Abstract
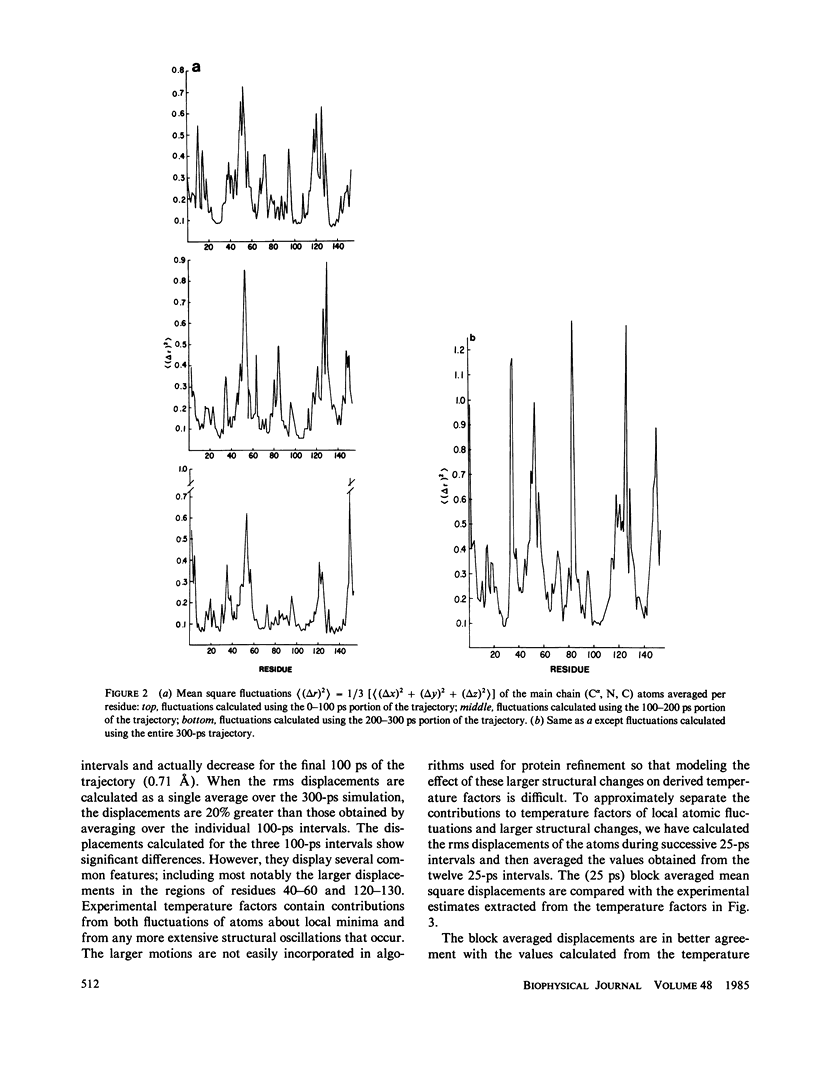
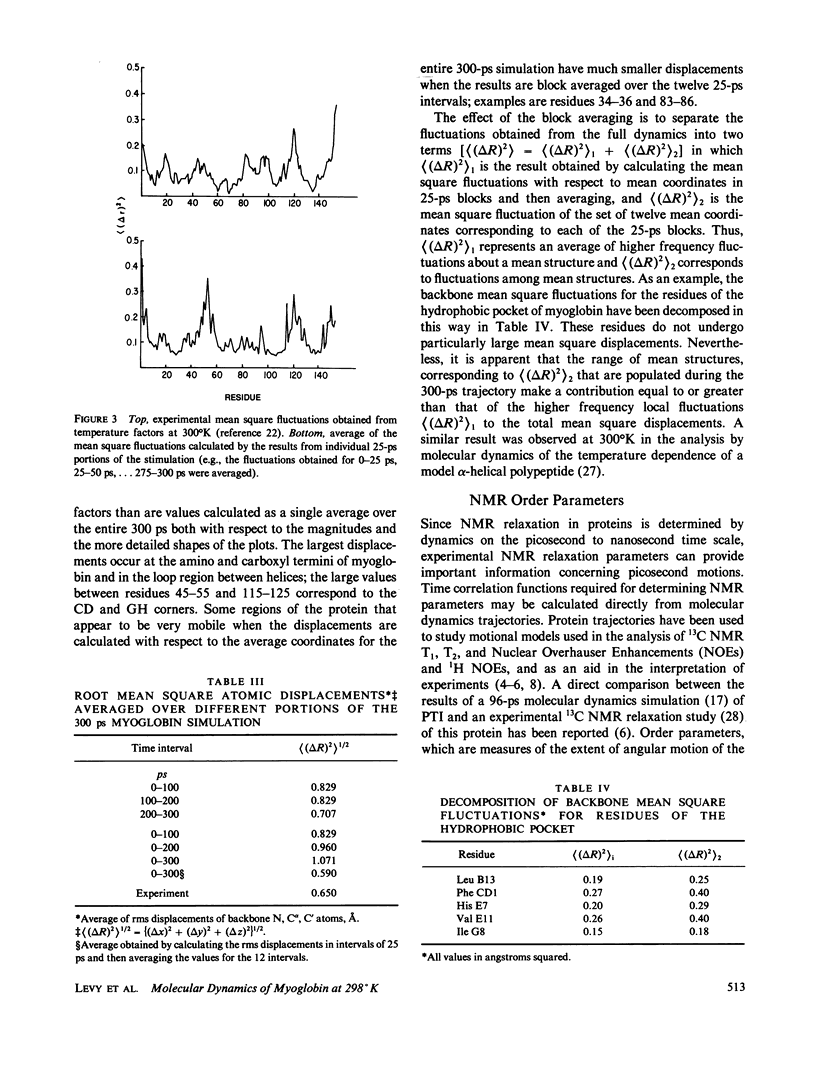
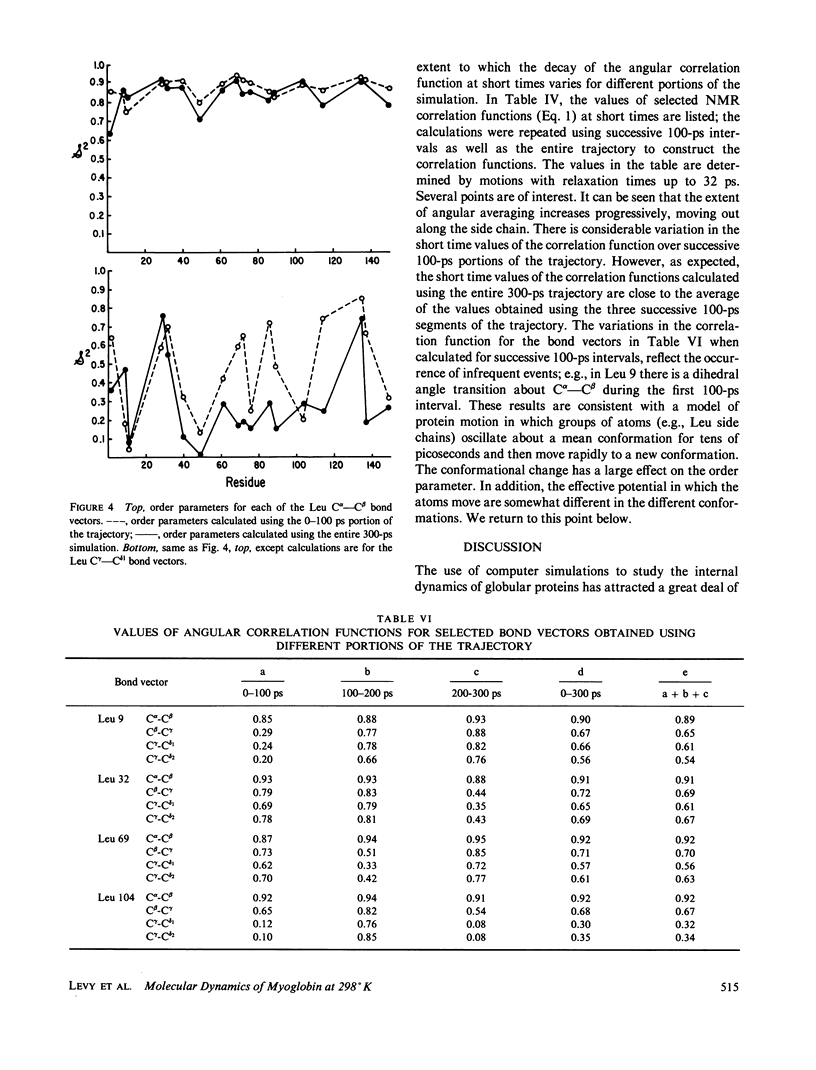
We have carried out a very long (300 ps) molecular dynamics simulation of the protein myoglobin. This trajectory is approximately three times longer than the longest previous molecular dynamics simulation of a protein, and ten times longer than protein simulations of comparable size (1,423 atoms in our model). Here we report results from this long simulation concerning the average structure, the mean square fluctuations of atoms about the average structure, and the nuclear magnetic resonance order parameters for various groups in myoglobin. The results demonstrate that the average coordinates change very slowly during the simulation. The relative atomic mobilities are well described by the simulation. For both the mean square atomic fluctuations and the order parameters, however, there are significant quantitative differences when values calculated using shorter portions of the trajectory are compared with results obtained for the entire 300-ps simulation. The implications of this result for obtaining converged properties from protein molecular dynamics simulations for comparison with experiment are discussed.
Full text
PDF






Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Artymiuk P. J., Blake C. C., Grace D. E., Oatley S. J., Phillips D. C., Sternberg M. J. Crystallographic studies of the dynamic properties of lysozyme. Nature. 1979 Aug 16;280(5723):563–568. doi: 10.1038/280563a0. [DOI] [PubMed] [Google Scholar]
- Frauenfelder H., Petsko G. A., Tsernoglou D. Temperature-dependent X-ray diffraction as a probe of protein structural dynamics. Nature. 1979 Aug 16;280(5723):558–563. doi: 10.1038/280558a0. [DOI] [PubMed] [Google Scholar]
- Hartmann H., Parak F., Steigemann W., Petsko G. A., Ponzi D. R., Frauenfelder H. Conformational substates in a protein: structure and dynamics of metmyoglobin at 80 K. Proc Natl Acad Sci U S A. 1982 Aug;79(16):4967–4971. doi: 10.1073/pnas.79.16.4967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoch J. C., Dobson C. M., Karplus M. Fluctuations and averaging of proton chemical shifts in the bovine pancreatic trypsin inhibitor. Biochemistry. 1982 Mar 16;21(6):1118–1125. doi: 10.1021/bi00535a002. [DOI] [PubMed] [Google Scholar]
- Karplus M., McCammon J. A. Protein structural fluctuations during a period of 100 ps. Nature. 1979 Feb 15;277(5697):578–578. doi: 10.1038/277578a0. [DOI] [PubMed] [Google Scholar]
- Karplus M., McCammon J. A. The internal dynamics of globular proteins. CRC Crit Rev Biochem. 1981;9(4):293–349. doi: 10.3109/10409238109105437. [DOI] [PubMed] [Google Scholar]
- Levitt M. Molecular dynamics of native protein. I. Computer simulation of trajectories. J Mol Biol. 1983 Aug 15;168(3):595–617. doi: 10.1016/s0022-2836(83)80304-0. [DOI] [PubMed] [Google Scholar]
- Levitt M. Molecular dynamics of native protein. II. Analysis and nature of motion. J Mol Biol. 1983 Aug 15;168(3):621–657. doi: 10.1016/s0022-2836(83)80306-4. [DOI] [PubMed] [Google Scholar]
- Levy R. M., Dobson C. M., Karplus M. Dipolar NMR relaxation of nonprotonated aromatic carbons in proteins. Structural and dynamical effects. Biophys J. 1982 Jul;39(1):107–113. doi: 10.1016/S0006-3495(82)84496-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy R. M., Karplus M., McCammon J. A. Molecular dynamics studies of NMR relaxation in proteins. Biophys J. 1980 Oct;32(1):628–630. doi: 10.1016/S0006-3495(80)84998-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy R. M., Perahia D., Karplus M. Molecular dynamics of an alpha-helical polypeptide: Temperature dependence and deviation from harmonic behavior. Proc Natl Acad Sci U S A. 1982 Feb;79(4):1346–1350. doi: 10.1073/pnas.79.4.1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao B., Pear M. R., McCammon J. A., Northrup S. H. Molecular dynamics of ferrocytochrome c: anharmonicity of atomic displacements. Biopolymers. 1982 Oct;21(10):1979–1989. doi: 10.1002/bip.360211005. [DOI] [PubMed] [Google Scholar]
- McCammon J. A., Gelin B. R., Karplus M. Dynamics of folded proteins. Nature. 1977 Jun 16;267(5612):585–590. doi: 10.1038/267585a0. [DOI] [PubMed] [Google Scholar]
- Northrup S. H., Pear M. R., Morgan J. D., McCammon J. A., Karplus M. Molecular dynamics of ferrocytochrome c. Magnitude and anisotropy of atomic displacements. J Mol Biol. 1981 Dec 25;153(4):1087–1109. doi: 10.1016/0022-2836(81)90469-1. [DOI] [PubMed] [Google Scholar]
- Petsko G. A., Ringe D. Fluctuations in protein structure from X-ray diffraction. Annu Rev Biophys Bioeng. 1984;13:331–371. doi: 10.1146/annurev.bb.13.060184.001555. [DOI] [PubMed] [Google Scholar]
- Richards F. M. Areas, volumes, packing and protein structure. Annu Rev Biophys Bioeng. 1977;6:151–176. doi: 10.1146/annurev.bb.06.060177.001055. [DOI] [PubMed] [Google Scholar]
- Richarz R., Nagayama K., Wüthrich K. Carbon-13 nuclear magnetic resonance relaxation studies of internal mobility of the polypeptide chain in basic pancreatic trypsin inhibitor and a selectively reduced analogue. Biochemistry. 1980 Nov 11;19(23):5189–5196. doi: 10.1021/bi00564a006. [DOI] [PubMed] [Google Scholar]
- Tainer J. A., Getzoff E. D., Alexander H., Houghten R. A., Olson A. J., Lerner R. A., Hendrickson W. A. The reactivity of anti-peptide antibodies is a function of the atomic mobility of sites in a protein. Nature. 1984 Nov 8;312(5990):127–134. doi: 10.1038/312127a0. [DOI] [PubMed] [Google Scholar]
- Westhof E., Altschuh D., Moras D., Bloomer A. C., Mondragon A., Klug A., Van Regenmortel M. H. Correlation between segmental mobility and the location of antigenic determinants in proteins. Nature. 1984 Sep 13;311(5982):123–126. doi: 10.1038/311123a0. [DOI] [PubMed] [Google Scholar]
- Wittebort R. J., Rothgeb T. M., Szabo A., Gurd F. R. Aliphatic groups of sperm whale myoglobin: 13C NMR study. Proc Natl Acad Sci U S A. 1979 Mar;76(3):1059–1063. doi: 10.1073/pnas.76.3.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Gunsteren W. F., Berendsen H. J., Hermans J., Hol W. G., Postma J. P. Computer simulation of the dynamics of hydrated protein crystals and its comparison with x-ray data. Proc Natl Acad Sci U S A. 1983 Jul;80(14):4315–4319. doi: 10.1073/pnas.80.14.4315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Gunsteren W. F., Karplus M. Protein dynamics in solution and in a crystalline environment: a molecular dynamics study. Biochemistry. 1982 May 11;21(10):2259–2274. doi: 10.1021/bi00539a001. [DOI] [PubMed] [Google Scholar]


