Abstract
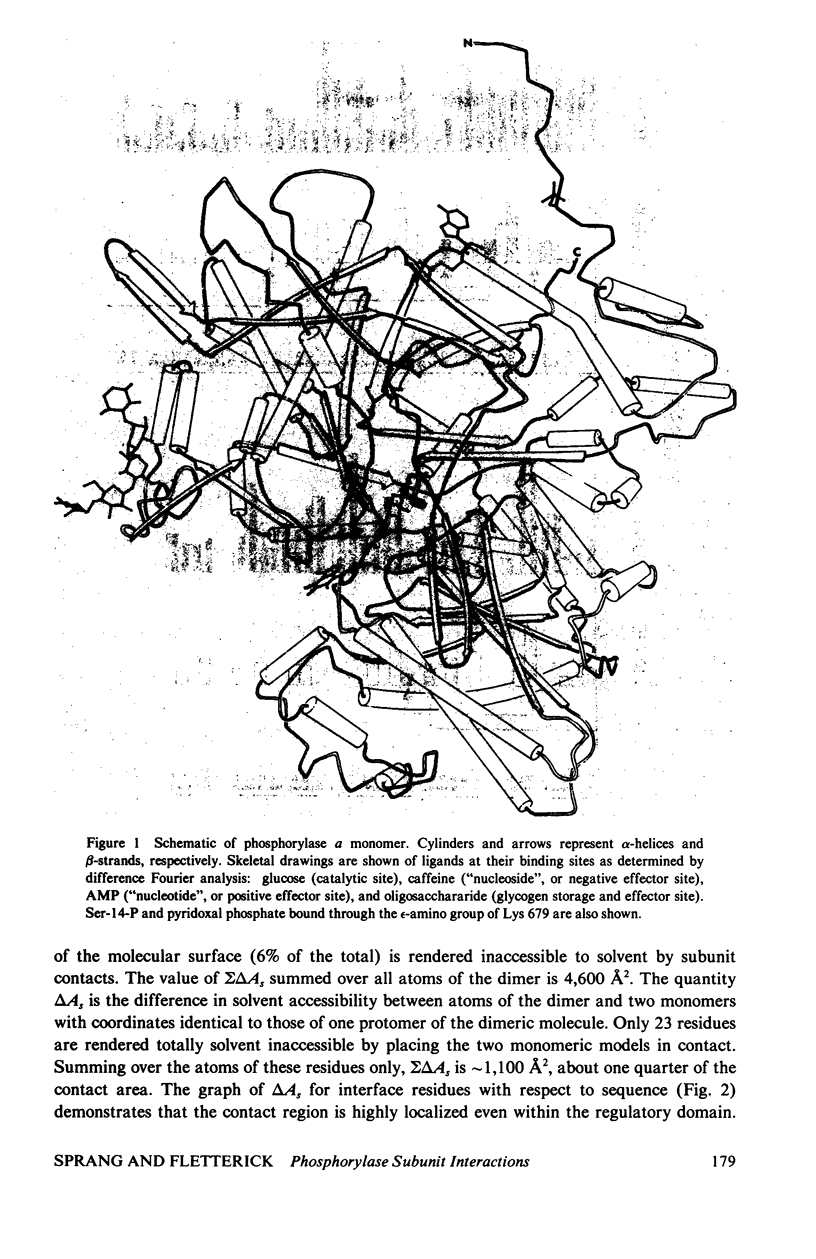
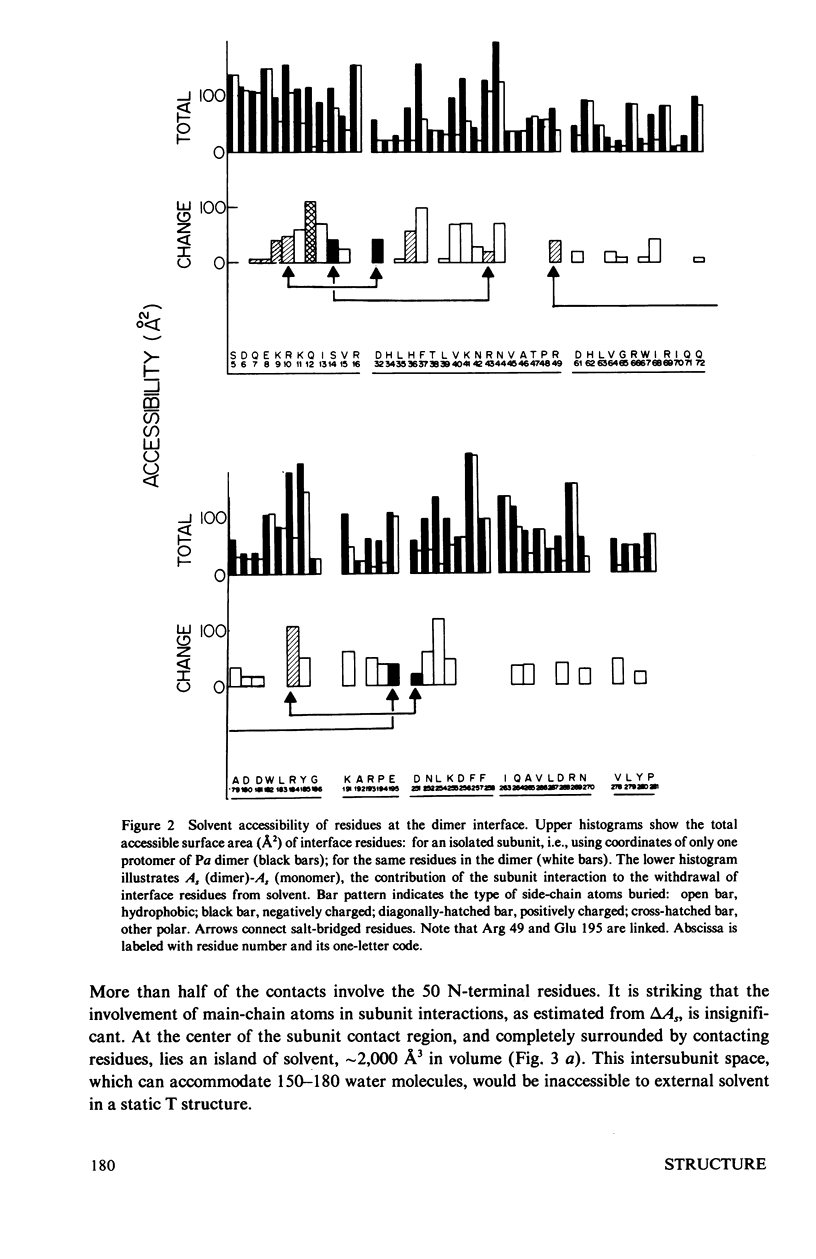
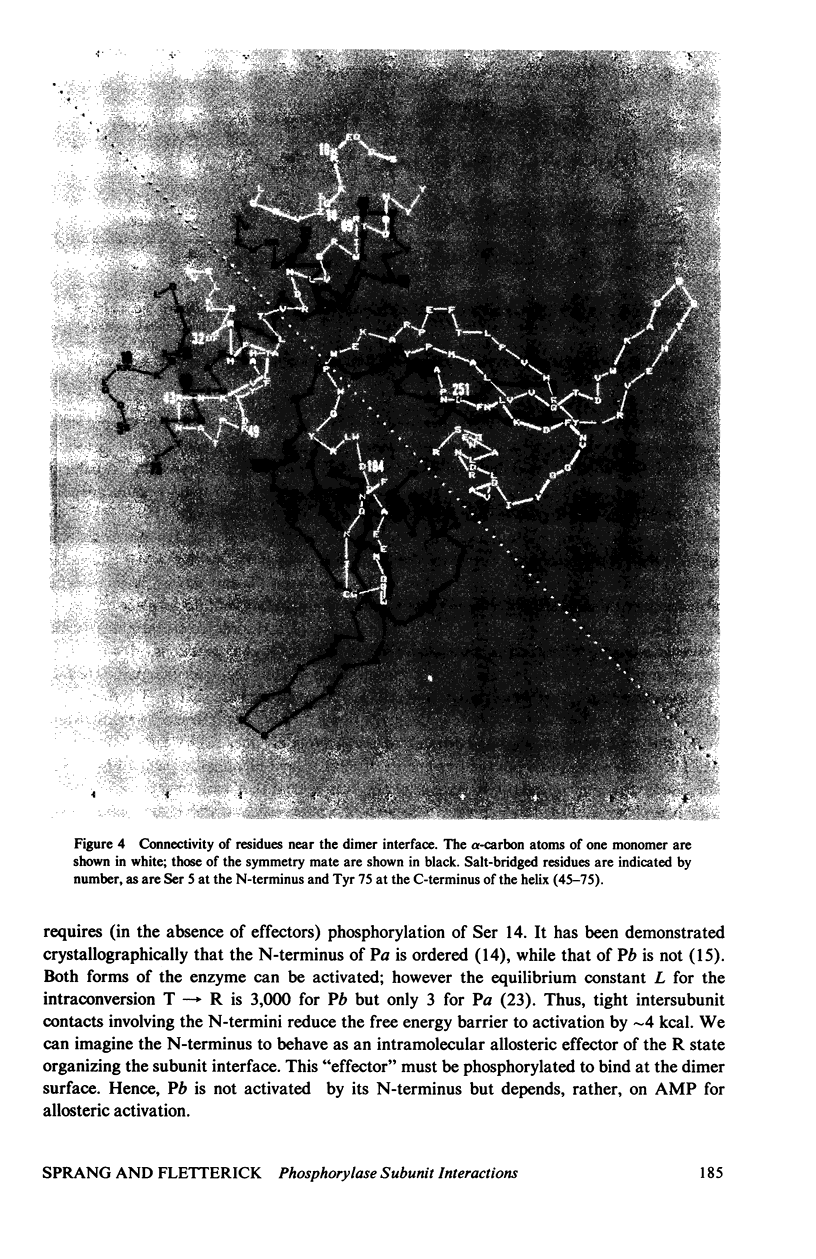
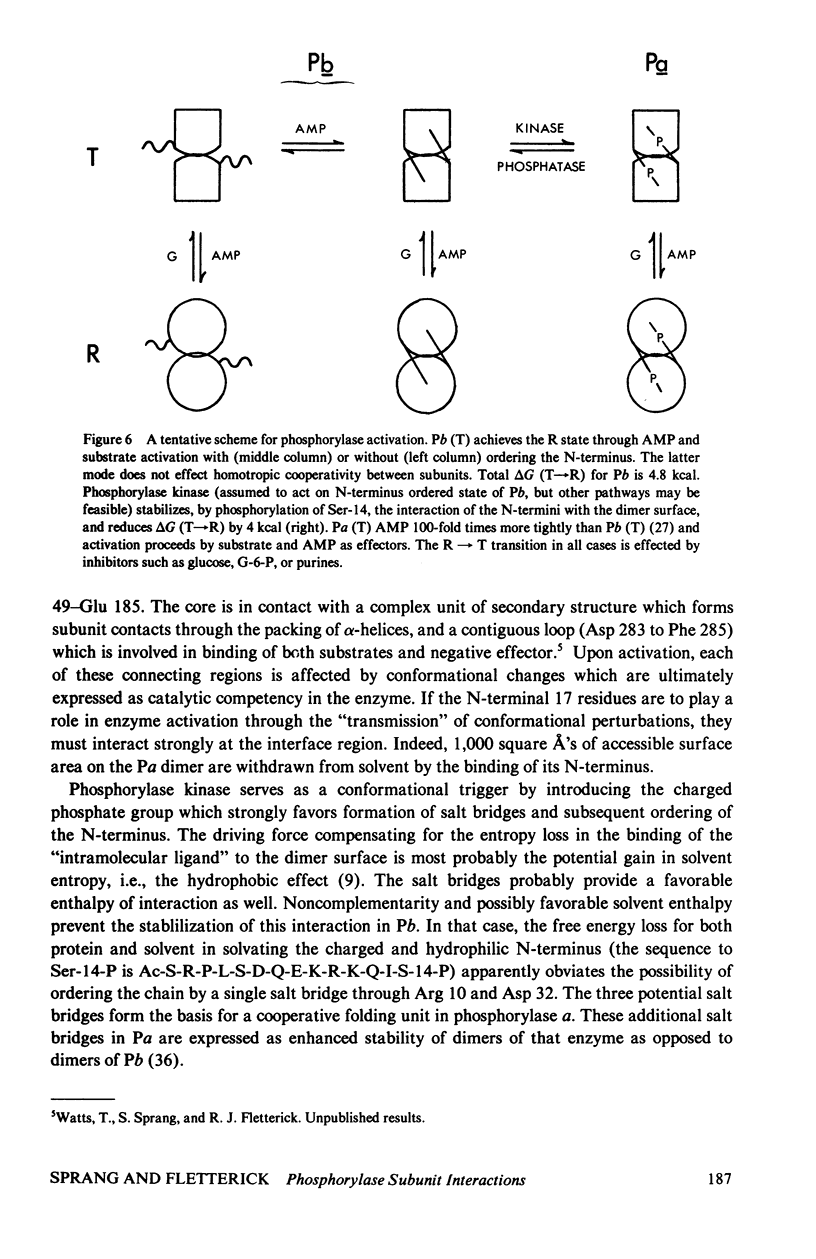
The contribution of intersubunit interactions to allosterically induced conformational changes in phosphorylase are considered. Phosphorylase a, Pa (phosphorylated at Ser-14), is significantly in the active (R) conformation, while phosphorylase b, Pb (nonphosphorylated), is predominantly in the inactive (T) conformation. The structure of glucose-inhibited (T) Pa has been determined at 2.5-A resolution and atomic coordinates have been measured. These data have been used to calculate the solvent accessible surface area at the subunit interface and map noncovalent interactions between protomers. The subunit contact involves only 6% of the Pa monomer surface, but withdraws an area of 4,600 A2 from solvent. The contact region is confined to the N-terminal (regulatory) domain of the subunit. Half of the residues involved are among the 70 N-terminal peptides. A total of approximately 100 atoms take part in polar or nonpolar contacts of less than 4.0 A with atoms of the symmetry-related monomer. The contact surface surrounds a central cavity at the core of the interface of sufficient volume to accommodate 150-180 solvent molecules. There are four intersubunit salt bridges. Two of these (Arg 10/Asp 32, Ser-14-P/Arg 43) are interactions between the N-terminus of one protomer with an alpha-helix loop segment near the N-terminus of the symmetry-related molecule. These two are relatively solvent accessible. The remainder (Arg 49/Glu 195, Arg 184/Asp 251) are nearer the interface core and are less accessible. The salt bridges at the N-terminus are surrounded by the polar and nonpolar contacts which may contribute to their stability. Analysis of the difference electron density between the isomorphous Pa and Pb crystal structures reveals that the N-terminal 17 residues of Pb are disordered. Pb thus lacks two intermolecular and one intersubunit (Ser-14-P/Arg 69) salt linkage present in Pa. The absence of these interactions in Pb is manifested in the difference in the free energy of T leads to R activation, which is 4 kcal more than that for Pa. Difference Fourier analysis of the T leads to R transition in substrate-activated crystals of Pa suggests that the 70 N-terminal residues undergo a concerted shift towards the molecular core; salt bridges are probably conserved in the transition. It is proposed that the N-terminus, when "activated" by phosphorylation (via a specific kinase) behaves as an intramolecular "effector" of the R state in phosphorylase and serves as the vehicle of homotropic cooperativity between subunits of the dimer.
Full text
PDF










Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baldwin J., Chothia C. Haemoglobin: the structural changes related to ligand binding and its allosteric mechanism. J Mol Biol. 1979 Apr 5;129(2):175–220. doi: 10.1016/0022-2836(79)90277-8. [DOI] [PubMed] [Google Scholar]
- Chothia C. Structural invariants in protein folding. Nature. 1975 Mar 27;254(5498):304–308. doi: 10.1038/254304a0. [DOI] [PubMed] [Google Scholar]
- Cohen P. The role of cyclic-AMP-dependent protein kinase in the regulation of glycogen metabolism in mammalian skeletal muscle. Curr Top Cell Regul. 1978;14:117–196. doi: 10.1016/b978-0-12-152814-0.50008-3. [DOI] [PubMed] [Google Scholar]
- FISCHER E. H., GRAVES D. J., CRITTENDEN E. R. S., KREBS E. G. Structure of the site phosphorylated in the phosphorylase b to a reaction. J Biol Chem. 1959 Jul;234(7):1698–1704. [PubMed] [Google Scholar]
- Fersht A. R. Conformational equilibria and the salt bridge in chymotrypsin. Cold Spring Harb Symp Quant Biol. 1972;36:71–73. doi: 10.1101/sqb.1972.036.01.012. [DOI] [PubMed] [Google Scholar]
- Fletterick R. J., Sprang S., Madsen N. B. Analysis of the surface topography of glycogen phosphorylase a: implications for metabolic interconversion and regulatory mechanisms. Can J Biochem. 1979 Jun;57(6):789–797. doi: 10.1139/o79-098. [DOI] [PubMed] [Google Scholar]
- Fletterick R. J., Sygusch J., Murray N., Madsen N. B. Low-resolution structure of the glycogen phosphorylase alpha monomer and comparison with phosphorylase beta. J Mol Biol. 1976 May 5;103(1):1–13. doi: 10.1016/0022-2836(76)90048-6. [DOI] [PubMed] [Google Scholar]
- Fletterick R. J., Sygusch J., Semple H., Madsen N. B. Structure of glycogen phosphorylase a at 3.0 A resolution and its ligand binding sites at 6 A. J Biol Chem. 1976 Oct 10;251(19):6142–6146. [PubMed] [Google Scholar]
- Graves D. J., Mann S. A., Philip G., Oliveira R. J. A probe into the catalytic activity and subunit assembly of glycogen phosphorylase. Desensitization of allosteric control by limited tryptic digestion. J Biol Chem. 1968 Dec 10;243(23):6090–6098. [PubMed] [Google Scholar]
- Greer J., Bush B. L. Macromolecular shape and surface maps by solvent exclusion. Proc Natl Acad Sci U S A. 1978 Jan;75(1):303–307. doi: 10.1073/pnas.75.1.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmreich E., Michaelides M. C., Cori C. F. Effects of substrates and a substrate analog on the binding of 5'-adenylic acid to muscle phosphorylase a. Biochemistry. 1967 Dec;6(12):3695–3710. doi: 10.1021/bi00864a012. [DOI] [PubMed] [Google Scholar]
- Hers H. G. The control of glycogen metabolism in the liver. Annu Rev Biochem. 1976;45:167–189. doi: 10.1146/annurev.bi.45.070176.001123. [DOI] [PubMed] [Google Scholar]
- Janski A. M., Graves D. J. An antibody probe for the amino-terminal region of glycogen phosphorylase. FEBS Lett. 1977 Oct 15;82(2):232–236. doi: 10.1016/0014-5793(77)80591-7. [DOI] [PubMed] [Google Scholar]
- Johnson L. N., Madsen N. B., Mosley J., Wilson K. S. The crystal structure of phosphorylase beta at 6 A resolution. J Mol Biol. 1974 Dec 25;90(4):703–717. doi: 10.1016/0022-2836(74)90534-8. [DOI] [PubMed] [Google Scholar]
- Kasvinsky P. J., Madsen N. B., Fletterick R. J., Sygusch J. X-ray crystallographic and kinetic studies of oligosaccharide binding to phosphorylase. J Biol Chem. 1978 Feb 25;253(4):1290–1296. [PubMed] [Google Scholar]
- Kasvinsky P. J., Madsen N. B., Sygusch J., Fletterick R. J. The regulation of glycogen phosphorylase alpha by nucleotide derivatives. Kinetic and x-ray crystallographic studies. J Biol Chem. 1978 May 10;253(9):3343–3351. [PubMed] [Google Scholar]
- Kasvinsky P. J., Shechosky S., Fletterick R. J. Synergistic regulation of phosphorylase a by glucose and caffeine. J Biol Chem. 1978 Dec 25;253(24):9102–9106. [PubMed] [Google Scholar]
- Koshland D. E., Jr, Némethy G., Filmer D. Comparison of experimental binding data and theoretical models in proteins containing subunits. Biochemistry. 1966 Jan;5(1):365–385. doi: 10.1021/bi00865a047. [DOI] [PubMed] [Google Scholar]
- Krebs E. G., Beavo J. A. Phosphorylation-dephosphorylation of enzymes. Annu Rev Biochem. 1979;48:923–959. doi: 10.1146/annurev.bi.48.070179.004423. [DOI] [PubMed] [Google Scholar]
- Lee B., Richards F. M. The interpretation of protein structures: estimation of static accessibility. J Mol Biol. 1971 Feb 14;55(3):379–400. doi: 10.1016/0022-2836(71)90324-x. [DOI] [PubMed] [Google Scholar]
- Levitt M., Chothia C. Structural patterns in globular proteins. Nature. 1976 Jun 17;261(5561):552–558. doi: 10.1038/261552a0. [DOI] [PubMed] [Google Scholar]
- Madsen N. B., KasvinskyPJ, Fletterick R. J. Allosteric transitions of phosphorylase a and the regulation of glycogen metabolism. J Biol Chem. 1978 Dec 25;253(24):9097–9101. [PubMed] [Google Scholar]
- Perutz M. F. Electrostatic effects in proteins. Science. 1978 Sep 29;201(4362):1187–1191. doi: 10.1126/science.694508. [DOI] [PubMed] [Google Scholar]
- Perutz M. F. Regulation of oxygen affinity of hemoglobin: influence of structure of the globin on the heme iron. Annu Rev Biochem. 1979;48:327–386. doi: 10.1146/annurev.bi.48.070179.001551. [DOI] [PubMed] [Google Scholar]
- Sprang S., Fletterick R. J. The structure of glycogen phosphorylase alpha at 2.5 A resolution. J Mol Biol. 1979 Jul 5;131(3):523–551. doi: 10.1016/0022-2836(79)90006-8. [DOI] [PubMed] [Google Scholar]
- Sprang S., Yang D., Fletterick R. J. Solvent accessibility properties of complex proteins. Nature. 1979 Jul 26;280(5720):333–335. doi: 10.1038/280333a0. [DOI] [PubMed] [Google Scholar]
- Sygusch J., Madsen N. B., Kasvinsky P. J., Fletterick R. J. Location of pyridoxal phosphate in glycogen phosphorylase a. Proc Natl Acad Sci U S A. 1977 Nov;74(11):4757–4761. doi: 10.1073/pnas.74.11.4757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber I. T., Johnson L. N., Wilson K. S., Yeates D. G., Wild D. L., Jenkins J. A. Crystallographic studies on the activity of glycogen phosphorylase b. Nature. 1978 Aug 3;274(5670):433–437. doi: 10.1038/274433a0. [DOI] [PubMed] [Google Scholar]