Abstract
In Saccharomyces cerevisiae at least five genes, EST1, EST2, EST3, TLC1 and CDC13, are required for telomerase activity in vivo. The telomerase catalytic subunit Est2p and telomerase RNA subunit Tlc1 constitute the telomerase core enzyme. Est1p and Est3p are the other subunits of telomerase holoenzyme. In order to dissect the function of Est3p in telomere replication, we over-expressed and purified recombinant wild-type and mutant Est3 proteins. The wild-type protein, as well as the K71A, E104A and T115A mutants were able to dimerize in vitro, while the Est3p-D49A, -K68A or -D166A mutant showed reduced ability to dimerize. Mutations in Est3p that decreased dimerization also appeared to cause telomere shortening in vivo. Double point mutation of Est3p-D49A-K68A and single point mutation of Est3p-K68A showed similar telomere shortening, suggesting that the K68 residue might be more important for telomerase activity. The ectopic co-expression of K71A or T115A mutant with wild-type Est3p using centromere plasmids caused telomere shortening, while co-expression of the D49A, K68A, D86A or F103A mutants with wild-type Est3p had no effect on telomere length regulation. These data suggested that dimerization is important for Est3p function in vivo.
INTRODUCTION
Telomeres are the physical ends of chromosomes and are composed of simple repeated telomeric DNA and its associated proteins (1). Telomeres protect chromosome ends from nucleolytic digestion, distinguish normal chromosome ends from DNA breaks and facilitate complete chromosomal replication (2–4). In Saccharomyces cerevisiae, the telomeric DNA is ∼300 ± 75 bp of C1–3A/TG1–3 sequence (5). Telomeric DNA elongation is mediated by telomerase, a specialized reverse transcriptase that catalyzes the addition of telomeric DNA sequence to chromosome ends by using Tlc1, the RNA component as template (6,7). In budding yeast S.cerevisiae, at least five genes, EST2, TLC1, EST1, EST3 and CDC13, are required for telomerase activity in vivo. Deficiency of any one of these genes leads to a common phenotype of initial viability with gradual telomere shortening and most cells dying after ∼100 generations (8–10).
EST2 encodes the catalytic subunit of telomerase with distinctive motifs common to all reverse transcriptases as well as motifs that are specific to telomerase (11–13). TLC1 encodes the 1.3 kb RNA subunit which contains a template region for the elongation of telomeric DNA (10). Est2p and Tlc1 are essential and sufficient for elongation activity of oligo primer in vitro and are believed to constitute the telomerase catalytic core (6,7). Cdc13p binds single-stranded telomeric DNA in vitro and in vivo, and appears to be a component of the telomere (14–17). Studies of different alleles of CDC13 reveal that Cdc13p is involved in both telomere protection and telomerase recruitment (16,18,19). Est1p binds the single-stranded G-rich telomeric DNA in vitro (20) and interacts with Tlc1 in vivo (21–23). In addition, Est1p interacts with Cdc13p in yeast two-hybrid analysis and glutathione S-transferase (GST)-pull down assay (24), and the Est1p-Cdc13p interaction recruits telomerase to telomeres (25,26). Recently, Est1p has been suggested to convert an inactive telomerase into an active form (17).
EST3 encodes two forms of protein, one is a truncated protein resulting from translation of only the first open reading frame and the other is the full-length Est3 protein which has been translated through the frame-shift site (27). Only the full-length Est3p (181 amino acid) with a molecular weight of 20.5 kDa is required for its in vivo telomere maintenance function (27). The est3 deletion strain gives rise to the same telomerase-deficient phenotypes observed in est2 and tlc1 mutants (8). Immunoprecipitation of either Est3p or Est1p allows for the isolation of telomerase activity that is attributed to Est2p and Tlc1 (21,28), and Est3p is associated with Tlc1 through interactions with Est2p (28). These observations suggest that telomerase holoenzyme contains Est1p and Est3p subunits in addition to the Est2p/Tlc1 core enzyme. Genetic studies suggest that Est3p interacts with the N-terminal domain of Est2p because over-expression of Est3p is able to rescue the temperature sensitivity of est2ts alleles containing mutations in the N-terminal Region I (13). To date, the mechanism for the function of Est3p in telomere replication is poorly understood.
To investigate the function(s) of Est3p in telomerase activity and potential interactions of Est3p with other subunits of telomerase, we carried out biochemical and genetic studies of Est3p. Wild-type and 13 mutant Est3 proteins were purified, and gel filtration analysis revealed that Est3p forms homodimer. Mutations that disrupted protein dimerization in vitro also showed telomere shortening in vivo. Our data suggest that the dimerization of Est3p contributes to telomere maintenance and may have implications for understanding the mechanism of how Est3p and the telomerase holoenzyme function in telomere replication.
MATERIALS AND METHODS
Yeast strains
Unless otherwise stated, all the yeast strains used in this work were derived from the YPH499 background (29). The est3 deletion strain was retrieved by tetrad dissection of a diploid strain with one wild-type EST3 allele, and selected on the synthetic glucose (SD) medium with different nutritional requirements and Southern blot.
Plasmids
The coding sequence of EST3 gene was amplified by a two-step PCR of yeast genomic DNA with primers flanked with BamHI and EcoRI sites and was cloned into over-expression vectors pGEX-4T-1 (from Invitrogen™ life technologies) and pYES3/CT (from Invitrogen™ life technologies). The nucleotide at position 277 in genomic EST3 gene was deleted during this two-step PCR cloning. The pRS316-EST3 centromere plasmid was constructed by cloning a 1278 bp fragment, which contains the Est3p coding region, and its upstream 493 bp and downstream 238 bp regions into the BamHI–HindIII sites of pRS316. The EST3 alleles for either over-expression or endogenous level expression were confirmed by DNA sequencing. Mutants of EST3 were created by site-directed mutagenesis with primer-specific two-step PCR using wild-type pRS316-EST3 plasmid as template to change the desired amino acids into Alanine (A) or Glutamate acid (E). All the mutants were cloned in yeast centromere plasmid pRS315 and confirmed by DNA sequencing. Two independent colonies of each mutant were isolated and tested for its function on telomere length regulation. All the mutants were also cloned into both pGEX-4T-1 and pYES3/CT over-expression vector.
Recombinant Est3p over-expression, purification and gel filtration
The pGEX-4T-1 plasmid that contained either wild-type or mutant EST3 was transformed into Escherichia coli strain BL21, and over-expression of Est3p was induced at an OD600 of 0.5 with 0.5 mM IPTG (Isopropyl-β-d-thiogalactopyranosid) at 16°C for 12–16 h. The GST–Est3 fusion proteins were purified according to the manufacturer's instructions using 250 µl glutathione beads per liter culture. All the procedures of purification were carried out at 4°C. The cells were collected and resuspended in 10 bed volumes of ice-cold phosphate-buffered saline (PBS) containing 1% Triton X-100. The cells were disrupted by two passages through a homogenizer (EmulsiFlex-C5, AVESTIN), and the supernatant was loaded on a glutathione column. The column was washed with PBS containing 1% Triton X-100 and PBS containing both 1% Triton X-100 and 1 M NaCl. The purified recombinant GST–Est3 proteins that were bound to GST beads were digested with thrombin in 5 bed volumes of thrombin-digestion buffer (20 mM Tris, pH 8.5, 100 mM NaCl, 0.33 mM CaCl2 and 1 mM DTT) at 4°C for 3 h under the condition of 1 U of thrombin/mg of GST–Est3p. The released Est3 proteins were dialyzed and aliquoted for further use. For Gel filtration, 100 µg of purified wild-type or mutant Est3 proteins in 100 µl buffer (20 mM Tris, pH 8.5, 50 mM NaCl) was loaded on an analytical size exclusion superdex 75 column (Pharmacia Biotech), which was calibrated with protein standard markers including BSA (67 kDa), ovalbumin (43 kDa), chymotrypsinogen A (25 kDa), ribonuclease A (13.7 kDa) according to the vendor's protocol. The column was developed with elution buffer (20 mM Tris, pH 8.5, 50 mM NaCl) at 0.3 ml/min, and 1 ml fractions were collected. The eluate of Est3p was analyzed by SDS–PAGE and western blot.
Telomere blot
Genomic DNA prepared from saturated yeast cultures of each yeast strain was digested with XhoI, separated by 1% agarose gel and transferred to HyBond N+ membranes (Amersham Pharmacia Biotech), cross-linked by UV and then probed with C1–3A/TG1–3 specific telomeric probe as previously documented (30).
Preparation of polyclonal antibodies against Est3p
Polyclonal antibody against Est3p was generated by immunization of rabbits with a GST–Est3 fusion protein. The antisera were affinity purified by antigen cross-linked column as previously documented (30,31).
RESULTS
Over-expression and purification of recombinant Est3p
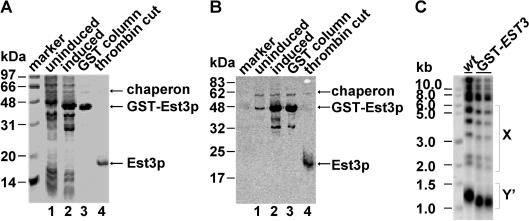
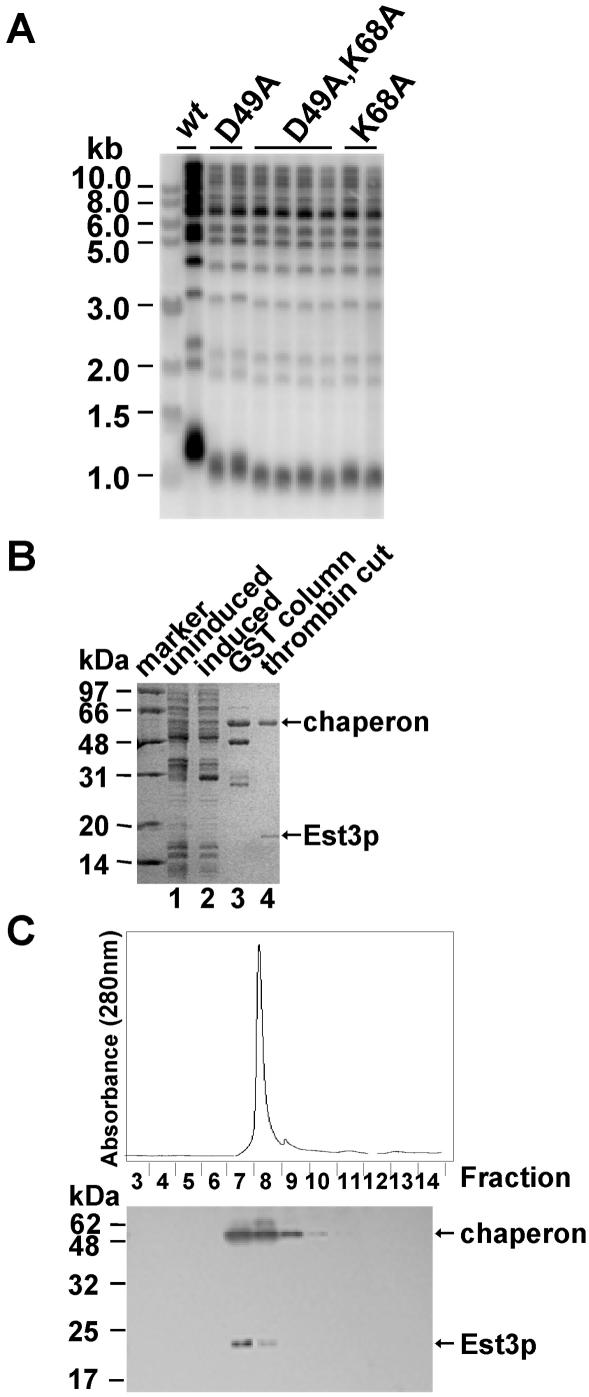
To study the function of Est3p in telomere replication, we started with purification of recombinant Est3p in vitro. Since a programmed frame-shift occurs during the translation of full-length Est3p of 181 amino acids in S.cerevisiae (27), the nucleotide adenine (A) at position 277 in EST3 gene was deleted to ensure the production of full-length Est3p when it is expressed in E.coli and S.cerevisiae. The modified gene was still considered to be EST3 because it encodes full-length wild-type Est3 protein. The EST3 gene was inserted into pGEX-4T-1 plasmid to express the GST–Est3p fusion in E.coli. The over-expression and purification of recombinant Est3p were as described in Materials and Methods. The Est3p was purified to near homogeneity as examined by Coomassie blue staining of an SDS–PAGE gel (Figure 1A) and confirmed by western blot with affinity purified antibodies against Est3p (Figure 1B). The GST fusion Est3p was over-expressed in E.coli and purified with a GST column (Figure 1A and B, lanes 1–3). Telomere Southern blot analysis revealed that the GST–Est3p fusion was not able to fully complement the wild-type Est3p in vivo as the telomeres in the cells that expressed GST–Est3p were shorter than those in EST3 wild-type cells (Figure 1C). For this reason the GST-tag was cleaved with thrombin before it was further characterized in vitro. The thrombin digestion released untagged Est3p from the glutathione beads (Figure 1A and B, lane 4). A 60 kDa protein was co-purified with Est3p and detected with a faint signal in the western blot analysis (Figure 1B). Other groups have also co-purified a similar 60 kDa protein and according to previous studies by Thain et al., this protein is likely to be the E.coli molecular chaperone GroEL, which plays an important role in helping protein folding in bacteria (32–34). The reason why the antibody raised against Est3p could cross-react with the chaperone is that the antigen, recombinant GST–Est3p used for immunizing rabbits, was likely contaminated with the 60 kDa chaperone (Figure 1A and B, lane 3).
Figure 1.
The purification and characterization of recombinant Est3p. (A) The GST–Est3p fusion was purified with glutathione Sepharose™ 4B beads (Amersham Biosciences) and the GST-tag was removed with thrombin digestion, the proteins were further analyzed with SDS–PAGE gel followed by Coomassie blue staining. Lane 1, uninduced total extract; lane 2, induced total extract; lane 3, purified GST–Est3p on glutathione beads; lane 4, Est3p after the GST-tag cleft by thrombin. Protein size standards are indicated in kilodaltons (kDa). (B) The purification of Est3p was examined by western blot with affinity purified primary antibody against Est3p. Prestained protein size standards (NEB) are indicated in kilodaltons (kDa). (C) GST–Est3p complementation experiment. Genomic DNA from each cell culture was prepared, digested with XhoI, separated on a 1% agarose gel and probed to a poly(GT) telomere specific probe. Isogenetic strains were as follows: wt (wild-type strain: YPH499), GST–EST3 (YPH499 background). Both X and Y′ telomere fragments are indicated.
Est3p forms homodimer
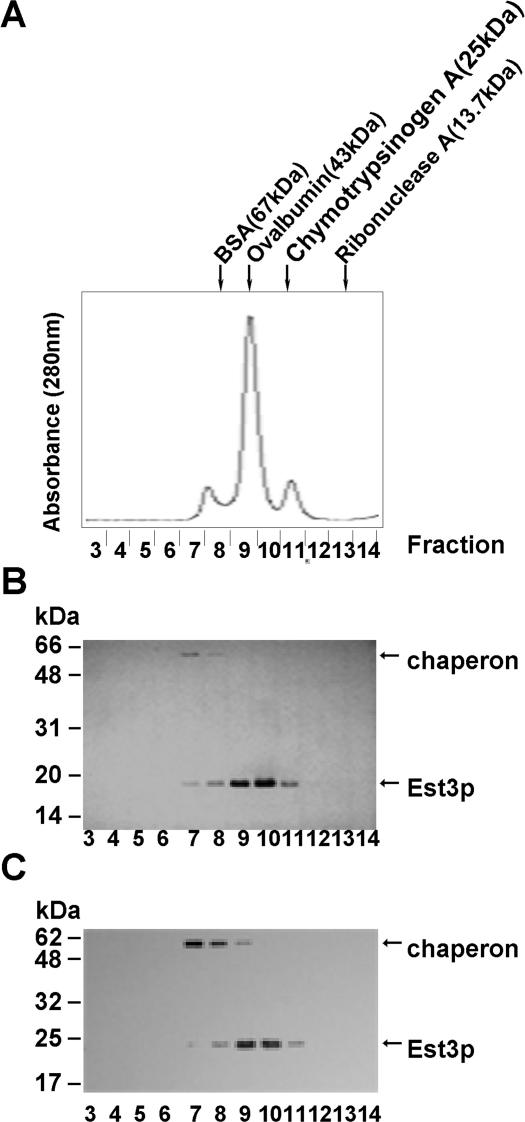
Telomerase has been suggested to function as a dimer or oligomer during the synthesis of telomeres (35–38). Since Est3p is a subunit of telomerase holoenzyme, we wanted to know whether Est3p could form a dimer or oligomer in vitro. The purified un-tagged recombinant Est3p (Figure 1A) was loaded onto an analytical size exclusion column that was calibrated with standard markers including BSA (67 kDa), ovalbumin (43 kDa), chymotrypsinogen A (25 kDa) and ribonuclease A (13.7 kDa) (Figure 2A). The elution profile of Est3p revealed three peaks corresponding to molecular weights of about 80, 40 and 20 kDa, respectively, with the 40 kDa peak being the most prominent (Figure 2A). Both silver staining and the western blot using specific primary antibody against Est3p showed that the 40 and 20 kDa peaks represented the Est3p while the 80 kDa peak was the molecular chaperone combined with Est3p (Figure 2B and C). These data suggest that the purified recombinant Est3p is able to form a homodimer in vitro.
Figure 2.
The purified wild-type Est3p forms dimer with gel filtration analysis. (A) Elution profile of gel filtration with an analytical size exclusion column superdex 75. The standard markers are as indicated: BSA (67 kDa), Ovalbumin (43 kDa), chymotrypsinogen A (25 kDa) and Ribonuclease A (13.7 kDa). The fractions near or within the peaks (from Fraction 3 to 14) were examined by silver staining (B) and western blot (C). Affinity-purified anti-Est3p antibody was used for western blot.
The telomeres were shortened in est3 mutants
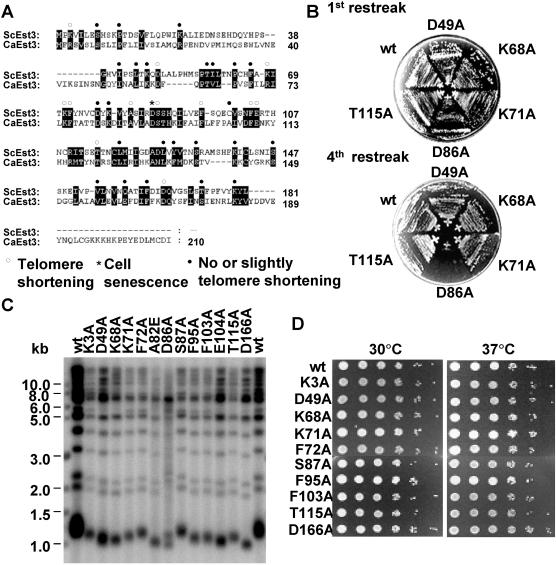
Besides biochemical characterization of Est3p, we also used genetic approach to screen for mutants of Est3p that would cause a change in telomere length. To facilitate our site-directed mutagenesis, we compared the sequences of S.cerevisiae Est3p (ScEst3p) and Candida albicans Est3p (CaEst3p) (39), and found out that they share 21% identity and 35% similarity (Figure 3A). The conserved amino acids were subjected to site-directed mutagenesis. Because little information about the structure or function of Est3p was available, we arbitrarily generated point mutations in 37 amino acids spanning nearly the full-length of Est3p (Figure 3A). In each mutant construct, a single amino acid was changed. The conserved amino acid [e.g. Lys (K), Ser (S), Pro (P), Asp (D), Thr (T), Phe (F), Glu (E), Leu (L), Tyr (Y), Gln (Q)] was changed to Ala (A), and the conserved Ala was changed to Glu (E) (see Figure 3A and Table 1). The mutations in Est3p will be referred to using the following abbreviation format of K3A, which stands for Ala (A) replacing the Arg (K) at amino acid position 3 of Est3p.
Figure 3.
Mutations of conserved residues in EST3 result in telomere shortening and do not show temperature sensitivity. (A) Sequence comparison of ScEst3p and CaEst3p. Sequences of EST3 in S.cerevisiae and C.albicans were obtained from SGD (Saccharomyces Genome Database: http://db.yeastgenome.org/cgi-bin/locus.pl?locus=Est3) and aligned with GENEDOC program. Positions that were mutated are indicated above the sequence of ScEst3p. Open circles indicate that mutation at this position caused telomere shortening; the asterisk indicates that the cell went senescence, while the solid circles mean little or no telomere shortening. (B) Growth of est3 mutant cells. The wild-type and mutant yeast cells (D49A, K68A, K71A, D86A and T115A) were streaked on selective SD plates four times, with each streak being counted as 25 generations. Only the first and fourth streaks are presented here. (C) Telomere Southern blot of est3 mutants. The mutant icons are labeled on the top. (D) Temperature sensitivity test of est3 mutants. The 30°C overnight cultured yeast cells were diluted to OD600 = 0.3, 3 µl of 10-fold serially diluted cells were spotted on the Leu− SD plates and the plates were incubated at 30 and 37°C, respectively. Isogenic strains tested are noted at the left.
Table 1.
est3 mutants and telomere phenotype
| Type | Telomere phenotype | Amino acid substitution |
|---|---|---|
| I | Cell senescence | D86A |
| II | Telomere shortening | K3A, D49A, K68A |
| K71A, F72A, A82E | ||
| S87A, F95A, F103A | ||
| E104A, T115A, D166A | ||
| III | Little or no telomere defect | S8A, P12A, K23A |
| I42E, L45E, K47A | ||
| T58A, I59E, P63A | ||
| F66A, D77A, K79A | ||
| C99E, L119E, L127A | ||
| Y129A, S133A, K140A | ||
| S147A, V154E, Q159A | ||
| F163A, T173A, K179A |
To determine the consequence of these single amino acid substitutions, the mutated est3 alleles were introduced into the est3Δ yeast strain. Each strain carrying a different mutant allele of est3 was restreaked on Leu− plates four times, with each restreak allowing for ∼25 generations of growth during colony formation. We found out that the substitution of Ala for Asp at position 86 (D86A) caused cell senescence, while other 36 mutants shown in Figure 3A and Table 1 did not display the similar senescence phenotype (Figure 3B and data not shown). To confirm the senescence of mutant D86A and to characterize other est3 mutants, the telomere length of each strain was determined by Southern blot analysis using a specific telomeric TG1–3 probe (Figure 3C and data not shown). Of the 37 mutants, 24 mutants showed little or no change of telomere length (Table 1 and data not shown). Twelve mutants (K3A, D49A, K68A, K71A, F72A, A82E, S87A, F95A, F103A, E104A, T115A and D166A) showed telomere shortening in the range of 50–200 bp (Figure 3C). One mutant, D86A, showed progressive telomere shortening and amplification of TG1–3 repeat telomeres (Figure 3C), a result consistent with telomerase-independent type II survival phenotype after senescence (40,41). None of the mutants showed telomere lengthening (Figure 3C and Table 1). Most of the mutations that resulted in telomere shortening were located in the N-terminal two-thirds of EST3 (amino acids 1–120), with only one mutation (D166A) located in the C-terminus. The mutants with shortened telomeres showed no temperature sensitivity when cultured at 37°C (Figure 3D and data not shown).
Est3p dimerization in vitro is affected in some of the Est3 mutants
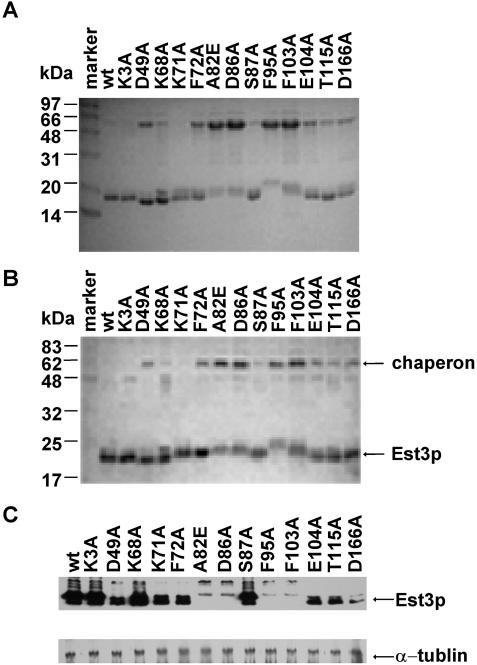
Because we observed that purified recombinant Est3p could form a homodimer in vitro (Figure 2), we wondered whether there was a functional connection between dimerization and telomere length regulation. Therefore, we purified 13 mutant proteins alongside wild-type Est3p in E.coli. The SDS–PAGE gel stained with Coomassie blue and western blot analysis indicated that the mutant proteins were purified to near homogeneity (Figure 4A and B). However, for the Est3p mutants D49A, F72A, A82E, D86A, F95A, F103A, E104A or T115A, much more molecular chaperone was co-purified (Figure 4A). Additionally, for unknown reasons, mutant proteins, Est3p-A82E, Est3p-D86A, Est3p-F95A, Est3p-F103A showed slower mobility in the SDS–PAGE gel than wild-type Est3p (Figure 4A). It is possible that some mutations of Est3p would cause conformational changes, and formation of a complex with molecular chaperone might help to stabilize these mutant proteins.
Figure 4.
Purification of wild-type and mutant Est3 proteins. The purified proteins were analyzed with SDS–PAGE stained by (A) Coomassie blue and (B) western blot using affinity purified polyclonal anti-Est3p antibody. (C) The over-expression of Est3 mutants in S.cerevisiae. The upper panel is a western blot probed with affinity purified polyclonal anti-Est3p antibody. The lower panel shows a western blot of the α-tubulin in the same membrane as used in the upper panel (loading control) probed with a monoclonal anti-α-tubulin antibody.
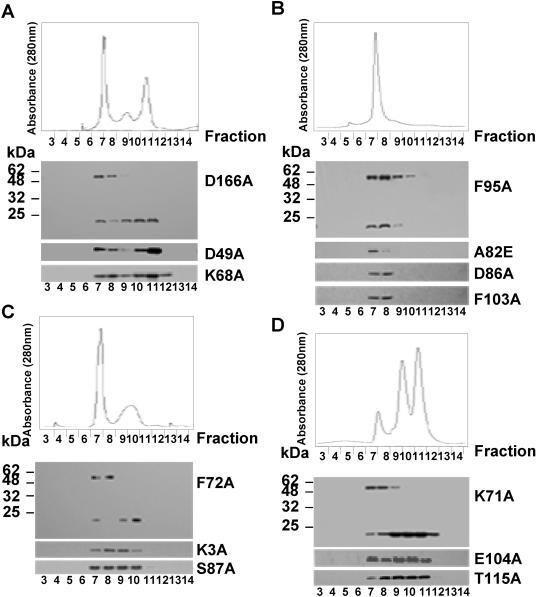
Each purified mutant Est3p was assayed on the same gel filtration column and western blots were performed to follow the elution of mutant Est3 proteins (Figure 5). The elution profiles of purified mutant Est3 proteins seemed to fall into several typical patterns. Within the D49A, K68A and D166A mutants, the 40 kDa-dimer peak was much less prominent compared with the wild-type Est3 protein (Figure 5A). In the A82E, D86A, F95A and F103A mutants, the 80 kDa peak was the major one, while both the 40 and 20 kDa peaks seemed to have disappeared (Figure 5B). In the K3A, F72A, S87A mutants (Figure 5C), the 40 kDa peak was observed, but was reduced in abundance when compared with wild-type Est3p. In the K71A, E104A and T115A mutants (Figure 5D), the 40 kDa peak was comparable with the 20 kDa. From these data, we concluded the following. The K3A, D49A, K68A, F72A, A82E, D86A, F95A, S87A, F103A and D166A mutant proteins have reduced (or lost) the ability to form stable homodimer in vitro. The A82E, D86A, F95A and F103A mutant proteins form a stable complex with the bacterial molecular chaperone, while the K71A, E104A and T115A mutant proteins were able to dimerize as well as wild-type Est3p.
Figure 5.
The gel filtration analysis of 13 mutant Est3 proteins. The top panels are the elution profiles whereas the y-axis is the relative UV absorbance at 280 nm, and the x-axis stands for the fraction number from 3 to 14. The panels under the elution profile are the results of western analysis of gel filtration fractions for each mutant indicated on the right of each panel.
Over-expression of wild-type and mutant Est3 proteins in S.cerevisiae
The correlation between the defect in dimer formation in vitro (Figure 5A) and the telomere length shortening in mutants, D49A, K68A or D166A in vivo (Figure 3C) suggests that dimer formation is important for Est3p function in vivo. For the mutants A82E, D86A, F95A, F103A, it remains possible that telomere shortening in vivo might be a result of Est3p instability rather than loss of dimerization. To determine whether or not the mutant proteins were stable when expressed in S.cerevisiae, we examined the Est3 proteins by western blot. We used conditions of Est3p over-expression because the endogenous expression level of Est3p was not detectable by western blot (data not shown). The over-expression of the wild-type and various mutant Est3 proteins is shown in Figure 4C, with α-tubulin used as protein loading control. Interestingly, the expression pattern of individual mutant in S.cerevisiae is quite similar to that of what we observed in E.coli. The expression level of the D49A, K71A, F72A, E104A, T115A and D166A mutant proteins was about 2- to 3-fold less than that of the wild-type protein (Figure 4C). The expression level of the K3A, K68A and S87A mutant proteins was comparable with that of wild-type (Figure 4C). The expression level of the A82E, D86A, F95A and F103A mutant proteins was much lower than that of wild-type (Figure 4C). These results suggested that the telomere shortening seen in the K3A, D49A, K68A, F72A, S87A and D166A strains (Figure 3C) might have been caused by a reduced ability of the Est3p mutants to dimerize, while telomere defect observed in the A82E, D86A, F95A and F103A mutants might have been caused by instability of the mutant proteins. The K71A, E104A and T115A mutant Est3 proteins seemed to be stable in both E.coli and S.cerevisiae (Figure 4A–C) and were able to dimerize in vitro (Figure 5D), indicating that the telomere shortening phenotype was due to something other than loss of dimerization. Taken together, these data suggest that dimerization of Est3p is important for maximum telomerase activity, but not essential for the minimal telomerase activity.
The telomere length in est3-D49A-K68A double mutation
The observations that the D49A or K68A amino acid change reduced dimer formation of Est3p in vitro and resulted in telomere shortening in vivo suggested that the two residues are important for Est3p function. To examine further whether Est3p dimerization is important for telomerase function in vivo, we made the double mutant Est3p-D49A-K68A. As shown in Figure 6A, the telomeres of est3-D49A-K68A double mutant cells were shorter than D49A, but similar to that of K68A, suggesting that the K68A mutation might have a greater affect on telomerase activity in vivo. We further purified the double mutant Est3p-D49A-K68A, and the Coomassie blue stained SDS–PAGE gel showed that much more molecular chaperone was co-purified during the purification of Est3p-D49A-K68A (Figure 6B) than that of the mutant Est3p-D49A or Est3p-K68A (Figure 5A). The elution profile of Est3p-D49A-K68A in a size exclusion column (Figure 6C) appeared to be different from that of Est3p-D49A or Est3p-K68A (Figure 5A), but similar to that of the Est3p-A82E, -D86A, -F95A and -F103A (Figure 5B). These data suggest that Est3p-D49A-K68A is unstable in vivo, and the loss of Est3p-D49A-K68A dimerization might be a secondary consequence of its instability.
Figure 6.
Characterization of double mutant Est3p-D49A-K68A. (A) Telomere Southern blot. The isogenic strains are as following: wild-type (YPH499), D49A mutant, D49AK68A double mutant, K68A mutant. (B) Commassie blue staining of purified Est3p-D49A-K68A. Lane 1, uninduced total extract; lane 2, induced total extract; lane 3, GST-Est3p-D49A-K68A fusion protein on glutathione beads; lane 4, Est3p-D49A-K68A after thrombin digestion. (C) Analysis of the Est3p-D49A-K68A double mutant by gel filtration with superdex 75 column. The upper panel is the elution profile, and the lower panel is western blot using affinity purified polyclonal anti-Est3p antibodies.
Functional interactions between Est3p dimers
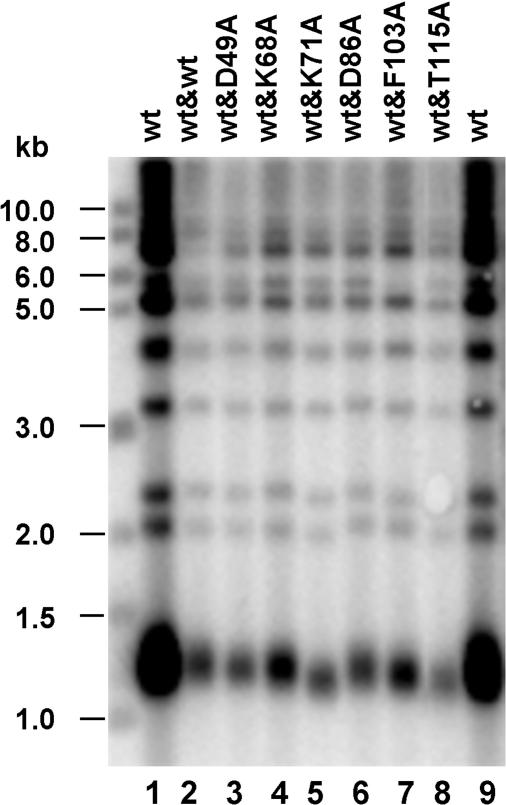
If dimerization is important for Est3p function in vivo, a heterodimer of mutant Est3p-K71A or Est3p-T115A with wild-type Est3p would cause a defect in telomere replication, while the Est3p-D49A and Est3p-K68A mutations would not be able to form a heterodimer with wild-type Est3p to enhance telomere replication. Thus, we introduced these mutant est3 alleles into yeast cells with the low-copy-number centromere plasmid pRS315 and introduced the wild-type EST3 allele with pRS316, another low-copy-number centromere plasmid. The co-expression of different mutant and wild-type Est3 proteins resulted in the different telomere lengths shown in Figure 7. Little or no telomere length change was detected in the cells where we co-expressed the wild-type Est3p and the Est3p mutants D49A (lane 3), K68A (lane 4), D86A (lane 6) or F103A (lane 7). In contrast, telomere shortening was seen in the cells where we co-expressed the wild-type Est3p and the Est3p mutants K71A (lane 5) or T115A (lane 8). These results suggest that the wild-type Est3p is not able to form a heterodimer with and is not affected by the D49A, K68A, D86A or F103A mutants, which are also unable to efficiently form a homodimer on their own (Figure 5A and B). These data also suggest that the wild-type Est3p is able to form a heterodimer with and is affected by the K71A or T115A mutants, which are able to form a homodimers by themselves (Figure 5D). The functional interaction of mutant and wild-type Est3p further supported the idea that Est3p forms a dimer in vivo, and the dimer form is more efficient in promoting telomere maintenance.
Figure 7.
Est3 mutants retaining the ability to dimerize show telomere shortening in ectopic co-expression with wild-type. Both wild-type EST3 (in pRS316 CEN plasmid) and mutant est3 (in pRS315 CEN plasmid) were co-introduced into est3 deletion cells, telomere Southern blot was performed, the isogenic transformants were as following: wt (wild-type), wt&wt, wt&est3-D49A, wt&est3-K68A, wt&est3-K71A, wt&est3-D86A, wt&est3-F103A, wt&est3-T115A.
DISCUSSION
Est3p is required for telomerase activity in vivo and is believed to be a subunit of yeast telomerase holoenzyme, but its function in telomere replication is not yet clear. We purified recombinant Est3p to near homogeneity (Figure 1A and B). Our affinity purification strategy was straightforward and efficient for the purification of wild-type Est3p. This method, however, was not as robust in the preparation of some of the Est3 mutant proteins (A82E, D86A, F95A and F103A) and a large amount of chaperone was co-purified with the Est3p (Figure 4A) although further purification step was employed (Figure 5B). We have also attempted to purify 6-histidine tagged Est3p from E.coli and similar difficulties were encountered (data not shown). Removing GST-tag from Est3p by thrombin digestion was a required procedure for in vitro characterization (Figure 1C); however, extensive digestion lowered the yield of recombinant Est3p (data not shown).
The purified Est3p was able to form a dimer when it was fractionated in a size exclusion column (Figure 2). The dimerization proficiency of Est3p observed in vitro correlated well with its ability to function in vivo. The substitution of alanine (A) for amino acid D49 or K68 in Est3p did not affect the stability of either of the mutant proteins (Figure 4), but did result in a reduction in their ability to dimerize in vitro (Figure 5A) and led to telomere shortening in vivo (Figure 3C). Co-expression of dimer-deficient D49A, K68A, D86A or F103A mutants with wild-type Est3p did not affect telomere replication (Figure 5A and B and Figure 7), but co-expression of dimer-proficient K71A or T115A mutant and wild-type Est3p caused telomere shortening (Figures 5D and 7). These lines of evidence strongly suggest that the dimerization of Est3p is important for its function in vivo.
It was previously reported that the telomerase RNA and the catalytic reverse transcriptase subunit in S.cerevisiae, Euplotes crassus and humans might form a dimer or multimer as a functional unit (35–38). Since Est3p is one of the subunits of the yeast telomerase holoenzyme, the dimerization of Est3p may play a role in helping telomerase form a dimer to elongate the telomere efficiently. This would explain our in vitro and in vivo observations for some of the Est3p mutants, such as D49A, K68A or D166A, but could not address the essential role of Est3p to telomerase activity, or the down regulation of telomerase activity in the Est3p mutant K3A, K71A, F72A, A82E, D86A, S87A, F95A, F103A, E104A and T115A, which also showed shortened telomeres (Figure 3C and Table 1). In other words, the dimerization of Est3p could be important for maximum telomerase activity, but is not essential for minimum telomerase activity. Therefore, it was unclear how the Est3 mutant proteins that retained the ability to homodimerize were affecting telomerase activity.
One possibility was that the mutant Est3 proteins were unstable and could not function efficiently. For example, the recombinant proteins carrying the mutations A82E, D86A, F95A or F103A showed slower mobility than wild-type in the SDS–PAGE gel (Figure 4A and C), and the over-expression of the A82E, D86A, F95A or F103A mutants were present at lower levels than that of over-expressed wild-type protein (Figure 4C). A possible explanation for this was that mutation in this region caused a conformational change of Est3p, and the telomere shortening observed in these mutants might be caused by the Est3p instability rather than loss of the ability to dimerize. The fact that mutant proteins form complex with the bacterial molecular chaperone also indicated that they were less stable (Figure 4A and C). The D86A mutation of Est3p caused progressive telomere shortening and cell senescence, while over-expression of Est3p-D86A in the est3Δ cells could stabilize shortened telomeres and prevent the cells from senescence (data not shown), suggesting that Est3p-D86A is not stable enough to maintain the minimum telomerase activity required for cell viability when it is present at the wild-type level.
Previous genetic studies by Friedman and co-workers (13) showed that over-expression of Est3p rescued the temperature sensitivity of est2ts specific alleles where the mutations were located in Region I, suggesting that Est3p interacts with N-terminal domain of Est2p. Thus, a second, but not exclusive possibility is that mutation of Est3p affects its interaction with the other subunits of telomerase holoenzyme, e.g. Est1p, Est2p and/or Tlc1. For the K71A, E104A and T115A mutants, the protein appeared to be stable in yeast (Figure 4C), and they were able to form dimers (Figure 5D). The shortened telomeres in these cells might be due to the loss of interaction between Est3p and one of the other telomerase holoenzyme components like Est1p, Est2p and/or Tlc1. However, due to the very low abundance of Est1, Est2 and Est3 proteins, we were unable to detect the interaction between them at the endogenous level (data not shown). Since reasonable amounts of the recombinant wild-type Est3p and the Est3p mutants K3A, K71A, F72A, S87A, E104A and T115A (which did not affect its dimerization, but caused telomere shortening in vivo) were successfully purified, it becomes possible to study direct interaction between Est3p and Est2p or Est1p when purified recombinant Est2p or Est1p becomes available.
The telomerase holoenzyme of S.cerevisiae has the ability to processively add more than one repeat per binding event in vivo (42–44), yet the purified recombinant telomerase core enzyme from S.cerevisiae can only add one-round of repetitive telomeric DNA in vitro (45). A third possibility would be that Est3p functions in the holoenzyme as a telomerase processivity factor, and hence mutation of Est3p causes telomere shortening in vivo. It has been proposed that the telomerase might switch templates during telomere synthesis (36). It remains possible that Est3p is a translocator for the template switching. The findings that Est3p forms a dimer, and the dimerization is important for its in vivo function seemed to be trivial with regard to uncovering the essential role(s) of Est3p in telomerase activity, but the constructed mutant yeast strains and the purified recombinant wild-type and mutant Est3 proteins will greatly facilitate the future genetic and biochemical studies for the function of Est3p in telomere replication.
Acknowledgments
We thank Dr Brian A. Lenzmeier (Buena Vista University, Storm Lake, IA, USA) for his critical reading on the manuscript. We thank anonymous reviewers for their critical suggestions on our work. This work is supported by a Chinese Academy of Sciences-Max Planck Society Professorship, a National Natural Science Foundation of China (NSFC 30125010) award to distinguished young scientist, and a NSFC grant (30270295). Funding to pay the Open Access publication charges for this article was provided by Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences.
Conflict of interest statement. None declared.
REFERENCES
- 1.Greider C.W. Telomere length regulation. Annu. Rev. Biochem. 1996;65:337–365. doi: 10.1146/annurev.bi.65.070196.002005. [DOI] [PubMed] [Google Scholar]
- 2.Blackburn E.H. Switching and signaling at the telomere. Cell. 2001;106:661–673. doi: 10.1016/s0092-8674(01)00492-5. [DOI] [PubMed] [Google Scholar]
- 3.de Lange T. Protection of mammalian telomeres. Oncogene. 2002;21:532–540. doi: 10.1038/sj.onc.1205080. [DOI] [PubMed] [Google Scholar]
- 4.Kass-Eisler A., Greider C.W. Recombination in telomere-length maintenance. Trends Biochem. Sci. 2000;25:200–204. doi: 10.1016/s0968-0004(00)01557-7. [DOI] [PubMed] [Google Scholar]
- 5.Zakian V.A. Structure, function, and replication of Saccharomyces cerevisiae telomeres. Annu. Rev. Genet. 1996;30:141–172. doi: 10.1146/annurev.genet.30.1.141. [DOI] [PubMed] [Google Scholar]
- 6.Counter C.M., Meyerson M., Eaton E.N., Weinberg R.A. The catalytic subunit of yeast telomerase. Proc. Natl Acad. Sci. USA. 1997;94:9202–9207. doi: 10.1073/pnas.94.17.9202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lingner J., Hughes T.R., Shevchenko A., Mann M., Lundblad V., Cech T.R. Reverse transcriptase motifs in the catalytic subunit of telomerase. Science. 1997;276:561–567. doi: 10.1126/science.276.5312.561. [DOI] [PubMed] [Google Scholar]
- 8.Lendvay T.S., Morris D.K., Sah J., Balasubramanian B., Lundblad V. Senescence mutants of Saccharomyces cerevisiae with a defect in telomere replication identify three additional EST genes. Genetics. 1996;144:1399–1412. doi: 10.1093/genetics/144.4.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lundblad V., Szostak J.W. A mutant with a defect in telomere elongation leads to senescence in yeast. Cell. 1989;57:633–643. doi: 10.1016/0092-8674(89)90132-3. [DOI] [PubMed] [Google Scholar]
- 10.Singer M.S., Gottschling D.E. TLC1: template RNA component of Saccharomyces cerevisiae telomerase. Science. 1994;266:404–409. doi: 10.1126/science.7545955. [DOI] [PubMed] [Google Scholar]
- 11.Nakamura T.M., Morin G.B., Chapman K.B., Weinrich S.L., Andrews W.H., Lingner J., Harley C.B., Cech T.R. Telomerase catalytic subunit homologs from fission yeast and human. Science. 1997;277:955–959. doi: 10.1126/science.277.5328.955. [DOI] [PubMed] [Google Scholar]
- 12.Peng Y., Mian I.S., Lue N.F. Analysis of telomerase processivity: mechanistic similarity to HIV-1 reverse transcriptase and role in telomere maintenance. Mol. Cell. 2001;7:1201–1211. doi: 10.1016/s1097-2765(01)00268-4. [DOI] [PubMed] [Google Scholar]
- 13.Friedman K.L., Heit J.J., Long D.M., Cech T.R. N-terminal domain of yeast telomerase reverse transcriptase: recruitment of Est3p to the telomerase complex. Mol. Biol. Cell. 2003;14:1–13. doi: 10.1091/mbc.E02-06-0327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hughes T.R., Weilbaecher R.G., Walterscheid M., Lundblad V. Identification of the single-strand telomeric DNA binding domain of the Saccharomyces cerevisiae Cdc13 protein. Proc. Natl Acad. Sci. USA. 2000;97:6457–6462. doi: 10.1073/pnas.97.12.6457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lin J.J., Zakian V.A. The Saccharomyces CDC13 protein is a single-strand TG1-3 telomeric DNA-binding protein in vitro that affects telomere behavior in vivo. Proc. Natl Acad. Sci. USA. 1996;93:13760–13765. doi: 10.1073/pnas.93.24.13760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nugent C.I., Hughes T.R., Lue N.F., Lundblad V. Cdc13p: a single-strand telomeric DNA-binding protein with a dual role in yeast telomere maintenance. Science. 1996;274:249–252. doi: 10.1126/science.274.5285.249. [DOI] [PubMed] [Google Scholar]
- 17.Taggart A.K., Teng S.C., Zakian V.A. Est1p as a cell cycle-regulated activator of telomere-bound telomerase. Science. 2002;297:1023–1026. doi: 10.1126/science.1074968. [DOI] [PubMed] [Google Scholar]
- 18.Pennock E., Buckley K., Lundblad V. Cdc13 delivers separate complexes to the telomere for end protection and replication. Cell. 2001;104:387–396. doi: 10.1016/s0092-8674(01)00226-4. [DOI] [PubMed] [Google Scholar]
- 19.Garvik B., Carson M., Hartwell L. Single-stranded DNA arising at telomeres in cdc13 mutants may constitute a specific signal for the RAD9 checkpoint. Mol. Cell. Biol. 1995;15:6128–6138. doi: 10.1128/mcb.15.11.6128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Virta-Pearlman V., Morris D.K., Lundblad V. Est1 has the properties of a single-stranded telomere end-binding protein. Genes Dev. 1996;10:3094–3104. doi: 10.1101/gad.10.24.3094. [DOI] [PubMed] [Google Scholar]
- 21.Zhou J., Hidaka K., Futcher B. The Est1 subunit of yeast telomerase binds the Tlc1 telomerase RNA. Mol. Cell. Biol. 2000;20:1947–1955. doi: 10.1128/mcb.20.6.1947-1955.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Livengood A.J., Zaug A.J., Cech T.R. Essential regions of Saccharomyces cerevisiae telomerase RNA: separate elements for Est1p and Est2p interaction. Mol. Cell. Biol. 2002;22:2366–2374. doi: 10.1128/MCB.22.7.2366-2374.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Seto A.G., Livengood A.J., Tzfati Y., Blackburn E.H., Cech T.R. A bulged stem tethers Est1p to telomerase RNA in budding yeast. Genes Dev. 2002;16:2800–2812. doi: 10.1101/gad.1029302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Qi H., Zakian V.A. The Saccharomyces telomere-binding protein Cdc13p interacts with both the catalytic subunit of DNA polymerase alpha and the telomerase-associated est1 protein. Genes Dev. 2000;14:1777–1788. [PMC free article] [PubMed] [Google Scholar]
- 25.Evans S.K., Lundblad V. Est1 and Cdc13 as comediators of telomerase access. Science. 1999;286:117–120. doi: 10.1126/science.286.5437.117. [DOI] [PubMed] [Google Scholar]
- 26.Peterson S.E., Stellwagen A.E., Diede S.J., Singer M.S., Haimberger Z.W., Johnson C.O., Tzoneva M., Gottschling D.E. The function of a stem-loop in telomerase RNA is linked to the DNA repair protein Ku. Nature Genet. 2001;27:64–67. doi: 10.1038/83778. [DOI] [PubMed] [Google Scholar]
- 27.Morris D.K., Lundblad V. Programmed translational frameshifting in a gene required for yeast telomere replication. Curr. Biol. 1997;7:969–976. doi: 10.1016/s0960-9822(06)00416-7. [DOI] [PubMed] [Google Scholar]
- 28.Hughes T.R., Evans S.K., Weilbaecher R.G., Lundblad V. The Est3 protein is a subunit of yeast telomerase. Curr. Biol. 2000;10:809–812. doi: 10.1016/s0960-9822(00)00562-5. [DOI] [PubMed] [Google Scholar]
- 29.Sikorski R.S., Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen Y.B., Yang C.P., Li R.X., Zeng R., Zhou J.Q. Def1p is involved in telomere maintenance in budding yeast. J. Biol. Chem. 2005;280:24784–24791. doi: 10.1074/jbc.M413562200. [DOI] [PubMed] [Google Scholar]
- 31.Zhou J.Q., Tan C.K., So A.G., Downey K.M. Purification and characterization of the catalytic subunit of human DNA polymerase delta expressed in baculovirus-infected insect cells. J. Biol. Chem. 1996;271:29740–29745. doi: 10.1074/jbc.271.47.29740. [DOI] [PubMed] [Google Scholar]
- 32.Thain A., Gaston K., Jenkins O., Clarke A.R. A method for the separation of GST fusion proteins from co-purifying GroEL. Trends Genet. 1996;12:209–210. doi: 10.1016/s0168-9525(96)90022-0. [DOI] [PubMed] [Google Scholar]
- 33.Maier G., Dietrich U., Panhans B., Schroder B., Rubsamen-Waigmann H., Cellai L., Hermann T., Heumann H. Mixed reconstitution of mutated subunits of HIV-1 reverse transcriptase coexpressed in Escherichia coli—two tags tie it up. Eur. J. Biochem. 1999;261:10–18. doi: 10.1046/j.1432-1327.1999.00304.x. [DOI] [PubMed] [Google Scholar]
- 34.Panne D., Raleigh E.A., Bickle T.A. McrBs, a modulator peptide for McrBC activity. EMBO J. 1998;17:5477–5483. doi: 10.1093/emboj/17.18.5477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Prescott J., Blackburn E.H. Functionally interacting telomerase RNAs in the yeast telomerase complex. Genes Dev. 1997;11:2790–2800. doi: 10.1101/gad.11.21.2790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wenz C., Enenkel B., Amacker M., Kelleher C., Damm K., Lingner J. Human telomerase contains two cooperating telomerase RNA molecules. EMBO J. 2001;20:3526–3534. doi: 10.1093/emboj/20.13.3526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Beattie T.L., Zhou W., Robinson M.O., Harrington L. Functional multimerization of the human telomerase reverse transcriptase. Mol. Cell. Biol. 2001;21:6151–6160. doi: 10.1128/MCB.21.18.6151-6160.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang L., Dean S.R., Shippen D.E. Oligomerization of the telomerase reverse transcriptase from Euplotes crassus. Nucleic Acids Res. 2002;30:4032–4039. doi: 10.1093/nar/gkf513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Singh S.M., Steinberg-Neifach O., Mian I.S., Lue N.F. Analysis of telomerase in Candida albicans: potential role in telomere end protection. Eukaryot. Cell. 2002;1:967–977. doi: 10.1128/EC.1.6.967-977.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen Q., Ijpma A., Greider C.W. Two survivor pathways that allow growth in the absence of telomerase are generated by distinct telomere recombination events. Mol. Cell. Biol. 2001;21:1819–1827. doi: 10.1128/MCB.21.5.1819-1827.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Signon L., Malkova A., Naylor M.L., Klein H., Haber J.E. Genetic requirements for RAD51- and RAD54-independent break-induced replication repair of a chromosomal double-strand break. Mol. Cell. Biol. 2001;21:2048–2056. doi: 10.1128/MCB.21.6.2048-2056.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Marcand S., Brevet V., Gilson E. Progressive cis-inhibition of telomerase upon telomere elongation. EMBO J. 1999;18:3509–3519. doi: 10.1093/emboj/18.12.3509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Diede S.J., Gottschling D.E. Telomerase-mediated telomere addition in vivo requires DNA primase and DNA polymerases alpha and delta. Cell. 1999;99:723–733. doi: 10.1016/s0092-8674(00)81670-0. [DOI] [PubMed] [Google Scholar]
- 44.Teixeira M.T., Arneric M., Sperisen P., Lingner J. Telomere length homeostasis is achieved via a switch between telomerase-extendible and -nonextendible states. Cell. 2004;117:323–335. doi: 10.1016/s0092-8674(04)00334-4. [DOI] [PubMed] [Google Scholar]
- 45.Liao X.H., Zhang M.L., Yang C.P., Xu L.X., Zhou J.Q. Characterization of recombinant Saccharomyces cerevisiae telomerase core enzyme purified from yeast. Biochem. J. 2005;390:169–176. doi: 10.1042/BJ20050208. [DOI] [PMC free article] [PubMed] [Google Scholar]