Abstract
Magnifying Genomes (MaGe) is a microbial genome annotation system based on a relational database containing information on bacterial genomes, as well as a web interface to achieve genome annotation projects. Our system allows one to initiate the annotation of a genome at the early stage of the finishing phase. MaGe's main features are (i) integration of annotation data from bacterial genomes enhanced by a gene coding re-annotation process using accurate gene models, (ii) integration of results obtained with a wide range of bioinformatics methods, among which exploration of gene context by searching for conserved synteny and reconstruction of metabolic pathways, (iii) an advanced web interface allowing multiple users to refine the automatic assignment of gene product functions. MaGe is also linked to numerous well-known biological databases and systems. Our system has been thoroughly tested during the annotation of complete bacterial genomes (Acinetobacter baylyi ADP1, Pseudoalteromonas haloplanktis, Frankia alni) and is currently used in the context of several new microbial genome annotation projects. In addition, MaGe allows for annotation curation and exploration of already published genomes from various genera (e.g. Yersinia, Bacillus and Neisseria). MaGe can be accessed at http://www.genoscope.cns.fr/agc/mage.
INTRODUCTION
During the last few years, the genomes of ∼280 bacteria have been completely sequenced, leading to an enormous demand for fast and accurate analysis of the resulting biological sequences. The information obtained from a genome depends largely on the quality of the annotation of its complete sequence [mainly, CoDing Sequence (CDS) identification and function prediction]. The quality of the annotation itself depends on the implemented bioinformatics tools and on the work and time dedicated to it by the annotators (1). A common annotation process starts with the use of highly automatic prediction of genes and biological functions of their product. Because of the large number of genomes currently annotated, part of this data from automatic methods is often directly stored in public databanks, leading to the propagation of existing annotation errors (2). Actually, validation of the first set of automatic data involves tedious manual work in which an expert performs additional searches and analyses. The first steps of such annotation processes obviously require powerful automatic tools. The recent publication of BaSys, a web software which permits a complete automatic annotation process for a new bacterial genome, highlights this need (3). In addition, databases for storage and management of heterogeneous data, together with complex but user-friendly interfaces, are also essential to manually annotate a genome efficiently.
To achieve the annotation of a complete genome, a number of annotation tools have been designed, with the first, strictly automatic systems focusing on human readable HTML reports (4–6). Since then, many efforts have been made in terms of project management (i.e. complex biological data models and integrated databases), spectrum of bioinformatics tools applied (including multiple genome comparison-based annotation strategies), sophistication of the user interfaces (extensive visualizations, fully interactive graphical interfaces) and the presence of convenient features such as data editors. Examples of commonly used annotation platforms are given by commercial systems, such as ERGO (7) or Pedant-Pro (successor of PEDANT), and open source systems, such as Artemis (8), GenDB (9) or Manatee (TIGR, unpublished). In addition, some newly developed tools perform automatic tasks for contig-assembly analysis together with automatic annotation of the successive assembly updates, thus allowing annotation of a genome to start during the finishing phase of the sequencing process (10,11).
In the study of microbial genomes, the increasing number and the diversity of sequenced genomes have led to the development of novel methods for the contextual analysis of genes and proteins, to detect functional constraints on genome evolution (12–15). Although results from these methods clearly demonstrate the added-value of genomic context analysis in the process of prokaryotic genome annotation (16), no existing annotation systems, except perhaps ERGO, systematically integrates them. To address this problem, we have developed a new microbial genome annotation system, called MaGe (Magnifying Genomes), which shares several functionalities with existing systems, mainly (i) an automatic annotation process including syntactic and functional annotations together with classification inferences, (ii) a relational database used to store the sequences and the analysis results, (iii) a web interface allowing multiple users to simultaneously annotate a genome and to query the database (e.g. search for functionalities and/or gene content between related species) and (iv) several connectivities and/or integration of other systems and databases. Since MaGe has been developed by people who are involved in manual expert annotation themselves, it offers original features such as a graphical gene context exploration. In order to detect gene groups that share locally conserved chromosomal organization, the annotated genome is compared with publicly available other ones. Synteny map visualization is then useful to quickly pinpoint genome rearrangements between related bacterial species. In addition, a customizable user-friendly gene editor has been developed to take into account the specificities of each bacterial annotation project (functional classifications, comparisons to reference genome data, etc.). In the context of the expert validation of the automatic predictions, the MaGe cartographic representations have already been shown to improve notably the final annotation quality (17,18). Our system is currently being used for the annotation or re-annotation of more than 16 microbial genomes (http://www.genoscope.cns.fr/agc/mage).
The MaGe system consists of three main components which are described in the following sections: (i) a set of bioinformatics methods currently implemented in the system, (ii) a relational database which contains sequence data and the results from the set of analysis methods and (iii) a graphical web interface. The setup and the management of a new annotation project are described in the last section of this paper.
BIOINFORMATICS METHODS
Prediction of genes and functional annotation tools
The annotation process begins with the FASTA formatted contig(s) on which appropriate algorithms for the identification of coding regions and various genetic elements are executed. A preliminary and essential step for a new genome annotation (anonymous DNA sequence) consists of construction of appropriate gene models. Our procedure is based on codon usage analysis and leads to the construction of gene models fitting well with the input genomic data (19). These models are then used in the core of the AMIGene gene-finding program (19), leading to more accurate prediction of small genes and/or atypical gene composition (20). In order to increase the reliability of the AMIGene results in terms of start codon positions, we have integrated the RBSfinder program into our software, which searches for ribosome-binding sites in the extragenic regions (21). tRNAscan-SE (22) has also been included in the annotation pipeline for the prediction of tRNA-encoding genes. In addition, other RNA structures, such as small RNAs and riboswitches, are identified using the Rfam database (23). We have also integrated the Petrin program (24) to identify putative rho-independent transcription termination sites. Other genetic elements such as intrachromosomal repeats are detected using the method described by Achaz et al. (25).
Extracted gene products are subjected to exhaustive bioinformatics analysis, including the gapped blastP algorithm (26) for general-purpose homology searches against the full non-redundant protein sequence databank UniProt (27). Queries are also submitted to more sensitive sequence similarity search tools, using motif/pattern/protein families compiled in the InterPro database (28) and the COG databank (29). In addition, genes coding for enzymes are classified using the PRIAM software (30), the results of which are used for metabolic pathway reconstruction (see below). Finally, functional assignations are also made using the HAMAP (High quality Automated and Manual Annotation of Microbial Proteomes) web server (http://www.expasy.org/sprot/hamap) (31). In terms of predicted structural features, we search for alpha-helical transmembrane regions with the tmHMM program (32), and signal peptides with SignalP (33). Highly sensitive comparison of each predicted protein with the SCOP database of known structural domains (34,35) is also carried out. Finally, to predict probable subcellular localization of the annotated protein in the cell (Integral Inner Membrane Proteins, IIMPs), another original approach developed by our group is applied (36).
Along with the fast growing number of sequenced prokaryotic genomes, an additional method that relies on gene context rather than on sequence similarity only has been developed in our group: synteny computation, which is undoubtedly one of the most original components of the MaGe system.
Comparative genomics through synteny analysis
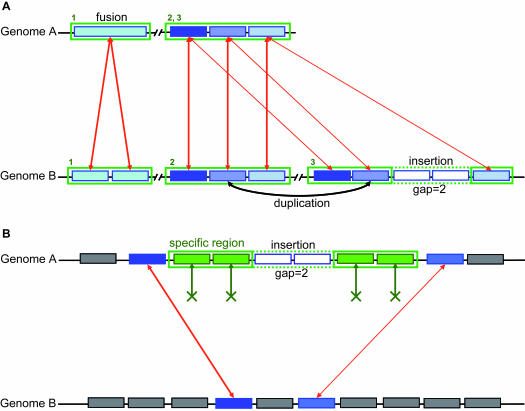
For assigning function to novel proteins, gene context approaches can complement the classical homology-based gene annotation. These ‘nonhomology-based’ inference methods rely on the fact that functionally associated proteins are encoded by genes that share similar selection pressures. In most of the proposed methods (14,37,38), orthologous pairs of proteins satisfy the bi-directional best hit (BBH) criterion, based on blast and/or Smith–Waterman (39) comparisons of complete genomes with one another. An innovative aspect of our approach is that we offer the possibility of retaining more than one homologous gene. Pairwise comparisons between predicted protein sequences of the studied genome and the proteins of another genome allow computation of ranked hits and BBH (for each protein, the three best hits are kept). Putative orthologous relations between two genomes are defined as gene couples satisfying the BBH criterion or an alignment threshold (generally, a minimum of 30% sequence identity on 80% of the length of the smallest protein). These relations are subsequently used to search for conserved gene clusters, e.g. synteny groups among several bacterial genomes. Our method, called the Syntonizer, is based on an exact graph-theoretical approach (40). This method allows for multiple correspondences between genes and, thus, paralogy relations and/or gene fusions are easily detected. All possible kinds of chromosomal rearrangements are allowed (inversion, insertion/deletion, see Figure 1A). A ‘gap’ parameter, representing the maximum number of consecutive genes which are not involved in a synteny group, is generally set to five genes. Comparative annotations of a new bacterial genome involve the computation of these synteny groups across all available microbial proteomes [NCBI databank, RefSeq section (41)].
Figure 1.
Synteny group and specific region detection. (A) Example of synteny groups (rectangles with green borders) between two genomes A and B. Syntonizer software allows multiple correspondences between genes (red arrows, e.g. blastP similarity results) to detect duplications and gene fusion/fission events. Local rearrangements (inversion; insertion/deletion) are allowed in our method. The gap parameter defines the number of consecutive genes not involved in synteny. The first synteny group shows a gene fusion event in genome A. The second synteny group shows a perfect gene order conservation in the two compared genomes. The third one is the result of a duplication in genome B together with the insertion of two genes (the gap parameter is then equal to 2). (B) Example of a specific region (rectangle with green border) in the genome A. Co-localized genes (plain green rectangles in genome A) have no ortholog in the compared genome B. Lack of correspondence relations (green arrows) are explicitly represented. A gap parameter represents the maximum number of consecutive genes with homologies in the compared genome. In this example, two genes are inserted (the gap parameter is then equal to 2).
From these comparison results, we define a species-specific gene as a gene having no ortholog in the compared species (significant similarities were not detected). This allowed us to compute specific regions between the genome under analysis, and a set of genomes selected for their phylogenetic proximity. Such regions are defined by at least two consecutive specific genes (Figure 1B). Insertion of genes which have homologies in the compared species is allowed in a specific region. A ‘gap’ parameter, representing the maximum number of consecutive genes which are not involved in a specific region (i.e. which have homologies), is generally set to two genes.
The predictive power of chromosomal clustering, which has already been demonstrated in several recent publications [see for example, (16,42,43)], may help the expert annotators to assign putative functions, even in the absence of relevant sequence similarity.
Automatic functional assignations
The computational methods described above form the core of the MaGe processing pipeline. This fully automated first round of annotation ends with a functional assignation procedure. The main purpose of this step is to infer, as precisely as possible, specific function(s) for each individual gene by the completeness of gene products, gene names, Enzyme Commission (EC) numbers and functional classes when possible (Supplementary Figure 1). Assignation of Gene Ontology terms (44) is directly obtained from the InterProScan results (28). Our procedure starts with the evaluation of the similarity results and gives a priority to the reference annotations of model organism(s), then InterPro domains and blast results against UniProt (Swiss-Prot curated annotations are preferably kept). At each step (Supplementary Figure 1), pairwise genome comparisons are evaluated taking synteny results into account (i.e. if two homologous genes are involved in a synteny group, the similarity threshold is lower). When a gene is member of more than one synteny group (i.e. in the case of multiple correspondences, see below), we assign its putative function using the corresponding gene which is involved in the synteny group sharing the most genes. These automatic assignments are just suggestions for functional role of the annotated genes; with the help of the MaGe graphical interface, the final decision is obviously up to the expert annotator.
Sequence and annotation updates
The finishing phase term is highly variable depending on the genome coverage by the DNA libraries, the number of clones sequenced during the random phase and the number of repeated sequences present in the genome. To give researchers a quicker access to genome information, it is therefore important to start the annotation of a genome during the finishing phase of a project. Progression of this phase can involve the alteration of numerous CDSs due to sequence gap closure and the addition, deletion or modification of one or more bases. We have therefore developed a procedure which maps annotated features from an earlier version to the updated version of the genome sequence assembly. For each update, newly predicted genes are compared with the previous set of annotated genes. Only corresponding genes which align perfectly are mapped, taking into account a possible modification of the start codon position. In the case of multiple correspondences (e.g. duplicated genes), the genomic context is explored to map only genes having a conserved neighborhood. At the end of the process, expert annotations of the mapped genes are transferred to the new version of the database. Then, a report allows one to retrieve locus name (i.e. label) correspondences between mapped genes, genes that no longer exist and newly predicted genes (after the gap closure).
The synteny results can be used in an alternative way to identify one (or several) possible supercontig organization on the final chromosome by comparison with a phylogenetically related complete genome (hereafter, reference genome) (Supplementary Figure 2). This process, which may be very helpful for the progression of the finishing phase, is achieved in two ways: (i) finding the best supercontig order (and orientation) which maximizes a global conservation of the co-linearity between the reference genome and the draft of the sequenced genome (ii) looking for significant synteny groups on the supercontig ends which are neighbors on the reference genome (Supplementary Figure 2A and B). All proposed results must be experimentally validated by PCR analysis.
Metabolic pathway reconstruction
The set of annotated EC numbers provides an access to the chemical repertoire of the organism and allows for reconstruction of metabolic pathways. Two sets of reference metabolic pathways are used and linked to the MaGe annotations (Table 1). A dynamic request to the KEGG server (45) allows one to visualize colored EC numbers on the metabolic diagrams (see ‘Metabolic pathway visualization’). In addition, for each prokaryotic genome being annotated in MaGe, an instance of the BioCyc scheme (built on an object database system, Ocelot) is created (46,47). The Pathway Tools software analyzes the list of predicted EC numbers and the product name of the CDSs, to identify a set of possible reactions which are subsequently matched against all pathways from MetaCyc (48). Each pathway is then evaluated and retained or not for the studied organism. At the end of the process, a PGDB is built (this new database is usually named organismCyc, i.e. AcinetoCyc, FrankiaCyc, etc.) and connected to the MaGe interface. In a second step, the Pathway Hole Filler program (49) is executed in order to find putative gene candidates for missing enzymes in the previously predicted metabolic pathways. KEGG and BioCyc metabolic network tools are clearly complementary, both in terms of metabolic datasets and of metabolic pathways graphical representation (see below). However, these two homology-based metabolic pathway reconstruction systems cannot predict novel pathways. For this purpose, the MaGe system is connected to the Pathway Hunter Tool (PHT) web server (50). Starting from the set of MaGe annotated EC numbers, and a source/destination metabolite pair selected by the user, the shortest metabolic pathways (k-shortest pathways) are computed by PHT (Table 1). Alternative routes can then be evaluated for biological significance. Used together, these three methods are helpful to infer functional coupling of genes which participate in the same cellular process.
Table 1.
Main features of the metabolic data sets used in MaGe
| KEGG | BioCyc | PHT | |
|---|---|---|---|
| Enzyme, reaction data | Ligand | Enzyme database + in-house curation | Ligand and Brenda databases |
| Pathway data | Multi-organisms, generic representation | Organism specific, experimentally validated (MetaCyc) | No |
| Gene/reaction correspondences | EC numbers | EC numbers + product names | EC numbers |
| Pathway reconstruction | Homology based (EC number mapping) | Homology based (pathway selection algorithm) | Ab initio reconstruction (k-shortest pathways) |
| Hole detection | No | Yes + Pathway Hole Filler | No |
| Data management | Flat files | Object Database, Ocelot | ? |
| MaGe integration | Web service | Local installation | Web service |
| MaGe annotation updates | Dynamic | Re-execution of Pathway Tools | Dynamic |
This table shows the main features of the three metabolic pathway reconstruction systems integrated in MaGe: KEGG (45), BioCyc (47) and PHT (Pathway Hunter Tool) (50). KEGG and BioCyc use the homology method to reconstruct metabolic pathways. PHT, which uses an ab initio algorithm to compute the shortest pathways between two metabolites, helps the user to find alternative routes.
Both KEGG and BioCyc predict pathways by comparing the enzymes within a given genome against the known set of reference pathways. However, while the very large KEGG metabolic maps are mosaics that combine pathways and reactions from many organisms (45), MetaCyc pathways describe single metabolic routes that have been experimentally elucidated in specific organisms (48) (Table 1). This latter metabolic resource is obviously more accurate than KEGG. However, in terms of completeness, the KEGG maps are sometimes useful to make hypotheses on potential alternative metabolic pathways. Another main difference between the two systems relies on the pathway prediction algorithm, which simply consists of coloring a set of static map diagrams to indicate the presence of enzymes within a KEGG map, whereas BioCyc uses the PathoLogic software which can predict whether a MetaCyc pathway is present or absent in the analyzed organism. While expert annotation is going on in the MaGe system, there is an automatic update of the metabolic pathway reconstruction in KEGG, but the corresponding BioCyc PGDB needs to be recomputed (Table 1). Finally, the Enzyme Commission system is widely used to assign enzymatic activity to gene products, but some limitations exist. Based on the experimental enzyme characterization, assignment of novel EC numbers is manually performed by an expert commission. Unfortunately, numerous new reactions are not fully characterized and are unlikely to receive EC numbers in a short time. Furthermore, an EC number may correspond to multiple reaction formulae and can then cause ambiguities in distinguishing substrate specificity. Partial EC numbers, such as 1.1.1.-, may also cause imprecision because they are used with two different meanings: (i) the substrate specificity of the enzyme is ‘unknown’, (ii) the exact activity of the enzyme is known but an EC number is ‘not yet available’. Therefore, the use of partial EC numbers may lead to erroneous assignment of enzymes to pathway reactions, resulting in incorrect enzyme-reaction associations (51). In BioCyc, only complete EC numbers are used in the process of metabolic pathways reconstruction.
THE RELATIONAL DATABASE
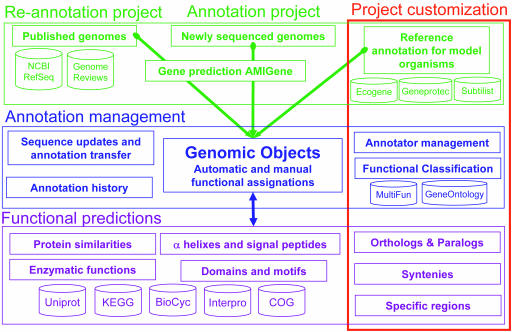
The MaGe system uses a relational database called PkGDB (Prokaryotic Genome DataBase) for storing, modifying and accessing very large datasets. A simplified view of the PkGDB data model is depicted in Figure 2. The core tables store information on organisms, sequences and genomic objects (RNA genes, CDSs, etc.). These annotations are coming from three main origins. First, in the case of a newly sequenced genome, gene prediction tools are run and their results are compiled as new genomic objects to be annotated (see ‘Bioinformatics Methods’). Second, the complete bacterial proteomes are extracted from the NCBI RefSeq (41) and EBI Genome Reviews (52) databanks and stored in PkGDB. Third, annotations of several interesting complete bacterial genomes (i.e. which could be improved in the context of a new MaGe genome project) are submitted to a human computer-assisted process, in order to improve their qualities [correction of inconsistencies, re-annotation of pseudogenes, searching for putative missing genes or wrongly annotated genes (20)]. These enriched sets of annotation data are subsequently used to search for synteny groups in the genome(s) to be annotated. Around this core structure, additional tables store functional prediction results (see ‘Bioinformatics Methods’). To retrieve and query results, each databank (e.g. UNIPROT, InterPro, COG, BioCyc, KEGG/LIGAND) used by a method is indexed in one or several tables (Figure 2). The system architecture permits easy integration of new method results. Finally, the database architecture supports integration of automatic and manual annotations and records a history of all the modifications. Automatic annotation can be updated at any stage of the project. Furthermore, sequence updates and annotation transfer are stored in the database, allowing users to check mapped genes, new genes and genes that do not exist anymore. Three user groups are defined: ‘curator’, ‘annotator’ and ‘guest’. Users having an ‘annotator’ status cannot directly save a novel annotation but instead, a mail is automatically sent to the ‘curators’ for a final review. In case of a public project, a ‘guest’ login status is activated and annotations are immediately made available. Anonymous users can then query and browse the data using the MaGe's functionalities.
Figure 2.
Simplified PkGDB relational model. PkGDB is made of three main components: sequence and annotation data (in green), annotation management (in blue) and functional predictions (in purple). Sequences and annotations come from three sources namely public databanks, sequencing centers and specialized databases focused on model organisms. For genomes of interest, a (re)-annotation process is performed using AMIGene (19) and leads to the creation of new ‘Genomic Objects’. Each ‘Genomic Object’ and associated functional prediction results are stored in PkGDB. The database architecture supports integration of automatic and manual annotations, and management of a history of annotations and sequence updates. The core of PkGDB can be supplemented by other tables to take into account genome project specificities (‘Project customization’, red rectangle).
These main components of PkGDB can be supplemented by other relational tables which take the specificities of each annotation project into account (tables surrounded by a red rectangle in Figure 2). For each MaGe project, a set of reference organism annotations can be defined and integrated in the automatic and manual annotation process. For this purpose, continuously updated annotations (often using experimental evidence) from specialized databases [e.g. GenprotEC (53) and Ecogene (54) for Escherichia coli, PseudoCAP for Pseudomonas aeruginosa (55) and Subtilist for Bacillus subtilis (56)] can be stored in PkGDB. Depending on the organism properties, various functional classifications can be integrated in the thematic database: either an already defined classification [e.g. MultiFun (57), B.subtilis (56), TIGR (58), COG (29) or FunCat (59) classifications] or a completely new one. Finally, parameters used to compute similarities, synteny groups and specific regions take into account the phylogenetic proximity of the newly annotated genome to the available bacterial proteomes. The corresponding results are stored in the database (Figure 2) and then explored in the MaGe graphical representation of the synteny maps and/or the ‘PhyloProfile and Synteny’ and ‘Specific regions’ functionalities (see ‘Genome browser and synteny maps’).
FUNCTIONALITIES OF THE WEB INTERFACE
The MaGe web interface consists of numerous dynamic web pages containing textual and graphical representations for accessing and querying data (Supplementary Figure 3). A specific effort has been made in terms of graphical representations of available analysis results, to make the manual expert annotation easier and more efficient.
Genome browser and synteny maps
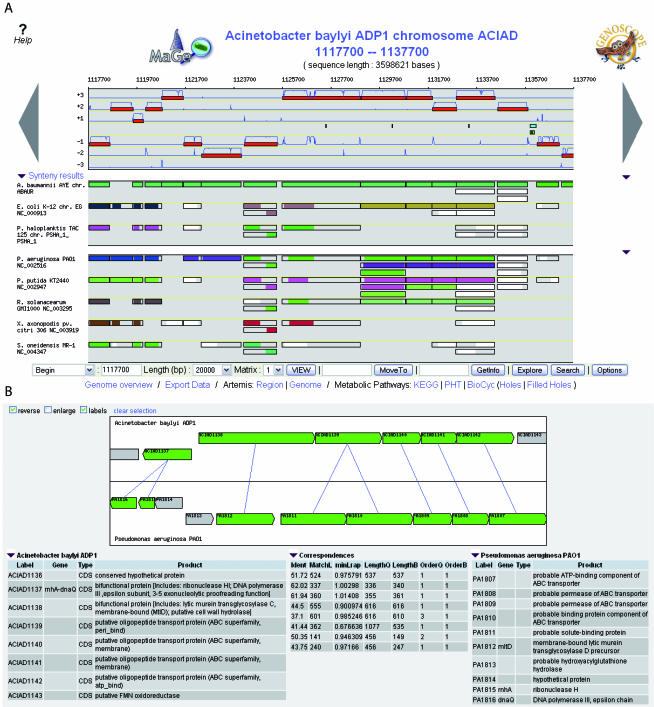
MaGe's main innovative functionality is a cartographic gene context exploration of the studied genome compared against all the available microbial genomes. This comparative genomics environment provides quality checks for both the automatic annotations and manual analysis. In Figure 3A, the first graphic map (genome browser) contains the complete Acinetobacter baylyi ADP1 chromosome, over which the user can navigate with complete freedom (moving and zooming functionalities). The predicted coding genes are drawn, on the six reading frames, in red rectangles together with the coding prediction curves which are computed with the selected gene model.
Figure 3.
MaGe's genome browser and synteny maps. (A) The Acinetobacter ADP1 chromosomal segment, extending between positions 1 117 700 and 1 137 700 bp, is represented on this graphical map of the MaGe interface developed on our database. Annotated CDSs are represented in the six reading frames of the sequence by red rectangles, and coding prediction curves are superimposed on the predicted CDSs (blue curves). The synteny maps, calculated on a set of selected genomes (three from PkGDB database and five from NCBI databank), are displayed below. In contrast with the graphic interface of the Acinetobacter ADP1 genome, there is no notion of scale on the synteny map: a rectangle has the same size of the CDS which is exactly opposite in the ADP1 genome, and it represents a putative ortholog between one CDS of the compared genome and one CDS of the Acinetobacter ADP1 genome. In addition, rectangles are colored depending on the part of the protein which aligns with the corresponding ADP1 protein. If, for several CDSs co-localized on the ADP1 genome, there are several co-localized orthologs in the compared genome, the rectangles will all be of the same color; otherwise, the rectangle is white. A group of rectangles of the same color thus indicates synteny between Acinetobacter ADP1 and the compared genome. (B) This second graphical representation of synteny has been obtained by clicking on one rectangle of the synteny maps (here one of the eight P.aeruginosa green genes). It allows the user to see how homologous genes, in a synteny group, are organized: here, one fusion event in Acinetobacter ADP1 (ACIAD1137: rnhA+dnaQ), a duplication of two genes (PA1810 and PA1811) and an insertion of two genes (PA1814 and PA1813) in P. aeruginosa. In addition, ACIAD1138 is similar to the mtlD gene of P.aeruginosa only in its N-terminal part, the second part of the protein sharing similarity with a COG family annotated as ‘LysM-repeat proteins and domains’ (COG1388).
The two following maps are representations of the synteny results (Figure 3A): each line shows the similarity results between the genome being annotated (i.e. Acinetobacter ADP1) and a given genome (i.e. the first three lines of the second synteny map are with three Pseudomonas species). The first synteny map is a selection of the hundred curated genomes in PkGDB to date (see ‘The relational database’), and the second one is a selection of the 280 complete prokaryotic proteomes available in public databanks. On these maps, a rectangle flags the existence of a gene in a compared organism which is similar to the opposite gene in the annotated genome. If, for several co-localized CDSs on the annotated genome, there are several co-localized homologs on the compared genome, the rectangles will all be of the same color; otherwise, the rectangle is white. A group of rectangles of the same color thus indicates a synteny group. This graphical representation allows the user to quickly see if the part of the genome being annotated shares similarities and locally conserved organization with the selected bacterial sequences (‘Options’ functionality). As shown in Figure 3A, this is the case with Acinetobacter baumannii, and the two selected Pseudomonas species, with the P.aeruginosa genome sharing the most important number of synteny groups in this part of the ADP1 genome.
In contrast with the genome browser, there is no notion of scale on the synteny maps: to see how homologous genes are organized in a synteny group, the user can simply interact on one gene in a given synteny group. For example, by clicking on one rectangle of the green synteny group between P.aeruginosa and Acinetobacter ADP1, both corresponding genome regions of the compared organisms are shown and orthologs are linked, allowing the user to explore fusion/fission, duplication, inversion and insertion/deletion of genes (Figure 3B). In our example, one interesting rearrangement appears clearly: the two P.aeruginosa homologs of the ADP1 CDS named ACIAD1137 are co-localized and transcribed on the same strand, showing that the corresponding biological functions (i.e. ribonuclease H and epsilon subunit of the DNA polymerase III) have been fused in the genome of Acinetobacter ADP1. Actually, the graphical representation of the synteny maps itself is also useful for detecting this kind of interesting feature: on each line, a rectangle has the same size as the corresponding annotated CDS in the studied genome. In addition, rectangles are colored depending on the part of the protein which aligns with the corresponding ADP1 protein (Figure 3A). It then becomes easy to see that ACIAD1137 has always two homologous genes in all the selected compared genomes (except with A.baumannii). However, the corresponding ADP1 protein aligns only on its N-terminal part with the first corresponding genes (rnhA gene), and on its C-terminal part with the second corresponding genes (dnaQ gene). Finally, these two homologous genes are involved in a synteny group containing eight genes in Pseudomonas species, six genes in Ralstonia solanacearum, three genes in E.coli, Pseudoalteromonas haloplanktis and Xanthomonas axonopodis, and only two genes in Shewanella oneidensis. In these last four bacteria, dnaQ and rnhA genes are transcribed anti-clockwise and in R.solanacearum, dnaQ gene is not co-localized with the rnhA gene (white rectangle). This raises interesting evolutionary questions concerning the fusion of these two biological functions involved in DNA replication.
Just below the three maps, several functionalities are available, such as the exploration of synteny results or annotated data using keywords (‘Explore’), the search for similarities using blast functionalities (26), or for patterns in DNA or protein sequences (‘Search’). At any time the user can download data in different common file formats (FASTA, EMBL, GenBank, etc.) or extract part of its DNA sequence (‘Export Data’). He/she can work with Artemis software (8) which is very useful for modifying erroneous start codon positions, for example, or explore KEGG (45), BioCyc (47) or PHT (50) metabolic pathways with MaGe annotations as input (see ‘Metabolic pathway reconstruction’ and ‘Metabolic pathway visualization’).
Automatic versus manual annotation
In spite of the continuous improvement in the overall quality of bioinformatic methods, some difficulties in gene functional assignment can hardly be addressed in a completely automatic way. Most notably, the problem of error propagation in databases (60), which is today very strong in the context of common ‘industrial’ production of genome data, can only be solved with human intervention. Thus, the set of automatic annotations produced by any system should be considered only as a useful first approximation.
In MaGe, automatic annotation is always available in the gene editor (‘Automatic annotation’, Supplementary Figure 4). This information is updated each time a new version of the complete genome sequence becomes available. Improvement of the annotation data quality can be made in the ‘Gene Validation’ section of the gene editor, which allows the user to modify, delete and add information. Annotation homogenization is achieved via a procedure which is automatically launched when gene annotations are saved in the database. This allows for a minimal checking of the annotation coherence. For instance, ‘ProductType’ field must be equal to enzyme if an EC number is given (Supplementary Figure 4). A further advantage of MaGe's manual annotation system is that it enables a group of users, possibly at different locations, to easily co-operate on specific annotations: email addresses of either the last annotator (in the gene editor) or all the different annotators for a specific gene (in the ‘History’ functionality, data not shown) are available. To help the user in the manual annotation of a gene, a summary of available method results are visualized in a completely customizable list (Supplementary Figure 4). This part of the gene editor is essentially a workbench for curation and analysis of a single gene or its protein family. It contains information on gene prediction (AMIGene) and duplication results, similarity results against annotation data from reference genomes, Swiss-Prot curated annotations and TrEMBL databank, synteny results using PkGDB curated proteomes and complete prokaryotic genomes stored in the NCBI RefSeq section (about 280 to date). These comprehensive synteny results are useful to update, if necessary, the list of currently selected genomes which are visualized in the synteny maps. Other tables include enzymatic function predictions (PRIAM results), similarity results against COG (COGnitor), protein domain databanks (InterProScan) and HAMAP families. Finally, clues on the probable protein localization are given by the SignalP and tmHMM results (Supplementary Figure 4). For each set of results, external links, if any, are provided (NiceProt, NiceEnzyme, InterPro and COG databases, HAMAP families). In addition, direct interaction with PubMed (only if the field ‘PubMedID’ is filled), and with KEGG (external link) or BioCyc (internal link) metabolic pathway(s), is available. This integrative strategy allows annotators to quickly browse functional evidence, tracking the history of a function and checking the gene context conservation with an orthologous gene having an experimentally demonstrated biological function.
Metabolic pathway visualization
Using MaGe, metabolic pathway exploration is accessible through three different tools: KEGG, BioCyc and PHT. Starting from the set of predicted and/or validated EC numbers, metabolic maps are dynamically drawn via a request to the KEGG web server. A color-based code enables comparison of the studied organism enzyme content with a selected related organism, with enzymes encoded by genes localized on the current MaGe genome region highlighted in yellow (Figure 4C). The useful representation of KEGG interconnected metabolic pathways is supplemented by the organism-specific PGDB built with the BioCyc system and an access to a PHT web form (see ‘Metabolic pathway reconstruction’).
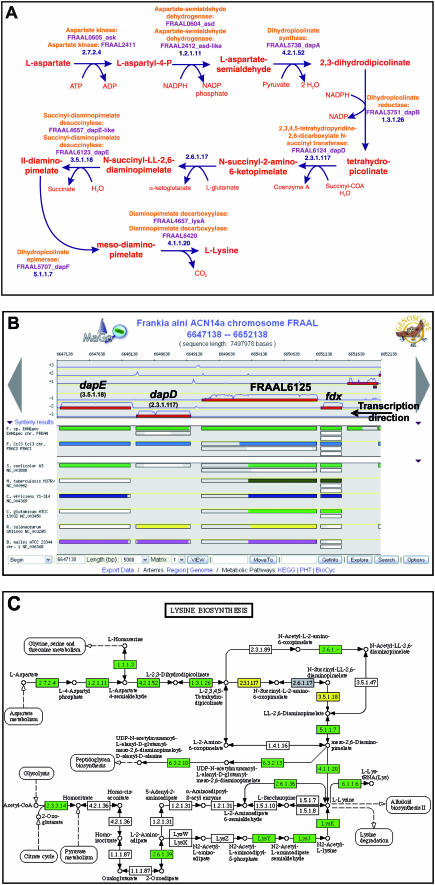
Figure 4.
Lysine biosynthesis in F.alni genome through the MaGe interfaces. Three screenshots showing lysine biosynthesis in F.alni. The FrankiaCyc Pathway/Genome DataBase (PGDB) is available through MaGe via a BioCyc web server (A). In addition, the user can obtain KEGG maps by comparison with E.coli (C). Yellow rectangles symbolize enzymes encoded by genes in the selected MaGe region (B) while green rectangles represent enzymes encoded by genes localized elsewhere in the studied genome. Gray boxes correspond to known enzymes in E.coli that are not present in the genome under study. Lastly, white boxes are enzymatic activities missing in both organisms. The BioCyc pathway selection algorithm reports only one possible pathway for lysine biosynthesis (A) in F.alni. The reported pathway apparently lacks the gene(s) encoding the succinyldiaminopimelate amino transferase activity (EC number 2.6.1.17). The lysine biosynthesis map from KEGG (C) also reports the lack of succinyldiaminopimelate amino transferase activity which has been detected in E.coli. Furthermore, genomic context exploration of the genes involved in this pathway, via the MaGe genome browser (B), reveals that the gene FRAAL6125 is co-localized with the characterized dapE and dapD genes. FRAAL6125 is a good candidate for dapC, a gene coding the missing activity and experimentally described in other species.
Exploration of metabolic pathways could be enhanced through gene context analysis. For example, in the case of lysine biosynthesis, three alternative routes are described in the literature: the succinylase, dehydrogenase and acetylase branches (61). During the study of Frankia alni genome, MaGe annotations combined with the FrankiaCyc PGDB revealed only one possible pathway involving the succinylase branch (Figure 4A). All of the genes coding for the enzymes of this pathway (ask, asd, dapA, dapB, dapD, dapE, dapF and lysA) have been found, except for the dapC gene which encodes a succinyldiaminopimelate amino transferase activity. In E.coli, the dapC gene does not exist, but the ArgD protein possesses both an acetylornithine and a succinyldiaminopimelate aminotransferase activity for arginine and lysine biosynthesis, respectively (62). In F.alni, the argD gene has been identified and its presence could explain the absence of dapC. Actually, studying the F.alni genomic context of the genes involved in lysine biosynthesis, we found a gene (FRAAL6125) described as a putative aminotransferase. This gene is co-localized with the characterized dapE and dapD genes which encode two of the three steps of the succinylase branch (Figure 4B). In addition, the corresponding KEGG map reveals the apparent lack of DapC activity and a co-localization of dapE and dapD genes (Figure 4C). Furthermore, the synteny results among thirty organisms show a chromosomal conservation of this three-gene organization. All these evidence leads us to assume that FRAAL6125 is a good candidate for dapC. These assumptions were confirmed by sequence comparison with experimentally demonstrated dapC genes in Corynebacterium glutamicum (63) and Bordetella pertussis (64) (52 and 32% amino acid identity, respectively). In contrast to the dapC homolog in other organisms, in F.alni the protein encoded by FRAAL6125 possesses an additional C-terminal domain of unknown function which is characterized by a glutamine- and glycine-rich content. This is shown, in Figure 4B, by the uncolored part of the rectangles in the synteny maps corresponding to the dapC homologs in the selected organisms. Two other strains of the Frankia genus (Cci3 and EAN1pec), sequenced by the United States Department Of Energy, show a similar genomic organization of the dapCDE gene cluster. But only the strain EAN1pec possesses this C-terminal domain (first synteny map in Figure 4B). This Frankia-specific C-terminal domain of DapC calls for more experimental investigation. This example shows that MaGe integration of gene context methods is a powerful tool for experts in metabolic analysis.
Data exploration
Although the notion of multigenome comparisons is omnipresent in the graphical interface of our system, the exploration functionality developed in MaGe is linked to the genome being selected for expert annotation only (‘Display organism’ in the ‘Options’ functionality). A simple keyword search enables the user to quickly retrieve genes of the annotated genome having a particular function. Several sets of data can be queried, such as automatic and validated annotations (expert work), or a specific set of annotated CDSs corresponding, for example, to conserved hypothetical proteins which are in synteny with other organisms. In addition, each kind of computed result (PRIAM, InterPro, blast similarities in reference genome annotation data, and in Swiss-Prot or TrEMBL databanks) can be retrieved. The result output is a list of candidate genes, the genomic contexts of which can be easily visualized (automatic displacement of the genome browser centered on a gene of interest).
In a second section, called ‘PhyloProfile and Synteny’ (Supplementary Figure 5), the user can search for genes of the studied organism which are homologs of genes in certain organisms and exclude those that are homologs of genes in other organisms. The phylogenetic profile method is designed to infer functional relationships between genes: proteins involved in the same biological process are likely to evolve in a correlated fashion (15). This method, combined with the integration of synteny results, allows one to detect a coevolution of gene groups which have a similar chromosomal organization. Integration of chromosomal proximity and gene content information has been reported to be more accurate than the single-gene phylogenetic profiles (65).
Using the synteny results stored in our database (see ‘The relational database’), the fusion/fission events can easily be computed. Our procedure detects synteny groups having two genes from a compared genome corresponding to a single annotated CDS in the target genome (Figure 1A). BlastP correspondences are evaluated to exclude the detection of tandem duplications by keeping only non-overlapping side-by-side alignments. These events are listed in the ‘Fusion/Fission’ item of the ‘Explore’ functionality (Supplementary Figure 5) and split into two tables: one containing the list of putative fused genes, and the other for fission events. Annotators can then browse results by checking for possible pseudogenes or for true functional evidence leading to the annotation of a multifunctional protein (see above, the case of rnhA and dnaQ gene fusion in Acinetobacter ADP1).
In a fourth section of the ‘Explore’ functionality, specific regions between the genome under analysis and a set of genomes selected for their phylogenetic proximity can be browsed (Supplementary Figure 5). Data are represented in a table listing gene clusters that have no correspondences in one or more compared organisms. One application of this comparative genomic analysis is the detection of genomic islands. A comparative study between two A.baumannii strains, AYE a multi-drug resistant strain and SDF a fully susceptible one, led us to decipher a 86 kb AYE-specific region where more than 40 resistance genes are clustered (P.-E. Fournier et al., manuscript in preparation).
SETTING UP A NEW ANNOTATION PROJECT
The MaGe system can be used either for the annotation of novel genomes or for curation of already annotated genomes available in public databanks (re-annotation projects). To start a new project, we first work on the integration, in PkGDB, of available bacterial genomes which are of interest in the context of the new thematic database (Supplementary Figure 6). Both complete and unfinished bacterial genomes are integrated in our database. The sequence(s) of the novel genome(s) are then submitted to the complete annotation pipeline analysis, including computation of synteny results with all the available proteomes in PkGDB and in the NCBI RefSeq databank. As explained in the ‘Metabolic pathway reconstruction’, a Pathway/Genome DataBase (PGDB) is built using the BioCyc software (47), and the corresponding database is made available from the MaGe interface. Some changes in the gene editor are made to take into account the specificity of each project. For example, the E.coli functional classification which is the default can be changed, or additional ‘BioProcess’ classes can be added (for the RhizoScope project shown in Supplementary Figure 6, three additional processes were added: Nitrogen fixation, Photosynthesis and Symbiosis). Finally, the new thematic database is made available to the research teams involved in the project (via a secure connection). In addition, the portion of the database information corresponding to bacterial genomes available in public databanks is made freely accessible via the MaGe interface (Supplementary Figure 6).
The MaGe system, initially developed and used in the context of the Acinetobacter ADP1 genome annotation (17), has also been used for the analysis of Pseudoalteromonas haloplanktis (18), Frankia alni and Pseudomonas entomophila. In the context of the MicroScope project which aims to build thematic databases for the (re)-annotation of prokaryotic genomes (http://www.genoscope.cns.fr/agc/microscope), a number of microbial genomes are currently being annotated using the MaGe system (16 projects to date): this includes pathogenic species (such as Leptospira biflexa, Neisseria meningitidis NEM8013 and E.coli strains) or environmental bacteria (such as Cenibacterium arsenoxidans and Bradyrhizobium sp. ORS278). In addition, our group is involved in a metagenomic project which aims to produce an inventory of the microorganisms present at two main stages of waste water treatment. Several large genomic regions from yet uncultured microorganisms have already been annotated and analyzed, giving us the opportunity to propose specific culture media for enrichment cultures for the corresponding bacteria.
The PkGDB database scheme and the MaGe web frontend are available upon request for a local installation. Furthermore, on demand, we can customize the MaGe system for a specific genome project (http://www.genoscope.cns.fr/agc/microscope).
IMPLEMENTATION OF MaGe
UNIX shell and perl scripts manage data integration and computations. Program executions are dispatched on a multi-processor computer system (40 Alpha 1 GHz CPUs) by the Platform LSF software (a batch application workload processing). Pattern search and sequence alignments are performed with the Biofacet package (66). The free MySQL database management system which is used by PkGDB provides a fast and a reliable access to data. For the MaGe web server, the Apache system and the PHP (Hypertext Preprocessor) language are used. PHP is a HTML-embedded scripting language allowing dynamic generation of the HTML page contents. Associated with the GD graphics library, web interface images are dynamically generated in PNG (Portable Network Graphics) format.
CONCLUSION
The MaGe annotation platform (i.e. a software suite with a multigenomes relational database and a web graphical interface) has proved to be a useful tool for expert annotation, mainly because it avoids most of the main automatic sequence annotation pitfalls. In the process of the expert annotation, our graphical representation of synteny results is obviously invaluable to highlight interesting features. Owing to the dynamic nature of the bioinformatics field, constant efforts are made to keep the set of computational techniques (i.e. additional methods and/or links to useful web sites are regularly added) and the integrated databases up-to-date. In this way, the recently published annotation environment SEED will be integrated into the MaGe system (42). Based on the notion of populated subsystem (i.e. a set of functional roles that together implement a specific biological process or structural complex), the SEED system should bring additional clues as far as biological function of uncalled genes is concerned. Our automatic annotation procedure takes into account the spurious function assignments caused by multidomain proteins and exploits functional coupling between genes located in adjacent positions on the chromosome. However, we plan to improve some decision rules, mainly by introducing data from predicted metabolic pathways obtained with the BioCyc software (46,47), and by combining co-localization results with phylogenetic profiles. In addition, MaGe is often used to annotate several closely related genomes, and a novel functionality is clearly required, which will permit a manual refinement of annotation on several related species at the same time. Other planned developments include new features in the genome browser (i.e. representation of global DNA and protein statistical tendencies), new features in the gene editor (i.e. graphical representation of functional annotations on the corresponding protein) and an improved interface for queries in the PkGDB database.
The growing availability of expression profiles (from microarray data and proteomics), supplemented with gene essentiality and regulation, protein–protein interaction and metabolomics data, brings a major source of clues for the clarification of gene function. The Genostar exploratory genomics platform offers a unified way of representing and managing data of various types and origins through a set of software modules which can exchange information (http://www.genostar.org). This system has already been connected to our PkGDB database: in the context of some annotation projects, MaGe high quality annotations are imported into Genostar and linked to various types of experimental data modeled in the GenoLink module (67). The MaGe functionalities combined with the advanced query interface of this module should also contribute to the characterization of the functions of orphan genes.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Supplementary Material
Acknowledgments
The authors would like to thank all MaGe users for their feedback that helped greatly in optimizing and improving many functionalities of the system. This work was supported by the French Centre National de la Recherche Scientifique (CNRS-URA8030), the GENOPOLE of Evry and the French Ministry of Research (funds allocated by the ACI IMPBio2004). The authors thank Susan Cure and Denis Bayada for their help in writing the manuscript. A particular thanks for François Le Fèvre for his help in setting up the BioCyc system. The authors thank the entire system network team of Genoscope for its essential contribution to the efficiency of the MaGe web interface. Funding to pay the Open Access publication charges for this article was provided by CNRG-composante Genoscope.
Conflict of interest statement. None declared.
REFERENCES
- 1.Galperin M.Y., Koonin E.V. Sources of systematic error in functional annotation of genomes: domain rearrangement, non-orthologous gene displacement and operon disruption. In Silico Biol. 1998;1:55–67. [PubMed] [Google Scholar]
- 2.Brenner S.E. Errors in genome annotation. Trends Genet. 1999;15:132–133. doi: 10.1016/s0168-9525(99)01706-0. [DOI] [PubMed] [Google Scholar]
- 3.Van Domselaar G.H., Stothard P., Shrivastava S., Cruz J.A., Guo A., Dong X., Lu P., Szafron D., Greiner R., Wishart D.S. BASys: a web server for automated bacterial genome annotation. Nucleic Acids Res. 2005;33:W455–W459. doi: 10.1093/nar/gki593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Riley M.L., Schmidt T., Wagner C., Mewes H.W., Frishman D. The PEDANT genome database in 2005. Nucleic Acids Res. 2005;33:D308–D310. doi: 10.1093/nar/gki019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gaasterland T., Sensen C.W. MAGPIE: automated genome interpretation. Trends Genet. 1996;12:76–78. doi: 10.1016/0168-9525(96)81406-5. [DOI] [PubMed] [Google Scholar]
- 6.Hoersch S., Leroy C., Brown N.P., Andrade M.A., Sander C. The GeneQuiz web server: protein functional analysis through the Web. Trends Biochem. Sci. 2000;25:33–35. doi: 10.1016/s0968-0004(99)01510-8. [DOI] [PubMed] [Google Scholar]
- 7.Overbeek R., Larsen N., Walunas T., D'Souza M., Pusch G., Selkov E., Jr, Liolios K., Joukov V., Kaznadzey D., Anderson I., et al. The ERGO genome analysis and discovery system. Nucleic Acids Res. 2003;31:164–171. doi: 10.1093/nar/gkg148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berriman M., Rutherford K. Viewing and annotating sequence data with Artemis. Brief Bioinform. 2003;4:124–132. doi: 10.1093/bib/4.2.124. [DOI] [PubMed] [Google Scholar]
- 9.Meyer F., Goesmann A., McHardy A.C., Bartels D., Bekel T., Clausen J., Kalinowski J., Linke B., Rupp O., Giegerich R., et al. GenDB—an open source genome annotation system for prokaryote genomes. Nucleic Acids Res. 2003;31:2187–2195. doi: 10.1093/nar/gkg312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frangeul L., Glaser P., Rusniok C., Buchrieser C., Duchaud E., Dehoux P., Kunst F. CAAT-Box, contigs-assembly and annotation tool-box for genome sequencing projects. Bioinformatics. 2004;20:790–797. doi: 10.1093/bioinformatics/btg490. [DOI] [PubMed] [Google Scholar]
- 11.Almeida L.G., Paixao R., Souza R.C., Costa G.C., Barrientos F.J., Santos M.T., Almeida D.F., Vasconcelos A.T. A System for Automated Bacterial (genome) Integrated Annotation—SABIA. Bioinformatics. 2004;20:2832–2833. doi: 10.1093/bioinformatics/bth273. [DOI] [PubMed] [Google Scholar]
- 12.Dandekar T., Snel B., Huynen M., Bork P. Conservation of gene order: a fingerprint of proteins that physically interact. Trends Biochem. Sci. 1998;23:324–328. doi: 10.1016/s0968-0004(98)01274-2. [DOI] [PubMed] [Google Scholar]
- 13.Marcotte E.M., Pellegrini M., Ng H.L., Rice D.W., Yeates T.O., Eisenberg D. Detecting protein function and protein–protein interactions from genome sequences. Science. 1999;285:751–753. doi: 10.1126/science.285.5428.751. [DOI] [PubMed] [Google Scholar]
- 14.Overbeek R., Fonstein M., D'Souza M., Pusch G.D., Maltsev N. Use of contiguity on the chromosome to predict functional coupling. In Silico Biol. 1999;1:93–108. [PubMed] [Google Scholar]
- 15.Pellegrini M., Marcotte E.M., Thompson M.J., Eisenberg D., Yeates T.O. Assigning protein functions by comparative genome analysis: protein phylogenetic profiles. Proc. Natl Acad. Sci. USA. 1999;96:4285–4288. doi: 10.1073/pnas.96.8.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Doerks T., von Mering C., Bork P. Functional clues for hypothetical proteins based on genomic context analysis in prokaryotes. Nucleic Acids Res. 2004;32:6321–6326. doi: 10.1093/nar/gkh973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barbe V., Vallenet D., Fonknechten N., Kreimeyer A., Oztas S., Labarre L., Cruveiller S., Robert C., Duprat S., Wincker P., et al. Unique features revealed by the genome sequence of Acinetobacter sp. ADP1, a versatile and naturally transformation competent bacterium. Nucleic Acids Res. 2004;32:5766–5779. doi: 10.1093/nar/gkh910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Medigue C., Krin E., Pascal G., Barbe V., Bernsel A., Bertin P.N., Cheung F., Cruveiller S., D'Amico S., Duilio A., et al. Coping with cold: the genome of the versatile marine Antarctica bacterium Pseudoalteromonas haloplanktis TAC125. Genome Res. 2005;15:1325–1335. doi: 10.1101/gr.4126905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bocs S., Cruveiller S., Vallenet D., Nuel G., Medigue C. AMIGene: Annotation of MIcrobial Genes. Nucleic Acids Res. 2003;31:3723–3726. doi: 10.1093/nar/gkg590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cruveiller S., Le Saux J., Vallenet D., Lajus A., Bocs S., Medigue C. MICheck: a web tool for fast checking of syntactic annotations of bacterial genomes. Nucleic Acids Res. 2005;33:W471–W479. doi: 10.1093/nar/gki498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Suzek B.E., Ermolaeva M.D., Schreiber M., Salzberg S.L. A probabilistic method for identifying start codons in bacterial genomes. Bioinformatics. 2001;17:1123–1130. doi: 10.1093/bioinformatics/17.12.1123. [DOI] [PubMed] [Google Scholar]
- 22.Lowe T.M., Eddy S.R. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Griffiths-Jones S., Moxon S., Marshall M., Khanna A., Eddy S.R., Bateman A. Rfam: annotating non-coding RNAs in complete genomes. Nucleic Acids Res. 2005;33:D121–D124. doi: 10.1093/nar/gki081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.d'Aubenton Carafa Y., Brody E., Thermes C. Prediction of rho-independent Escherichia coli transcription terminators. A statistical analysis of their RNA stem-loop structures. J. Mol. Biol. 1990;216:835–858. doi: 10.1016/s0022-2836(99)80005-9. [DOI] [PubMed] [Google Scholar]
- 25.Achaz G., Coissac E., Viari A., Netter P. Analysis of intrachromosomal duplications in yeast Saccharomyces cerevisiae: a possible model for their origin. Mol. Biol. Evol. 2000;17:1268–1275. doi: 10.1093/oxfordjournals.molbev.a026410. [DOI] [PubMed] [Google Scholar]
- 26.Altschul S.F., Madden T.L., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bairoch A., Apweiler R., Wu C.H., Barker W.C., Boeckmann B., Ferro S., Gasteiger E., Huang H., Lopez R., Magrane M., et al. The Universal Protein Resource (UniProt) Nucleic Acids Res. 2005;33:D154–D159. doi: 10.1093/nar/gki070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mulder N.J., Apweiler R., Attwood T.K., Bairoch A., Bateman A., Binns D., Bradley P., Bork P., Bucher P., Cerutti L., et al. InterPro, progress and status in 2005. Nucleic Acids Res. 2005;33:D201–D205. doi: 10.1093/nar/gki106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Natale D.A., Galperin M.Y., Tatusov R.L., Koonin E.V. Using the COG database to improve gene recognition in complete genomes. Genetica. 2000;108:9–17. doi: 10.1023/a:1004031323748. [DOI] [PubMed] [Google Scholar]
- 30.Claudel-Renard C., Chevalet C., Faraut T., Kahn D. Enzyme-specific profiles for genome annotation: PRIAM. Nucleic Acids Res. 2003;31:6633–6639. doi: 10.1093/nar/gkg847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gattiker A., Michoud K., Rivoire C., Auchincloss A.H., Coudert E., Lima T., Kersey P., Pagni M., Sigrist C.J., Lachaize C., et al. Automated annotation of microbial proteomes in SWISS-PROT. Comput. Biol. Chem. 2003;27:49–58. doi: 10.1016/s1476-9271(02)00094-4. [DOI] [PubMed] [Google Scholar]
- 32.Krogh A., Larsson B., von Heijne G., Sonnhammer E.L. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 33.Bendtsen J.D., Nielsen H., von Heijne G., Brunak S. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 2004;340:783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- 34.Gough J., Karplus K., Hughey R., Chothia C. Assignment of homology to genome sequences using a library of hidden Markov models that represent all proteins of known structure. J. Mol. Biol. 2001;313:903–919. doi: 10.1006/jmbi.2001.5080. [DOI] [PubMed] [Google Scholar]
- 35.Andreeva A., Howorth D., Brenner S.E., Hubbard T.J., Chothia C., Murzin A.G. SCOP database in 2004: refinements integrate structure and sequence family data. Nucleic Acids Res. 2004;32:D226–D229. doi: 10.1093/nar/gkh039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pascal G., Medigue C., Danchin A. Universal biases in protein composition of model prokaryotes. Proteins. 2005;60:27–35. doi: 10.1002/prot.20475. [DOI] [PubMed] [Google Scholar]
- 37.von Mering C., Jensen L.J., Snel B., Hooper S.D., Krupp M., Foglierini M., Jouffre N., Huynen M.A., Bork P. STRING: known and predicted protein–protein associations, integrated and transferred across organisms. Nucleic Acids Res. 2005;33:D433–D437. doi: 10.1093/nar/gki005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Janga S.C., Collado-Vides J., Moreno-Hagelsieb G. Nebulon: a system for the inference of functional relationships of gene products from the rearrangement of predicted operons. Nucleic Acids Res. 2005;33:2521–2530. doi: 10.1093/nar/gki545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smith T.F., Waterman M.S. Identification of common molecular subsequences. J. Mol. Biol. 1981;147:195–197. doi: 10.1016/0022-2836(81)90087-5. [DOI] [PubMed] [Google Scholar]
- 40.Boyer F., Morgat A., Labarre L., Pothier J., Viari A. Syntons, metabolons and interactons: an exact graph-theoretical approach for exploring neighbourhood between genomic and functional data. Bioinformatics. 2005;21:4209–4215. doi: 10.1093/bioinformatics/bti711. [DOI] [PubMed] [Google Scholar]
- 41.Pruitt K.D., Tatusova T., Maglott D.R. NCBI Reference Sequence (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2005;33:D501–D504. doi: 10.1093/nar/gki025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Overbeek R., Begley T., Butler R.M., Choudhuri J.V., Chuang H.Y., Cohoon M., de Crecy-Lagard V., Diaz N., Disz T., Edwards R., et al. The subsystems approach to genome annotation and its use in the project to annotate 1000 genomes. Nucleic Acids Res. 2005;33:5691–5702. doi: 10.1093/nar/gki866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Enault F., Suhre K., Claverie J.M. Phydbac ‘Gene Function Predictor’: a gene annotation tool based on genomic context analysis. BMC Bioinformatics. 2005;6:247. doi: 10.1186/1471-2105-6-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Harris M.A., Clark J., Ireland A., Lomax J., Ashburner M., Foulger R., Eilbeck K., Lewis S., Marshall B., Mungall C., et al. The Gene Ontology (GO) database and informatics resource. Nucleic Acids Res. 2004;32:D258–D261. doi: 10.1093/nar/gkh036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kanehisa M., Goto S., Kawashima S., Okuno Y., Hattori M. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004;32:D277–280. doi: 10.1093/nar/gkh063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Karp P.D., Paley S., Romero P. The Pathway Tools software. Bioinformatics. 2002;18(Suppl. 1):S225–S232. doi: 10.1093/bioinformatics/18.suppl_1.s225. [DOI] [PubMed] [Google Scholar]
- 47.Krummenacker M., Paley S., Mueller L., Yan T., Karp P.D. Querying and computing with BioCyc databases. Bioinformatics. 2005;21:3454–3455. doi: 10.1093/bioinformatics/bti546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Krieger C.J., Zhang P., Mueller L.A., Wang A., Paley S., Arnaud M., Pick J., Rhee S.Y., Karp P.D. MetaCyc: a multiorganism database of metabolic pathways and enzymes. Nucleic Acids Res. 2004;32:D438–D442. doi: 10.1093/nar/gkh100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Green M.L., Karp P.D. A Bayesian method for identifying missing enzymes in predicted metabolic pathway databases. BMC Bioinformatics. 2004;5:76. doi: 10.1186/1471-2105-5-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rahman S.A., Advani P., Schunk R., Schrader R., Schomburg D. Metabolic pathway analysis web service (Pathway Hunter Tool at CUBIC) Bioinformatics. 2005;21:1189–1193. doi: 10.1093/bioinformatics/bti116. [DOI] [PubMed] [Google Scholar]
- 51.Green M.L., Karp P.D. Genome annotation errors in pathway databases due to semantic ambiguity in partial EC numbers. Nucleic Acids Res. 2005;33:4035–4039. doi: 10.1093/nar/gki711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kersey P., Bower L., Morris L., Horne A., Petryszak R., Kanz C., Kanapin A., Das U., Michoud K., Phan I., et al. Integr8 and Genome Reviews: integrated views of complete genomes and proteomes. Nucleic Acids Res. 2005;33:D297–D302. doi: 10.1093/nar/gki039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Serres M.H., Goswami S., Riley M. GenProtEC: an updated and improved analysis of functions of Escherichia coli K-12 proteins. Nucleic Acids Res. 2004;32:D300–D302. doi: 10.1093/nar/gkh087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rudd K.E. EcoGene: a genome sequence database for Escherichia coli K-12. Nucleic Acids Res. 2000;28:60–64. doi: 10.1093/nar/28.1.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Winsor G.L., Lo R., Sui S.J., Ung K.S., Huang S., Cheng D., Ching W.K., Hancock R.E., Brinkman F.S. Pseudomonas aeruginosa Genome Database and PseudoCAP: facilitating community-based, continually updated, genome annotation. Nucleic Acids Res. 2005;33:D338–D343. doi: 10.1093/nar/gki047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Moszer I., Jones L.M., Moreira S., Fabry C., Danchin A. SubtiList: the reference database for the Bacillus subtilis genome. Nucleic Acids Res. 2002;30:62–65. doi: 10.1093/nar/30.1.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Serres M.H., Riley M. MultiFun, a multifunctional classification scheme for Escherichia coli K-12 gene products. Microb. Comp. Genomics. 2000;5:205–222. doi: 10.1089/omi.1.2000.5.205. [DOI] [PubMed] [Google Scholar]
- 58.Haft D.H., Selengut J.D., White O. The TIGRFAMs database of protein families. Nucleic Acids Res. 2003;31:371–373. doi: 10.1093/nar/gkg128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ruepp A., Zollner A., Maier D., Albermann K., Hani J., Mokrejs M., Tetko I., Guldener U., Mannhaupt G., Munsterkotter M., et al. The FunCat, a functional annotation scheme for systematic classification of proteins from whole genomes. Nucleic Acids Res. 2004;32:5539–5545. doi: 10.1093/nar/gkh894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bork P., Bairoch A. Go hunting in sequence databases but watch out for the traps. Trends Genet. 1996;12:425–427. doi: 10.1016/0168-9525(96)60040-7. [DOI] [PubMed] [Google Scholar]
- 61.Velasco A.M., Leguina J.I., Lazcano A. Molecular evolution of the lysine biosynthetic pathways. J. Mol. Evol. 2002;55:445–459. doi: 10.1007/s00239-002-2340-2. [DOI] [PubMed] [Google Scholar]
- 62.Ledwidge R., Blanchard J.S. The dual biosynthetic capability of N-acetylornithine aminotransferase in arginine and lysine biosynthesis. Biochemistry. 1999;38:3019–3024. doi: 10.1021/bi982574a. [DOI] [PubMed] [Google Scholar]
- 63.Hartmann M., Tauch A., Eggeling L., Bathe B., Mockel B., Puhler A., Kalinowski J. Identification and characterization of the last two unknown genes, dapC and dapF, in the succinylase branch of the L-lysine biosynthesis of Corynebacterium glutamicum. J. Biotechnol. 2003;104:199–211. doi: 10.1016/s0168-1656(03)00156-1. [DOI] [PubMed] [Google Scholar]
- 64.Fuchs T.M., Schneider B., Krumbach K., Eggeling L., Gross R. Characterization of a Bordetella pertussis diaminopimelate (DAP) biosynthesis locus identifies dapC, a novel gene coding for an N-succinyl-L,L-DAP aminotransferase. J. Bacteriol. 2000;182:3626–3631. doi: 10.1128/jb.182.13.3626-3631.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zheng Y., Roberts R.J., Kasif S. Genomic functional annotation using co-evolution profiles of gene clusters. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-11-research0060. RESEARCH0060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Glemet E., Codani J.J. LASSAP, a LArge Scale Sequence compArison Package. Comput. Appl. Biosci. 1997;13:137–143. doi: 10.1093/bioinformatics/13.2.137. [DOI] [PubMed] [Google Scholar]
- 67.Durand P., Labarre L., Meil A., Wojcik J. GenoLink: discovering drug target proteins by exploring networks of heterogeneous biological data. ERCIM. 2005;60:31–32. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.