Abstract
Histone acetyltransferases and histone deacetylases regulate the acetylation of histones and transcription factors, and in doing so have major roles in the control of cell fate. Many recent results have indicated that their function is strictly regulated in cells through the modulation of their levels, activity and availability for interaction with specific transcription factors. In this review, we present the various molecular mechanisms that bring about this tight regulation and discuss how these regulatory events influence cellular responses to environmental changes.
Introduction
In eukaryotes, the packaging of DNA in chromatin interferes with DNA metabolic processes such as transcription, replication and DNA repair. Chromatin structure and function can be affected by various post-translational modifications of the amino-terminal tails of nucleosomal histones, of which lysine acetylation is the best characterized. Acetylation is thought to increase DNA accessibility through the neutralization of the positive charge of lysine residues. This modification correlates largely with transcriptional activation, but it is also involved in DNA replication, histone deposition and DNA repair. Histone acetylation also regulates protein–protein interactions, as some acetylated lysines are recognized by bromodomains, which are found in many proteins that regulate chromatin function (Strahl & Allis, 2000).
Histone acetylation is catalysed by histone acetyl transferases (HATs), whereas the reverse reaction is performed by histone deacetylases (HDACs). HATs and HDACs are classified into many families that are often conserved from yeast to humans (Marmorstein & Roth, 2001; Thiagalingam et al., 2003). For example, human class I, class II and class III HDACs are homologous to the yeast Rpd3, Hda1 and Sir2 HDACs, respectively. HATs and HDACs are usually embedded in large multimolecular complexes (Fig. 1), in which the other subunits function as cofactors for the enzyme, and they have a strict specificity for acetylation sites. HATs and HDACs participate in the genome-wide turnover of acetyl groups on histones and, in addition, some also modify other factors. Through their physical interaction with sequence-specific transcription factors, they are also targeted to specific promoters (Fig. 1), where they locally modify histones or transcription factors and thus regulate gene transcription.
Figure 1.
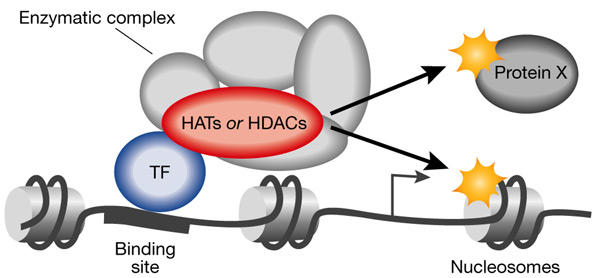
Model of local action of histone acetyltransferases and histone deacetylases. Histone acetyltransferases (HATs) and histone deacetylases (HDACs) are recruited to their target promoters through a physical interaction with a sequence-specific transcription factor (TF). They usually function within a multimolecular complex ('enzymatic complex'), in which the other subunits are necessary for them to modify nucleosomes around the binding site. These enzymes can also modify factors other than histones (protein X) to regulate transcription. Note that the position of the modified nucleosome that is shown has been chosen at random for this figure.
Because of the importance of histone acetylation in chromatin function, HATs and HDACs have major roles in the control of cell fate and their misregulation is involved in the development of some human tumours (Timmermann et al., 2001). Moreover, they are targeted by many viral proteins, which often affect their activity (Caron et al., 2003). Consistent with the importance of HATs and HDACs, they are tightly regulated in living cells and their activity is modulated by signalling pathways.
Although many reviews have focused on the various HAT and HDAC families and their roles in chromatin function, transcriptional regulation or cell fate (for example, see Marmorstein & Roth, 2001; Thiagalingam et al., 2003), none has extensively explored how their function can be regulated. Here, we describe the various mechanisms by which mammalian cells control HAT and HDAC activity, grouping them into three main classes that regulate the amount of enzyme, their enzymatic activity, or their availability for interaction with specific transcription factors.
Regulating enzyme quantity
As for all other proteins, an obvious way to regulate the activity of HATs and HDACs is to regulate their expression, and indeed the transcription of some of these enzymes is known to be tightly regulated during development. Although the molecular mechanisms of this regulation and the signals involved have, in most cases, not been defined, some interesting insights into the transcriptional regulation of the Hdac1 gene promoter have been obtained. For example, Hdac1 messenger RNA expression is induced by histone hyperacetylation, which suggests that a feedback loop controls histone acetylation levels in vivo (Hauser et al., 2002). In addition, Hdac1 mRNAs levels increase in a mouse lymphocyte cell line that has been stimulated with growth factors (for example, interleukin-2 (IL-2); Bartl et al., 1997), and in Swiss 3T3 cells that have been stimulated by serum (Hauser et al., 2002). This induction might be of crucial importance for cell growth, as the inactivation of Hdac1 causes proliferation defects in mice (Lagger et al., 2002).
The intracellular level of a protein depends on the rate of its translation and its half-life, and so far, one example of the regulation of HAT stability has been described. Tip60, a HAT that is involved in apoptosis and DNA repair after double-stranded breaks, is degraded by the proteasome after ubiquitylation by the ubiquitin ligase Mdm2 (Legube et al., 2002). Interestingly, DNA-damage-induced signalling leads to Tip60 accumulation, which suggests that the response to DNA damage involves the precise control of Tip60 expression. Similarly, Wiper-Bergeron and collaborators found that Hdac1 is also subject to proteasome-dependent degradation and that this process is important for glucocorticoid-induced preadipocyte differentiation (Wiper-Bergeron et al., 2003).
Regulating enzyme activity
Two main mechanisms that regulate the enzymatic activity of HATs and HDACs have been described: post-translational modifications (see supplementary information online for a more complete list of HAT and HDAC post-translational modifications) and protein–protein interactions. In addition, the availability of metabolic cofactors might also influence acetylation levels.
Post-translational modifications. The activities of many HATs and HDACs have been shown to be regulated through phosphorylation. For example, the HAT activity of CREB-binding protein (CBP) is stimulated on phosphorylation by cyclin E/cyclin-dependent kinase 2 (Aitsi-Ali et al., 1998). This event could be important for cell-cycle progression, as the HAT activity of CBP is required for progression to S phase.
The cellular response to DNA damage also involves changes in the HAT activity of some enzymes as a result of their phosphorylation. For example, activating transcription factor 2 (ATF2) is a sequence-specific transcription factor that has HAT activity and is phosphorylated in response to irradiation with ultraviolet light (Kawasaki et al., 2000). This phosphorylation increases ATF2's HAT activity, which leads to the transcriptional activation of promoters containing cAMP-responsive elements, the DNA elements recognized by ATF2.
The enzymatic activities of HATs and HDACs can also be regulated by other modifications; for example, HDAC4 has been shown to be sumoylated by the SUMO (small ubiquitin-related modifier) E3 ligase RANBP2 (Kirsh et al., 2002). This seems to be important for enzyme function, as a point mutation in the sumoylated residue reduces HDAC4 activity.
Protein–protein interactions. Both HATs and HDACs are usually part of large, multimolecular complexes, which contain other components that are often required for enzyme activity. However, the modulation of this activity through the regulation of complex assembly has not been seen so far. By contrast, many examples of the control of HAT activity by factors that are not bona fide components of the complex have been described.
The activity of CBP or the closely related p300 HAT has been shown to be stimulated in cis by a variety of sequence-specific transcription factors such, as HNF1-α, HNF4, Sp1, Zta, NF-E2, C/EBP-α and phosphorylated Elk1 (Chen et al., 2001; Li et al., 2003; Soutoglou et al., 2001). Through this stimulation, these sequence-specific transcription factors are thought to increase the acetylation of histones (or other transcription factors) at their target promoters. By contrast, other transcription factors, such as Msx3, Hox proteins and Twist, block HAT activity (Hamamori et al., 1999; Mehra-Chaudhary et al., 2001; Shen et al., 2001), and this property is shared by the Rsk2 kinase (Merienne et al., 2001). Interestingly, some members of this latter class have been shown to exert an effect in trans: for example, Twist can repress the activity of many transcription factors that function through CBP or p300/CBP-associated factor (pCAF; Hamamori et al., 1999). Thus, binding of a transcription factor to a HAT can affect gene expression through other transcription factors.
The activity of acetylation-controlling enzymes can also be modulated through the recruitment to the complex of an enzyme that catalyses the reverse reaction. Indeed, some complexes containing both HAT and HDAC activities have been characterized. For example, sumoylated p300 can interact with HDAC6, and this interaction brings about transcriptional repression (Girdwood et al., 2003).
Availability of metabolic cofactors. The activity of enzymes can also be regulated by the availability of cofactors. HAT activity is dependent on the presence of acetyl-coenzyme A and it is therefore possible that intracellular levels of this cofactor might be used to control acetylation levels. However, this has not been described so far. The deacetylation reaction catalysed by class I as well as class II HDACs does not require any cofactors, whereas deacetylation by HDACs of the Sir2 family is dependent on the presence of NAD+ (Denu, 2003). Interestingly, a genetic link has been established between Sir2 and some enzymes belonging to the NAD+ metabolic pathways, which suggests that metabolic networks and acetylation levels are coupled.
Regulating enzyme availability
Many transcription factors regulate transcription by physically recruiting HATs and HDACs to promoters (Fig. 1), and the modulation of this interaction by signalling pathways has been widely documented. In many cases, this regulation is achieved by post-translational modifications of the transcription factors, but this is outside the scope of this review. However, some examples have been described in which the enzyme itself is directly targeted. The availability of HATs or HDACs can be regulated by changing either their subcellular localization or their capacity to interact with specific transcription factors.
The former is an attractive way to control acetylation levels, as cytoplasmic enzymes cannot modify chromatin-incorporated histones. The best-characterized example of such a mode of regulation is that of the class II HDACs (see below). Another example is HDAC3, which can be relocated to the cytoplasm by its physical interaction with the adaptor TAB2 protein in the presence of IL-1β (Baek et al., 2002).
The availability of HATs and HDACs for a given signalling event can also be modulated in a more specific manner by changing their ability to be recruited by specific transcription factors. For example, CBP is phosphorylated in its GF box, and this phosphorylation is required for the recruitment of CBP by the AP1 transcription factor (Zanger et al., 2001). The methylation of CBP within its CREB-binding KIX domain by coactivator arginine methyltransferase 1 (CARM1) decreases its affinity for phosphorylated CREB, so that it is then available to interact with other transcription factors (Xu et al., 2001). Together, these results suggest that the range of transcription factors that can recruit CBP is affected significantly by its post-translational modifications.
What happens during signalling?
In general, the extent to which the molecular events described above affect histone or protein acetylation levels in response to a signal is still poorly understood. However, recent studies have described in detail some molecular mechanisms that might represent paradigms of what happens during signalling.
One example is the phosphorylation of class II HDACs that occurs during muscle differentiation. Some type II HDACs, such as HDAC5, can bind to and repress the activity of the myocyte enhancer factor 2 (MEF2) transcription factor, which is important for muscle differentiation (McKinsey et al., 2001). On induction of muscle differentiation, these class II enzymes are phosphorylated by Ca2+/calmodulin-dependent protein kinases (CaMKs) and are thereby relocalized to the cytoplasm. This regulation is of crucial importance for muscle differentiation. Indeed, an HDAC5 that is mutated in its phosphorylation site is constitutively localized to the nucleus and is a potent inhibitor of myogenesis. Conversely, an HDAC5 mutant that is retained in the cytoplasm is unable to inhibit muscle differentiation. These results have led to a model in which muscle-specific genes are repressed in proliferating myoblasts through the actions of class II HDACs associated with MEF2 (McKinsey et al., 2001). On the signal to differentiate, phosphorylation of these HDACs creates a binding site for the 14-3-3 chaperone proteins, which leads to their nuclear export, and muscle-specific genes are expressed. Consistent with this model, an increase in histone acetylation of MEF2-dependent muscle-specific genes can be detected during myogen-esis (Lu et al., 2000). Thus, during the process of muscle differentiation, phosphorylation of class II HDACs at the endpoints of signal transduction pathways seems to be directly responsible for the regulation of histone acetylation on specific promoters.
Another interesting example is the activation of immediate-early genes after the stimulation of resting cells to proliferate. On growth factor treatment, the signal is transduced through the mitogen-activated protein kinase (MAPK) pathway to the nucleus. Activation of some immediate-early genes, such as cIL8, relies on the phosphorylation of the transcription factor Elk1 by Mapk. The involvement of p300 or CBP in immediate-early gene transactivation has been widely documented, but the molecular mechanisms underlying their action have remained largely unknown. Li and colleagues recently reported that p300 was constantly present on the cIL8 promoter, probably through its interaction with unphosphorylated Elk1 (Li et al., 2003). After signalling, Elk1 phosphorylation changes the Elk1/p300 contacts and p300 HAT activity is induced. Thus, in this case, signal-induced gene transactivation is likely to be due to the stimulation of p300 HAT activity after changes in its interactions with a transcription factor.
Perspectives
Besides the regulatory events described above, HATs and HDACs are subject to a variety of post-translational modifications, the molecular roles of which remain largely uncharacterized (see supplementary information online). In some cases, these modifications are likely to have an important role, as they affect the transcriptional activity of HATs and HDACs. For example, phosphorylation of p300 at its carboxy terminus increases its ability to coactivate the CEBP-β transcription factor (Schwartz et al., 2003), whereas phosphorylation of CBP by CaMK favours its ability to mediate CREB transcriptional activation (Impey et al., 2002). In addtition, CBP methylation by CARM1 outside the KIX domain is important for its ability to function as a coactivator for the oestrogen receptor (Chevillard-Briet et al., 2002). In all these cases, it would be interesting to investigate whether these modifications affect the acetylation status of specific promoters.
All of the studies described in this review have focused on the molecular events that target the HATs and HDACs themselves. However, bona fide enzymes are usually multimolecular complexes in which cofactors are required for the catalytic subunit to modify nucleosomes. It will be informative to investigate to what extent these cofactors are subject to regulation, and whether signalling can affect their activity. In addition, according to the 'histone code' hypothesis (Strahl & Allis, 2000), which states that the various histone modifications act interdependently to specify a given function for chromatin, acetylation is linked to other histone modifications. Furthermore, HATs and HDACs function in a concerted way with ATP-dependent chromatin-remodelling complexes. Regulation of any of these enzymes, therefore, could add another layer of complexity to the acetylation status of specific promoters.
Finally, a striking proportion of the molecular events described above lead to the regulation of the CBP/p300 enzymes (see supplementary information online; Fig. 2). CBP and p300 have important roles in opposing processes such as cell proliferation and terminal differentiation (Goodman & Smolik, 2000). Moreover, they can co-activate many different transcription factors. Their levels are thought to be limiting in cells, and so their activity and availability are likely to be tightly regulated. Post-translational modifications of these proteins might act in concert to specify a functional state and, as already suggested (Gamble & Freedman, 2002), this could establish the 'CBP/p300 code' in a manner reminiscent of the popular histone code. The binding of proteins that affect CBP/p300 functions (see above) could also be involved in the establishment of the code. It will be interesting to determine whether such a code is restricted to CBP/p300 or applies more generally to the other HATs or HDACs that are important for cell fate.
Figure 2.
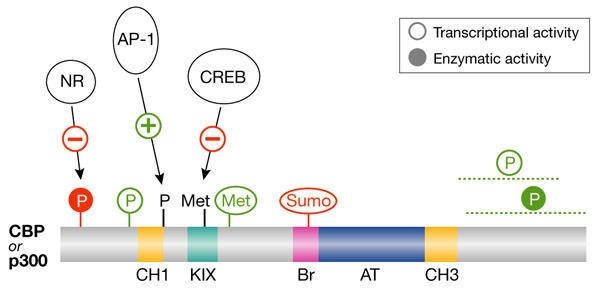
Functional 'code' of CREB-binding protein/p300 post-translational modifications. Representation of the CREB-binding protein (CBP) and p300 histone acetyl transferases including their functional domains (cysteine/histidine-rich domains (CH1 and CH3); CREB-binding domain (KIX); bromodomain (Br); and acetyl transferase domain (AT)) and some of their post-translational modifications (phosphorylation (P); methylation (Met); and sumoylation (Sumo)). The post-translational modifications activating (in green) or inhibiting (in red) CBP or p300 enzymatic activity (filled circles) or transcriptional activity (empty circles) are shown. The transcription factors for which binding to CBP or p300 is affected by CBP/p300 modifications are also shown. Note that some modifications have only been documented for CBP or for p300. The lines below some modifications indicate the region in which the modification occurs for sites that have not been fully characterized. NR, nuclear receptor superfamily.
Supplementary information is available at EMBO reports online (http://www.nature.com/embor/journal/vaop/ncurrent/extref/4-embor941-s1.pdf).
Supplementary Material
Supplementary information


D.T. is the recipient of an EMBO Young Investigator Award
Acknowledgments
We thank L. Vandel and V. Régnier for critical reading of the manuscript. We apologize to our colleagues for important studies that have not been cited due to space limitations. D.T. is supported by a grant from the Ligue Nationale Contre le Cancer as an Equipe Labellisée. G.L. is the recipient of a studentship from the Ligue Nationale Contre le Cancer.
References
- Ait-Si-Ali S. et al. ( 1998) Histone acetyltransferase activity of CBP is controlled by cycle-dependent kinases and oncoprotein E1A. Nature, 396, 184–186. [DOI] [PubMed] [Google Scholar]
- Baek S.H., Ohgi K.A., Rose D.W., Koo E.H., Glass C.K. & Rosenfeld M.G. ( 2002) Exchange of N-CoR corepressor and Tip60 coactivator complexes links gene expression by NF-κB and β-amyloid precursor protein. Cell, 110, 55–67. [DOI] [PubMed] [Google Scholar]
- Bartl S., Taplick J., Lagger G., Khier H., Kuchler K. & Seiser C. ( 1997) Identification of mouse histone deacetylase 1 as a growth factor-inducible gene. Mol. Cell. Biol., 17, 5033–5043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caron C., Col E. & Khochbin S. ( 2003) The viral control of cellular acetylation signaling. Bioessays, 25, 58–65. [DOI] [PubMed] [Google Scholar]
- Chen C.J., Deng Z., Kim A.Y., Blobel G.A. & Lieberman P.M. ( 2001) Stimulation of CREB binding protein nucleosomal histone acetyltransferase activity by a class of transcriptional activators. Mol. Cell. Biol., 21, 476–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevillard-Briet M., Trouche D. & Vandel L. ( 2002) Control of CBP co-activating activity by arginine methylation. EMBO J., 21, 5457–5466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denu J.M. ( 2003) Linking chromatin function with metabolic networks: Sir2 family of NAD+-dependent deacetylases. Trends Biochem. Sci., 28, 41–48. [DOI] [PubMed] [Google Scholar]
- Gamble M.J. & Freedman L.P. ( 2002) A coactivator code for transcription. Trends Biochem. Sci., 27, 165–167. [DOI] [PubMed] [Google Scholar]
- Girdwood D., Bumpass D., Vaughan O.A., Thain A., Anderson L.A., Snowden A.W., Garcia-Wilson E., Perkins N.D. & Hay R.T. ( 2003) p300 transcriptional repression is mediated by SUMO modification. Mol. Cell, 11, 1043–1054. [DOI] [PubMed] [Google Scholar]
- Goodman R.H. & Smolik S. ( 2000) CBP/p300 in cell growth, transformation, and development. Genes Dev., 14, 1553–1577. [PubMed] [Google Scholar]
- Hamamori Y., Sartorelli V., Ogryzko V., Puri P.L., Wu H.Y., Wang J.Y., Nakatani Y. & Kedes L. ( 1999) Regulation of histone acetyltransferases p300 and PCAF by the bHLH protein twist and adenoviral oncoprotein E1A. Cell, 96, 405–413. [DOI] [PubMed] [Google Scholar]
- Hauser C., Schuettengruber B., Bartl S., Lagger G. & Seiser C. ( 2002) Activation of the mouse histone deacetylase 1 gene by cooperative histone phosphorylation and acetylation. Mol. Cell. Biol., 22, 7820–7830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Impey S., Fong A.L., Wang Y., Cardinaux J.R., Fass D.M., Obrietan K., Wayman G.A., Storm D.R., Soderling T.R. & Goodman R.H. ( 2002) Phosphorylation of CBP mediates transcriptional activation by neural activity and CaM kinase IV. Neuron, 34, 235–244. [DOI] [PubMed] [Google Scholar]
- Kawasaki H., Schiltz L., Chiu R., Itakura K., Taira K., Nakatani Y. & Yokoyama K.K. ( 2000) ATF-2 has intrinsic histone acetyltransferase activity which is modulated by phosphorylation. Nature, 405, 195–200. [DOI] [PubMed] [Google Scholar]
- Kirsh O. et al. ( 2002) The SUMO E3 ligase RanBP2 promotes modification of the HDAC4 deacetylase. EMBO J., 21, 2682–2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagger G. et al. ( 2002) Essential function of histone deacetylase 1 in proliferation control and CDK inhibitor repression. EMBO J., 21, 2672–2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legube G., Linares L.K., Lemercier C., Scheffner M., Khochbin S. & Trouche D. ( 2002) Tip60 is targeted to proteasome-mediated degradation by Mdm2 and accumulates after UV irradiation. EMBO J., 21, 1704–1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q.J., Yang S.H., Maeda Y., Sladek F.M., Sharrocks A.D. & Martins-Green M. ( 2003) MAP kinase phosphorylation-dependent activation of Elk-1 leads to activation of the co-activator p300. EMBO J., 22, 281–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J., McKinsey T.A., Zhang C.L. & Olson E.N. ( 2000) Regulation of skeletal myogenesis by association of the MEF2 transcription factor with class II histone deacetylases. Mol. Cell, 6, 233–244. [DOI] [PubMed] [Google Scholar]
- Marmorstein R. & Roth S.Y. ( 2001) Histone acetyltransferases: function, structure, and catalysis. Curr. Opin. Genet. Dev., 11, 155–161. [DOI] [PubMed] [Google Scholar]
- McKinsey T.A., Zhang C.L. & Olson E.N. ( 2001) Control of muscle development by dueling HATs and HDACs. Curr. Opin. Genet. Dev., 11, 497–504. [DOI] [PubMed] [Google Scholar]
- Mehra-Chaudhary R., Matsui H. & Raghow R. ( 2001) Msx3 protein recruits histone deacetylase to down-regulate the Msx1 promoter. Biochem. J., 353, 13–22. [PMC free article] [PubMed] [Google Scholar]
- Merienne K., Pannetier S., Harel-Bellan A. & Sassone-Corsi P. ( 2001) Mitogen-regulated RSK2–CBP interaction controls their kinase and acetylase activities. Mol. Cell. Biol., 21, 7089–7096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz C., Beck K., Mink S., Schmolke M., Budde B., Wenning D. & Klempnauer K.H. ( 2003) Recruitment of p300 by C/EBPβ triggers phosphorylation of p300 and modulates coactivator activity. EMBO J., 22, 882–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen W.F., Krishnan K., Lawrence H.J. & Largman C. ( 2001) The HOX homeodomain proteins block CBP histone acetyltransferase activity. Mol. Cell. Biol., 21, 7509–7522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soutoglou E., Viollet B., Vaxillaire M., Yaniv M., Pontoglio M. & Talianidis I. Transcription factor-dependent regulation of CBP and P/CAF histone acetyltransferase activity. EMBO J., 20, 1984–1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl B.D. & Allis C.D. ( 2000) The language of covalent histone modifications. Nature, 403, 41–45. [DOI] [PubMed] [Google Scholar]
- Thiagalingam S., Cheng K.H., Lee H.J., Mineva N., Thiagalingam A. & Ponte J.F. ( 2003) Histone deacetylases: unique players in shaping the epigenetic histone code. Ann. NY Acad. Sci., 983, 84–100. [DOI] [PubMed] [Google Scholar]
- Timmermann S., Lehrmann H., Polesskaya A. & Harel-Bellan A. ( 2001) Histone acetylation and disease. Cell. Mol. Life Sci., 58, 728–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiper-Bergeron N., Wu D., Pope L., Schild-Poulter C. & Hache R.J. ( 2003) Stimulation of preadipocyte differentiation by steroid through targeting of an HDAC1 complex. EMBO J., 22, 2135–2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W., Chen H., Du K., Asahara H., Tini M., Emerson B.M., Montminy M. & Evans R.M. ( 2001) A transcriptional switch mediated by cofactor methylation. Science, 294, 2507–2511. [DOI] [PubMed] [Google Scholar]
- Zanger K., Radovick S. & Wondisford F.E. ( 2001) CREB binding protein recruitment to the transcription complex requires growth factor-dependent phosphorylation of its GF box. Mol. Cell, 7, 551–558. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary information


