Abstract
X-ray crystallography can reveal the magnitudes and principal directions of the mean-square displacements of every atom in a protein. This structural information is complementary to the temporal information obtainable by spectroscopic techniques such as nuclear magnetic resonance. Determination of the temperature dependence of the mean-square displacements makes it possible to separate large conformational motions from simple thermal vibrations. The contribution of crystal lattice disorder to the overall apparent displacement can be estimated by Mössbauer spectroscopy. This technique has been applied to high resolution x-ray diffraction data from sperm whale myoglobin in its Met iron and oxy cobalt forms. Both crystal structures display regions of large conformational motions, particularly at the chain termini and in the region of the proximal histidine. Overall, the mean-square displacement increases with increasing distance from the center of gravity of the molecule. Some regions of the heme pocket in oxy cobalt myoglobin are more rigid than the corresponding regions in Met myoglobin.
Full text
PDF




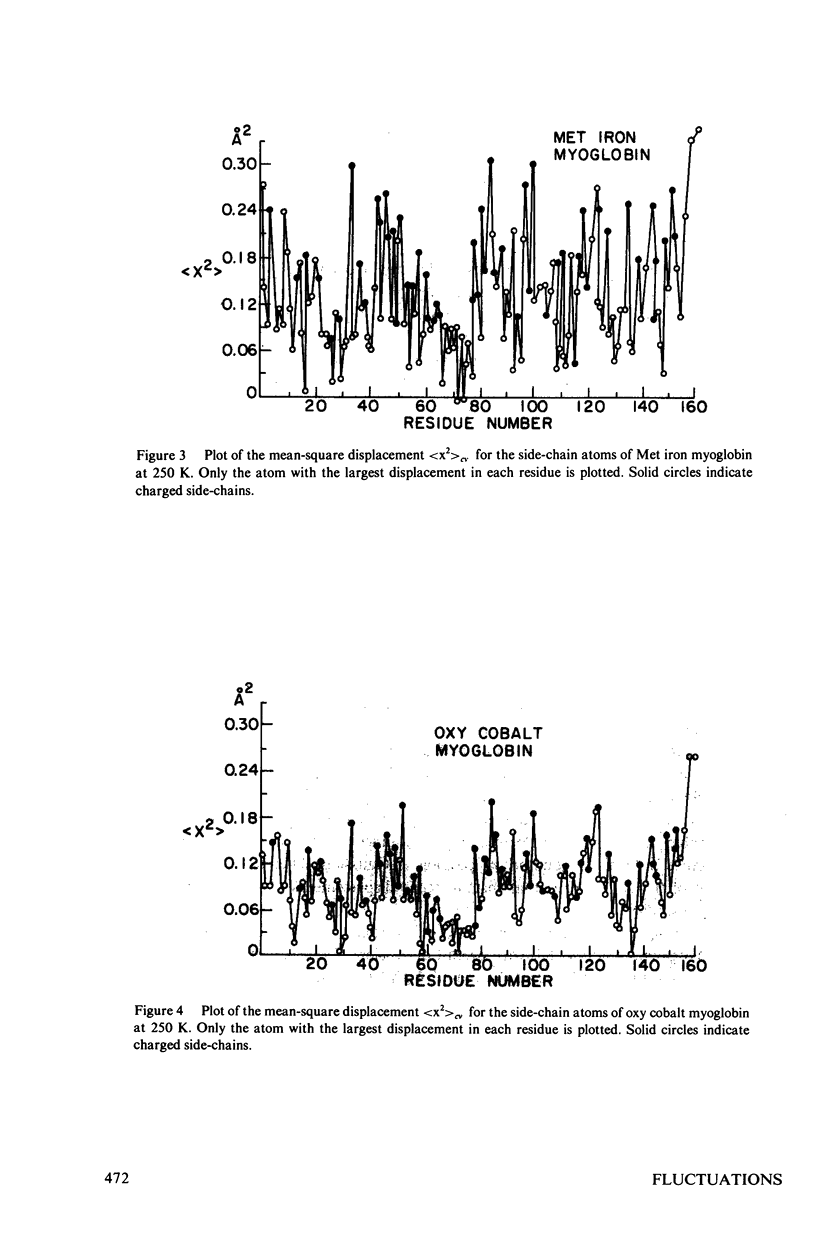

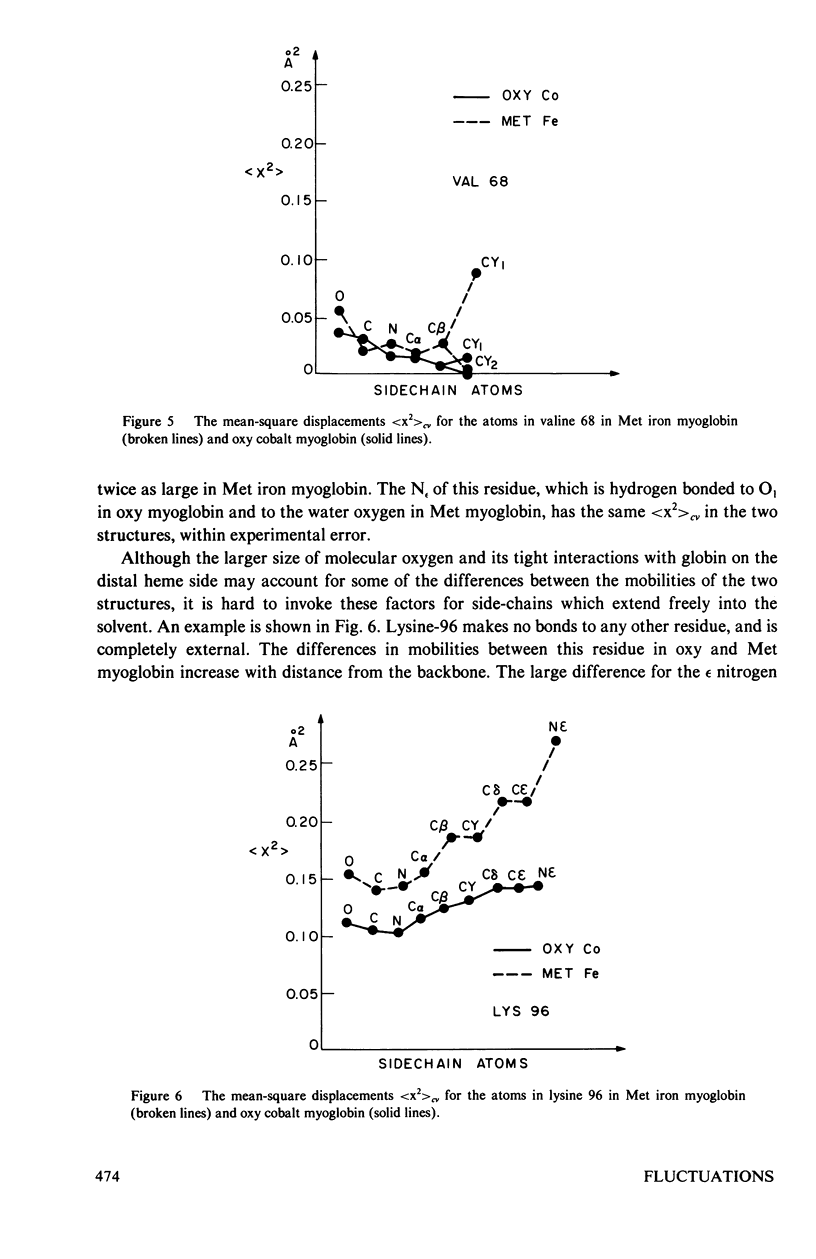




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Artymiuk P. J., Blake C. C., Grace D. E., Oatley S. J., Phillips D. C., Sternberg M. J. Crystallographic studies of the dynamic properties of lysozyme. Nature. 1979 Aug 16;280(5723):563–568. doi: 10.1038/280563a0. [DOI] [PubMed] [Google Scholar]
- Austin R. H., Beeson K. W., Eisenstein L., Frauenfelder H., Gunsalus I. C. Dynamics of ligand binding to myoglobin. Biochemistry. 1975 Dec 2;14(24):5355–5373. doi: 10.1021/bi00695a021. [DOI] [PubMed] [Google Scholar]
- Eftink M. R., Ghiron C. A. Dynamics of a protein matrix revealed by fluorescence quenching. Proc Natl Acad Sci U S A. 1975 Sep;72(9):3290–3294. doi: 10.1073/pnas.72.9.3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Englander S. W., Downer N. W., Teitelbaum H. Hydrogen exchange. Annu Rev Biochem. 1972;41:903–924. doi: 10.1146/annurev.bi.41.070172.004351. [DOI] [PubMed] [Google Scholar]
- Feldmann R. J. The design of computing systems for molecular modeling. Annu Rev Biophys Bioeng. 1976;5:477–510. doi: 10.1146/annurev.bb.05.060176.002401. [DOI] [PubMed] [Google Scholar]
- Frauenfelder H., Petsko G. A., Tsernoglou D. Temperature-dependent X-ray diffraction as a probe of protein structural dynamics. Nature. 1979 Aug 16;280(5723):558–563. doi: 10.1038/280558a0. [DOI] [PubMed] [Google Scholar]
- Hampton D. A., Brill A. S. Crystalline state disorder and hyperfine component line widths in ferric hemoglobin chains. Biophys J. 1979 Feb;25(2 Pt 1):301–311. doi: 10.1016/s0006-3495(79)85293-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopfield J. J. Relation between structure, co-operativity and spectra in a model of hemoglobin action. J Mol Biol. 1973 Jun 25;77(2):207–222. doi: 10.1016/0022-2836(73)90332-x. [DOI] [PubMed] [Google Scholar]
- Karplus M., McCammon J. A. Protein structural fluctuations during a period of 100 ps. Nature. 1979 Feb 15;277(5697):578–578. doi: 10.1038/277578a0. [DOI] [PubMed] [Google Scholar]
- McCammon J. A., Gelin B. R., Karplus M. Dynamics of folded proteins. Nature. 1977 Jun 16;267(5612):585–590. doi: 10.1038/267585a0. [DOI] [PubMed] [Google Scholar]
- Norvell J. C., Nunes A. C., Schoenborn B. P. Neutron diffraction analysis of myoglobin: structure of the carbon monoxide derivative. Science. 1975 Nov 7;190(4214):568–570. doi: 10.1126/science.1188354. [DOI] [PubMed] [Google Scholar]
- Petsko G. A. Protein crystallography at sub-zero temperatures: cryo-protective mother liquors for protein crystals. J Mol Biol. 1975 Aug 15;96(3):381–392. doi: 10.1016/0022-2836(75)90167-9. [DOI] [PubMed] [Google Scholar]
- Phillips S. E. Structure of oxymyoglobin. Nature. 1978 May 18;273(5659):247–248. doi: 10.1038/273247a0. [DOI] [PubMed] [Google Scholar]
- Sternberg M. J., Grace D. E., Phillips D. C. Dynamic information from protein crystallography. An analysis of temperature factors from refinement of the hen egg-white lysozyme structure. J Mol Biol. 1979 May 25;130(3):231–252. doi: 10.1016/0022-2836(79)90539-4. [DOI] [PubMed] [Google Scholar]
- Wittebort R. J., Rothgeb T. M., Szabo A., Gurd F. R. Aliphatic groups of sperm whale myoglobin: 13C NMR study. Proc Natl Acad Sci U S A. 1979 Mar;76(3):1059–1063. doi: 10.1073/pnas.76.3.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodward C. K., Hilton B. D. Hydrogen exchange kinetics and internal motions in proteins and nucleic acids. Annu Rev Biophys Bioeng. 1979;8:99–127. doi: 10.1146/annurev.bb.08.060179.000531. [DOI] [PubMed] [Google Scholar]
- Wyckoff H. W., Doscher M., Tsernoglou D., Inagami T., Johnson L. N., Hardman K. D., Allewell N. M., Kelly D. M., Richards F. M. Design of a diffractometer and flow cell system for X-ray analysis of crystalline proteins with applications to the crystal chemistry of ribonuclease-S. J Mol Biol. 1967 Aug 14;27(3):563–578. doi: 10.1016/0022-2836(67)90059-9. [DOI] [PubMed] [Google Scholar]