Abstract
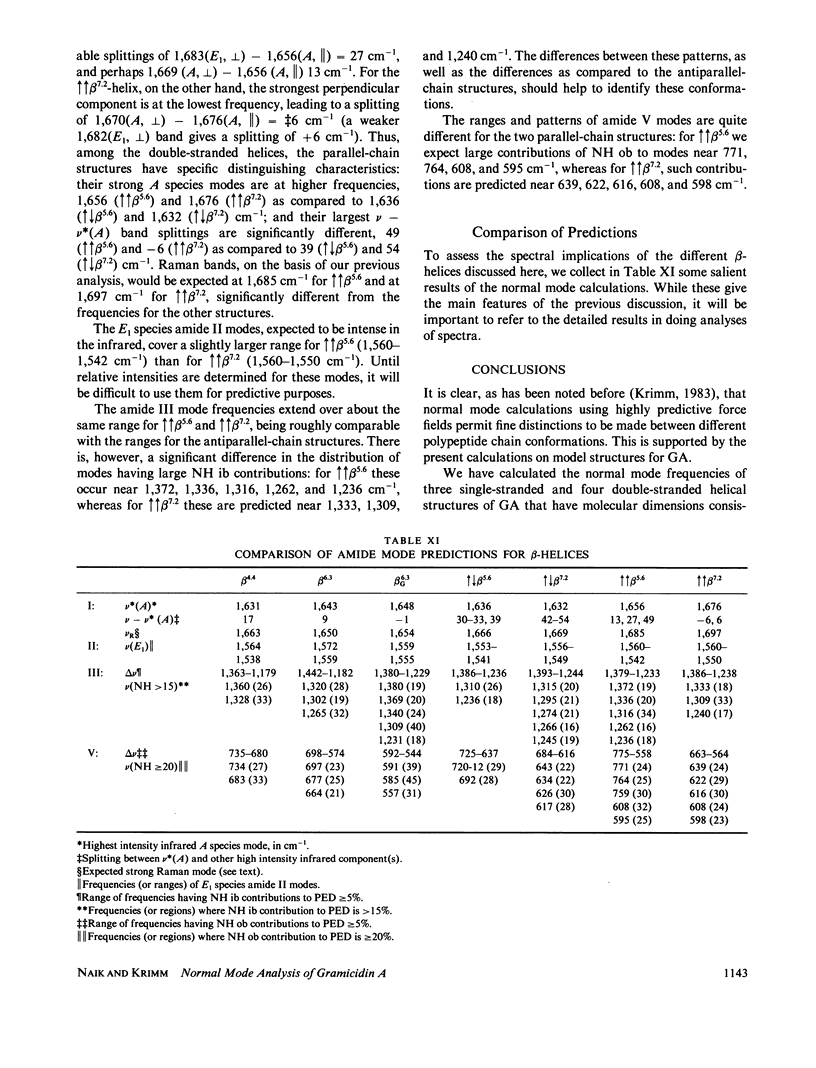
Normal mode frequencies have been calculated for single-stranded beta 4.4 and beta 6.3 and for double-stranded increases decreases beta 5.6, increases decreases beta 7.2, increases increases beta 5.6, and increases increases beta 7.2 helices that are possible models for the structure of gramicidin A. The force field used in the calculations is one that reproduces the frequencies of model polypeptide chain structures to about +/- 5 cm-1, and is therefore expected to provide meaningful distinctions between these conformations. The calculations predict significant differences in the infrared and Raman spectra of these beta-helices, suggesting that they should be identifiable from their spectra (which is shown in the following paper to be the case). The most sensitive region is that of the amide I frequencies, where the predicted patterns of intense infrared mode, infrared splittings, and intense Raman mode provide a characteristic identification of each of the above structures.
Full text
PDF

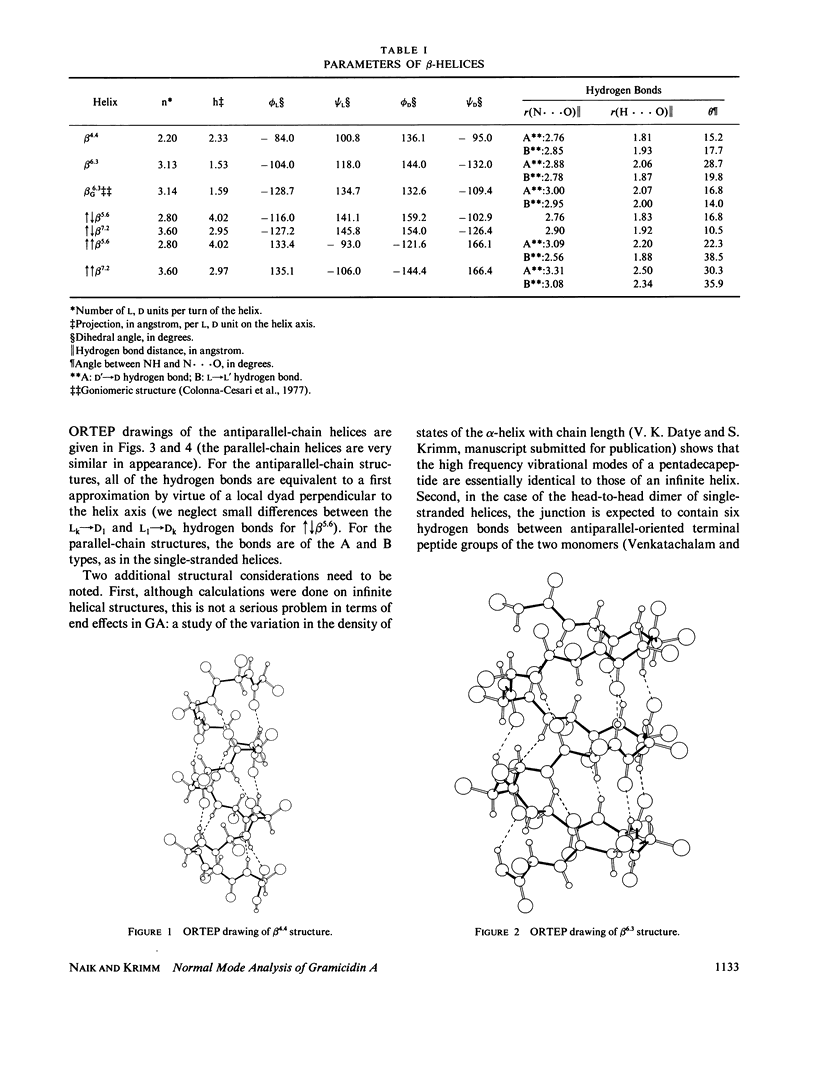
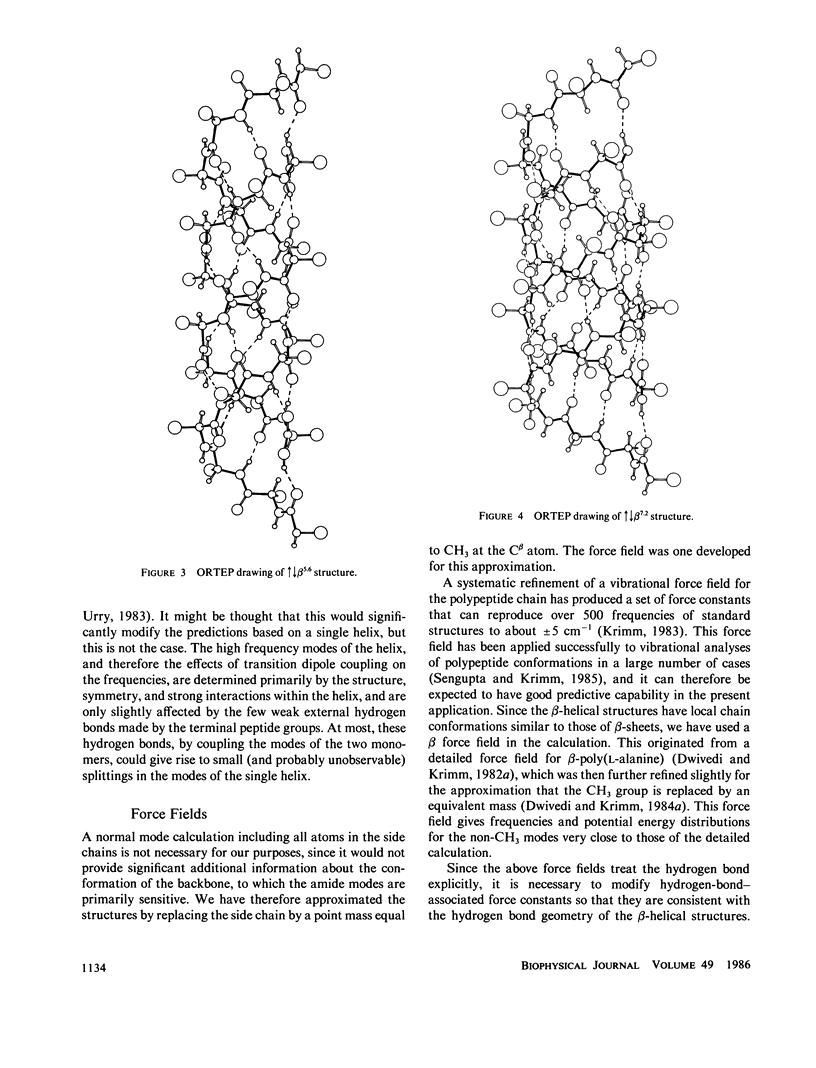

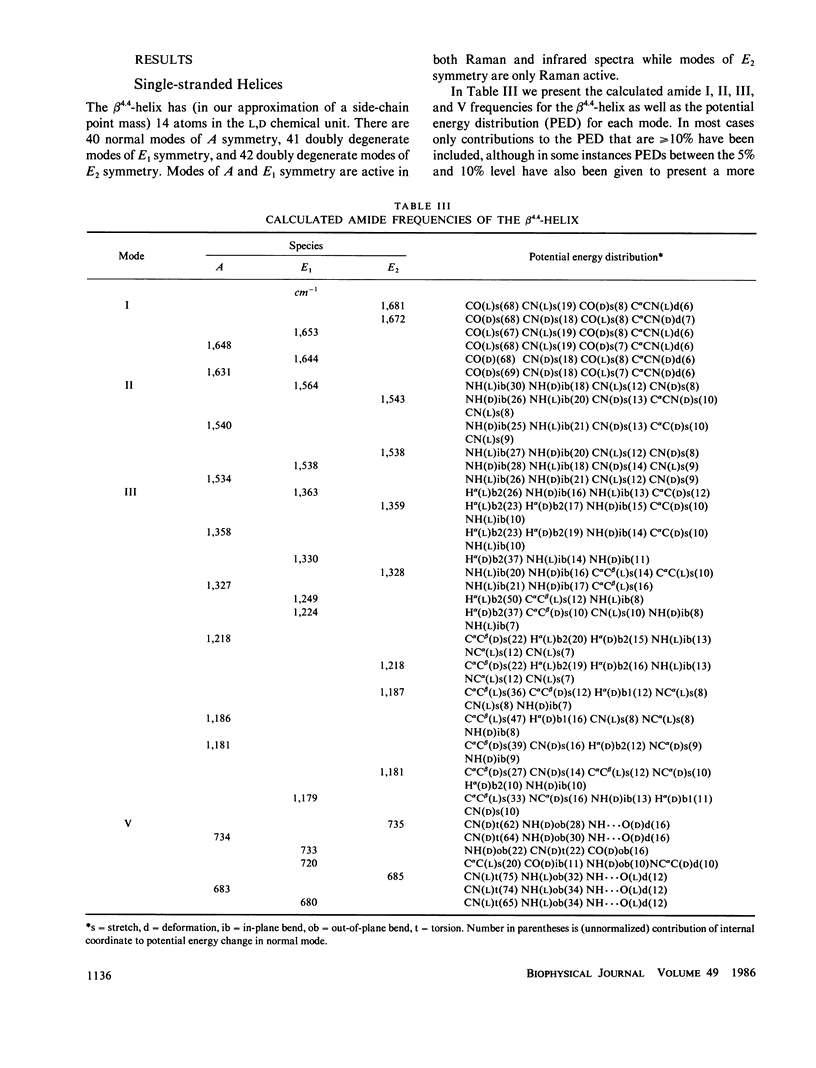
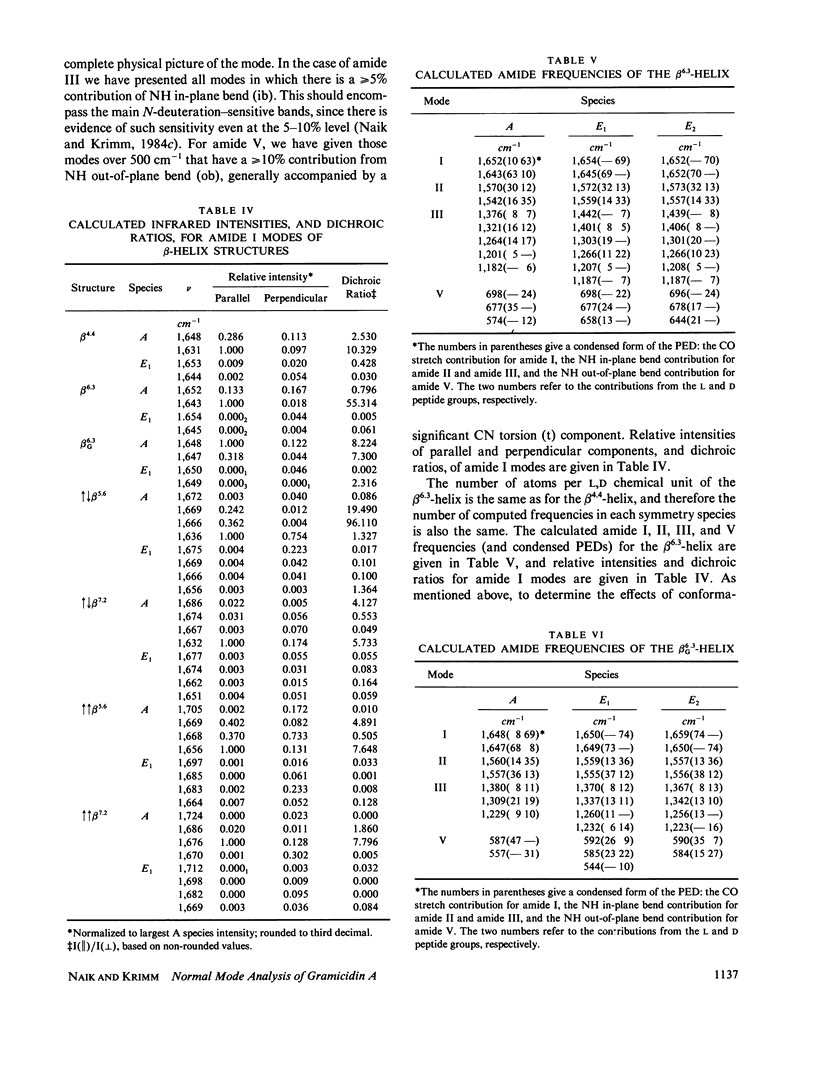





Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abe Y., Krimm S. Normal vibrations of crystalline polyglycine I. Biopolymers. 1972;11(9):1817–1839. doi: 10.1002/bip.1972.360110905. [DOI] [PubMed] [Google Scholar]
- Apell H. J., Bamberg E., Alpes H., Läuger P. Formation of ion channels by a negatively charged analog of gramicidin A. J Membr Biol. 1977 Feb 24;31(1-2):171–188. doi: 10.1007/BF01869403. [DOI] [PubMed] [Google Scholar]
- Bamberg E., Läuger P. Channel formation kinetics of gramicidin A in lipid bilayer membranes. J Membr Biol. 1973;11(2):177–194. doi: 10.1007/BF01869820. [DOI] [PubMed] [Google Scholar]
- Benedetti E., Di Blasio B., Pedone C., Lorenzi G. P., Tomasic L., Gramlich V. A double-stranded beta-helix with antiparallel chains in a crystalline oligo-L-D-peptide. Nature. 1979 Dec 6;282(5739):630–630. doi: 10.1038/282630a0. [DOI] [PubMed] [Google Scholar]
- Colonna-Cesari F., Premilat S., Heitz F., Spach G., Lotz B. Helical structures of poly(D-L-peptides). A conformational energy analysis. Macromolecules. 1977 Nov-Dec;10(6):1284–1288. doi: 10.1021/ma60060a023. [DOI] [PubMed] [Google Scholar]
- Dwivedi A. M., Krimm S. Vibrational analysis of peptides, polypeptides, and proteins. XVIII. Conformational sensitivity of the alpha-helix spectrum: alpha I- and alpha II-poly(L-alanine). Biopolymers. 1984 May;23(5):923–943. doi: 10.1002/bip.360230509. [DOI] [PubMed] [Google Scholar]
- Haydon D. A., Hladky S. B. Ion transport across thin lipid membranes: a critical discussion of mechanisms in selected systems. Q Rev Biophys. 1972 May;5(2):187–282. doi: 10.1017/s0033583500000883. [DOI] [PubMed] [Google Scholar]
- Heitz F., Lotz B., Spach G. AlphaDL and piDL helices of alternating poly-gamma-benzyl-D-L-glutamate. J Mol Biol. 1975 Feb 15;92(1):1–13. doi: 10.1016/0022-2836(75)90088-1. [DOI] [PubMed] [Google Scholar]
- Iqbal Z., Weidekamm E. Raman and infrared spectroscopic study of gramicidin A conformations. Arch Biochem Biophys. 1980 Jul;202(2):639–649. doi: 10.1016/0003-9861(80)90472-5. [DOI] [PubMed] [Google Scholar]
- Koeppe R. E., 2nd, Berg J. M., Hodgson K. O., Stryer L. Gramicidin A crystals contain two cation binding sites per channel. Nature. 1979 Jun 21;279(5715):723–725. doi: 10.1038/279723a0. [DOI] [PubMed] [Google Scholar]
- Koeppe R. E., 2nd, Hodgson K. O., Stryer L. Helical channels in crystals of gramicidin A and of a cesium--gramicidin A complex: an x-ray diffraction study. J Mol Biol. 1978 May 5;121(1):41–54. doi: 10.1016/0022-2836(78)90261-9. [DOI] [PubMed] [Google Scholar]
- Koeppe R. E., 2nd, Schoenborn B. P. 5-A Fourier map of gramicidin A phased by deuterium-hydrogen solvent difference neutron diffraction. Biophys J. 1984 Mar;45(3):503–507. doi: 10.1016/S0006-3495(84)84186-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolb H. A., Läuger P., Bamberg E. Correlation analysis of electrical noise in lipid bilayer membranes: kinetics of gramicidin A channels. J Membr Biol. 1975;20(1-2):133–154. doi: 10.1007/BF01870632. [DOI] [PubMed] [Google Scholar]
- Krasne S., Eisenman G., Szabo G. Freezing and melting of lipid bilayers and the mode of action of nonactin, valinomycin, and gramicidin. Science. 1971 Oct 22;174(4007):412–415. doi: 10.1126/science.174.4007.412. [DOI] [PubMed] [Google Scholar]
- Krimm S., Abe Y. Intermolecular interaction effects in the amide I vibrations of polypeptides. Proc Natl Acad Sci U S A. 1972 Oct;69(10):2788–2792. doi: 10.1073/pnas.69.10.2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krimm S. Vibrational analysis of conformation in peptides, polypeptides, and proteins. Biopolymers. 1983 Jan;22(1):217–225. doi: 10.1002/bip.360220130. [DOI] [PubMed] [Google Scholar]
- Lotz B., Colonna-Cesari F., Heitz F., Spach G. A family of double helices of alternating poly(gamma-benzyl-D-L-glutamate), a stereochemical model for gramicidin A. J Mol Biol. 1976 Oct 5;106(4):915–942. doi: 10.1016/0022-2836(76)90343-0. [DOI] [PubMed] [Google Scholar]
- Moore W. H., Krimm S. Transition dipole coupling in Amide I modes of betapolypeptides. Proc Natl Acad Sci U S A. 1975 Dec;72(12):4933–4935. doi: 10.1073/pnas.72.12.4933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore W. H., Krimm S. Vibrational analysis of peptides, polypeptides, and proteins. I. Polyglycine I. Biopolymers. 1976 Dec;15(12NA-NA-770103-770104):2439–2464. doi: 10.1002/bip.1976.360151210. [DOI] [PubMed] [Google Scholar]
- Nabedryk E., Gingold M. P., Breton J. Orientation of gramicidin A transmembrane channel. Infrared dichroism study of gramicidin in vesicles. Biophys J. 1982 Jun;38(3):243–249. doi: 10.1016/S0006-3495(82)84555-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naik V. M., Krimm S., Denton J. B., Némethy G., Scheraga H. A. Vibrational analysis of peptides, polypeptides and proteins. XXVII. Structure of gramicidin S from normal mode analyses of low-energy conformations. Int J Pept Protein Res. 1984 Dec;24(6):613–626. [PubMed] [Google Scholar]
- Naik V. M., Krimm S. The structure of crystalline and membrane-bound gramicidin A by vibrational analysis. Biochem Biophys Res Commun. 1984 Dec 28;125(3):919–925. doi: 10.1016/0006-291x(84)91371-8. [DOI] [PubMed] [Google Scholar]
- Naik V. M., Krimm S. Vibrational analysis of peptides, polypeptides, and proteins. XVII. Normal modes of crystalline Pro-Leu-Gly-NH2, a type II beta-turn. Int J Pept Protein Res. 1984 Jan;23(1):1–24. [PubMed] [Google Scholar]
- Naik V. M., Krimm S. Vibrational analysis of the structure of crystalline gramicidin a. Biophys J. 1984 Jan;45(1):109–112. doi: 10.1016/S0006-3495(84)84129-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naik V. M., Krimm S. Vibrational analysis of the structure of gramicidin A. II. Vibrational spectra. Biophys J. 1986 Jun;49(6):1147–1154. doi: 10.1016/S0006-3495(86)83743-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popov E. M., Lipkind G. M. Konformatsionnoe sostoianie i mekhanizm funktsionirovaniia gramitsidina A. Mol Biol (Mosk) 1979 Mar-Apr;13(2):363–376. [PubMed] [Google Scholar]
- Prasad B. V., Chandrasekaran R. Conformation of polypeptide chains containing both L- and D-residues. II. Double-helical structures of poly-LD-peptides. Int J Pept Protein Res. 1977;10(2):129–138. doi: 10.1111/j.1399-3011.1977.tb02786.x. [DOI] [PubMed] [Google Scholar]
- Ramachnandran G. N., Chandrasekaran R. Conformation of peptide chains containing both L- & D-residues. I. Helical structures with alternating L- & D-residues with special reference to the LD-ribbon & the LD-helices. Indian J Biochem Biophys. 1972 Mar;9(1):1–11. [PubMed] [Google Scholar]
- Rothschild K. J., Stanley H. E. Raman spectroscopic investigation of gramicidin A' conformations. Science. 1974 Aug 16;185(4151):616–618. doi: 10.1126/science.185.4151.616. [DOI] [PubMed] [Google Scholar]
- Sengupta P. K., Krimm S. Vibrational analysis of peptides, polypeptides, and proteins. XXXII. alpha-Poly(L-glutamic acid). Biopolymers. 1985 Aug;24(8):1479–1491. doi: 10.1002/bip.360240805. [DOI] [PubMed] [Google Scholar]
- Urry D. W. A molecular theory of ion-conductng channels: a field-dependent transition between conducting and nonconducting conformations. Proc Natl Acad Sci U S A. 1972 Jun;69(6):1610–1614. doi: 10.1073/pnas.69.6.1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urry D. W., Goodall M. C., Glickson J. D., Mayers D. F. The gramicidin A transmembrane channel: characteristics of head-to-head dimerized (L,D) helices. Proc Natl Acad Sci U S A. 1971 Aug;68(8):1907–1911. doi: 10.1073/pnas.68.8.1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urry D. W., Shaw R. G., Trapane T. L., Prasad K. U. Infrared spectra of the Gramicidin A transmembrane channel: the single-stranded-beta 6-helix. Biochem Biophys Res Commun. 1983 Jul 18;114(1):373–379. doi: 10.1016/0006-291x(83)91637-6. [DOI] [PubMed] [Google Scholar]
- Urry D. W., Spisni A., Khaled A. Characterization of micellar-packaged gramicidin A channels. Biochem Biophys Res Commun. 1979 Jun 13;88(3):940–949. doi: 10.1016/0006-291x(79)91499-2. [DOI] [PubMed] [Google Scholar]
- Urry D. W. The gramicidin A transmembrane channel: a proposed pi(L,D) helix. Proc Natl Acad Sci U S A. 1971 Mar;68(3):672–676. doi: 10.1073/pnas.68.3.672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urry D. W., Trapane T. L., Prasad K. U. Is the gramicidin a transmembrane channel single-stranded or double-stranded helix? A simple unequivocal determination. Science. 1983 Sep 9;221(4615):1064–1067. doi: 10.1126/science.221.4615.1064. [DOI] [PubMed] [Google Scholar]
- Veatch W. R., Fossel E. T., Blout E. R. The conformation of gramicidin A. Biochemistry. 1974 Dec 17;13(26):5249–5256. doi: 10.1021/bi00723a001. [DOI] [PubMed] [Google Scholar]
- Veatch W. R., Mathies R., Eisenberg M., Stryer L. Simultaneous fluorescence and conductance studies of planar bilayer membranes containing a highly active and fluorescent analog of gramicidin A. J Mol Biol. 1975 Nov 25;99(1):75–92. doi: 10.1016/s0022-2836(75)80160-4. [DOI] [PubMed] [Google Scholar]
- Veatch W., Stryer L. The dimeric nature of the gramicidin A transmembrane channel: conductance and fluorescence energy transfer studies of hybrid channels. J Mol Biol. 1977 Jun 15;113(1):89–102. doi: 10.1016/0022-2836(77)90042-0. [DOI] [PubMed] [Google Scholar]
- Wallace B. A. Ion-bond forms of the gramicidin a transmembrane channel. Biophys J. 1984 Jan;45(1):114–116. doi: 10.1016/S0006-3495(84)84131-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace B. A., Veatch W. R., Blout E. R. Conformation of gramicidin A in phospholipid vesicles: circular dichroism studies of effects of ion binding, chemical modification, and lipid structure. Biochemistry. 1981 Sep 29;20(20):5754–5760. doi: 10.1021/bi00523a018. [DOI] [PubMed] [Google Scholar]
- Weinstein S., Wallace B. A., Blout E. R., Morrow J. S., Veatch W. Conformation of gramicidin A channel in phospholipid vesicles: a 13C and 19F nuclear magnetic resonance study. Proc Natl Acad Sci U S A. 1979 Sep;76(9):4230–4234. doi: 10.1073/pnas.76.9.4230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstein S., Wallace B. A., Morrow J. S., Veatch W. R. Conformation of the gramicidin A transmembrane channel: A 13C nuclear magnetic resonance study of 13C-enriched gramicidin in phosphatidylcholine vesicles. J Mol Biol. 1980 Oct 15;143(1):1–19. doi: 10.1016/0022-2836(80)90121-7. [DOI] [PubMed] [Google Scholar]
- Zingsheim H. P., Neher E. The equivalence of fluctuation analysis and chemical relaxation measurements: a kinetic study of ion pore formation in thin lipid membranes. Biophys Chem. 1974 Oct;2(3):197–207. doi: 10.1016/0301-4622(74)80045-1. [DOI] [PubMed] [Google Scholar]


