisation of microtubules and microfilaments in flax cells during the resistance response to flax rust infection. Planta 195 237–247. [Google Scholar]
- Lahaye, T., and Bonas, U. (2001). Molecular secrets of bacterial type III effector proteins. Trends Plant Sci. 6 479–485. [DOI] [PubMed] [Google Scholar]
- Lawrence, G.J. (1989). Flax rust from Linum marginale: Pathogenicity reactions on the Linum usitatissimum set of differential varieties. Can. J. Bot. 67 3187–3191. [Google Scholar]
- Lawrence, G.J., Finnegan, E.J., Ayliffe, M.A., and Ellis, J.G. (1995). The L6 gene for flax rust resistance is related to the Arabidopsis bacterial resistance gene Rps2 and the tobacco viral resistance gene N. Plant Cell 7 1195–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence, G.J., Mayo, G.M.E., and Shepherd, K.W. (1981. a). Interactions between genes controlling pathogenicity in the flax rust fungus. Phytopathology 71 12–19. [Google Scholar]
- Lawrence, G.J., Shepherd, K.W., and Mayo, G.M.E. (1981. b). Fine structure of genes controlling pathogenicity in flax rust, Melampsora lini. Heredity 46 297–313. [Google Scholar]
- Luderer, R., and Joosten, M.H.A.J. (2001). Avirulence proteins of plant pathogens: Determinants of victory and defeat. Mol. Plant Pathol. 6 355–364. [DOI] [PubMed] [Google Scholar]
- Luderer, R., Takken, F.W., de Wit, P.J.G.M., and Joosten, M.H.A.J. (2002). Cladosporium fulvum overcomes Cf-2-mediated resistance by producing truncated AVR2 elicitor proteins. Mol. Microbiol. 45 875–884. [DOI] [PubMed] [Google Scholar]
- Marti, M., Good, R.T., Rug, M., Knuepfer, E., and Cowman, A.F. (2004). Targeting malaria virulence and remodeling proteins to the host erythrocyte. Science 306 1930–1933. [DOI] [PubMed] [Google Scholar]
- Murdoch, L.J., Kobayashi, I., and Hardham, A.R. (1998). Production and characterisation of monoclonal antibodies to cell wall components of the flax rust fungus. Eur. J. Plant Pathol. 104 331–346. [Google Scholar]
- Nakai, K., and Horton, P. (1999). PSORT: A program for detecting the sorting signals of proteins and predicting their subcellular localization. Trends Biochem. Sci. 24 34–35. [DOI] [PubMed] [Google Scholar]
- Orth, K., Xu, Z., Mudgett, M.B., Bao, Z.Q., Palmer, L.E., Bliska, J.B., Mangel, W.F., Staskawicz, B.J., and Dixon, J.E. (2000). Disruption of signalling by Yersinia effector YopJ, a ubiquitin-like protein protease. Science 290 1594–1597. [DOI] [PubMed] [Google Scholar]
- Pallaghy, P.K., Nielsen, K.J., Craik, D.J., and Norton, R.S. (1994). A common structural motif incorporating a cystine knot and a triple-stranded β-sheet in toxic and inhibitory polypeptides. Protein Sci. 3 1833–1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panstruga, R. (2003). Establishing compatibility between plants and obligate biotrophic pathogens. Curr. Opin. Plant Biol. 6 320–326. [DOI] [PubMed] [Google Scholar]
- Perfect, S.E., and Green, J.R. (2001). Infection structures of biotrophic and hemibiotrophic fungal plant pathogens. Mol. Plant Pathol. 2 101–108. [DOI] [PubMed] [Google Scholar]
- Rehmany, A.P., Gordon, A., Rose, L.E., Allen, R.L., Armstrong, M.R., Whisson, S.C., Kamoun, S., Tyler, B.M., Birch, P.R.J., and Beynon, J.L. (2005). Differential recognition of highly divergent downy mildew avirulence gene alleles by RPP1 resistance genes from two Arabidopsis lines. Plant Cell 17 1839–1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rep, M., van der Does, H.C., Meijer, M., van Wijk, R., Houterman, P.M., Dekker, H.L., de Koster, C.G., and Cornelissen, B.J.C. (2004). A small, cysteine-rich protein secreted by Fusarium oxysporum during colonization of xylem vessels is required for I-3-mediated resistance in tomato. Mol. Microbiol. 53 1373–1383. [DOI] [PubMed] [Google Scholar]
- Rohe, M., Gierlich, A., Hermann, H., Hahn, M., Schmidt, B., Rosahl, S., and Knogge, W. (1995). The race-specific elicitor, NIP1, from the barley pathogen, Rhynchosporium secalis, determines avirulence on host plants of the Rrs1 resistance genotype. EMBO J. 14 4168–4177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rooney, H.C.E., van't Klooster, J.W., van der Hoorn, R.A.L., Joosten, M.H.A.J., Jones, J.D.G., and de Wit, P.J.G.M. (2005). Cladosporium Avr2 inhibits tomato Rcr3 protease for Cf-2-dependent disease resistance. Science 308 1783–1786. [DOI] [PubMed] [Google Scholar]
- Shan, W., Cao, M., Leung, D., and Tyler, B.M. (2004). The Avr1b locus of Phytophthora sojae encodes an elicitor and a regulator required for avirulence on soybean plants carrying resistance gene Rps1b. Mol. Plant Microbe Interact. 17 394–403. [DOI] [PubMed] [Google Scholar]
- Shao, F., Golstein, C., Ade, J., Stoutemyer, M., Dixon, J.E., and Innes, R.W. (2003). Cleavage of Arabidopsis PBS1 by a bacterial type III effector. Science 301 1230–1233. [DOI] [PubMed] [Google Scholar]
- Tague, B.W., Dickinson, C.D., and Chrispeels, M.J. (1990). A short domain of the plant vacuolar protein phytohemagglutinin targets invertase to the yeast vacuole. Plant Cell 2 533–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian, M., Benedetti, B., and Kamoun, S. (2005). A second Kazal-like protease inhibitor from Phytophthora infestans inhibits and interacts with the apoplastic pathogenesis-related protease P69B of tomato. Plant Physiol. 138 1785–1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian, M., Huitema, E., Da Cunha, L., Torto-Alalibo, T., and Kamoun, S. (2004). A Kazal-like extracellular serine protease inhibitor from Phytophthora infestans targets the tomato pathogenesis-related protease P69B. J. Biol. Chem. 279 26370–26377. [DOI] [PubMed] [Google Scholar]
- van den Ackerveken, G.F.J.M., van Kan, J.A.L., and de Wit, P.J.G.M. (1992). Molecular analysis of the avirulence gene avr9 of the fungal tomato pathogen Cladosporium fulvum fully supports the gene-for-gene hypothesis. Plant J. 2 359–366. [DOI] [PubMed] [Google Scholar]
- van den Ackerveken, G.F.J.M., Vossen, P., and de Wit, J.G.M.P. (1993). The AVR9 race-specific elicitor of Cladosporium fulvum is processed by endogenous and plant proteases. Plant Physiol. 103 91–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Burg, H.A., Westerink, N., Francoijs, K., Roth, R., Woestenenk, E., Boeren, S., de Wit, P.J.G.M., Joosten, M.H.A.J., and Vervoort, J. (2003). Natural disulfide bond-disrupted mutants of AVR4 of the tomato pathogen Cladosporium fulvum are sensitive to proteolysis, circumvent Cf-4-mediated resistance, but retain their chitin binding ability. J. Biol. Chem. 278 27340–27346. [DOI] [PubMed] [Google Scholar]
- van den Hooven, H.W., van den Burg, H.A., Vossen, P., Boeren, S., de Wit, P.G.M., and Vervoort, J. (2001). Disulfide bond structure of the AVR9 elicitor of the fungal tomato pathogen Cladosporium fulvum: Evidence for a cystine knot. Biochemistry 40 3458–3466. [DOI] [PubMed] [Google Scholar]
- van der Hoorn, R.A.L., Laurent, F., Roth, R., and de Wit, P.J.G.M. (2000). Agroinfiltration is a versatile tool that facilitates comparative analyses of Avr9/Cf-9-induced and Avr4/Cf-4-induced necrosis. Mol. Plant Microbe Interact. 13 439–446. [DOI] [PubMed] [Google Scholar]
- van't Slot, K.A.E., van den Burg, H.A., Kloks, C.P.A.M., Hilbers, C.W., Knogge, W., and Papavoine, C.H.M. (2003). Solution structure of the plant disease resistance-triggering protein NIP1 from the fungus Rhynchosporium secalis shows a novel β-sheet fold. J. Biol. Chem. 278 45730–45736. [DOI] [PubMed] [Google Scholar]
- Voegele, R.T., Hahn, M., Lohaus, G., Link, T., Heiser, I., and Mendgen, K. (2005). Possible roles for mannitol and mannitol dehydrogenase in the biotrophic plant pathogen Uromyces fabae. Plant Physiol. 137 190–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voegele, R.T., and Mendgen, K. (2003). Rust haustoria: Nutrient uptake and beyond. New Phytol. 159 93–100. [DOI] [PubMed] [Google Scholar]
- Voegele, R.T., Struck, C., Hahn, M., and Mendgen, K. (2001). The role of haustoria in sugar supply during infection of broad bean by the rust fungus Uromyces fabae. Proc. Natl. Acad. Sci. USA 98 8133–8138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westerink, N., Brandwagt, B.F., de Wit, P.J.G.M., and Joosten, M.H.A.J. (2004). Cladosporium fulvum circumvents the second functional resistance gene homologue at the Cf-4 locus (Hcr9-E) by secretion of a stable avr4E isoform. Mol. Microbiol. 54 533–545. [DOI] [PubMed] [Google Scholar]
- Wilson, R.J., Baillie, B.K., and Jones, D.A. (2005). ER retrieval of Avr9 compromises its elicitor activity consistent with perception of Avr9 at the plasma membrane. Mol. Plant Pathol. 6 193–197. [DOI] [PubMed] [Google Scholar]
- Zimmer, D.E., and Comstock, V.E. (1973). New genes for rust resistance in flax. Phytopathology 63 777–780. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.