Abstract
Translation initiation represents a key step during regulation of gene expression in chloroplasts. Here, we report on the identification and characterization of three suppressor point mutations which overcome a translational defect caused by the deletion of a U-rich element in the 5′-untranslated region (5′-UTR) of the psbD mRNA in the green alga Chlamydomonas reinhardtii. All three suppressors affect a secondary RNA structure encompassing the psbD AUG initiation codon within a double-stranded region as judged by the analysis of site-directed chloroplast mutants as well as in vitro RNA mapping experiments using RNase H. In conclusion, the data suggest that these new element serves as a negative regulator which mediates a rapid shut-down of D2 synthesis.
INTRODUCTION
The control of mRNA translation represents a key regulatory step for gene expression in both prokaryotes and eukaryotes. Despite the fundamental differences between their translation machineries, in both types of organisms the initial steps of translation are known to play an essential role (1). These early events of protein synthesis depend on a variety of cis- and trans-acting determinants which usually mediate their functions via the 5′- and 3′-untranslated regions (5′ and 3′-UTR) of protein-coding transcripts (2–6). In chloroplasts, the typical endosymbiotic organelles of photoautotrophic plants and algae—a special situation developed during evolution. A basic translational system, which is of prokarytic phylogenetic origin, is embedded in a eukaryotic cellular context. This results in a novel hybrid apparatus for protein synthesis which is controlled by nucleus-encoded eukaryotic-type factors (7–9). During the last years, substantial work has been invested to identify these factors and to elucidate the molecular mechanisms which underlie regulatory translational processes within this specialized cellular compartment.
Genetic analyses in the green alga Chlamydomonas reinhardtii and vascular plants have recently led to the identification and cloning of some of the trans-acting translation factors involved (10–14). Moreover, the genes for RNA-binding proteins which associate with chloroplast 5′-UTRs in vitro [for a review see (15,16) and in vivo (17,18)] were identified by biochemical means. However, apart from the target regions on chloroplast mRNAs, relatively little is known about the precise molecular working mode of the respective factors.
Initially, the cis-acting RNA targets were mapped by analyzing reporter gene constructs carrying mutated versions of distinct chloroplast 5′-UTRs (19–22) or by site-directed mutagenesis of the endogenous chloroplast gene regions (16,23,24). Alternatively, a chloroplast in vitro translation system from tobacco was used to define cis-acting translational elements within various plastid 5′-UTRs in higher plants (25,26). These analyses revealed that the typical prokaryotic signal for translation initiation, i.e. a Shine–Dalgarno element located 4–12 nt upstream of the AUG start codon, is functional in some but not all chloroplast mRNAs (25,27–29). Furthermore, sequences surrounding the AUG start codon were shown to significantly affect translational efficiency (30–32).
More systematic mutagenesis approaches then identified additional regions within 5′-UTRs as well as downstream regions (33,34) which affect chloroplast protein synthesis. In both C.reinhardtii and tobacco, stem–loop structures within the psbA 5′-UTR have been shown to be critical for determining translational efficiency (27,35). RNA secondary structure elements within 5′-UTRs were also found to affect protein synthesis from the psbC, petD and rps7 mRNAs in C.reinhardtii (20,22,24,36) and the atpB mRNA in tobacco (25).
We have previously demonstrated that the psbD 5′-UTR in C.reinhardtii contains the target site for the nucleus-encoded RNA stability factor Nac2 (3,37) which connects processes of psbD RNA stabilization and translation initiation [for a recent review see (38)]. Nac2 guides the RNA-binding protein RBP40 to its cognate target site which is located 15 nt upstream of the psbD AUG start codon. This cis-acting element comprises a stretch of multiple U residues whose deletion in the mutant ΔU completely abolished the synthesis of the psbD gene product, i.e. the D2 protein of the photosystem II reaction centre (23), and, furthermore, led to the loss of RBP40-binding. A cis-acting suppressor of the ΔU mutation was isolated and shown to harbour a 5 bp duplication within the mutated region which partially restored both photosynthetic growth and RNA recognition by RBP40 (39).
Here, we report on the identification and characterization of three novel, independent second-site suppressor mutations of the ΔU mutation which are all located further downstream of the U-element close to the AUG start codon. Site-directed mutagenesis studies demonstrated that these mutations affect a secondary RNA structure including the AUG start codon. The data suggest that this structure serves as a negative regulatory element for D2 synthesis.
MATERIALS AND METHODS
Algal strains, suppressor isolation and genetic crosses
C.reinhardtii strains were grown on tris-acetate-phosphate medium at 25°C (40). Suppressors of the ΔU mutation were isolated as described (39). In brief, ΔU cells were plated on HS medium and kept in the dark for 24 h, exposed to ultraviolet (UV)-light (7.5 mJ and 254 nm) in a stratalinker (Stratagene) and transfered to darkness for another 24 h-period to prevent photoreactivation. Suppressors were selected in bright light (100 µE m−2 s−1) for a period of up to 6 weeks. To test whether the suppressor mutations reside within the nuclear or chloroplast genome, all three suppressor strains (mt+) were genetically crossed to the wild-type (mt−). All 4 members out of 33 (suΔU+9), 31 (suΔU−3) or 18 (suΔU+10) analyzed tetrads from these crosses were able to grow photoautotrophically indicating a chloroplast localization of the respective suppressor mutations. Photoautotrophic growth rates were followed by measurement of the OD700 of cell cultures.
Plasmid construction and chloroplast transformation
Plasmids containing psbD 5′-UTR mutations were generated via mutagenesis PCR as described (23) with oligonucleotides 1963 and 1365 as well as oligonucleotides including the mutation, i.e. su2-a: 5′-gcaatgacaatttcgatcgg-3′; su2-b: 5′-ccgatcgaaattgtcattgc-3′; su4-a: 5′-gcaatgacaatggcgatcgg-3′; su4-b: 5′-ccgatcgccattgtcattgc-3′; su5-a: 5′-gagatacacacaatgacaat-3′; su5-b: 5′-attgtcattgtgtgtatctc-3′; revsu2-a: 5′-ggagatacacgaaatgacaa-3′; revsu2-b: 5′-ttgtcatttcgtgtatctcc-3′; revsu4-a: 5′-gagatacacgccatgacaat-3′; revsu4-b: 5′-attgtcatggcgtgtatctc-3′; revsu5-a: 5′-atgacaattgtgatcggtac-3′; revsu5-b: 5′-gtaccgatcacaattgtcat-3′; mutsu-a: 5′-ggagatacacgccatgacaa-3′; mutsu-b: 5′-ttgtcatggcgtgtatctcc-3′. Chloroplasts were then transformed with these plasmids using a helium-driven particle gun (41). The resultant strains were selected for photoautotrophic growth on HS medium plates. Plasmid 72.1 containing the wild-type psbD 5′-UTR was used as a positive control (23).
Analysis of nucleic acids and proteins
Total DNA from C.reinhardtii was isolated using the DNeasy Plant Kit (Qiagen, Hilden). Algal RNA was prepared with hot phenol (42). RNA secondary structures were calculated by using the RNAdraw software (43). Northern analysis, primer extension assays and western analysis were performed exactly as described (44). Radioactive labelling of RNAs and UV cross-linking with proteins were also performed as described (39).
RNase H mapping of RNA secondary structure
Templates comprising 134 bp (wt) or 127 bp (suΔU+10, suΔU+9 and suΔU−3) for in vitro synthesis of the various psbD RNA probes were PCR-amplified from appropriate DNAs with the oligonucleotide su3131 : 5′-tgtgcgtttctcttgatatgtaccg-3′, complementary to the coding region of psbD from position +39 to +15 relative to the ATG and oligonucleotide 2126: 5′-taatacgactcactatagggacacaatgattaaaattaaa-3′ spanning the psbD 5′ region from position −74, as well as the T7 promotor sequence (39). In vitro transcription reactions and radioactive labelling of the RNAs were performed as described (23). RNA probes (15 fmol) were diluted in cacodylate buffer (50 mM Na-cacodylate, 20 mM CaCl2 and 10 mM KCl) and incubated with 10 pmol of the oligonucleotide RH-1: 5′-aattgtcattgcgtgtatct-3′ which is complementary to position −11 to +9 relatively to the AUG start codon. The samples were heated to 60°C for 5 min and cooled down with a rate of 1°C per min to 25°C. After addition of one volume 2× cacodylate buffer with MgCl2 (50 mM Na-cacodylate, 10 mM KCl, 20 mM CaCl2 and 20 mM MgCl2), samples were incubated with 0.5 U RNase H (Ambion, Cambridgeshire) for 2 min or 5 min and loaded on 12% Polyacrylamide TBE gels (45). After electrophoresis, gels were sealed in plastic bags and exposed to Fuji X-ray films at −20°C.
Pulse labelling of membrane proteins
For pulse labelling of proteins, cells were grown as described (23). After measurement of chlorophyll content, cells were harvested and resuspended to 80 µg chlorophyll/ml. A total of 500 µl of the cells were incubated with cycloheximide (10 µg/ml) for 10 min. Subsequently, cells were fed with 50 µCi 35S-sulphate (Amersham, Freiburg) for 20 min in bright light (100 µE m−2 s−1). After centrifugation, sedimented cells were frozen in liquid nitrogen. Broken-cell preparation and gel electrophoresis were carried out as described (44). Signals were quantitated by using the Scion Image software and standardized to the internal Cytf control.
RESULTS
Three independent point mutations suppress the ΔU mutation
Previously, it was shown that a striking U-rich element within the 5′-UTR of the chloroplast psbD mRNA is involved in its translation (Figure 1). This element is recognized by the RNA-binding protein RBP40, a process that is dependent on the presence of the nucleus-encoded RNA stability factor Nac2 (39). Replacement of the U-track by a BamHI restriction site in the mutant ΔU resulted in complete loss of both D2 synthesis as well as RBP40-binding (23). Furthermore, the photosynthetic revertant suΔU containing a 5 bp duplication immediately upstream of the BamHI site restored both D2 synthesis and RBP40-binding to almost wild-type levels. These findings together with a more comprehensive mutational analysis underlined the significance of the U-element/RBP40 interaction for translation of the psbD message (39).
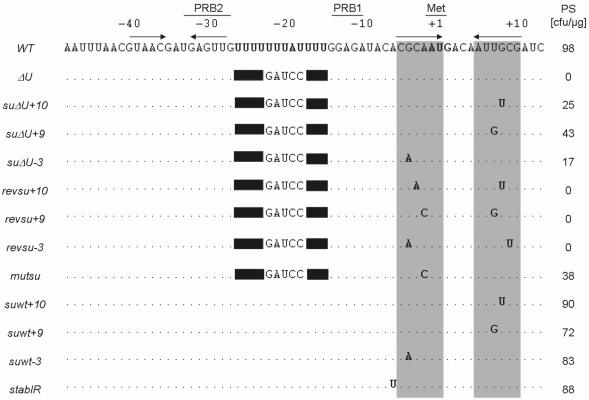
Figure 1.
Sequence alignment of the psbD 5′ region from wild-type (WT) and analyzed mutant strains. Positions relative to the initiation codon (Met) and the formerly described PRB1 and PRB2 boxes (23) are indicated above the sequences. Dots and solid boxes mark conserved residues and deletions, respectively. The sequence of the U-rich region and the AUG start codon are given in boldface and horizontal arrows represent computer-predicted stem–loop structures of the wild-type region. PS, representative number of photoautotrophically growing colonies after transformation of the mutant ΔU with 1 µg of indicated DNAs.
We have now isolated three novel, independent phenotypic revertants of the ΔU mutation, namely suΔU+10, suΔU+9 and suΔU−3. Genetic crosses revealed that each suppressor mutation resides within the chloroplast genome (see Materials and Methods). However, in contrast to the previously characterized suΔU strain, sequencing of the psbD region revealed that in the suppressors the immediate vicinity of the mutated U-tract is not altered. Instead, each suppressor strain harbours a single point muation which is localized downstream of the U-stretch at positions +10 (suΔU+10), +9 (suΔU+9) and −3 (suΔU−3) with regard to the AUG start codon (Figure 1). Biolistic back-transformations of chloroplasts with constructs containing the psbD leader region of either suΔU+10, suΔU+9 or suΔU−3 showed that all three constructs were able to complement the ΔU mutant verifying that indeed each mutation is sufficient to cause the suppressor phenotype on its own (Figure 1).
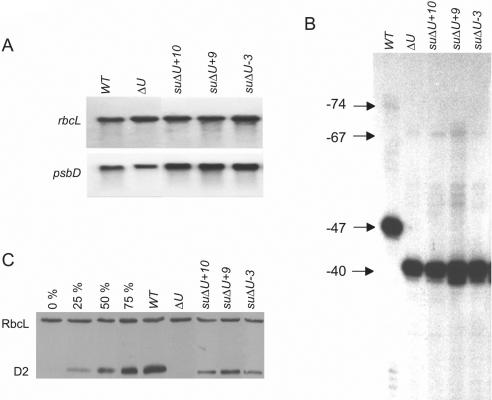
All three suppressors accumulated almost wild-type levels of psbD mRNA (Figure 2A; Table 1) indicating that the respective point mutations have no effect on RNA stabilization. The psbD mRNA has previously been shown to exist in two forms. The difference lies in the length of their 5′-UTR and is most likely due to a 5′ processing event (23,38). In neither of the three suppressor strains, this 5′ maturation was compromised (Figure 2B). However, D2 protein accumulation was found to be restored to varying extent. Whilst the suppressor suΔU+9 accumulated almost 50% of D2 protein as compared to the wild-type, suΔU+10 and suΔU−3 reached only levels of 20 and 15%, respectively (Figure 2C; Table 1). This was in agreement with reduced photoautotrophic doubling times of 37 h for suΔU+9, 38 h for suΔU+10 and 45 h for suΔU−3 as compared to 35 h for the wild-type (Table 1).
Figure 2.
Molecular characterization of suppressor strains suΔU+10, +9, and −3. (A) Northern analysis of total RNA (10 µg) from the strains indicated at the top was carried out with a radiolabelled psbD- or, as an internal standard, a rbcL-specific probe. (B) Indicated RNAs were assayed by primer extension analysis using oligonucleotide 3131. The arrows mark the 5′ ends of the longer form starting at position −74 relative to the AUG start codon and the shorter form starting at position −47 of the psbD 5′-UTR from the wild-type. In the ΔU mutant and the suppressors, psbD leaders are shorter by 7 nt due to the deletion at the poly(U) region (see Figure 1). (C) Western analysis of total proteins (10 µg) from the same strains was performed by immunolabelling with antibodies directed against either the D2 protein or, as an internal standard, the Rubisco holoenzyme from spinach. A serial dilution of wild-type proteins (0–75%) in ΔU proteins was co-analyzed.
Table 1.
Molecular characteristics of analyzed strains
| Strains | WT | ΔU | suΔU+10 | suΔU+9 | suΔU−3 | mutsu | suwt+10 | suwt+9 | suwt−3 | stabIR |
|---|---|---|---|---|---|---|---|---|---|---|
| Photoautotrophic growtha | 35 ± 2 | – | 38 ± 1.5 | 37 ± 1 | 45 ± 2 | 40 ± 2 | 31 ± 1.5 | 30 ± 2 | 32 ± 1 | 39 ± 2 |
| RNA | 100 | 80 ± 5 | 108 ± 6 | 112 ± 4 | 115 ± 3 | 114 ± 5 | 104 ± 3 | 99 ± 4 | 105 ± 4 | 112 ± 5 |
| Proteinb | 100 | 0 | 20 ± 6 | 46 ± 5 | 15 ± 7 | 42 ± 6 | 117 ± 8 | 116 ± 9 | 118 ± 9 | 61 ± 6 |
Values display the mean of three independent experiments except for b.
aDoubling times in HS medium at 100 µE m−2 s−1.
bValues display the mean of 10 independent experiments.
Suppressor mutations do not restore RBP40-binding
As mentioned above, in the previously described cis-acting suppressor suΔU, a 5 bp duplication immediately upstream of the U-rich translation element resulted in restored binding of RBP40 to the psbD 5′-UTR thereby explaining the observed suppressor phenotype (39). Therefore, we tested whether the suppressor mutations suΔU+10, +9 and −3 also affect RBP40-binding in vitro. Radiolabelled RNA probes from each 5′-UTR were incubated with stromal protein extract from the wild-type, and the RBP40 signal was monitored after UV cross-linking of RNAs and proteins. As expected, a strong binding signal was observed with a wild-type psbD 5′ probe but not with a mutant ΔU probe. The same held for the three suppressor RNAs, none of which led to a RBP40-binding signal (Figure 3A). To further confirm the different RNA-binding properties of RBP40, competition binding experiments were carried out by using radiolabelled wild-type RNA and increasing excess of now unlabelled probes. As shown in Figure 3B, only the homologous wild-type 5′-UTR efficiently competed with the wild-type probe while excess of the suppressor RNAs had only a minor effect on RBP40-binding. Taken together, these data strongly suggest that in contrast to the known suppressor suΔU, the suppressor mutations suΔU+10, suΔU+9 and suΔU−3 do not restore the binding of RBP40 to the psbD U-track but instead appear to act via a RBP40-independent mechanism.
Figure 3.
Binding of RBP40 to suppressor 5′-UTRs. (A) Radiolabelled psbD 5′-UTR probes from the strains indicated at the top and chloroplast stromal protein extract were analysed by UV cross-linking (39). (B) In competition binding experiments, radiolabelled wild-type psbD 5′ probe was pre-incubated with 5-, 50- or 500-fold molar excess of the marked unlabeled competitor RNAs. (C) The diagram displays the intensities of the RBP40-binding signal in relation to that of the signal without competitor (0×).
The psbD AUG start codon is part of a secondary RNA structure
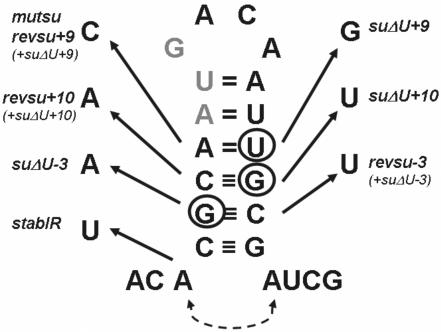
Bioinformatic inspection of the psbD 5′ RNA secondary structure revealed that all three supressor mutations are located in a putative stem-structure of a RNA hairpin which partly encompasses also the AUG start codon (Figure 4). To test whether this RNA secondary structure or the RNA sequence context close to the AUG codon is important for mediating suppression, compensatory mutations were introduced into the psbD leader by site-directed mutagenesis (23). The mutations revsu+10, revsu+9 and revsu−3 each restored the putative stem-region in the respective suppressor backgrounds by an appropriate exchange of the mismatching nucleotides on the opposite strand of the stem (Figure 4). When mutant ΔU chloroplasts were transformed with these constructs, no complementations were observed for neither of them (Figure 1). This suggested that indeed the restoration of the RNA stem–loop structure inhibited the suppression effect independent of the sequence context.
Figure 4.
RNA stem–loop structure of the psbD 5′ region. The wild-type sequence of the psbD region around the initiation codon (grey) is delineated with nucleotide exchanges in the various mutants marked by arrows. All mutants harbour single point mutations in the stem-region except revsu+10, +9 and −3 which in addition contain the respective suppressor point mutations (see also Figure 1). The additional base pairing in stabIR is indicated by a dotted line.
To further substantiate that the suppressor mutations affect a critical structural rather than a sequence element, the secondary structure of the wild-type and the suppressor psbD 5′-UTRs were determined by comparative RNase H mapping (45). Radiolabelled RNA probes were hybridized with oligonucleotide RH-1 which is complementary to the respective AUG regions. RNA/DNA hybrid formation is assumed to be possible only in unpaired RNA stretches which then become a substrate for RNase H. Consequently, unstructured RNA regions will cause an efficient cleavage of the RNA into defined smaller products. As shown in Figure 5, the wild-type psbD 5′-UTR was almost resistant against RNase H digestion over a time period of 5 min suggesting that a double-stranded RNA structure prevents stable base pairing with the oligonucleotide. In contrast, all three suppressor RNAs were efficiently cleaved into expected major fragments of 59 and 30 nt being indicative of a more or less unstructured RNA region in the vicinity of the start codon.
Figure 5.
In vitro mapping of RNA secondary structure. 100 fmol of indicated radiolabelled RNA probes of 116 nt (wt) and 109 nt (suΔU+10, suΔU+9 and suΔU−3) were incubated with oligonucleotide RH-1 complementary to the AUG regions and 0.5 U RNase H for 2 or 5 min. Major RNase H cleavage products of 59 and 30 nt (arrows) were visualized on autoradiograms after separation by denaturating PAGE. Computer-predicted secondary RNA structures with a boxed AUG initiation codon are given at the top. Position of oligonucleotide RH-1 is indicated by bold characters.
These findings strongly suggested that the psbD 5′-UTR forms a stable RNA stem–loop structure which partially includes the AUG codon in its stem-region. Furthermore, this structure appears to act as a negatively regulating element for D2 synthesis at least in a ΔU genetic background (Figure 1) probably by preventing access of the small ribosomal subunit to the initiation codon. If this scenario were true, then, a suppression of the ΔU mutation should be achievable by altering other nucleotides of the stem-region. Thus, an A to C mutation at position −1 relative to the start codon was introduced into the ΔU 5′-UTR to test whether this stem-destabilizing mutation, similar to the genetically selected suppressors suΔU+10, +9 and −3, would also lead to a suppression effect. A corresponding construct indeed was capable of complementing the ΔU mutant (Figure 1). In the resulting transformant mutsu, D2 accumulation was found to be restored to 42% of the wild-type level (Figure 6B; Table 1) further supporting the idea of a repressive RNA structure located at the psbD translation initiation codon.
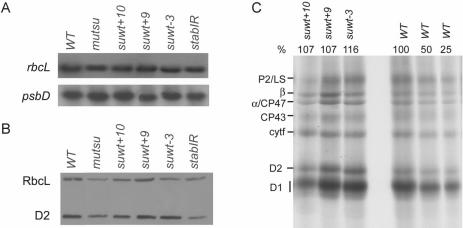
Figure 6.
Molecular characterization of site-directed psbD mutants. For northern (A) and western (B) analyses see legend of Figure 2. (C) Proteins from indicated strains were pulse labelled with 35S-sulphate for 20 min and separated by SDS–PAGE.
Analysis of suppressor mutations in the presence of the psbD U-rich element
The above mentioned data provided good evidence that the psbD stem–loop structure is a negative determinant for psbD gene expression in the absence of the U-rich translation element and, as a consequence, the absence of RBP40 activity. Thus, the next question concerned the phenotype of mutants which contain both the U-element and any of the suΔU+10, +9 and −3 mutations. Adequate site-directed mutant versions, named suwt+10, suwt+9 and suwt−3, were constructed and transformed into ΔU chloroplasts. As expected, all three constructs complemented the U-stretch mutation (Figure 1). Interestingly, resulting transformants were shown to accumulate slightly increased levels of D2 protein (Figure 6B; Table 1). This was in agreement with a moderate enhancement of D2 synthesis in suwt+10, +9 and −3 as compared to the wild-type in protein pulse labelling experiments using radioactive sulphate (Figure 6C). This increase in D2 accumulation in suwt+10, +9 and −3 suggested that also other subunits of the PS II complex accumulated to enhanced levels because unassembled PS II proteins are rapidly degraded by a chloroplast proteolytic ‘clearing system’ (23). Consistently, all three strains exhibited a slightly accelerated photoautotrophic growth rate as compared to the wild-type (Table 1). These findings supported the idea that the RNA secondary structure serves as a negative element for psbD gene expression and, similar to the situation in cyanobacteria, D2 protein accumulation represents a key regulatory step for PS II assembly (46).
In conclusion, these results suggested that the RNA stem–loop structure containing the psbD AUG start codon negatively regulates synthesis of the D2 protein and as a consequence PS II accumulation. As an additional control for this hypothesis, the site-directed mutant stabIR was constructed by introducing a U residue at position −5 of the wild-type psbD leader (Figure 4). This alteration led to a stem-region which was extended by 1 bp and thus, a RNA structure of enhanced stability should be formed. Although this construct was still able to complement the ΔU mutant (Figure 1), the subsequent molecular characterization of the resulting transformant stabIR revealed a significant reduction of D2 accumulation to nearly 60% compared to the wild-type level (Figure 6B; Table 1). This finding provided an additional piece of evidence for the RNA conformation at the translation initiation codon playing a critical role for psbD gene expression.
DISCUSSION
In this report, we describe the identification and characterization of a structural RNA element which serves as a negative regulatory determinant for the synthesis of the D2 protein in C.reinhardtii chloroplasts. The analysis of genetically selected suppressors, various site-directed chloroplast mutants as well as in vitro mapping studies using RNase H showed that the AUG start codon of the psbD mRNA is located in a double-stranded RNA region which has to be resolved before translation initiation. It should be noted, however, that in suppressor strains suΔU+9 and suΔU+10 the amino acid sequence of the D2 protein was changed at position 3 from isoleucine to methionine and at position 4 from alanine to valine, respectively (Figure 4). These alterations might have influenced overall D2 protein stability and thus D2 accumulation. For instance, it was reported previously that the replacement of the threonine at position 2 of D2 with acidic—but not neutral—amino acid residues abolished D2 synthesis/accumulation at a post-initiation level in C.reinhardtii (47,48). However, similar effects are not likely to play a significant role for the D2 accumulation rates in suppressors suΔU+9 and suΔU+10 since they represent gain of function mutants. Additionally, when placed in an otherwise wild-type background, both point mutations apparently had no negative effect on PSII activity as documented by the fitness of the strains suwt+9 and suwt+10 (Table 1). Hence, it seems rather unlikely that the amino acid changes in suΔU+9 and suΔU+10 add a substantial effect to the suppression mechanism.
The influences of RNA secondary structure elements on chloroplast protein synthesis rates have been noticed before. In principle these structures can have two different functions (49). First, they might serve as recognition sites for trans-acting, translation-activating factors as has been suggested for several chloroplast 5′-UTRs (21,22,24,27,35,36). Secondly and like the element described here, secondary RNA structures might serve as negative regulatory elements which block access of the small ribosomal subunit to the translation initiation region resembling the situation which is frequently found in prokaryotes (1). In chloroplasts for instance, it has been proposed that the processing of petD precursor RNAs in maize results in the release of the initiation codon from base pairing within a secondary structure and only thereby allows efficient synthesis of the petD gene product (50). Also in barley chloroplasts, methyl jasmonate-dependent processing of the rbcL 5′-UTR was proposed to influence translation initiation rates (51). By using a chloroplast in vitro translation system from tobacco, it was recently shown that the RNA conformation around the AUG codon of the atpB mRNA negatively affects translation efficiency (25). Moreover, bioinformatic inspection of the close vicinity of translational initiation regions (each ranging from position −15 to +18) from all chloroplast genes in C.reinhardtii revealed that, potentially, nine of the respective initiation codons might be parts of double-stranded RNA structures with a thermodynamic stability in the range of the psbD mRNA one (ΔG = −8.53 kcal at 25°C). These regions include that of the psbN (−9.31 kcal), atpB (−7.53 kcal), psbC (−7.32 kcal), ORF2971 (−6.77 kcal), rpoA (−6.71 kcal), rpl2 (−6.4 kcal), psaA (−6.06 kcal) and petG (−5.3 kcal) genes. However, only additional experimental data will provide an answer to the question whether such ‘prokaryotic-like’, negative principles of regulation represent a more common theme of chloroplast gene expression.
Whilst the psbD stem–loop structure plays a crucial role for gene expression in the absence of the U-rich region and thus RBP40 activity, its destabilization in an otherwise wild-type background did not result in a dramatic but only moderate increase in D2 synthesis. This suggests that under optimal conditions the inhibition of D2 synthesis via the stem–loop is not strictly rate-limiting. This is reminiscent of the situation found in chloroplast mutants of the petA- or atpB-1 nt (31,52). In both cases nucleotide changes in the respective −1 triplets did not show any measurable effect on protein accumulation. Only if the AUG start codon was changed into the sub-optimal AUU codon, differences in protein accumulation became detectable (31,52). Under optimal conditions, i.e. the presence of the U-element, most probably the availability of positively regulating trans-acting factors such as the Nac2 protein and the Nac2-dependent RNA-binding protein RBP40 mainly determine D2 synthesis rates as has recently been proposed (39,44). In the absence or reduction of Nac2/RBP40 activity however, this element might be involved in a rapid shut-down of psbD translation thereby allowing a fast adjustment of protein synthesis rates to changing environmental conditions.
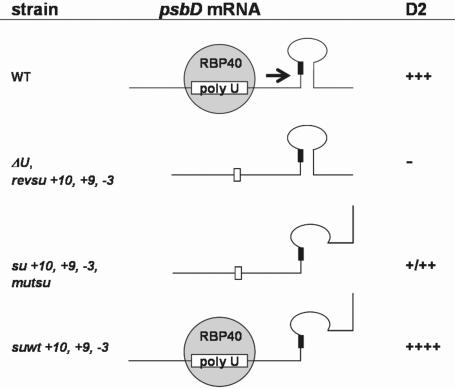
Figure 7 summarizes the main psbD gene expression modes which were characterized during the course of this work. In conclusion, the fact that the isolated suppressor mutations overcome a defect in RBP40-binding without restoring the interaction between this factor and the U-rich element strongly suggests that at least one function of RBP40 is to resolve the secondary structure in the wild-type situation. We have recently identified the RBP40 gene but its putative structure does not reveal a typical helicase-like activity (I. Elles and J. Nickelsen, unpublished data). Hence, it appears more likely that the binding of RBP40 alters the psbD RNA conformation thereby enabling access of the small ribosomal subunit to the initiation codon. Future work is focussing on the interactions between the involved trans-acting factors and the characterized RNA elements and, thus, their precise molecular working modes.
Figure 7.
Model of D2 synthesis modes. Analyzed strains and their relative D2 levels are indicated at the left and right margins, respectively. The psbD mRNA structure is given along with RBP40. Black boxes represent the AUG initiation codon. For further explanation see text.
Acknowledgments
We thank U. Aschke for skilled technical assistance, U. Kück for providing laboratory space and F. Narberhaus for critical reading of the manuscript. Antiserum against the Rubisco Holoenzyme from spinach was kindly provided by G. Wildner. This work was supported by a grant from the Deutsche Forschungsgemeinschaft to J.N. (SFB 480-TPB8). Funding to pay the Open Access publication charges for this article was provided by the DFG.
Conflict of interest statement. None declared.
REFERENCES
- 1.Kozak M. Initiation of translation in prokaryotes and eukaryotes. Gene. 1999;234:187–208. doi: 10.1016/s0378-1119(99)00210-3. [DOI] [PubMed] [Google Scholar]
- 2.Drager R.G., Girard-Bascou J., Choquet Y., Kindle K.L., Stern D.B. In vivo evidence for 5′→3′ exoribonuclease degradation of an unstable chloroplast mRNA. Plant J. 1998;13:85–96. doi: 10.1046/j.1365-313x.1998.00016.x. [DOI] [PubMed] [Google Scholar]
- 3.Nickelsen J., van Dillewijn J., Rahire M., Rochaix J.D. Determinants for stability of the chloroplast psbD RNA are located within its short leader region in Chlamydomonas reinhardtii. EMBO J. 1994;13:3182–3191. doi: 10.1002/j.1460-2075.1994.tb06617.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vaistij F.E., Boudreau E., Lemaire S.D., Goldschmidt-Clermont M., Rochaix J.D. Characterization of Mbb1, a nucleus-encoded tetratricopeptide-like repeat protein required for expression of the chloroplast psbB/psbT/psbH gene cluster in Chlamydomonas reinhardtii. Proc. Natl Acad. Sci. USA. 2000;97:14813–14818. doi: 10.1073/pnas.97.26.14813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Suay L., Salvador M.L., Abesha E., Klein U. Specific roles of 5′ RNA secondary structures in stabilizing transcripts in chloroplasts. Nucleic Acids Res. 2005;33:4754–4761. doi: 10.1093/nar/gki760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Del Campo E.M., Sabater B., Martin M. Post-transcriptional control of chloroplast gene expression. Accumulation of stable psaC mRNA is due to downstream RNA cleavages in the ndhD gene. J. Biol. Chem. 2002;277:36457–36464. doi: 10.1074/jbc.M204500200. [DOI] [PubMed] [Google Scholar]
- 7.Zerges W. Translation in chloroplasts. Biochimie. 2000;82:583–601. doi: 10.1016/s0300-9084(00)00603-9. [DOI] [PubMed] [Google Scholar]
- 8.Manuell A., Beligni M.V., Yamaguchi K., Mayfield S.P. Regulation of chloroplast translation: interactions of RNA elements, RNA-binding proteins and the plastid ribosome. Biochem. Soc. Trans. 2004;32:601–605. doi: 10.1042/BST0320601. [DOI] [PubMed] [Google Scholar]
- 9.Beligni M.V., Yamaguchi K., Mayfield S. The translational apparatus of Chlamydomonas reinhardtii chloroplast. Photosynth Res. 2004;82:315–325. doi: 10.1007/s11120-004-2440-5. [DOI] [PubMed] [Google Scholar]
- 10.Fisk D.G., Walker M.B., Barkan A. Molecular cloning of the maize gene crp1 reveals similarity between regulators of mitochondrial and chloroplast gene expression. EMBO J. 1999;18:2621–2630. doi: 10.1093/emboj/18.9.2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Auchincloss A.H., Zerges W., Perron K., Girard-Bascou J., Rochaix J.D. Characterization of Tbc2, a nucleus-encoded factor specifically required for translation of the chloroplast psbC mRNA in Chlamydomonas reinhardtii. J. Cell. Biol. 2002;157:953–962. doi: 10.1083/jcb.200201060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meurer J., Lezhneva L., Amann K., Godel M., Bezhani S., Sherameti I., Oelmuller R. A peptide chain release factor 2 affects the stability of UGA-containing transcripts in Arabidopsis chloroplasts. Plant Cell. 2002;14:3255–3269. doi: 10.1105/tpc.006809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dauvillee D., Stampacchia O., Girard-Bascou J., Rochaix J.D. Tab2 is a novel conserved RNA binding protein required for translation of the chloroplast psaB mRNA. EMBO J. 2003;22:6378–6388. doi: 10.1093/emboj/cdg591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Somanchi A., Barnes D., Mayfield S.P. A nuclear gene of Chlamydomonas reinhardtii, Tba1, encodes a putative oxidoreductase required for translation of the chloroplast psbA mRNA. Plant J. 2005;42:341–352. doi: 10.1111/j.1365-313X.2005.02378.x. [DOI] [PubMed] [Google Scholar]
- 15.Nickelsen J. Chloroplast RNA-binding proteins. Curr. Genet. 2003;43:392–399. doi: 10.1007/s00294-003-0425-0. [DOI] [PubMed] [Google Scholar]
- 16.Bruick R.K., Mayfield S.P. Light-activated translation of chloroplast mRNAs. Trends Plant Sci. 1999;4:190–195. doi: 10.1016/s1360-1385(99)01402-8. [DOI] [PubMed] [Google Scholar]
- 17.Schmitz-Linneweber C., Williams-Carrier R., Barkan A. RNAimmunoprecipitation and microarray analysis show a chloroplast pentatricopeptide repeat protein to be associated with the 5′ region of mRNAs whose translation it activates. Plant Cell. 2005;17:2791–2804. doi: 10.1105/tpc.105.034454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nakamura T., Ohta M., Sugiura M., Sugita M. Chloroplast ribonucleoproteins are associated with both mRNAs and intron-containing precursor tRNAs. FEBS Lett. 1999;460:437–441. doi: 10.1016/s0014-5793(99)01390-3. [DOI] [PubMed] [Google Scholar]
- 19.Staub J.M., Maliga P. Accumulation of D1 polypeptide in tobacco plastids is regulated via the untranslated region of the psbA mRNA. EMBO J. 1993;12:601–606. doi: 10.1002/j.1460-2075.1993.tb05692.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zerges W., Girard-Bascou J., Rochaix J.D. Translation of the chloroplast psbC mRNA is controlled by interactions between its 5′ leader and the nuclear loci TBC1 and TBC3 in Chlamydomonas reinhardtii. Mol. Cell Biol. 1997;17:3440–3448. doi: 10.1128/mcb.17.6.3440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stampacchia O., Girard-Bascou J., Zanasco J.L., Zerges W., Bennoun P., Rochaix J.D. A nuclear-encoded function essential for translation of the chloroplast psaB mRNA in Chlamydomonas. Plant Cell. 1997;9:773–782. doi: 10.1105/tpc.9.5.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fargo D.C., Boynton J.E., Gillham N.W. Mutations altering the predicted secondary structure of a chloroplast 5′ untranslated region affect its physical and biochemical properties as well as its ability to promote translation of reporter mRNAs both in the Chlamydomonas reinhardtii chloroplast and in Escherichia coli. Mol. Cell Biol. 1999;19:6980–6990. doi: 10.1128/mcb.19.10.6980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nickelsen J., Fleischmann M., Boudreau E., Rahire M., Rochaix J.D. Identification of cis-acting RNA leader elements required for chloroplast psbD gene expression in Chlamydomonas. Plant Cell. 1999;11:957–970. doi: 10.1105/tpc.11.5.957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Higgs D.C., Shapiro R.S., Kindle K.L., Stern D.B. Small cis-acting sequences that specify secondary structures in a chloroplast mRNA are essential for RNA stability and translation. Mol. Cell Biol. 1999;19:8479–8491. doi: 10.1128/mcb.19.12.8479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hirose T., Sugiura M. Multiple elements required for translation of plastid atpB mRNA lacking the Shine–Dalgarno sequence. Nucleic Acids Res. 2004;32:3503–3510. doi: 10.1093/nar/gkh682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hirose T., Sugiura M. Cis-acting elements and trans-acting factors for accurate translation of chloroplast psbA mRNAs: development of an in vitro translation system from tobacco chloroplasts. EMBO J. 1996;15:1687–1695. [PMC free article] [PubMed] [Google Scholar]
- 27.Mayfield S.P., Cohen A., Danon A., Yohn C.B. Translation of the psbA mRNA of Chlamydomonas reinhardtii requires a structured RNA element contained within the 5′ untranslated region. J. Cell. Biol. 1994;127:1537–1545. doi: 10.1083/jcb.127.6.1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sakamoto W., Chen X., Kindle K.L., Stern D.B. Function of the Chlamydomonas reinhardtii petD 5′ untranslated region in regulating the accumulation of subunit IV of the cytochrome b6/f complex. Plant J. 1994;6:503–512. doi: 10.1046/j.1365-313x.1994.6040503.x. [DOI] [PubMed] [Google Scholar]
- 29.Fargo D.C., Zhang M., Gillham N.W., Boynton J.E. Shine–Dalgarno-like sequences are not required for translation of chloroplast mRNAs in Chlamydomonas reinhardtii chloroplasts or in Escherichia coli. Mol. Gen. Genet. 1998;257:271–282. doi: 10.1007/s004380050648. [DOI] [PubMed] [Google Scholar]
- 30.Chen X., Kindle K., Stern D. Initiation codon mutations in the Chlamydomonas chloroplast petD gene result in temperature-sensitive photosynthetic growth. EMBO J. 1993;12:3627–3635. doi: 10.1002/j.1460-2075.1993.tb06036.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Esposito D., Fey J.P., Eberhard S., Hicks A.J., Stern D.B. In vivo evidence for the prokaryotic model of extended codon–anticodon interaction in translation initiation. EMBO J. 2003;22:651–656. doi: 10.1093/emboj/cdg072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hirose T., Sugiura M. Functional Shine–Dalgarno-like sequences for translational initiation of chloroplast mRNAs. Plant Cell. Physiol. 2004;45:114–117. doi: 10.1093/pcp/pch002. [DOI] [PubMed] [Google Scholar]
- 33.Rott R., Liveanu V., Drager R.G., Stern D.B., Schuster G. The sequence and structure of the 3′-untranslated regions of chloroplast transcripts are important determinants of mRNA accumulation and stability. Plant Mol. Biol. 1998;36:307–314. doi: 10.1023/a:1005943701253. [DOI] [PubMed] [Google Scholar]
- 34.Kuroda H., Maliga P. Complementarity of the 16S rRNA penultimate stem with sequences downstream of the AUG destabilizes the plastid mRNAs. Nucleic Acids Res. 2001;29:970–975. doi: 10.1093/nar/29.4.970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zou Z., Eibl C., Koop H.U. The stem–loop region of the tobacco psbA 5′-UTR is an important determinant of mRNA stability and translation efficiency. Mol. Genet. Genomics. 2003;269:340–349. doi: 10.1007/s00438-003-0842-2. [DOI] [PubMed] [Google Scholar]
- 36.Zerges W., Auchincloss A.H., Rochaix J.D. Multiple translational control sequences in the 5′ leader of the chloroplast psbC mRNA interact with nuclear gene products in Chlamydomonas reinhardtii. Genetics. 2003;163:895–904. doi: 10.1093/genetics/163.3.895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Boudreau E., Nickelsen J., Lemaire S.D., Ossenbuhl F., Rochaix J.D. The Nac2 gene of Chlamydomonas encodes a chloroplast TPR-like protein involved in psbD mRNA stability. EMBO J. 2000;19:3366–3376. doi: 10.1093/emboj/19.13.3366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Herrin D.L., Nickelsen J. Chloroplast RNA processing and stability. Photosynth Res. 2004;82:301–304. doi: 10.1007/s11120-004-2741-8. [DOI] [PubMed] [Google Scholar]
- 39.Ossenbühl F., Nickelsen J. Cis- and trans-acting determinants for translation of psbD mRNA in Chlamydomonas reinhardtii. Mol. Cell. Biol. 2000;20:8134–8142. [Google Scholar]
- 40.Gorman D.S., Levine R.P. Cytochrome f and plastocyanin: their sequence in the photosynthetic electron transport chain of Chlamydomonas reinhardtii. Proc. Natl Acad. Sci. USA. 1965;54:1665–1669. doi: 10.1073/pnas.54.6.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fischer N., Stampacchia O., Redding K., Rochaix J.D. Selectable marker recycling in the chloroplast. Mol. Gen. Genet. 1996;251:373–380. doi: 10.1007/BF02172529. [DOI] [PubMed] [Google Scholar]
- 42.Kück U., Choquet Y., Schneider M., Dron M., Bennoun P. Structural and transcriptional analysis of two homologous genes for the P700 chlorophyll a-apoproteins in Chlamydomonas reinhardtii: evidence for in vivo trans splicing. EMBO J. 1992;6:2185–2195. doi: 10.1002/j.1460-2075.1987.tb02489.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Matzura O., Wennborg A. RNAdraw: an integrated program for RNA secondary structure calculation and analysis under 32-bit Microsoft Windows. Comput. Appl. Biosci. 1996;12:247–249. doi: 10.1093/bioinformatics/12.3.247. [DOI] [PubMed] [Google Scholar]
- 44.Klinkert B., Schwarz C., Pohlmann S., Pierre Y., Girard-Bascou J., Nickelsen J. Relationship between mRNA levels and protein accumulation in a chloroplast promoter-mutant of Chlamydomonas reinhardtii. Mol. Genet. Genom. 2005;274:631–643. doi: 10.1007/s00438-005-0056-x. [DOI] [PubMed] [Google Scholar]
- 45.Chowdhury S., Ragaz C., Kreuger E., Narberhaus F. Temperature-controlled structural alterations of an RNA thermometer. J. Biol. Chem. 2003;278:47915–47921. doi: 10.1074/jbc.M306874200. [DOI] [PubMed] [Google Scholar]
- 46.Komenda J., Reisinger V., Muller B.C., Dobakova M., Granvogl B., Eichacker L.A. Accumulation of the D2 protein is a key regulatory step for assembly of the photosystem II reaction center complex in Synechocystis PCC 6803. J. Biol. Chem. 2004;279:48620–48629. doi: 10.1074/jbc.M405725200. [DOI] [PubMed] [Google Scholar]
- 47.Fleischmann M.M., Rochaix J.D. Characterization of mutants with alterations of the phosphorylation site in the D2 photosystem II polypeptide of Chlamydomonas reinhardtii. Plant Physiol. 1999;119:1557–1566. doi: 10.1104/pp.119.4.1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Andronis C., Kruse O., Deak Z., Vass I., Diner B.A., Nixon P.J. Mutation of residue threonine-2 of the D2 polypeptide and its effect on photosystem II function in Chlamydomonas reinhardtii. Plant Physiol. 1998;117:515–524. doi: 10.1104/pp.117.2.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Danon A. Translational regulation in the chloroplast. Plant Physiol. 1997;115:1293–1298. doi: 10.1104/pp.115.4.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Barkan A., Walker M., Nolasco M., Johnson D. A nuclear mutation in maize blocks the processing and translation of several chloroplast mRNAs and provides evidence for the differential translation of alternative mRNA forms. EMBO J. 1994;13:3170–3181. doi: 10.1002/j.1460-2075.1994.tb06616.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Reinbothe S., Reinbothe C., Heintzen C., Seidenbecher C., Parthier B. A methyl jasmonate-induced shift in the length of the 5′ untranslated region impairs translation of the plastid rbcL transcript in barley. EMBO J. 1993;12:1505–1512. doi: 10.1002/j.1460-2075.1993.tb05794.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Esposito D., Hicks A.J., Stern D.B. A role for initiation codon context in chloroplast translation. Plant Cell. 2001;13:2373–2384. doi: 10.1105/tpc.010236. [DOI] [PMC free article] [PubMed] [Google Scholar]