Abstract
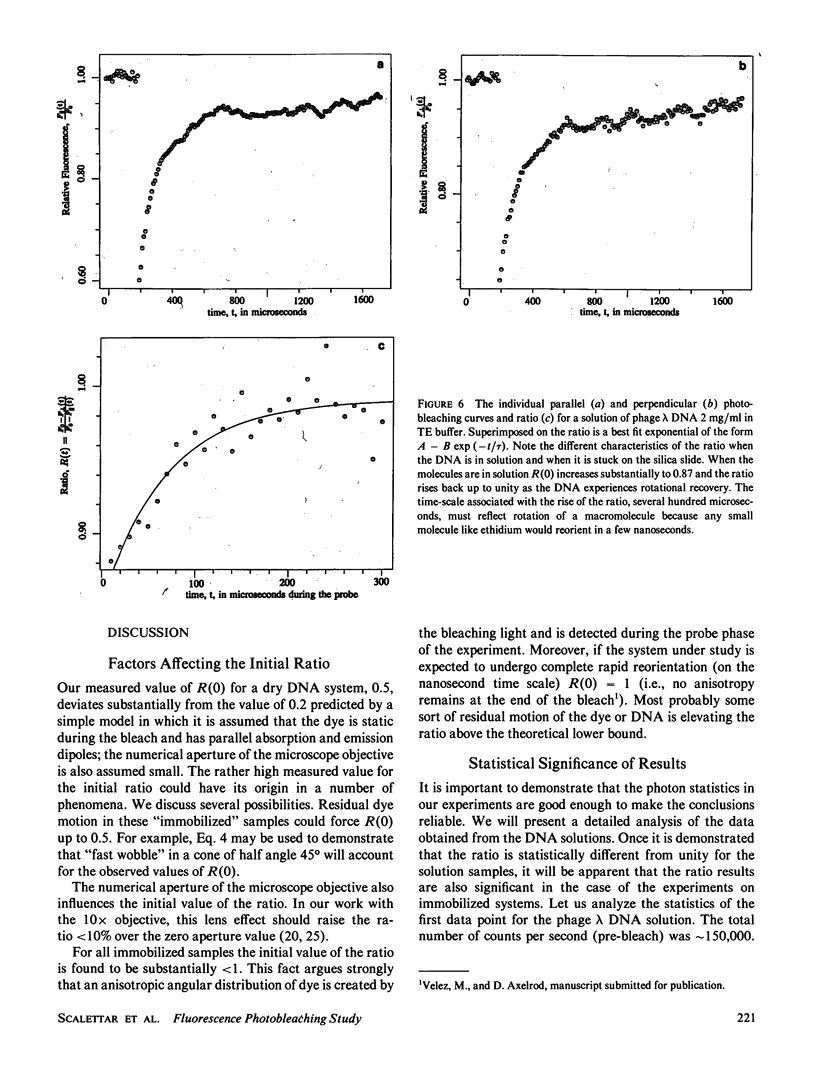
We have conducted a polarized fluorescence photobleaching recovery (FPR) study of the rotational dynamics of ethidium azide labeled DNA. Polarized photobleaching experiments provide data on microsecond and millisecond molecular reorientation that complement the information available from nanosecond fluorescence depolarization studies. In polarized FPR experiments an anisotropic angular concentration of fluorophore is created by bleaching dye molecules in a preferred orientation with a short, intense pulse of polarized light. The sample is then weakly illuminated, and the temporal variation in the emitted fluorescence is monitored. The fluorescence signal will systematically change as molecules undergo post-bleach reorientation and the angular distribution of dye tends toward isotropy. We have observed that the time dependence of our microsecond FPR curves is also determined in part by nonrotational phenomena. To isolate the reorientational recovery we conduct our FPR experiments in two modes (called parallel and perpendicular) that differ only in the polarization of the bleaching light. A quotient function, R(t), is constructed from the data obtained in these two modes; the variation with time of this new quantity is governed solely by processes that are sensitive to the polarization of the incident light (e.g., molecular rotation). It is found experimentally that R(t) remains constant, as expected, for rotationally restricted DNA systems despite a temporal recovery in the parallel and perpendicular FPR curves. We also follow the dynamics of solutions of phage lambda DNA as revealed in the temporal dependence of R(t). This DNA system rotationally relaxes after approximately 100 microseconds and the dye/DNA complex reorients substantially during the 10-microseconds bleach period. Our FPR data are interpreted in terms of dynamic models of DNA motion.
Full text
PDF
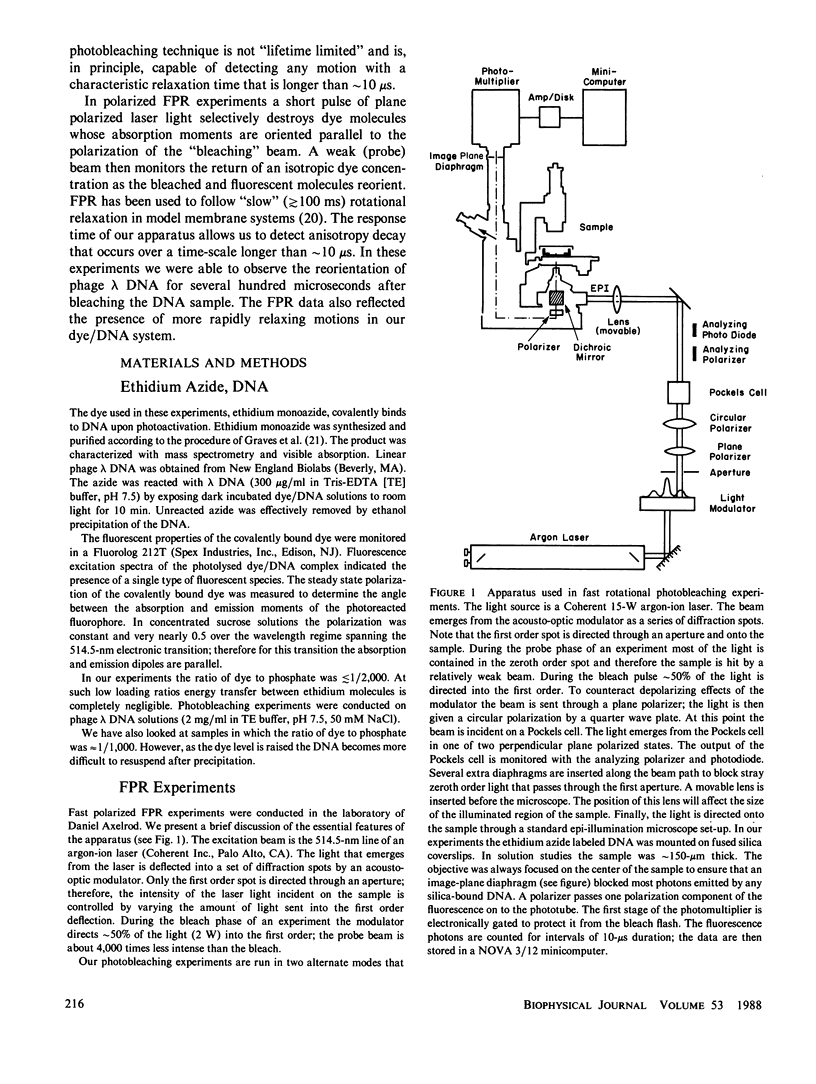
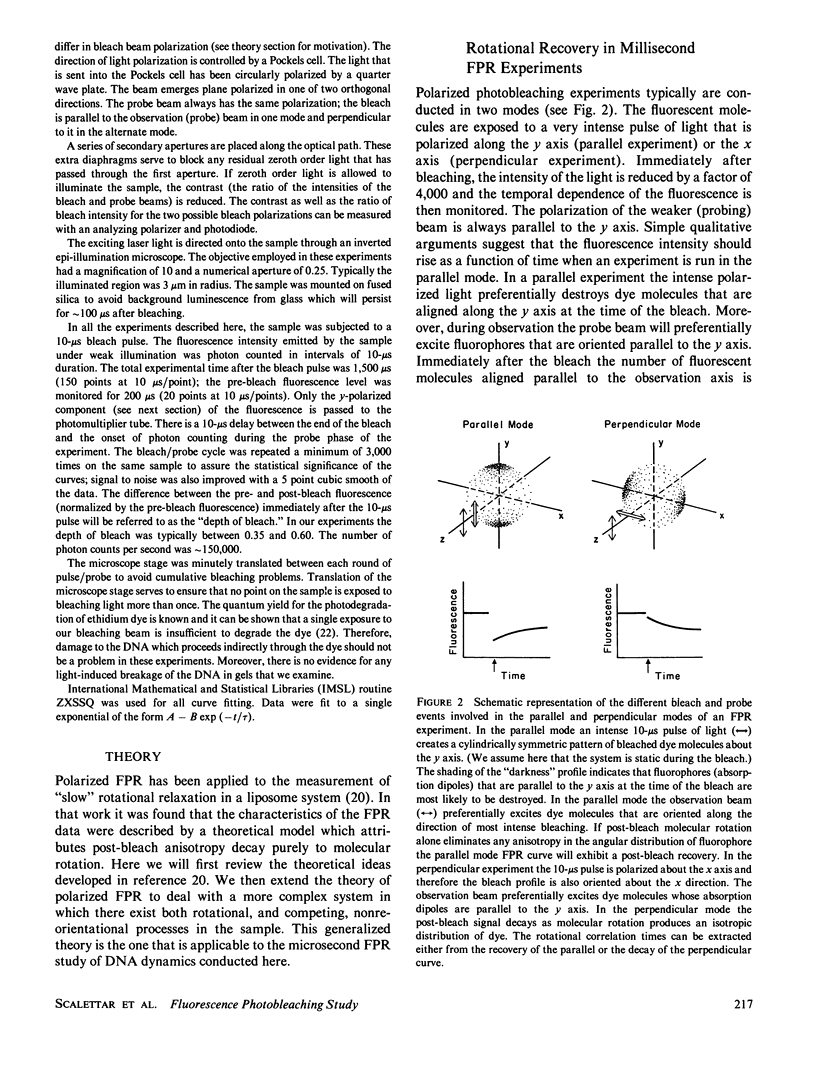

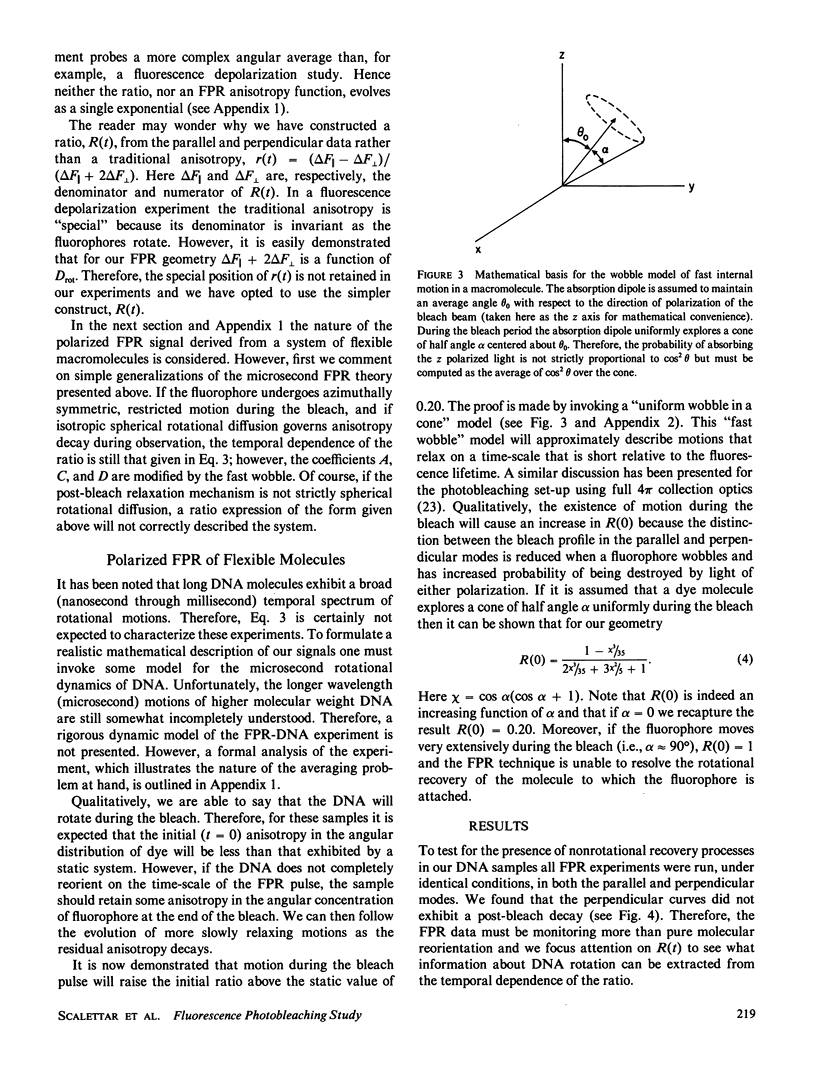
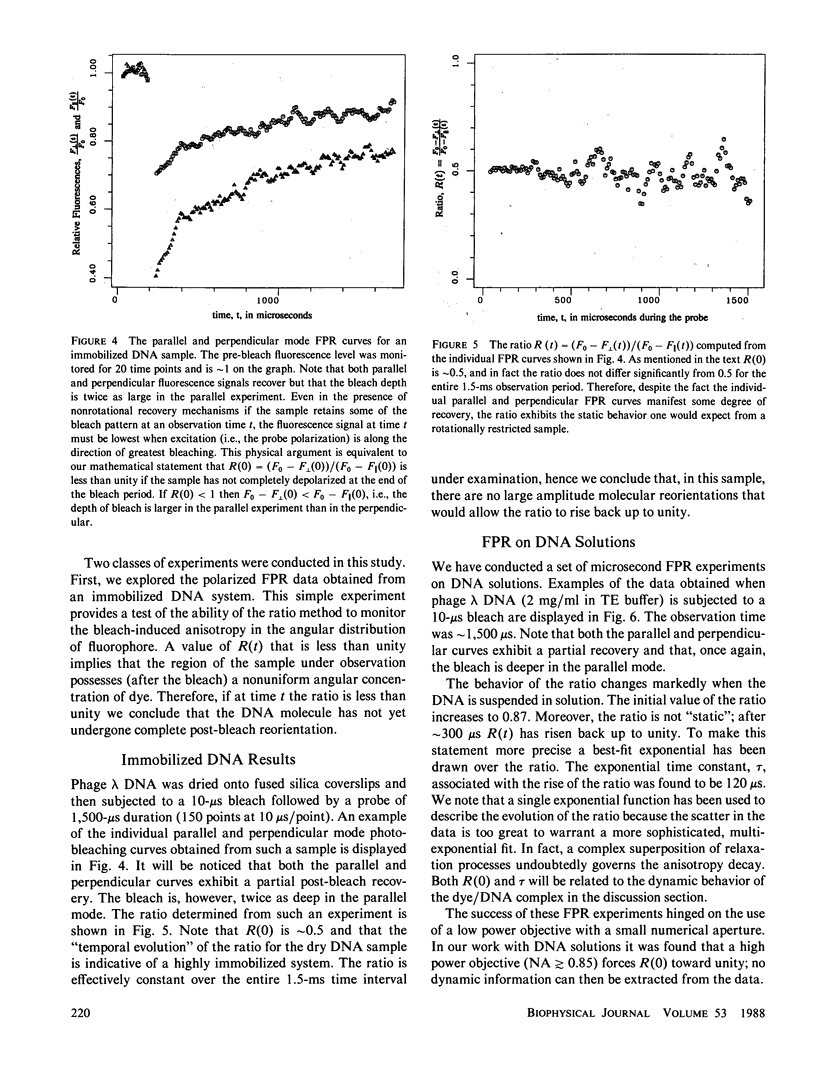





Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Allison S. A., Shibata J. H., Wilcoxon J., Schurr J. M. NMR relaxation in DNA. I. The contribution of torsional deformation modes of the elastic filament. Biopolymers. 1982 Apr;21(4):729–762. doi: 10.1002/bip.360210403. [DOI] [PubMed] [Google Scholar]
- Axelrod D. Carbocyanine dye orientation in red cell membrane studied by microscopic fluorescence polarization. Biophys J. 1979 Jun;26(3):557–573. doi: 10.1016/S0006-3495(79)85271-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark-Lewis I., Aebersold R., Ziltener H., Schrader J. W., Hood L. E., Kent S. B. Automated chemical synthesis of a protein growth factor for hemopoietic cells, interleukin-3. Science. 1986 Jan 10;231(4734):134–139. doi: 10.1126/science.3079915. [DOI] [PubMed] [Google Scholar]
- Diekmann S., Hillen W., Morgeneyer B., Wells R. D., Pörschke D. Orientation relaxation of DNA restriction fragments and the internal mobility of the double helix. Biophys Chem. 1982 Jul;15(4):263–270. doi: 10.1016/0301-4622(82)80009-4. [DOI] [PubMed] [Google Scholar]
- Ding D. W., Rill R., Van Holde K. E. The dichroism of DNA in electric fields. Biopolymers. 1972;11(10):2109–2124. doi: 10.1002/bip.1972.360111011. [DOI] [PubMed] [Google Scholar]
- Early T. A., Kearns D. R. 1H nuclear magnetic resonance investigation of flexibility in DNA. Proc Natl Acad Sci U S A. 1979 Sep;76(9):4165–4169. doi: 10.1073/pnas.76.9.4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genest D., Wahl P. Fluorescence anisotropy decay due to rotational brownian motion of ethidium intercalated in double strand DNA. Biochim Biophys Acta. 1978 Dec 21;521(2):502–509. doi: 10.1016/0005-2787(78)90292-7. [DOI] [PubMed] [Google Scholar]
- Graves D. E., Yielding L. W., Watkins C. L., Yielding K. L. Synthesis, separation and characterization of the mono- and diazide analogs of ethidium bromide. Biochim Biophys Acta. 1977 Nov 2;479(1):98–104. doi: 10.1016/0005-2787(77)90129-0. [DOI] [PubMed] [Google Scholar]
- Hagerman P. J. Investigation of the flexibility of DNA using transient electric birefringence. Biopolymers. 1981 Jul;20(7):1503–1535. doi: 10.1002/bip.1981.360200710. [DOI] [PubMed] [Google Scholar]
- Hogan M. E., Jardetzky O. Internal motions in DNA. Proc Natl Acad Sci U S A. 1979 Dec;76(12):6341–6345. doi: 10.1073/pnas.76.12.6341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan M., Wang J., Austin R. H., Monitto C. L., Hershkowitz S. Molecular motion of DNA as measured by triplet anisotropy decay. Proc Natl Acad Sci U S A. 1982 Jun;79(11):3518–3522. doi: 10.1073/pnas.79.11.3518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magde D., Elson E. L., Webb W. W. Fluorescence correlation spectroscopy. II. An experimental realization. Biopolymers. 1974 Jan;13(1):29–61. doi: 10.1002/bip.1974.360130103. [DOI] [PubMed] [Google Scholar]
- Millar D. P., Robbins R. J., Zewail A. H. Direct observation of the torsional dynamics of DNA and RNA by picosecond spectroscopy. Proc Natl Acad Sci U S A. 1980 Oct;77(10):5593–5597. doi: 10.1073/pnas.77.10.5593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz K. S., Schurr J. M. Rotational relaxation of macromolecules determined by dynamic light scattering. II. Temperature dependence for DNA. Biopolymers. 1973;12(7):1543–1564. doi: 10.1002/bip.1973.360120709. [DOI] [PubMed] [Google Scholar]
- Smith L. M., Weis R. M., McConnell H. M. Measurement of rotational motion in membranes using fluorescence recovery after photobleaching. Biophys J. 1981 Oct;36(1):73–91. doi: 10.1016/S0006-3495(81)84717-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas J. C., Allison S. A., Appellof C. J., Schurr J. M. Torison dynamics and depolarization of fluorescence of linear macromolecules. II. Fluorescence polarization anisotropy measurements on a clean viral phi 29 DNA. Biophys Chem. 1980 Oct;12(2):177–188. doi: 10.1016/0301-4622(80)80050-0. [DOI] [PubMed] [Google Scholar]
- Thompson D. S., Gill S. J. Polymer relaxation times from birefringence relaxation measurements. J Chem Phys. 1967 Dec 15;47(12):5008–5017. doi: 10.1063/1.1701752. [DOI] [PubMed] [Google Scholar]
- Wahl P., Paoletti J., Le Pecq J. B. Decay of fluorescence emission anisotropy of the ethidium bromide-DNA complex. Evidence for an internal motion in DNA. Proc Natl Acad Sci U S A. 1970 Feb;65(2):417–421. doi: 10.1073/pnas.65.2.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegener W. A., Rigler R. Separation of translational and rotational contributions in solution studies using fluorescence photobleaching recovery. Biophys J. 1984 Dec;46(6):787–793. doi: 10.1016/S0006-3495(84)84077-1. [DOI] [PMC free article] [PubMed] [Google Scholar]


