Abstract
High-risk human papillomaviruses (HPVs) (e.g., HPV-16) cause anogenital and head and neck cancers, and low-risk HPVs (e.g., HPV-6) cause benign hyperproliferative disease. The E7 protein of HPV-16 binds all retinoblastoma tumor suppressor protein (pRB) family members with higher affinity than HPV-6E7. The HPV-16 E7 protein has been reported to target pRB family members for degradation and to immortalize cells. Here we tested the hypothesis that the low-risk E7 protein has an intrinsic ability to decrease expression of pRB family members. First, we introduced a high-affinity pRB-binding site into HPV-6 E7 (6E7G22D) and showed that, in human foreskin keratinocytes, HPV-6 E7G22D decreased the level of pRB protein but not pRB mRNA. Second, we analyzed the ability of wild-type HPV-6 E7 to destabilize the other pRB family members, p107 and p130. HPV-6 E7, like HPV-16 E7, decreased the level of p130 protein. This decrease was inhibited by MG132, a proteasome inhibitor. Binding of HPV-6 E7 to p130 was necessary but not sufficient to decrease the level of p130. Furthermore, the destabilization of p130 correlated with a decrease in the expression of involucrin, a differentiation marker. We suggest that the shared activity of HPV-16 E7 and HPV-6 E7 to destabilize p130 and decrease or delay differentiation may be related to the role of E7 in the HPV life cycle. The added ability of HPV-16 E7 to regulate pRB and p107 may be related to oncogenic activity.
Keywords: keratinocytes, RB family members, human papillomaviruses, differentiation
Human papillomaviruses (HPVs) are nonenveloped viruses that contain an ≈8,000-bp circular DNA genome. HPVs infect mucosal and cutaneous stratified squamous epithelia and are divided into high-risk and low-risk viruses based on their pathogenicity (1). The low-risk viruses, e.g., HPV-6 and HPV-11, mainly cause benign hyperproliferative disease, including the most prevalent viral sexually transmitted disease, condyloma acuminata or genital warts. The high-risk viruses, e.g., HPV-16, HPV-18, and HPV-31, are normally found in malignant tumors, including cervical cancer and some head and neck cancers (1). The replication cycle of all HPVs is differentiation-dependent (1, 2). The virus presumably enters through a break in the epithelium and initially infects the basal cells, which are the proliferating epithelial cells (keratinocytes). In these cells, the virus undergoes the nonproductive stage of its life cycle, where it is maintained as a low-copy-number episome. When the infected cell moves to the suprabasal compartment, both the high-risk and low-risk viruses undergo the productive phase of their life cycle: amplifying their DNA, synthesizing structural proteins, and producing infectious virus. Uninfected cells in this suprabasal compartment normally have exited the cell cycle and begun to differentiate. Because HPVs require the host cell DNA replication machinery to replicate, the viral DNA must encode proteins able to generate an intracellular environment within this differentiated compartment appropriate for this replication. Although only the E7 protein encoded by high-risk HPVs can immortalize human epithelial cells and play a role in the development of cervical carcinoma, the E7 proteins of both high-risk and low-risk viruses can create this replication-competent environment (3). Also, both proteins are required for episomal maintenance in undifferentiated cells and/or genome amplification upon differentiation (4-6). This finding suggests that high-risk and low-risk E7 proteins share some activities that are important for virus replication.
Structurally, both high-risk and low-risk E7 proteins contain 98 aa and can be divided into three regions: conserved region 1 (CR1, amino acids 2-15), CR2 (amino acids 16-38), and the C-terminal region containing two zinc finger domains (amino acids 39-98) (7, 8). The E7 proteins of high-risk HPV-16 and low-risk HPV-6 share 50% amino acid sequence identity and 15% conservative changes (7). One of the main properties of E7 is that, through its LXCXE motif in CR2, it can bind and inactivate the retinoblastoma tumor suppressor protein (pRB) and its family members, resulting in the release of the E2F transcription factors and the transactivation of E2F-responsive genes necessary for cell-cycle progression (8-10). High-risk HPV-16 E7 also targets pRB, p107, and p130, the three RB family members, for degradation (11-14), an activity requiring sequences outside of the LXCXE motif in E7 (12-15). HPV DNA is not maintained as well when the LXCXE motif in E7 is disrupted (16), suggesting that E7 may play a role in the HPV life cycle by disrupting pRB family member-mediated transcriptional repression of certain genes involved in cell cycling.
Despite having the LXCXE motif, the low-risk HPV-6 E7 binds to pRB family members with lower affinity than the high-risk HPV-16 E7 (7, 17-19). HPV-6 E7 cannot target pRB for degradation (20), but its effect on p107 and p130 has not been reported. In this study we investigated the ability of HPV-6 E7 to destabilize pRB family members. We show that a mutation in HPV-6 E7 that significantly increases the binding affinity to pRB allows HPV-6 E7 to target pRB for degradation. In addition, we show that wild-type HPV-6 E7 can degrade p130 but neither p107 nor pRB. The ability of both high-risk and low-risk E7 to target p130 for degradation provides a unifying activity for high-risk and low-risk E7 proteins that might explain their shared involvement in the virus life cycle.
Results
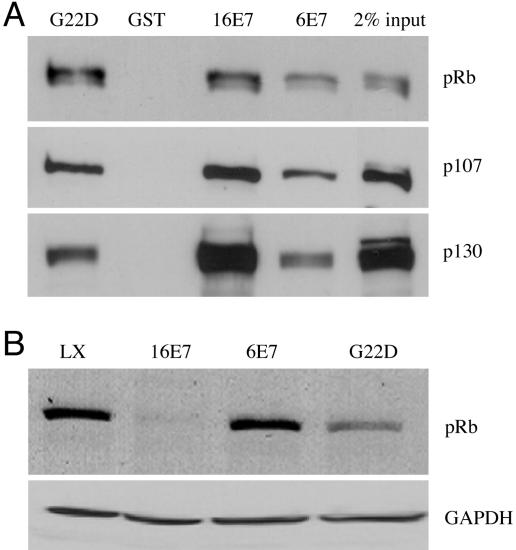
HPV-6 E7 Can Destabilize pRB When a High-Affinity pRB-Binding Site Is Introduced. Degradation of pRB by HPV-16 E7 requires at least two regions of the E7 protein in addition to the high-affinity pRB binding site (12-15). Because HPV-6 E7 binds pRB, p107, and p130 with lower affinity than does HPV-16 E7 (7, 17-19), and only the latter can target pRB for degradation, we wanted to determine whether increasing the affinity of HPV-6 E7 for pRB would result in gain of function with respect to pRB degradation. Both HPV-16 E7 and HPV-6 E7 have the LXCXE pRB binding motif; however, the amino acid residue preceding this motif in HPV-16 E7 is aspartic acid whereas in HPV-6 E7 it is glycine. Mutating the glycine to aspartic acid (G22D) results in a mutant protein with a high-affinity pRB binding site (21). Using the glutathione S-transferase (GST) pull-down assay, we confirmed that HPV-6 E7 bound pRB with lower affinity than did HPV-16 E7 whereas HPV-6 E7G22D bound pRB as efficiently as did HPV-16 E7 (Fig. 1A). In contrast, HPV-6 E7G22D showed a lesser increase in affinity for p107 and no increase in affinity for p130 compared with HPV-6 E7 (Fig. 1 A). To determine whether HPV-6 E7G22D gained the ability to destabilize pRB, human foreskin keratinocytes (HFKs) were infected with parental retrovirus LXSN, retrovirus encoding HPV-16 E7 [L(16E7)SN], HPV-6 E7 [L(6E7)SN], or HPV-6 E7G22D [L(6E7G22D)SN], and grown in C-K-SFM, keratinocyte serum-free medium supplemented with human recombinant epidermal growth factor and bovine pituitary extract. Western blot analysis of whole-cell lysates demonstrated that HPV-6 E7G22D gained function with respect to destabilizing pRB (Fig. 1B, compare G22D with 6E7), although it was not as efficient as HPV-16 E7 (Fig. 1B, compare G22D with 16E7). RT-PCR showed that E7 expression did not decrease the pRB mRNA level (data not shown).
Fig. 1.
Characterization of HPV-6 E7G22D with respect to binding of pRB family members and degradation of pRB. (A) The ability of HPV-6, HPV-16, and HPV-6 E7G22D to bind pRB family members. Jurkat nuclear extracts were incubated with GST, GST-16E7, GST-6E7, or GST-6E7G22D, and bound proteins were detected after separation by SDS/PAGE and transfer to nitrocellulose membrane. The membrane was probed with antibodies to pRB, p107, and p130. Ponceau red staining of the membrane before probing indicated that similar levels of GST or GST fusion proteins were present in the precipitate (data not shown). The average ± SD of bound pRB in three independent experiments, corrected for Ponceau red and relative to GST16E7 (set to 1.0), was 1.22 ± 0.15 (G22D) and 0.35 ± 0.06 (6E7); the average ± SD of bound p107 was 0.73 ± 0.08 (G22D) and 0.36 ± 0.01 (6E7); the average of bound p130 was 0.42 ± 0.07 (G22D) and 0.23 ± 0.05 (6E7). (B) Immunoblot of pRB in the presence and absence of E7. HFKs were infected with control retrovirus LXSN, L(16E7)SN, or L(6E7)SN and grown in C-K-SFM. Whole-cell lysates were analyzed by immunoblot by using antibodies to pRB and GAPDH, the latter as a loading control. The average ± SD of pRB in three independent experiments, corrected for GAPDH and relative to LXSN, was 0.24 ± 0.14 (16E7), 0.89 ± 0.32 (6E7), and 0.50 ± 0.19 (G22D).
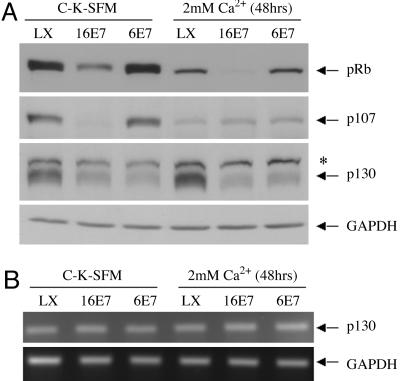
HPV-6 E7, Like HPV-16 E7, Can Destabilize p130 in both Undifferentiated and Differentiated Growth Conditions. The destabilization of pRB by HPV-6 E7G22D suggests that wild-type HPV-6 E7 may have the ability to target proteins for degradation. To test whether wild-type HPV-6 E7 can modulate the stability of the other two pRB family members, HFKs were infected with LXSN, L(16E7)SN, or L(6E7)SN and grown in C-K-SFM (undifferentiated conditions) or C-K-SFM containing 2 mM CaCl2 (differentiated conditions), keratinocyte growth conditions that support the nonproductive and productive phases of the virus life cycle, respectively. The steady-state level of each pRB family member in whole-cell lysates was determined by Western blot analysis. As expected, HPV-16 E7 destabilized p107 and p130 in undifferentiated keratinocytes (13, 14). In differentiated keratinocytes, HPV-16 E7 also destabilized p130 but not p107. HPV-6 E7 decreased the steady-state level of p130 in both undifferentiated and differentiated growth conditions, although it did not destabilize p107 under either growth condition (Fig. 2A). Neither HPV-16 E7 nor HPV-6 E7 expression decreased the p130 mRNA level (Fig. 2B).
Fig. 2.
The effect of HPV E7 proteins on p130 expression in undifferentiated and differentiated conditions. (A) Immunoblot of pRB family members in the presence and absence of E7 and in different growth conditions. HFKs were infected with control retrovirus LXSN, L(16E7)SN, or L(6E7)SN and grown in C-K-SFM or C-K-SFM containing 2 mM CaCl2 for 48 h. Whole-cell lysates were analyzed by immunoblot by using antibodies to pRB, p107, p130, and GAPDH. The average ± SD in three independent experiments, corrected for GAPDH and relative to LXSN in C-K-SFM, of pRB was 0.21 ± 0.09 (16E7) and 1.14 ± 0.20 (6E7) in C-K-SFM and 0.32 ± 0.13 (LXSN), 0.01 ± 0.02 (16E7), and 0.22 ± 0.08 (6E7) in CaCl2; the average ± SD of p107 was 0.27 ± 0.18 (16E7) and 1.14 ± 0.19 (6E7) in C-K-SFM and 0.25 ± 0.09 (LXSN), 0.26 ± 0.09 (16E7), and 0.27 ± 0.12 (6E7) in CaCl2; the average ± SD of p130 was 0.35 ± 0.04 (16E7) and 0.36 ± 0.08 (6E7) in C-K-SFM and 1.21 ± 0.06 (LXSN), 0.35 ± 0.05 (16E7), and 0.42 ± 0.04 (6E7) in CaCl2. *, Unknown protein cross-reacting with anti-p130. (B) Analysis of p130 mRNA levels. HFKs were infected and grown as in A, and total RNA was analyzed by RT-PCR with primers amplifying p130 and GAPDH. The results shown are for 26 cycles; similar results were obtained at 22 and 30 cycles. Data are representative of three independent experiments.
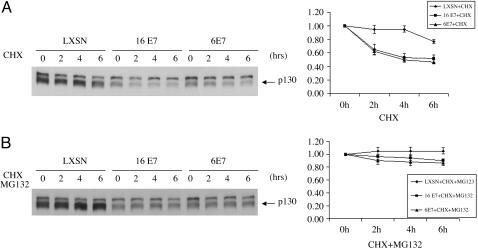
HPV-6 E7 Decreases p130 Half-Life Through the Proteasome Pathway. A previous study showed that HPV-16 E7 decreases p130 half-life through the proteasome pathway (13). To test whether HPV-6 E7 also destabilizes p130 by decreasing its half-life through the proteasome pathway, we infected HFKs with LXSN, L(16E7)SN, and L(6E7)SN and subsequently treated the cells with 0.25 mM cycloheximide, a protein synthesis inhibitor, or 0.25 mM cycloheximide plus 50 μM MG132, a proteasome pathway inhibitor. After 2, 4, and 6 h, the cells were harvested and the levels of p130 in the whole-cell lysates were determined by Western blot analysis. Densitometric analysis showed that HPV-6 E7, like HPV-16 E7, significantly decreased p130 half-life (Fig. 3A). The ability of both E7s to decrease p130 half-life was inhibited by MG132 (Fig. 3B).
Fig. 3.
The half life of p130 in the presence and absence of E7 protein and in the presence and absence of a proteasome inhibitor. (A) Decreased half-life of p130 in the presence of HPV E7 protein. HFKs were infected with control retrovirus LXSN, L(16E7)SN, or L(6E7)SN and treated with 0.25 mM cycloheximide for the indicated times. Immunoblots of whole-cell lysates are shown (Left), and densitometry is plotted for the average ± SD of three independent experiments (Right). (B) Inhibition of p130 degradation by MG132. HFKs were infected as in A and treated with 0.25 mM cycloheximide plus 50 μM MG132 for the indicated times. Immunoblots of whole-cell lysates are shown (Left), and densitometry is plotted for the average ± SD of three independent experiments (Right).
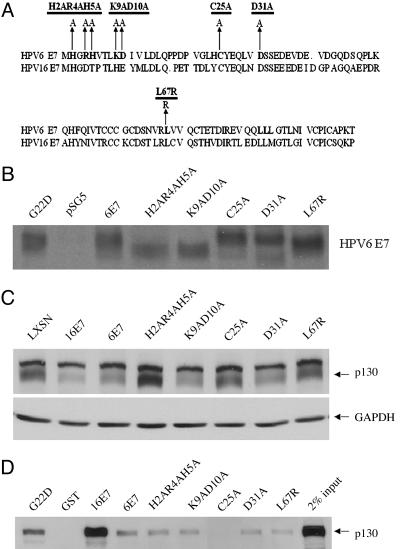
Binding of HPV-6 E7 to p130 Is Necessary but Not Sufficient for Destabilization of p130. To gain insight into which residues of HPV-6 E7 are responsible for destabilization of p130, mutants were generated, and recombinant retroviruses encoding these mutants were prepared (Fig. 4A). The stability of all mutants was verified (Fig. 4B). Differences in mobility of the proteins reflect charge differences (22, 23). HFKs were infected with parental retrovirus LXSN, L(16E7)SN, L(6E7)SN, or retroviruses encoding the HPV-6 E7 mutants. Whole-cell lysates were prepared and analyzed by Western blot using antibodies to p130. Mutants H2AR4AH5A in CR1, C25A in CR2, which disrupts the LXCXE motif, and L67R in the C terminus lost the ability to destabilize p130 (Fig. 4C). Mutants K9AD10A in CR1 and D31A in CR2 retained the ability to degrade p130.
Fig. 4.
Construction of HPV-6 E7 mutants and their characterization with respect to p130 binding and degradation. (A) Map of HPV-6 E7 mutants. (B) Stability of mutants. COS-7 cells were transfected with pSG5, pSG5(6E7), pSG5(H2AR4AH5A), pSG5(K9AD10A), pSG5(C25A), pSG5(D31A), and pSG5(L67R). Forty-eight hours after transfection, the cells were labeled with 500 μCi of 35S-labeled methionine and cysteine and lysed, and HPV-6 E7 was immunoprecipitated by a polyclonal antibody (anti-6E7). The immunoprecipitates were separated on 15% SDS/PAGE gel and exposed to x-ray film. (C) Amino acids required for destabilization of p130. HFKs were infected with LXSN, L(16E7)SN, L(6E7)SN, or retroviruses encoding the HPV-6 E7 mutants. Whole-cell lysates from transduced cells grown in C-K-SFM were analyzed by Western blot with antibodies to p130. The average ± SD of p130 in three independent experiments, corrected for GAPDH and relative to LXSN, was 0.33 ± 0.06 (16E7), 0.35 ± 0.06 (6E7), 1.03 ± 0.05 (H2AR4AH5A), 0.43 ± 0.06 (K9AD10A), 0.79 ± 0.15 (C25A), 0.37 ± 0.07 (D31A), and 0.89 ± 0.32 (L67R). (D) E7 amino acids required for binding to p130. GST pull-downs were conducted as in Fig. 1 with GST, GST-16E7, GST-6E7H2AR4AH5A, GST-6E7K9AD10A, GST-6E7C25A, GST-6E7D31A, and GST-6E7L67R, and the immunoblots were probed with antibodies to p130. Ponceau red staining indicated equal amounts of GST and GST fusion proteins in the precipitate (data not shown). The average ± SD of bound p130 in three independent experiments, corrected for Ponceau red and relative to GST6E7, was 6.86 ± 2.40 (16E7), 1.23 ± 0.76 (H2AR4AH5A), 0.51 ± 0.18 (K9AD10A), 0.16 ± 0.09 (C25A), 1.12 ± 0.12 (D31A), and 0.71 ± 0.05 (L67R).
To determine whether loss of ability to destabilize p130 was due to loss of the ability of HPV-6 E7 to bind p130, pull-down assays were performed using GST, GST-16E7, GST-6 E7, and GST-6 E7 mutants. Among all HPV-6 E7 mutants, only C25A did not bind p130 (Fig. 4D). Mutants H2AR4AH5A and L67R, which lost the ability to destabilize p130, retained the ability to bind p130. Furthermore, although mutants H2AR4AH5A and D31A bound p130 as efficiently as did wild-type HPV-6 E7, only mutant D31A reduced the level of p130. These data suggest that binding of HPV-6 E7 to p130 is necessary but not sufficient for destabilization of p130.
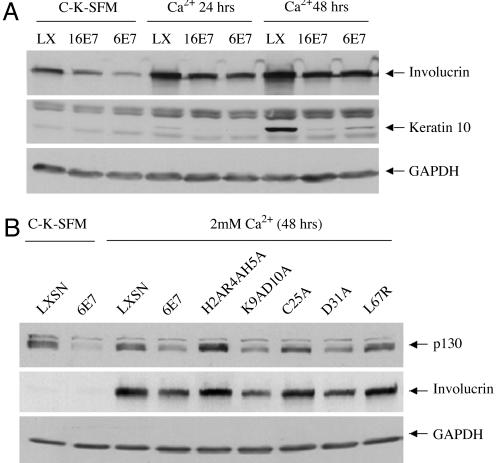
Decreasing/Delaying Differentiation Is a Shared Activity of HPV-16 E7 and HPV-6 E7 and Correlates with the Ability of HPV-6 E7 to Destabilize p130. pl30 is normally expressed in the G0 phase of the cell cycle and plays an important role in the maintenance of G0 arrest and regulation of differentiation (24-28). High-risk HPV E7 can decrease or delay differentiation (29, 30). Because both high-risk and low-risk E7s decrease p130 stability, they may also share the ability to decrease or delay differentiation. To test this, HFKs were infected with LXSN, L(16E7)SN, or L(6E7)SN and grown in C-K-SFM or C-K-SFM containing 2 mM CaCl2 for 24 and 48 h. Whole-cell lysates were analyzed by Western blot with antibodies to two differentiation markers, involucrin and keratin 10. After CaCl2 treatment, both HPV-16 E7 and HPV-6 E7 decreased/delayed the expression of involucrin and keratin 10 compared with control cells (Fig. 5A).
Fig. 5.
HPV E7-mediated decreased/delayed keratinocyte differentiation and correlation with the ability of HPV-6 E7 to destabilize p130. (A) Effect of HPV-16 E7 and HPV-6 E7 on differentiation. HFKs were transduced and grown as described in Fig. 1. Whole-cell lysates were analyzed on Western blots by using antibodies to involucrin, keratin 10, and GAPDH. The average ± SD of involucrin in three independent experiments, corrected for GAPDH, are as follows: relative to LXSN in C-K-SFM, 0.54 ± 0.17 (16E7) and 0.35 ± 0.18 (6E7); relative to LXSN in CaCl2 for 24 h, 0.46 ± 0.11 (16E7) and 0.45 ± 0.12 (6E7); relative to LXSN in CaCl2 for 48 h, 0.63 ± 0.15 (16E7) and 0.54 ± 0.19 (6E7). The average ± SD of keratin 10 in three independent experiments, corrected for GAPDH and relative to LXSN in CaCl2 for 48 h, was 0.04 ± 0.00 (16E7) and 0.15 ± 0.05 (6E7). (B) Effect of HPV-6 E7 and HPV-6 E7 mutants on differentiation. HFKs were infected and grown as in Fig. 1, and immunoblots were performed on whole-cell extracts by using antibodies to p130, involucrin, and GAPDH. The average ± SD of p130 in three independent experiments, corrected for GAPDH, was as follows: relative to LXSN in C-K-SFM, 0.31 ± 0.08 (6E7); relative to LXSN in CaCl2, 0.43 ± 0.02 (6E7), 1.08 ± 0.10 (H2AR4AH5A), 0.52 ± 0.13 (K9AD10A), 0.86 ± 0.09 (C25A), 0.48 ± 0.08 (D31A), and 0.88 ± 0.10 (L67R); the average ± SD of involucrin relative to LXSN in CaCl2 was 0.38 ± 0.19 (6E7), 0.89 ± 0.03 (H2AR4AH5A), 0.51 ± 0.14 (K9AD10A), 0.88 ± 0.14 (C25A), 0.43 ± 0.09 (D31A), and 0.79 ± 0.01 (L67R).
To determine whether the ability of HPV-6 E7 protein to decrease differentiation correlated with its ability to destabilize p130, HFKs were infected with LXSN, L(6E7)SN, or retroviruses encoding the HPV-6 E7 mutants, and whole-cell lysates were subjected to Western blot analysis with antibodies to p130 and involucrin. Those HPV-6 E7 mutants that destabilized p130 (K9AD10A and D31A) decreased involucrin expression, and those that lost the ability to destabilize p130 (mutants H2AR4AH5A, C25A, and L67R) also lost the ability to decrease differentiation (Fig. 5B).
Discussion
The pRB family members pRB, p107, and p130 play key roles in the regulation of cell growth, differentiation, and apoptosis (24-26, 31, 32). The three pRB family members are structurally related, with homologous highly conserved pocket regions that interact with the E2F family of transcription factors. pRB normally interacts with and inhibits the function of the activator E2Fs, E2F1-3, whereas p107 and p130 interact with the repressor E2Fs, E2F4 and E2F5 (32). HPV E7 protein, adenovirus E1A, and simian virus 40 large T antigen (SVLT), through the LXCXE motif in their CR2 domains, bind pRB family members, resulting in the release of E2F transcription factors and the transactivation of E2F-responsive genes (10, 33-35). pRB, p107, and p130 cooperate in cell-cycle control (24, 26). The pRB protein is highly expressed in proliferating cells and controls the G1-S transition, which is most critical for cell-cycle progression. p107 is abundant in S-phase cells and decreases in quiescent cells or terminally differentiated cells. In contrast, p130 is highly expressed in differentiated cells and quiescent cells and decreases when cells reach S-phase. Consistent with these reports, we found that p130 was increased and pRB and p107 were decreased in differentiated keratinocytes compared with cycling undifferentiated cells (Fig. 1B, compare LX lanes in C-K-SFM with LX lanes in 2 mM Ca2+).
In this study we report the finding that HPV-6 E7, like HPV-16 E7, can destabilize p130 in both undifferentiated and differentiated HFKs (Figs. 2, 3, 4, 5). Under the same conditions, HPV-6 E7 does not destabilize pRB or p107 (Figs. 1B and 2 A). Hence, the ratio of pRB family members upon differentiation would be changed in the presence of either low-risk or high-risk E7 protein compared with uninfected cells (Fig. 2 A). Because p130, E2F4, and HDAC form a corepressor complex on E2F-responsive promoters in quiescent cells and E2F4 is recruited to the nucleus by p130 (36-38), destabilization of p130 by HPV E7 may result in a replacement of the corepressor complex with E2F1, E2F2, and E2F3 on E2F-responsive promoters in quiescent cells or differentiated cells and induce the cells to reenter the cell cycle. The shared ability of low-risk and high-risk E7 to target p130 for degradation may be a critical common denominator among all HPVs, which allows them to replicate in the differentiated compartment.
HPV-6 E7 binds to p130 at least five times less efficiently than HPV-16 E7 (Figs. 1 A and 4D) but can destabilize p130 as efficiently as HPV-16 E7 (Figs. 2 A, 3, and 4C). The mechanism by which HPV-6 E7 and HPV-16 E7 destabilize p130 is unknown. Like HPV-16 E7, HPV-6 E7 decreases p130 half-life through the proteasome pathway (Fig. 3). Mutational analysis showed that HPV-6 E7H2AR4AH5A, an N-terminal mutant of HPV-6 E7, and mutant C25A, which disrupts the LXCXE motif in CR2, lost the ability to destabilize p130 (Figs. 4C and 5B). Similarly, HPV-16 E7H2P, an N-terminal mutant of HPV-16 E7, and Δ21-24, deleted in the LXCXE motif of HPV-16 E7, lose the ability to destabilize p130 (14), suggesting that HPV-16 E7 and HPV-6 E7 may share a common mechanism to destabilize p130. HPV-6 E7H2AR4AH5A and the C-terminal mutant L67R bind p130 as well as HPV-6 E7 but do not destabilize p130, suggesting that binding to p130 is necessary but not sufficient for destabilization of p130 by HPV-6 E7 (Fig. 4D). The HPV-6 E7L67R was generated because of a similar mutation in HPV-16 E7. HPV-16 E7L67R mutates a histone deacetylase binding site (39). This mutant, although it is able to bind pRB, is defective in its ability to abrogate pRB-mediated cell-cycle arrest, but its effect on p130 degradation has not been reported (39).
Like HPV-6 E7, SVLT also interacts with the three pRB family members through LXCXE but only destabilizes p130 (40). SVLT is a DnaJ chaperone with a J domain in its N-terminal region which recruits and activates the ATPase activity of the cellular chaperone protein Hsc70 (41). The J domain is required for p130 dephosphorylation and degradation (40, 42). HPV-16 E7-mediated transactivation of an E2F-responsive gene can be blocked by Hsj1, a human DnaJ protein (43). HPV E7 does not have J domain, but it does bind h-Tid-1, a member of the DnaJ family of proteins (44), and, therefore, may indirectly interact with chaperone proteins required for stabilization of p130. In addition, the phosphorylation status of p130 affects its stability. Cdk4/6 phosphorylation of p130 results in its proteasomal degradation in S phase (27), and phosphorylation by glycogen synthase kinase 3 stabilizes p130 in G0 (45). Thus, the mechanism of E7-induced degradation of p130 is likely to involve the interaction of E7 with cellular proteins that regulate p130 stability. This interaction might explain why differences in the affinity of high-risk and low-risk E7 for p130 are not reflected in differences in ability to target it for degradation.
Although HPV-6 E7 does not destabilize pRB in retrovirally transduced HFKs (Fig. 1B), HPV-6 E7G22D, a HPV-6 E7 mutant with a high-affinity pRB binding site, can destabilize pRB (Fig. 1B), indicating that HPV-6 E7 can destabilize the pRB protein upon sufficient physical interaction. However, binding of E7 to pRB is necessary but not sufficient for degradation of pRB, because E7 encoded by low-risk cutaneous HPV-1, adenovirus E1A, and SVLT bind pRB with high affinity but do not destabilize pRB (15, 35, 46, 47). Furthermore, HPV-16 E7 targets pRB for degradation via an activity that is in addition to pRB binding (13, 14). The similar activity of HPV-6 E7G22D and HPV-16 E7 with respect to pRB suggests that HPV-6 and HPV-16 E7 share the other sequences needed to target pRB for degradation. The fact that, in contrast to pRB interactions, both wild-type HPV-6 E7 and HPV-16 E7 can degrade p130 despite differences in affinity for p130 suggests that the mechanism involved in degrading pRB and p130 may be different. This difference may, in part, explain why HPV-6 E7 has only weak immortalizing activity and cannot alone immortalize HFKs (48).
p130 plays an important role in regulation of differentiation (25, 26, 28, 32). The ability of HPV-6 E7 to decrease differentiation correlates with its ability to destabilize p130 (Fig. 5B). Destabilization of p130 and decreasing differentiation are shared activities of HPV-16 E7 and HPV-6 E7 (Fig. 5A). These activities are important for the productive stage of HPV life cycle because the virus must either delay differentiation or induce differentiated cells to create an environment supportive of the amplification of viral DNA (2). Our results suggest a model whereby both low-risk and high-risk viruses solve the dilemma of amplifying their genome in the differentiation compartment of the epithelium, at least in part, by targeting p130 for degradation.
Materials and Methods
Cell Culture. HFKs were prepared as previously described (49) and kept in C-K-SFM (Invitrogen). Experiments on undifferentiated cells were conducted at 80% confluence. For differentiation conditions, cells were grown to confluence and incubated for 48 h in C-K-SFM containing 2 mM CaCl2. The retrovirus packaging cell lines PA317 and Phoenix-ampho cells (American Type Culture Collection) and COS-7 cells were grown in DMEM plus 10% FCS. Where noted, cells were treated with 0.25 mM cycloheximide (Sigma) or 50 μM MG132 (Sigma).
Retrovirus Infection. Retrovirus infections were performed as described in ref. 49. Third-passage HFKs were grown to ≈40% confluence in a 10-cm dish and infected with 5 ml of the recombinant retrovirus or parental virus (1 × 106 virus particles per ml). After G418 selection, the cells were expanded and used for experiments.
Mutational Analysis and Metabolic Labeling. The HPV-6 E7 mutants H2AR4AH5A, K9AD10A, C25A, D31A, and L67R were generated by using the QuikChange Site-Directed Mutagenesis kit (Stratagene) and confirmed by sequencing. HPV-6 E7 mutants were subcloned into pSG5 (Stratagene). DNA transfections for transient expression of HPV-6 E7 mutants were performed on COS-7 cells as described in ref. 50. Forty-eight hours after transfection, the cells were incubated in methionine and cysteine-free DMEM (Invitrogen) supplemented with 5% dialyzed FBS (HyClone) for 1 h, followed by incubation with 500 μCi (1 Ci = 37 GBq) of Trans 35S (MP Biomedical) in the above-described starvation medium. After 1.5 h, the cells were lysed in 0.1% Nonidet P-40, 0.25 M NaCl, 5 mM EDTA, and 50 mM Hepes (pH 7.5) to which 1% (vol/vol) Sigma protease inhibitor mixture and 0.5 mM DTT had been added (50). HPV-6 E7 was immunoprecipitated with a polyclonal antibody (anti-6E7), and the proteins were separated by SDS/PAGE. The gel was fixed in 10% glacial acetic acid and 30% methanol, treated with enlightening (PerkinElmer) for gel autoradiography enhancement, and dried before exposure to x-ray film.
Semiquantitative RT-PCR. Total RNA was extracted from HFKs by using Tri-Reagent (Molecular Research Center) as described in ref. 51. RT-PCR was performed following the protocol of the RT-PCR kit (Invitrogen). Briefly, 0.5 μg of RNA sample was digested with DNase I and incubated with reverse transcriptase at 50°C for 30 min. The samples were then subjected to PCR with primers as follows: p130, 5′TGTCACACCAGTTCCTGGAC and 3′GCAAAGTTGTTCCTGTCACC; GAPDH, 5′GAAGGTGAAGGTCGGAGTCA and 3′GAAGATGGTGATGGGATTTC. PCR parameters were as follows: 94°C for 5 min; 94°C for 30 sec, 55°C for 30 sec, 72°C for 30 sec for 22, 26, and 30 cycles, followed by extension at 72°C for 7 min. The amplified DNA products were separated on a 2% agarose gel and stained with ethidium bromide.
Western Blot Analysis. Whole-cell lysates were extracted by using SDS lysis buffer (20 mM Tris, pH 6.8/1% SDS/1 mM EDTA). Forty micrograms of the cell lysates was separated by SDS/PAGE. The protein was transferred to a nitrocellulose membrane (Bio-Rad) and probed with antibodies to involucrin (Sigma), pRB (BD Biosciences), p130 (BD Biosciences), p107 (Santa Cruz Biotechnology), or GAPDH (Chemi-Con).
Analysis of Protein Interactions. Jurkat nuclear extract (500 μg) was incubated with 10 μg of GST or GST fusion protein in binding buffer [20 mM Hepes/150 mM KCl/4 mM MgCl2/1 mM EDTA/0.02% Nonidet P-40/10% glycerol/0.035% 2-mercaptoethanol/1% (vol/vol) Sigma protease inhibitor mixture] and rocked for 1 h at 4°C. Glutathione-Sepharose beads (Amersham Pharmacia Biosciences) were added to each reaction and rocked for another 1 h. The beads were then washed with 1 ml of washing buffer (20 mM Hepes/150 mM KCl/4 mM MgCl2/1 mM EDTA/0.1% Nonidet P-40/10% glycerol/0.035% 2-mercaptoethanol/1% Sigma protease inhibitor mixture) five times and boiled with 2× SDS sample buffer, and the proteins were separated by SDS/PAGE. Proteins were transferred to the nitrocellulose membrane, stained with Ponceau red, and probed with antibodies to pRB, p107, and p130, as described under Western Blot Analysis.
Acknowledgments
We thank Grova Mae Lewis for technical support and Kathy Rundell for comments on the manuscript. This work was supported by National Institutes of Health Grant AI49254, the Lilly Center for Women's Health, and the Indiana Genomics Initiative of Indiana University, which is supported in part by Lilly Endowment Inc.
Author contributions: B.Z. and A.R. designed research; B.Z. and W.C. performed research; W.C. contributed new reagents/analytic tools; B.Z., W.C., and A.R. analyzed data; and B.Z. and A.R. wrote the paper.
Conflict of interest statement: No conflicts declared.
Abbreviations: HPV, human papillomavirus; CR, conserved region; HFK, human foreskin keratinocyte; GST, glutathione S-transferase.
References
- 1.zur Hausen, H. (2002) Nat. Rev. Cancer 2, 342-350. [DOI] [PubMed] [Google Scholar]
- 2.Longworth, M. S. & Laimins, L. A. (2004) Microbiol. Mol. Biol. Rev. 68, 362-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cheng, S., Schmidt-Grimminger, D. C., Murant, T., Broker, T. R. & Chow, L. T. (1995) Genes Dev. 9, 2335-2349. [DOI] [PubMed] [Google Scholar]
- 4.Thomas, J. T., Hubert, W. G., Ruesch, M. N. & Laimins, L. A. (1999) Proc. Natl. Acad. Sci. USA 96, 8449-8454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Flores, E. R., Allen-Hoffmann, B. L., Lee, D. & Lambert, P. F. (2000) J. Virol. 74, 6622-6631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Oh, S. T., Longworth, M. S. & Laimins, L. A. (2004) J. Virol. 78, 2620-2626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gage, J. R., Meyers, C. & Wettstein, F. O. (1990) J. Virol. 64, 723-730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Phelps, W. C., Munger, K., Yee, C. L., Barnes, J. A. & Howley, P. M. (1992) J. Virol. 66, 2418-2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dyson, N., Howley, P. M., Munger, K. & Harlow, E. (1989) Science 243, 934-937. [DOI] [PubMed] [Google Scholar]
- 10.Chellappan, S., Kraus, V. B., Kroger, B., Munger, K., Howley, P. M., Phelps, W. C. & Nevins, J. R. (1992) Proc. Natl. Acad. Sci. USA 89, 4549-4553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boyer, S. N., Wazer, D. E. & Band, V. (1996) Cancer Res. 56, 4620-4624. [PubMed] [Google Scholar]
- 12.Jones, D. L. & Munger, K. (1997) J. Virol. 71, 2905-2912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gonzalez, S. L., Stremlau, M., He, X., Basile, J. R. & Munger, K. (2001) J. Virol. 75, 7583-7591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Helt, A. M. & Galloway, D. A. (2001) J. Virol. 75, 6737-6747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Giarre, M., Caldeira, S., Malanchi, I., Ciccolini, F., Leao, M. J. & Tommasino, M. (2001) J. Virol. 75, 4705-4712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Longworth, M. S. & Laimins, L. A. (2004) J. Virol. 78, 3533-3541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Munger, K., Werness, B. A., Dyson, N., Phelps, W. C., Harlow, E. & Howley, P. M. (1989) EMBO J. 8, 4099-4105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Demers, G. W., Espling, E., Harry, J. B., Etscheid, B. G. & Galloway, D. A. (1996) J. Virol. 70, 6862-6869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hu, T. H., Ferril, S. C., Snider, A. M. & Barbosa, M. S. (1995) Int. J. Oncol. 6, 167-174. [DOI] [PubMed] [Google Scholar]
- 20.Demers, G. W., Foster, S. A., Halbert, C. L. & Galloway, D. A. (1994) Proc. Natl. Acad. Sci. USA 91, 4382-4386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heck, D. V., Yee, C. L., Howley, P. M. & Munger, K. (1992) Proc. Natl. Acad. Sci. USA 89, 4442-4446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Armstrong, D. J. & Roman, A. (1992) J. Gen. Virol. 73, 1275-1279. [DOI] [PubMed] [Google Scholar]
- 23.Armstrong, D. J. & Roman, A. (1993) Biochem. Biophys. Res. Commun. 192, 1380-1387. [DOI] [PubMed] [Google Scholar]
- 24.Nevins, J. R. (1998) Cell Growth Differ. 9, 585-593. [PubMed] [Google Scholar]
- 25.Lipinski, M. M. & Jacks, T. (1999) Oncogene 18, 7873-7882. [DOI] [PubMed] [Google Scholar]
- 26.Classon, M. & Dyson, N. (2001) Exp. Cell Res. 264, 135-147. [DOI] [PubMed] [Google Scholar]
- 27.Tedesco, D., Lukas, J. & Reed, S. I. (2002) Genes Dev. 16, 2946-2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ruiz, S., Segrelles, C., Bravo, A., Santos, M., Perez, P., Leis, H., Jorcano, J. L. & Paramio, J. M. (2003) Development 130, 2341-2353. [DOI] [PubMed] [Google Scholar]
- 29.Woodworth, C. D., Cheng, S., Simpson, S., Hamacher, L., Chow, L. T., Broker, T. R. & DiPaolo, J. A. (1992) Oncogene 7, 619-626. [PubMed] [Google Scholar]
- 30.Jones, D. L., Alani, R. M. & Munger, K. (1997) Genes Dev. 11, 2101-2111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cam, H. & Dynlacht, B. D. (2003) Cancer Cell 3, 311-316. [DOI] [PubMed] [Google Scholar]
- 32.Cobrinik, D. (2005) Oncogene 24, 2796-2809. [DOI] [PubMed] [Google Scholar]
- 33.Phelps, W. C., Bagchi, S., Barnes, J. A., Raychaudhuri, P., Kraus, V., Munger, K., Howley, P. M. & Nevins, J. R. (1991) J. Virol. 65, 6922-6930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ikeda, M. A. & Nevins, J. R. (1993) Mol. Cell. Biol. 13, 7029-7035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Helt, A. M. & Galloway, D. A. (2003) Carcinogenesis 24, 159-169. [DOI] [PubMed] [Google Scholar]
- 36.Takahashi, Y., Rayman, J. B. & Dynlacht, B. D. (2000) Genes Dev. 14, 804-816. [PMC free article] [PubMed] [Google Scholar]
- 37.Chestukhin, A., Litovchick, L., Rudich, K. & DeCaprio, J. A. (2002) Mol. Cell. Biol. 22, 453-468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rayman, J. B., Takahashi, Y., Indjeian, V. B., Dannenberg, J. H., Catchpole, S., Watson, R. J., te Riele, H. & Dynlacht, B. D. (2002) Genes Dev. 16, 933-947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brehm, A., Nielsen, S. J., Miska, E. A., McCance, D. J., Reid, J. L., Bannister, A. J. & Kouzarides, T. (1999) EMBO J. 18, 2449-2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stubdal, H., Zalvide, J., Campbell, K. S., Schweitzer, C., Roberts, T. M. & DeCaprio, J. A. (1997) Mol. Cell. Biol. 17, 4979-4990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Srinivasan, A., McClellan, A. J., Vartikar, J., Marks, I., Cantalupo, P., Li, Y., Whyte, P., Rundell, K., Brodsky, J. L. & Pipas, J. M. (1997) Mol. Cell. Biol. 17, 4761-4773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lin, J. Y. & DeCaprio, J. A. (2003) J. Biol. Chem. 278, 46482-46487. [DOI] [PubMed] [Google Scholar]
- 43.Sheng, Q., Denis, D., Ratnofsky, M., Roberts, T. M., DeCaprio, J. A. & Schaffhausen, B. (1997) J. Virol. 71, 9410-9416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schilling, B., De-Medina, T., Syken, J., Vidal, M. & Munger, K. (1998) Virology 247, 74-85. [DOI] [PubMed] [Google Scholar]
- 45.Litovchick, L., Chestukhin, A. & DeCaprio, J. A. (2004) Mol. Cell. Biol. 24, 8970-8980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ciccolini, F., Di Pasquale, G., Carlotti, F., Crawford, L. & Tommasino, M. (1994) Oncogene 9, 2633-2638. [PubMed] [Google Scholar]
- 47.Schmitt, A., Harry, J. B., Rapp, B., Wettstein, F. O. & Iftner, T. (1994) J. Virol. 68, 7051-7059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Halbert, C. L., Demers, G. W. & Galloway, D. A. (1992) J. Virol. 66, 2125-2134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang, B., Spandau, D. F. & Roman, A. (2002) J. Virol. 76, 220-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Armstrong, D. J. & Roman, A. (1997) Virology 239, 238-246. [DOI] [PubMed] [Google Scholar]
- 51.Zhang, B., Laribee, R. N., Klemsz, M. J. & Roman, A. (2004) Virology 329, 189-198. [DOI] [PubMed] [Google Scholar]