Abstract
Cdyl (chromodomain-Y-like) is a chromodomain-containing protein that is predominantly expressed during mouse spermiogenesis. In its carboxy-terminal portion, there is a domain with homology to the coenzyme A (CoA) pocket of the enoyl-CoA hydratase/isomerase, which is shown here to be able to bind CoA and histone deacetylases (HDACs). It also efficiently represses transcription. Moreover, the binding of Hdac1 represses the ability of Cdyl to bind CoA, and a Cdyl–CoA interaction only occurs in the absence of HDACs. These data suggest that Cdyl is primarily a transcriptional co-repressor. However, the degradation of cellular Hdac1 and Hdac2, as observed here in the elongating spermatids, may provide an HDAC-free environment in which Cdyl could bind CoA and participate in the global chromatin remodelling that occurs in these cells.
Introduction
A group of related genes encode the proteins Cdy (chromodomain Y) and Cdyl (Cdy-like), which both contain a canonical chromodomain in their amino-terminal region, as well as a domain with significant sequence homology to the enoyl-coenzyme A (CoA) hydratase/isomerase catalytic domain (Lahn & Page, 1999).
Recently, it has been shown that both Cdy and Cdyl have histone acetyltransferase (HAT) activity (Lahn et al., 2002) and that Cdyl is expressed abundantly in elongating spermatids during histone hyperacetylation and replacement. These observations establish a potential link between Cdy and Cdyl function and chromatin remodelling during spermatogenesis. Apart from the chromodomain, the other potentially important domain present in Cdy and Cdyl has significant sequence homology to the 'CoA pocket' of the enoyl-CoA hydratase/isomerase. Enoyl-CoA hydratase is one of the four enzymes that catalyse the degradation of fatty acids by the β-oxidation pathway in mitochondria. The role of enoyl-CoA hydratase is to catalyse the hydratation of 2-trans-enoyl-CoA to (s)-3-hydroxyacyl-CoA (Engel et al., 1996). From these data, Cdyl seems to be a potentially important protein that is expressed in somatic cells and overexpressed in germinal cells during spermiogenesis, but little is known about its function. Here, we report the functional characterization of Cdyl. Our data suggest that in somatic cells, as well as during the early stages of spermiogenesis, Cdyl is primarily a repressor of transcription that might potentially be converted into a HAT during the late stages of spermatid maturation.
Results
Cdyl is a potent repressor of transcription
We have identified a chromodomain protein that also contains a CoA channel, which is now known as Cdyl (Lahn & Page, 1999), as a highly conserved protein among vertebrates (see supplementary information online). The presence of these domains suggests a role for Cdyl in the control of transcription. We therefore investigated the ability of Cdyl to modulate the transcription of a reporter gene. The DNA-binding domain of Gal4 (Gal4DB; amino acids 1–147) was fused to Cdyl, and the fusion protein was expressed to target a luciferase reporter gene containing five Gal4 binding sites upstream of the simian virus 40 (SV40) early promoter (Fig. 1A). Figure 1B shows that the expression of increasing amounts of Gal4–Cdyl led to a marked repression of the SV40 promoter, whereas the expression of Gal4DB alone or the haemagglutinin (HA)-tagged Cdyl (not fused to Gal4DB) did not affect the expression of the reporter gene. To localize the repression domain of Cdyl, the chromodomain and the putative CoA pocket that constitutes half of the protein (called the CoA pocket domain in this study) were fused to Gal4DB, and their repressor activities were monitored as described above. Figure 1C shows that only the CoA-pocket domain of Cdyl represses transcription.
Figure 1.
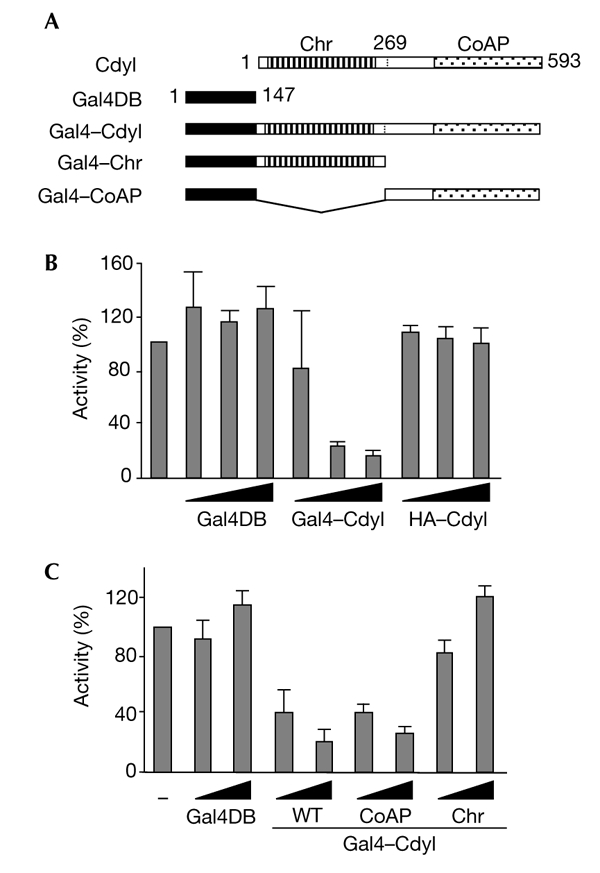
Cdyl is a transcriptional repressor. (A) The Gal4 DNA-binding domain (Gal4DB) was fused to either full-length Cdyl (Gal4–Cdyl), the Cdyl chromodomain (Chr; the Gal4–Chr construct), or the Cdyl carboxy-terminal half, which is homologous to the enoyl-coenzyme A (CoA) hydratase (the Gal4–CoA-pocket (CoAP) construct). (B,C) HeLa cells were cotransfected with 200 ng of a reporter plasmid containing five Gal4-binding sites cloned upstream of the simian virus 40 early promoter, and increasing amounts (10, 30 or 100 ng in (B) and 30 or 100 ng in (C)) of the indicated expression constructs. Gal4DB corresponds to the Gal4 DNA-binding domain alone, and haemagglutinin (HA)–Cdyl is Cdyl fused to an HA-tag (and does not contain Gal4DB). Luciferase activity was measured and normalized with respect to that of β-galactosidase. Mean values of at least three independent assays are shown. Cdyl, chromodomain-Y-like; WT, wild type.
Cdyl has a functional CoA pocket
Significant sequence homology between the CoA-binding channel of enoyl-CoA hydratase and a domain in the Cdyl C-terminal region (Fig. 2A) led us to investigate the ability of Cdyl to bind CoA. Agarose beads covered with covalently bound CoA were used for this. Figure 2B shows that wild-type Cdyl expressed in cells (Fig. 2B, lane 4), but not its chromodomain-containing region (Fig. 2B, lane 3), interacts efficiently with the CoA–agarose beads. In these experiments, the CoA-binding activity of the Cdyl CoA-pocket domain alone could not be investigated because this fragment of Cdyl remained tightly associated with the insoluble nuclear materials after the extraction of nuclear proteins.
Figure 2.
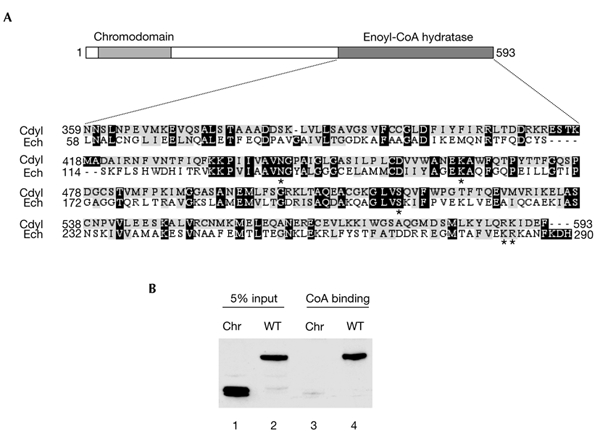
The domain of Cdyl that is homologous to enoyl-coenzyme-A hydratase binds coenzyme A. (A) Sequence homology between the coenzyme A (CoA) pocket of mouse enoyl-CoA hydratase (Ech) and mouse Cdyl. The asterisks indicate the amino acids that are replaced by alanine in the mutants shown in Fig. 3. (B) Whole-cell extracts from U2OS cells that express wild-type (WT) Cdyl and a Cdyl fragment that contains the chromodomain (Chr) were incubated with CoA–agarose. The beads were washed and the bound proteins were detected by western blotting using an anti-Cdyl antibody. Cdyl, chromodomain-Y-like.
To determine whether the transcriptional repressor activity of Cdyl is associated with the ability of the protein to bind CoA, we prepared four Cdyl mutants in which amino acids conserved between Cdyl and the catalytic domain of enoyl-CoA hydratase (Fig. 2A, asterisks) were replaced by alanines. The amino acids replaced were N441, K463, S516 and diamino acids R588 and K589. The mutant proteins were fused to Gal4DB, and their repressor activities were monitored as described above. Interestingly, all the mutants were as active as the wild-type protein in repressing transcription (Fig. 3A). To test the effect of these mutations on the CoA-binding activity of Cdyl, we used the CoA pull-down method described above. Figure 3B shows that two of the mutants have lost their ability to interact with CoA (Fig. 3B, lanes 4 and 5). As all the mutants were active in repressing transcription, these results show that the repressor activity of Cdyl is independent of its CoA-binding ability.
Figure 3.
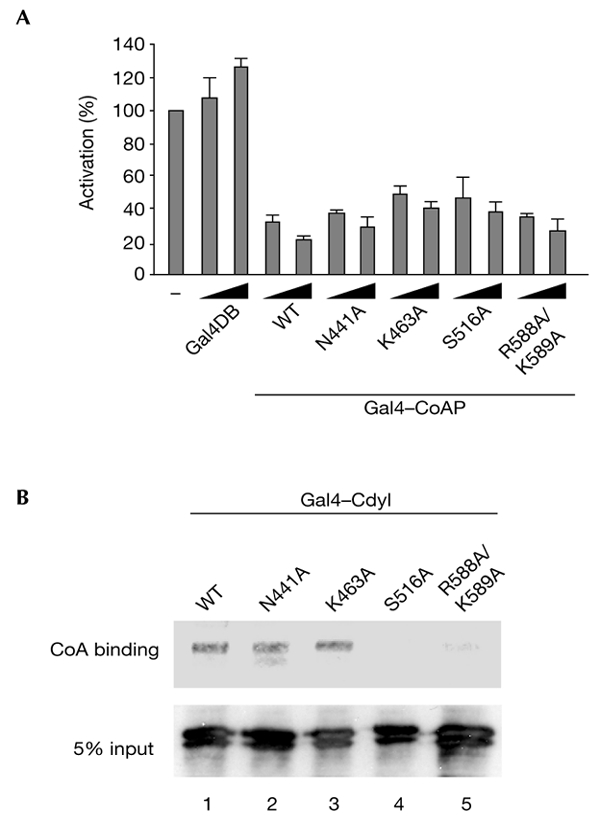
Cdyl mutated in its coenzyme A pocket is also a transcriptional repressor. (A) Amino acids N441, K463, S516, R588 and K589 of Cdyl, which are conserved between enoyl-coenzyme A (CoA) hydratase and Cdyl (see Fig. 2A), were replaced by alanine. The mutated proteins were fused to the Gal4 DNA-binding domain (Gal4DB), and their ability to repress transcription was monitored, as described in Fig. 1. Mean values from at least three independent assays are shown. (B) U2OS cells were transfected with vectors that express either wild-type (WT) Cdyl or one of the four mutants of Cdyl. Extracts from transfected cells were used to perform a CoA pull-down assay on CoA beads as described in Fig. 2. Beads were washed and the bound proteins were detected by western blotting using an anti-Cdyl antibody. Cdyl, chromodomain-Y-like.
The Cdyl CoA-pocket domain recruits histone deacetylases
As histone deacetylases (HDACs) are mainly involved in the repression of transcription, we wondered whether they interact with Cdyl. To investigate this possibility, cells were transfected with the expression vector FLAG–Cdyl. After transfection, extracts were prepared, the FLAG-tagged proteins were immunoprecipitated and the presence of endogenous HDACs was monitored using anti-Hdac1, anti-Hdac2 and anti-Hdac3 antibodies. Figure 4A shows that endogenous Hdac1 and Hdac2, but not Hdac3, were co-immunoprecipitated with full-length Cdyl. To demonstrate the association of endogenous Cdyl with endogenous HDACs, we took advantage of the enhanced expression of Cdyl in spermatid cells (Lahn et al., 2002). Mouse spermatogenic cells were separated according to their stage of differentiation using a sedimentation chamber. Extracts from pooled pachytene and spermatid-containing fractions were prepared, and immunoprecipitation was performed using either anti-Hdac1 or anti-Hdac2 antibodies. Figure 4B confirms that, as previously reported (Lahn et al., 2002), Cdyl expression is mainly detected in the spermatid cell population (also see later). It also shows that, in spermatid cells, Cdyl was associated with both HDACs (Fig. 4B, lanes 4 and 5).
Figure 4.
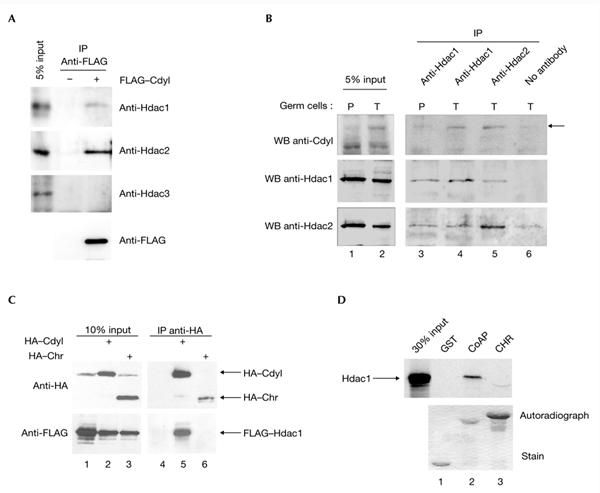
Cdyl interacts with histone deacetylases 1 and 2 through its coenzyme-A-pocket domain. (A) Cdyl recruits endogenous histone deacetylase 1 (Hdac1) and Hdac2. COS7 cells were transfected with 5 μg of expression vector for FLAG–Cdyl (+) or an empty vector (−), and Cdyl-containing complexes were immunoprecipitated from whole-cell extracts with an anti-FLAG antibody, followed by elution with a 3X FLAG peptide (Sigma). A western blot probed with an anti-FLAG antibody showed the efficiency of the immunoprecipitations (anti-FLAG panel). A western blot probed with anti-Hdac1, anti-Hdac2 and anti-Hdac3 antibodies showed the association of endogenous Hdac1 and Hdac2, but not Hdac3, with Cdyl. (B) Immunoprecipitations (IPs) were performed on extracts prepared from fractionated spermatogenic cells enriched in pachytene spermatocytes (P) and spermatids (T) with anti-Hdac1 (lanes 3 and 4), anti-Hdac2 (lane 5) covalently linked to protein-G–sepharose, or with no antibody as a negative control (lane 6). Cdyl (upper panels), Hdac1 (middle panels) and Hdac2 (lower panels) were detected using the corresponding antibodies. (C) COS7 cells were transfected with 5 μg of expression vectors for haemagglutinin (HA)–Cdyl (lane 2), HA–chromodomain (Chr; lane 3) or with an empty vector as a control (lane 1), and with 5 μg of expression vectors for FLAG-tagged Hdac1. IPs were performed on whole-cell extracts using an anti-HA antibody and were analysed by SDS–polyacrylamide gel electrophoresis (PAGE) on an 8% gel using anti-HA and anti-FLAG antibodies (lanes 4–6). (D) The CoA-pocket domain (CoAP) and the Chr of Cdyl fused to glutatione-S-transferase (GST), and GST alone as a control, were immobilized on gluthatione beads and incubated with 35S-radiolabelled Hdac1. Bound proteins were eluted and loaded onto a 10% SDS–PAGE gel, which was stained (lower panel) and used to expose an autoradiograph (upper panel). Cdyl, chromodomain-Y-like; WB, western blot.
The domain of Cdyl that is involved in the interaction with HDACs was mapped by transfection of the FLAG–Hdac1 construct either alone or with constructs encoding HA-tagged Cdyl wild-type or mutant proteins. Figure 4C shows that the chromodomain-containing half of Cdyl does not interact with Hdac1 (Fig. 4C, lane 6), which suggests that Hdac1 is recruited by the CoA-pocket domain of the protein. The same result was obtained using a FLAG–Hdac2 expression vector (not shown). The direct involvement of the CoA pocket of Cdyl in the interaction with Hdac1 was analysed by an in vitro binding assay: the chromodomain or CoA-pocket domains of Cdyl were fused to glutathione-S-transferase (GST) and incubated with 35S-labelled Hdac1. Figure 4D shows that Hdac1 specifically interacts with the CoA-pocket domain (Fig. 4D, lane 2) but not with the chromodomain (Fig. 4D, lane 3), which confirms our in vivo data.
Finally, the analysis of the intracellular localization of Cdyl and Hdac1/2 in overexpressing cells by immunofluorescence shows that, whereas Hdac1 and Hdac2 expressed alone are homogeneously distributed in the nucleus, the co-expression of Cdyl induces the recruitment of these HDACs into the Cdyl-specific nuclear spots (see supplementary information online), thereby confirming an efficient interaction between Cdyl and these two HDACs.
HDAC recruitment prevents Cdyl from binding CoA
The fact that Cdyl mutants that are incapable of CoA binding have retained the ability to repress transcription (Fig. 3) suggests that they could also be as active as wild-type Cdyl in recruiting HDACs. To confirm this hypothesis, all the Gal4–Cdyl mutants with mutations in the CoA pocket were expressed with FLAG–Hdac1. After an anti-Gal4 immunoprecipitation, the presence of FLAG–Hdac1 was confirmed in the immunoprecipitated materials that contained either wild-type or mutated Gal4–Cdyl (Fig. 5A). These data therefore show that Cdyl is able to bind Hdac1 irrespectively of its ability to bind CoA. We also used the CoA pull-down method to test the ability of the Cdyl–Hdac1 complex to interact with CoA. Cells were transfected with vectors expressing either Cdyl or the chromodomain-containing half of the protein in the absence or presence of Hdac1. Figure 5B shows that, as expected, wild-type Cdyl was efficiently retained on the CoA beads (Fig. 5B, lane 6), whereas, interestingly, the Cdyl–Hdac1 complex was unable to bind the CoA beads (Fig. 5B, lane 8). The control immunoprecipitation of a fraction of the cell extracts showed that both Cdyl and Hdac1 were together in a complex (Fig. 5B, lane 10). The immunoprecipitated Cdyl–Hdac1 complex was also incubated with an excess of free CoA to test whether a Cdyl–CoA interaction would lead to the release of Hdac1. However, even in the presence of millimolar concentrations of CoA, no release of Hdac1 was seen (data not shown). These experiments therefore show that the interaction of Hdac1 with the CoA pocket of Cdyl prevents the protein from binding CoA and that Cdyl can bind CoA only in an HDAC-free environment.
Figure 5.
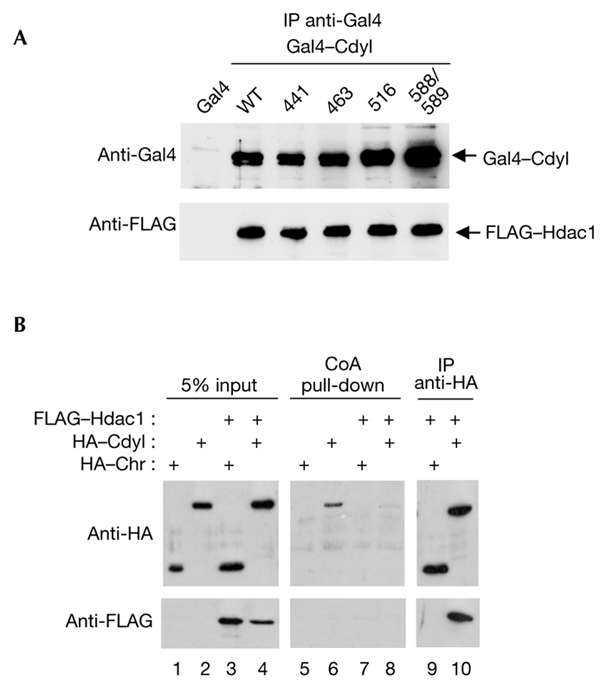
Coenzyme A and histone deacetylase binding by Cdyl. (A) Although they are unable to bind coenzyme A (CoA), Cdyl mutants can recruit histone deacetylase 1 (Hdac1). Wild-type (WT) Gal4–Cdyl and each of the four Gal4–Cdyl mutants described in the legend of Fig. 3 were co-expressed with FLAG–Hdac1. Twenty-four hours post-transfection, extracts were prepared and Gal4 fusion proteins were immunoprecipitated (IP) using an anti-Gal4 antibody. The association of the co-expressed Hdac1 was then monitored using an anti-FLAG antibody. (B) Hdac1 interaction inhibits CoA binding by Cdyl. COS7 cells were transfected with expression vectors for haemagglutinin (HA)–Cdyl full-length (lanes 2 and 4) or chromodomain (Chr; lanes 1 and 3; as negative control), with (lanes 3 and 4) or without (lanes 1 and 2) the expression vector for FLAG–Hdac1. Whole-cell extracts were prepared and subjected to a CoA pull-down (lanes 5–8; corresponding to inputs 1–4, respectively) and (on extracts corresponding to inputs 3 and 4 only) to an anti-HA immunoprecipitation (lanes 9 and 10). The beads were then washed and the bound proteins were analysed by western blotting using anti-HA (upper panels) or anti-FLAG (lower panels) antibodies. Cdyl, chromodomain-Y-like.
Involvement of Cdyl in repression of cellular genes
Recently, Cdyl, Hdac1 and Hdac2 were shown to be part of a C-terminal binding protein (CtBP) transcriptional co-repressor complex that is involved in the repression of expression of the E-cadherin gene (Shi et al., 2003). To investigate the specific function of Cdyl in the repression of E-cadherin, Cdyl-encoding constructs were transfected with a luciferase reporter plasmid driven by the E-cadherin core promoter (Comijn et al., 2001). Figure 6 shows that Cdyl has an important function in the repression of the E-cadherin promoter (Fig. 6A). It also confirms the participation of CtBP in this repression, as the co-transfection of a vector that expresses a small interfering RNA against CtBP (Shi et al., 2003) partially relieves the Cdyl-dependent repression of E-cadherin (Fig. 6B).
Figure 6.
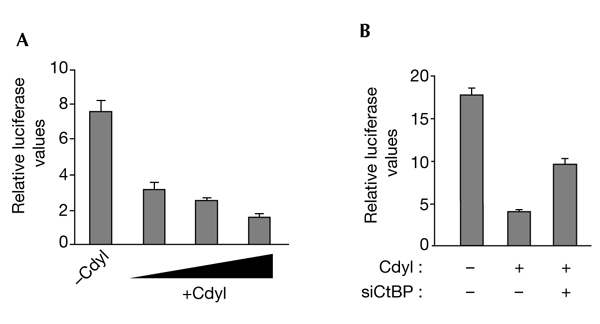
Cdyl is involved in the repression of the E-cadherin gene. (A) 293T cells were co-transfected with 20 ng of a reporter plasmid containing the E-cadherin promoter (positions −308 to +40) and increasing amounts (8, 16 or 50 ng) of a haemagglutinin (HA)–Cdyl expression vector. (B) 293T cells were co-transfected with 20 ng of a reporter plasmid that contains the E-cadherin promoter in the presence (+) or absence (−) of 12.5 ng of an HA–Cdyl expression vector and 40 ng of a C-terminal binding protein small interfering RNA plasmid (siCtBP; Shi et al., 2003). Luciferase activity was measured and normalized as described in the legend to Fig. 1. Cdyl, chromodomain-Y-like.
The disappearance of HDACs during mouse spermiogenesis
Our data, and those reported by Shi and colleagues, demonstrate the transcriptional repressor function of Cdyl in somatic cells. However, Cdyl is highly expressed in elongating spermatids at the same time as histone acetylation (Lahn et al., 2002), at a stage in which there is almost no transcription. In these cells, Cdyl should therefore have functions other than transcriptional repression, which probably involve its catalytic domain and CoA-binding activity. Our data suggest that for Cdyl CoA-binding activity to be activated, the HDACs must somehow change qualitatively or quantitatively during spermiogenesis. To investigate this possibility, extracts from cells enriched in pachytene spermatocytes, round and elongating (RE) spermatids, and elongating and condensing (EC) spermatids (Fig. 7A) were used to obtain a western blot, which was successively probed with anti-Hdac1, anti-Hdac2, anti-acetylated-H4 and anti-Cdyl antibodies. Figure 7B shows that Hdac1 and Hdac2 disappear in the elongating and condensing spermatids. Interestingly, these cells contain a significant amount of Cdyl and accumulate hyperacetylated histone H4. A northern blot confirmed that Cdyl is only expressed in spermatids and is present in RE and EC spermatid fractions before and after the disappearance of the HDACs (Fig. 7C). These observations suggest that Cdyl could be one of the proteins involved in inducing histone hyperacetylation during spermatid maturation, as previously suggested (Lahn et al., 2002).
Figure 7.
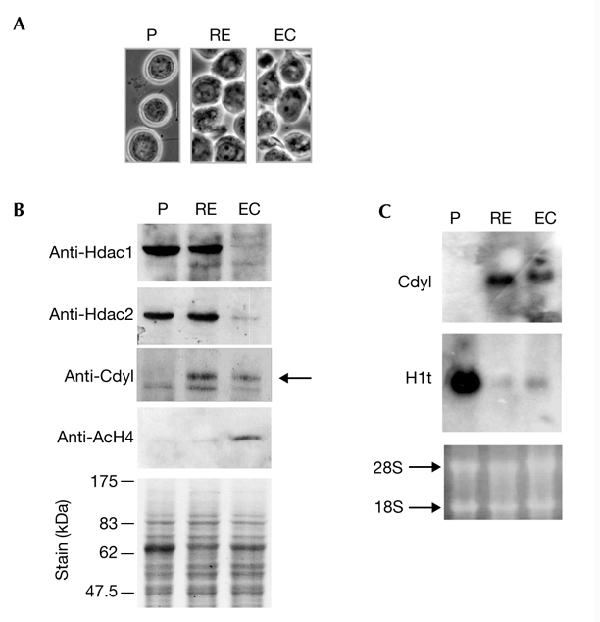
Histone hyperacetylation during spermiogenesis is associated with the disappearance of endogenous histone deacetylases 1 and 2. (A,B) Equivalent amounts of extracts were prepared from fractionated spermatogenic cells enriched in pachytene spermatocytes (P), round and elongating spermatids (RE) and elongating and condensing spermatids (EC; corresponding cell fractions shown in (A)). These were used to prepare western blots (B), which were successively probed with the following antibodies: anti-histone deacetylase 1 (Hdac1), anti-Hdac2, anti-Cdyl (chromodomain-Y-like) and anti-acetylated histone H4 (AcH4). The arrowhead indicates the Cdyl protein. (C) RNA prepared from the indicated fractions was used to obtain a northern blot, which was successively probed with Cdyl and testis H1 (H1t) probes. The ethidium-bromide-stained gel before transfer is shown (lower panel).
Discussion
Here, Cdyl is characterized as a new co-repressor of transcription that is able to recruit Hdac1 and Hdac2. Interestingly, while this manuscript was under consideration, Shi and colleagues published evidence for the association of endogenous Cdyl with endogenous Hdac1/Hdac2 in a CtBP-containing co-repressor complex, which functions in the repression of E-cadherin gene expression (Shi et al., 2003). We have confirmed these data and, moreover, have demonstrated the specific involvement of Cdyl in repressing the E-cadherin promoter.
Our data show that the repressor domain of Cdyl has homology with enoyl-CoA hydratase/isomerase, and could directly recruit HDACs. We have characterized this domain further, and show that the homology between Cdyl and enoyl-CoA hydratase/isomerases has a functional significance, as Cdyl can also bind CoA. However, the ability of this domain to bind CoA is repressed by HDAC recruitment, and Cdyl can only bind CoA in the absence of HDACs. Interestingly, we have found that mouse spermiogenesis is a situation in which the cellular content of Hdac1 and Hdac2 markedly decreases, whereas Cdyl levels do not decrease. In condensing spermatids, the absence of Hdac1 and Hdac2 would therefore allow Cdyl to bind CoA. Recently, recombinant Cdyl has been shown to have an in vitro HAT activity and was suggested to participate in histone acetylation during spermiogenesis (Lahn et al., 2002). Although we were unable to detect significant HAT activity associated with recombinant Cdyl (data not shown), our data suggest that the possibility of a participation of Cdyl in histone/protein acetylation in elongating spermatids remains open. Indeed, in these cells, the decrease in Hdac1 and Hdac2 levels could allow the protein to bind CoA or its derivatives and to catalyse an enzymatic reaction.
Methods
Fractionation of spermatogenic cells.
Spermatogenic cells of all stages were recovered from the testes of an adult mouse. Cell suspensions enriched in mouse spermatogenic cells at specific stages of their differentiation were obtained by velocity sedimentation at unit gravity on a BSA gradient, in accordance with a method described previously (Bellve, 1993; Romrell et al., 1976). This technique allows germ cells to be separated according to their size and density.
Reporter-gene assays.
HeLa cells were transfected by the calcium phosphate co-precipitation method. 293T cells were transfected using the Fugene transfection system (Roche). Total amounts of cytomegalovirus (CMV) promoter-containing plasmids were adjusted with pcDNA empty vectors. In each transfection, 100 ng (HeLa cells) or 2 ng (293T cells) of a pCMV–β-galactosidase (β-Gal) control plasmid was also used for normalization purposes. Cells were collected 36 h post-transfection and used for reporter-gene assays. Luciferase and β-Gal activities were measured using luciferase and β-Gal assay systems from Promega and Clontech, respectively, and luciferase activity was normalized to that of β-Gal.
Immunoprecipitation and coenzyme A and glutathione-S-transferase pull-down assays.
COS7 or U2OS cells were transfected with the appropriate expression vectors using the Fugene transfection system (Roche). Transfected cells or germ cells were collected and resuspended in 100 μl (per 4 × 106 cells) of lysis buffer (20% glycerol, 3 mM MgCl2, 50 mM Hepes, pH 7.9, 0.1% Nonidet P40, 1 mM dithiothreitol, 0.5 mM PMSF (phenylmethylsulphonyl fluoride) supplemented with 500 mM KCl. After 20 min at 4 °C, extracts were centrifuged for 10 min at 12,000 r.p.m. Immunoprecipitations were performed on supernatants by adding 1 μg (per 100 μl of extract) of the appropriate antibodies and 25 μl of protein-G–sepharose for 1 h at 4 °C. After three washes with the lysis buffer, the immunocomplexes were denatured in Laemmli buffer and analysed by SDS–polyacrylamide gel electrophoresis using 8% gels, followed by western blotting. For Hdac1 and Hdac2 immunoprecipitations from germ-cell extracts, antibodies were covalently linked to protein-G–sepharose before incubation with the extracts, and complexes were eluted with glycine (0.1 M, pH 2.6) for 15 min at 25 °C.
For CoA pull-down assays, supernatants were diluted in lysis buffer to 150 mM KCl and incubated for 90 min at 4 °C with 20 μl of CoA–agarose beads (Sigma). Beads were then washed three times with 150 mM KCl lysis buffer, and the retained proteins were denatured in Laemmli buffer and analysed by western blotting with a mouse polyclonal anti-Cdyl antibody. GST pull-down experiments were carried out as described previously (Lemercier et al., 2002).
Antibodies.
Antibodies used for immunoprecipitations were anti-FLAG M2 (Sigma), anti-HA (Clone 3F10; Roche), anti-Gal4, anti-Hdac1 and anti-Hdac2 (Santa Cruz). For western blotting, the detection of HA-tagged and FLAG-tagged proteins was performed using mouse anti-HA (Clone 12CA5; Roche) and anti-FLAG M2 antibodies, respectively. Anti-Hdac3 antibody was purchased from Upstate. Anti-Cdyl antibody was obtained in our laboratory from mice immunized by the injection of bacterially expressed Cdyl.
Supplementary information is available at EMBO reports online (http://www.nature.com/embor/journal/vaop/ncurrent/extref/4-embor917-s1.pdf).
Supplementary Material
Supplementary information
Acknowledgments
We are grateful to D. Allis, S. Khorasanizadeh and B. Lahn for helpful discussions, to E. Seto for the gift of FLAG–Hdac1 and FLAG–Hdac2, and to Y. Shi for the gift of the CtBP siRNA plasmid. We are also grateful to S. Curtet and R. Iratni for technical assistance. This work was supported by the INSERM Assistance Médicale à la Procréation and Région Rhône-Alpes 'émergence' programmes. C.C. is supported by the INSERM 'délégation' programme and L.A.v.G. by a postdoctoral fellowship from the Fund for Scientific Research Flanders, and he thanks D. Huylebroeck for support.
References
- Bellve A.R. ( 1993) Purification, culture, and fractionation of spermatogenic cells. Methods Enzymol., 225, 84–113. [DOI] [PubMed] [Google Scholar]
- Comijn J., Berx G., Vermassen P., Verschueren K., van Grunsven L.A., Bruyneel E., Mareel M., Huylebroek D. & van Roy F. ( 2001) The two handed E-box binding zinc finger protein SIP1 downregulates E-cadherin and induces invasion. Mol. Cell, 7, 1267–1278. [DOI] [PubMed] [Google Scholar]
- Engel C.K., Mathieu M., Zeelen J.P., Hiltunen J.K. & Wierenga R.K. ( 1996) Crystal structure of enoyl-coenzyme A (CoA) hydratase at 2.5 angstroms resolution: a spiral fold defines the CoA-binding pocket. EMBO J., 15, 5135–5145. [PMC free article] [PubMed] [Google Scholar]
- Lahn B.T. & Page D.C. ( 1999) Retroposition of autosomal mRNA yielded testis-specific gene family on human Y chromosome. Nature Genet., 21, 429–433. [DOI] [PubMed] [Google Scholar]
- Lahn B.T., Tang Z.L., Zhou J., Barndt R.J., Parvinen M., Allis C.D. & Page D.C. ( 2002) Previously uncharacterized histone acetyltransferases implicated in mammalian spermatogenesis. Proc. Natl Acad. Sci. USA, 99, 8707–8712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemercier C., Brocard M.P., Puvion-Dutilleul F., Kao H.Y., Albagli O. & Khochbin S. ( 2002) Class II histone deacetylases are directly recruited by BCL6 transcriptional repressor. J. Biol. Chem., 277, 22045–22052. [DOI] [PubMed] [Google Scholar]
- Romrell L.J., Bellve A.R. & Fawcett D.W. ( 1976) Separation of mouse spermatogenic cells by sedimentation velocity. Dev. Biol., 49, 119–131. [DOI] [PubMed] [Google Scholar]
- Shi J., Sawada J.I., Sui G., Affar E.B., Whetstine J.R., Lan F., Ogawa F., Po-Shan Luke M., Nakatani Y. & Shi Y. ( 2003) Coordinated histone modifications mediated by a CtBP co-repressor complex. Nature, 422, 735–738. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary information


