Abstract
Recently, we discovered an intriguing hemoprotein [aliphatic aldoxime dehydratase (OxdA)] that catalyzes the dehydration of aliphatic aldoximes [R–CH=N–OH] to the corresponding nitriles [R–C≡N] in the industrial Pseudomonas chlororaphis B23 strain. Unlike the utilization of H2O2 or O2 as a mediator of the catalysis by other heme-containing enzymes (e.g., P450), OxdA is notable for the direct binding of a substrate to the heme iron, experimental evidence of which was obtained here by means of resonance Raman (RR) analysis with an isotope technique. We found that the addition of a large amount of butyraldoxime (final concentration, 200 mM) to ferrous OxdA with a low enzyme concentration (final concentration, 5 μM) yields a long-lived OxdA–substrate complex (named OS-II), whose UV-vis spectrum is different from the corresponding spectra of the OxdA–substrate complex I and CO-bound, ferrous, and ferric forms of OxdA. Intriguingly, the RR analysis demonstrated that OS-II includes a highly oxidized heme with strong bonding between a substrate and the heme iron, as judged from the heme oxidation state marker ν4 band at 1,379 cm–1 and the 15N-isotope-substituted butyraldoxime sensitive band at 857 cm–1 in the RR spectra. It is noteworthy that OS-II has a highly oxidized heme like the ferryl-oxo heme species (e.g., compound II) formed by some general hemoproteins, although the function of OxdA is different from those (transport of electrons, transport of oxygen, sensing of oxygen or carbon monoxide, and catalysis of redox reactions) of general hemoproteins.
Keywords: hemoprotein, nitrile
We have extensively studied the biological metabolism of toxic compounds (that have a triple bond between carbon and nitrogen), such as nitriles [R–C≡N] (1–4) and isonitriles [R–N≡C] (5–7). The microbial degradation of nitriles proceeds through two different enzymatic pathways (8–9): (i) nitrilase catalyzes the hydrolysis of nitriles into acids [R–C(=O)–OH] and ammonia (10–12); and (ii) nitrile hydratase (NHase) catalyzes the hydration of nitriles to amides [R–C(=O)–NH2] (13–16), which are subsequently hydrolyzed to acids and ammonia by amidase (17–19). These enzymes have received much attention in applied (8, 20) as well as academic fields (12, 21). One of the fruits of our application-oriented nitrile studies is the current industrial production of acrylamide and nicotinamide using the NHase of Rhodococcus rhodochrous J1 (1, 9). On the other hand, the NHase of Pseudomonas chlororaphis B23 (22), which was previously used as a catalyst for acrylamide manufacture (8, 20, 23), is now used for the industrial production of 5-cyanovaleramide (24). Recently, we discovered a novel hemoprotein that catalyzes the dehydration of aliphatic aldoximes to the corresponding nitriles in P. chlororaphis B23 (25) and named it aliphatic aldoxime dehydratase (OxdA); it has been approved as a new enzyme by Nomenclature Committee of the International Union of Biochemistry and Molecular Biology (IUBMB): EC 4.99.1.5. (www.chem.qmul.ac.uk/iubmb/enzyme/EC4/99/1/5.html). The OxdA reaction not only is academically interesting but also is expected to be applicable to the practical production of nitriles, because it occurs under mild conditions, in contrast to the chemical dehydration of aldoximes under harsh conditions (26).
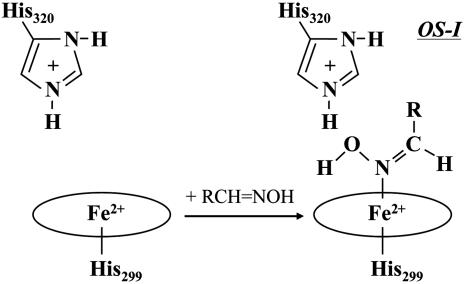
The reaction mechanism of OxdA has yet to be explored, because reaction intermediates in the catalytic cycle are not well understood. We previously reported that ferrous OxdA, containing a five-coordinated high-spin heme and His-299 as its proximal ligand, is the reactive form of the enzyme (27, 28), and that His-320 in the distal heme pocket would play a crucial role in donating a proton to a substrate during aldoxime dehydration (28). Using the stopped-flow rapid scan technique in combination with electronic absorption spectroscopy, we recently detected an initial reaction intermediate [named OxdA–substrate complex I (OS-I)], the heme coordination structure of which was suggestive of Fe(II)–N(OH)=CH–R (29), with butyraldoxime used as the substrate (Scheme 1). Although we have partially revealed the function and structure of OxdA, we have not so far answered the most interesting question regarding OxdA, “What hallmark of OxdA enables it to create a carbon–nitrogen triple bond from an aldoxime and to eliminate a water from the substrate even in an aqueous solution?” Here, we show that direct binding of a substrate followed by the formation of a highly oxidative state of OxdA is one possible answer.
Scheme 1.
Results and Discussion
The reaction intermediates of heme-containing enzymes, especially high-valent iron-oxo intermediates (referred to as compounds I and II) of cytochrome P450 and peroxidases, have received the close attention of a large number of researchers (30–34). This interest is due not only to their evident biological importance but also to their potential and appreciable economic value. The reaction intermediate of OxdA is also very attractive, because the OxdA reaction is very unique and intriguing in the following: (i) dehydration of the substrate in an aqueous solution; (ii) utilization of the favorable hydrophobic environment of the heme pocket on catalysis of dehydration; (iii) synthesis of a C–N triple bond, which is a highly toxic functional group; and (iv) direct binding of the substrate to the heme iron in contrast to other heme-containing enzymes (e.g., horseradish peroxidase). However, information on the reaction intermediate of OxdA has been quite limited and, thus, the reaction mechanism of OxdA is not yet well understood.
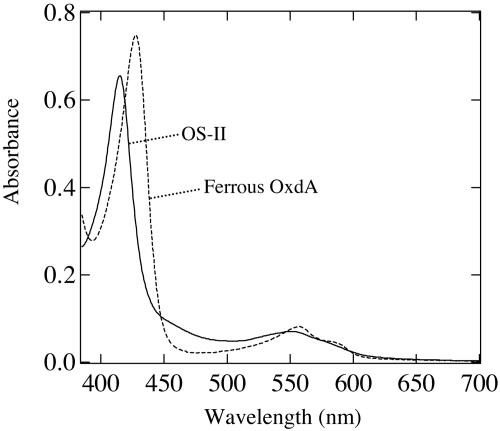
Here, we found that the addition of a large amount of butyraldoxime (final concentration, 200 mM) to a solution of ferrous OxdA with a low enzyme concentration (final concentration, 5 μM) yields a long-lived OxdA–substrate complex (named OS-II), whose UV-vis spectrum exhibits a Soret peak at 415.5 nm (Fig. 1). The UV-vis spectrum of OS-II was different from the corresponding spectra of OS-I and CO-bound, ferrous, and ferric forms of OxdA (25, 29). A gradual decrease in the Soret peak of OS-II and a concomitant increase in the Soret peak of ferrous OxdA occurred 10 min after the addition of butyraldoxime, the spectrum of ferrous OxdA finally being observed. Although the reaction rate on synthesis of butyronitrile was reduced, the stoichiometric conversion of butyraldoxime to butyronitrile was confirmed by gas chromatography.
Fig. 1.
Absorption spectra of ferrous OxdA and OS-II yielded after mixing ferrous OxdA (final concentration, 5 μM) with butyraldoxime (final concentration, 200 mM). The reaction buffer comprised 10 mM sodium dithionite and 100 mM potassium phosphate (pH 7.0).
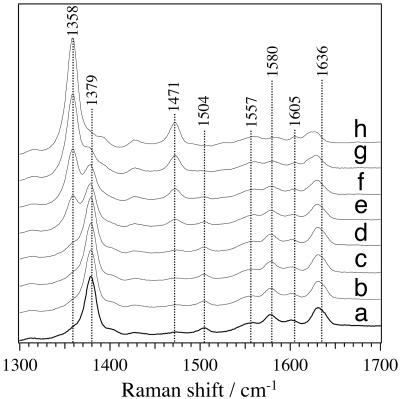
To obtain a more detailed insight into OS-II, we carried out resonance Raman (RR) spectroscopic studies on OS-II, exciting the species with a 413.1-nm Kr+ ion laser line. Fig. 2 shows the time-resolved RR spectra in the high-frequency region during the reaction process starting from OS-II immediately afforded upon the addition of excess butyraldoxime (final concentration, 200 mM) to ferrous OxdA (final concentration, 5 μM). The RR spectra of OS-II (Fig. 2 a, 5 min and b, 10 min) indicate there is no change for 10 min after the addition of excess butyraldoxime to ferrous OxdA. Then, there followed gradual decreases in the intensities of the heme marker bands of OS-II, such as the ν4 and ν3 bands at 1,379 and 1,504 cm–1, and concomitant increases in the intensities of the corresponding marker bands at 1,358 and 1,471 cm–1 of ferrous OxdA, as shown in Fig. 2 c–h. Intriguingly, the frequency of the ν4 band at 1,379 cm–1 of OS-II was even higher than that of ferric OxdA at 1,376 cm–1 (27). It has been established that the frequencies of the ν4 band are influenced by the conjugative π electron interaction between the central iron and the porphyrin ring, and that a decrease in the π electron density causes an upshift of the ν4 band (35). Thus, such a high ν4 band frequency as 1,379 cm–1 is reminiscent of that of a ferryl-oxo heme species observed in horseradish peroxidase (called compound II), which exhibits a ν4 band at 1,379 cm–1 (36). OS-II would be considered to contain a highly oxidized heme compared with that involved in ferric OxdA.
Fig. 2.
Time-resolved high-frequency RR spectra obtained after mixing ferrous OxdA (final concentration, 5 μM) with butyraldoxime (final concentration, 200 mM). The spectra were obtained at (a)5,(b) 10, (c) 15, (d) 20, (e) 25, (f) 30, (g) 35, and (h) 40 min, respectively. The RR spectrum of OS-II exhibited ν2, ν3, and ν10 bands at 1,580, 1,504, and 1,636 cm–1, respectively, which are characteristic of a six-coordinated low-spin heme (36, 42).
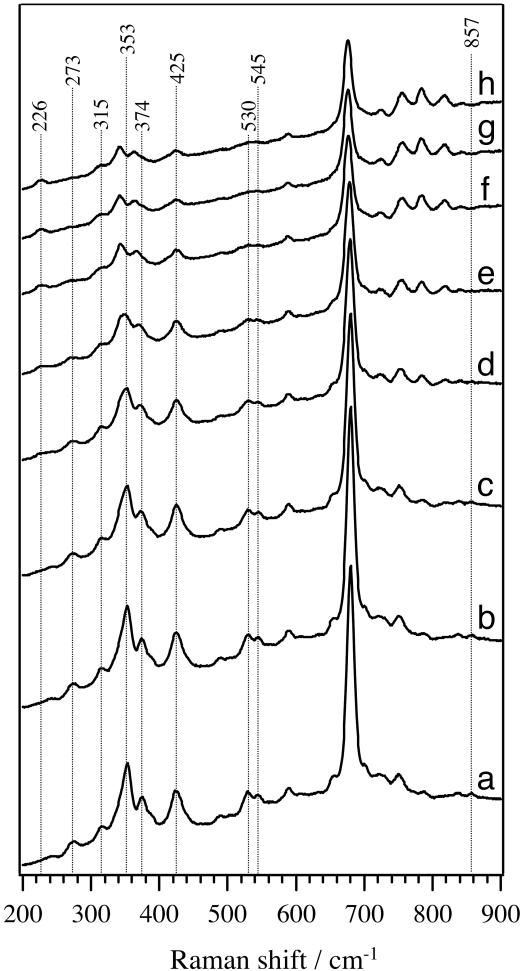
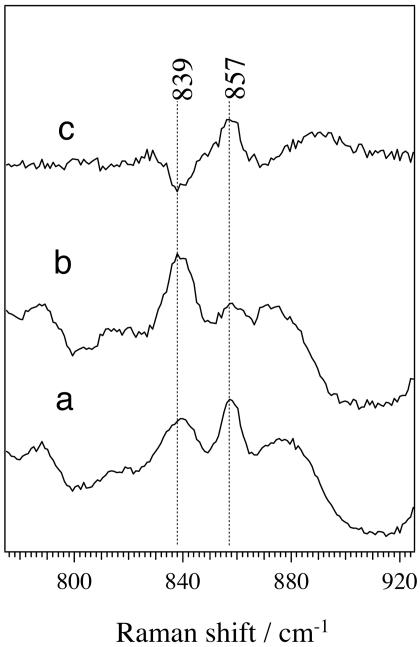
To obtain direct evidence of the binding of a substrate to the heme iron of OxdA, we examined the low-frequency region of the RR spectrum of OS-II. Fig. 3 shows the time-resolved RR spectra from OS-II to ferrous OxdA in the low-frequency region, with experimental conditions the same as those for the high-frequency spectra shown in Fig. 2. The RR spectra of OS-II exhibited no Fe-His band around 226 cm–1; this observation implies the direct binding of the substrate to the heme iron of OxdA, because it is known that the νFe-His line becomes very weak when a ligand is bound to the position trans to the histidine (37, 38). The low-frequency heme-ligand vibrational mode can be determined by using an isotope ligand, which causes an isotope shift of the ligand-heme iron vibrational mode due to the mass effect on oscillation (39, 40). Therefore, we used [15N]butyraldoxime (C3H7CH=15NOH) and observed the butyraldoxime-isotope dependence of the RR spectra of OS-II (Fig. 4). The difference spectrum (Fig. 4c) exhibited marked positive and negative peaks at 857 and 839 cm–1, respectively, suggesting that the modes are associated with oscillation of the N atom. There are two possibilities as to the origin of the isotope sensitive modes. One possibility is that the modes arise from N–OH stretching (νN–OH). However, this idea was rejected, because the 15N isotope-sensitive band at 857 cm–1 was insensitive to the solvent deuterium effect (data not shown). The other possibility is that the vibration involves stretching of a relatively strong Fe–N bond, the bond order of which may be more than one. The observed shift upon 15N substitution of as large as 18 cm–1 is in reasonable agreement with the theoretical value (15 cm–1) expected for an isolated diatomic harmonic stretching oscillator. In addition, our previous works also supported direct binding of the N atom of a substrate to the heme iron (28, 29). From these observations, we concluded that OS-II has the heme iron directly coordinated by the N atom of butyraldoxime. Intriguingly, the 857-cm–1 vibrational frequency is much higher than the Fe–N single bond stretching frequency (500–600 cm–1) observed for the NO adducts of hemoproteins (41, 42). Previously, Wagner and Nakamoto (43, 44) reported a similar high Fe–N stretching frequency (876 cm–1) in the RR spectra of the photolysis products of the tetraphenylporphyrin azide complex, for which the formation of a perferryliron-N triple bond [Fe(V)≡N] was proposed. In the OxdA reaction, however, the final product (nitrile) contains a carbon-nitrogen triple bond, and therefore the formation of Fe(V)≡N is unlikely to occur. Furthermore, density functional theory calculations have demonstrated that Fe(V)–N and Fe(IV)–N actually have a double bond characteristic with Fe–N distances of 1.722 and 1.698 Å, respectively (45). Thus, these findings suggest that the bond order between the heme iron and the nitrogen atom of the substrate part in OS-II is more than one; an Fe=N double bond may be formed.
Fig. 3.
Time-resolved low-frequency RR spectra obtained after mixing ferrous OxdA (final concentration, 5 μM) with butyraldoxime (final concentration, 200 mM). The spectra were obtained at (a)5,(b) 10, (c) 15, (d) 20, (e) 25, (f) 30, (g) 35, and (h) 40 min, respectively.
Fig. 4.
The RR spectra of OS-II obtained with natural abundant butyraldoxime (a) and [15N]butyraldoxime (C3H7CH=15NOH) (b). Spectrum c is the a–b difference spectrum.
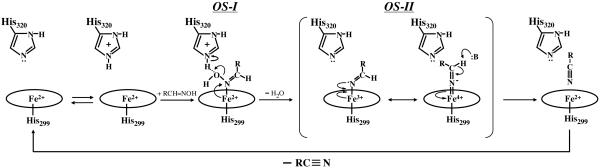
The question arises how the highly oxidized heme and the strong Fe–N bond are concomitantly formed in the reaction cycle. To answer this question, we here propose a possible mechanism for the creation of a carbon–nitrogen triple bond from aldoxime by OxdA (Fig. 5). First, OS-I is formed on mixing of ferrous OxdA with butyraldoxime (29). In our previous work (28), we suggested that the distal histidine (His-320) of OxdA donates a proton to the OH group of butyraldoxime. Second, after elimination of H2O from the heme-bound substrate, Fe(II)Por–N+=CH–R might be yielded, and electron transfer would occur from the ferrous iron to the cationic substrate. The resultant complex, Fe(III)Por–N=CH–R, may be in a resonance structure with Fe(IV)Por=N–=CH–R or Fe(III)Por•+=N–=CH–R, if the transfer of one more electron occurs from the heme to the substrate. The possibility that a cation radical porphyrin is formed in the OxdA reaction can be ruled out, because such a cation hole in porphyrin is known to cause a reduction of the absorbance in the Soret region (46); an upshift of ν2; and downshifts of ν3, ν4, and ν10 in the RR spectra (47), but the present experimental observation does not indicate such a trend. Thus, the mechanism of two-electron transfer from the ferrous iron to the substrate would result in the formation of a highly oxidized heme iron and an Fe=N double bond. The formation of OS-II would be followed by abstraction of the proton attached to the substrate carbon atom. Although it could be difficult to cleave the C–H bond of a neutral aldoxime (HO–N=CHR), the β-hydrogen of Fe(IV)Por=N–=CH–R may be prone to elimination by any weak base because of the intramolecular electron transfer from the substrate to the heme. The possible mechanism shown in Fig. 5 is largely in accordance with the results of studies on various iron porphyrin systems by Mansuy and coworkers (48, 49), suggesting that the ferryl state is formed in the aldoxime dehydration cycle.
Fig. 5.
A possible reaction mechanism for the synthesis of a nitrile by OxdA, where B indicates a basic amino acid residue of the OxdA active site.
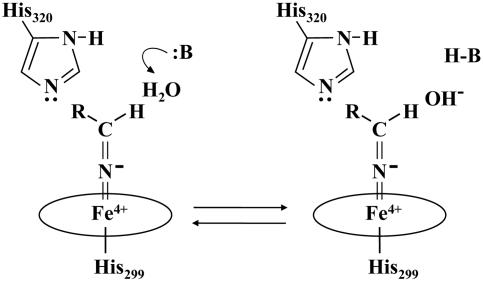
Previous analysis involving the stopped-flow rapid scan technique (29), in which a small amount of aldoxime (<50 mM) was added to ferrous OxdA, demonstrated that the rate-limiting step of the OxdA reaction is conversion of OS-I to the next species, but we report here that it is conversion of OS-II to ferrous OxdA when a large amount of aldoxime (200 mM) is added to ferrous OxdA, the final result being a marked increase in the lifetime of OS-II. We cannot give a solid reason for why the rate-limiting step of the OxdA reaction changes with the amount of added aldoxime. However, it has been occasionally reported that the rate-limiting step changes because of the effect of an exogenous compound (50), and it is reasonable to believe that OxdA has the ability to suppress the synthesis of excess nitrile, because excess nitrile is physiologically toxic for P. chlororaphis B23 itself. For example, one of the reasons for the change in the rate-limiting step may be the repression by protonation of B of OS-II, which would consequently diminish the ability of deprotonatation from the heme bound substrate (Fig. 6). It is speculated that a water molecule is attracted to the heme pocket of OxdA because of the accumulation of nitriles (and aldoximes), which are polar molecules, followed by the protonation of B, or that the water molecule yielded during the reaction from OS-I to OS-II could not reach an outer active site due to the accumulation of nitriles (and aldoximes). OS-II was also detected on the addition of butyraldoxime (final concentration, 40 mM) with butyronitrile (final concentration, 150 mM), but not on the addition of only butyraldoxime (final concentration, 40 mM).
Fig. 6.
A possible mechanism for the repression of the OxdA reaction on the addition of a large amount of aldoxime, where B indicates a basic amino acid residue of the OxdA active site.
Herein, we demonstrated a reaction intermediate of a heme-containing enzyme with a highly oxidized heme that is formed concomitantly upon direct binding of a substrate. It is noteworthy that the OxdA reaction is a rare example of a heme directly activating an organic substrate, unlike the utilization of H2O2 or O2 as a mediator of the catalysis by other heme-containing enzymes (e.g., P450).
Materials and Methods
Materials. Butyraldoxime was purchased from Tokyo Kasei Kogyo, Tokyo). 15N-labeled hydroxylamine hydrochloride was obtained from Sigma-Aldrich. All other biochemicals except for [15N]butyraldoxime were standard commercial preparations.
Plasmid, Strain, and Medium. Escherichia coli BL21-Codonplus(DE3)-RIL (Novagen) carrying pET-oxdA, which was included for the OxdA structural gene, was used for the expression of OxdA. E. coli transformants were grown in 2 × YT medium (1.6% tryptone/1% yeast extract/0.5% NaCl) containing 50 μg/ml kanamycin and 34 μg/ml chloramphenicol.
Expression and Purification of Recombinant OxdA. OxdA was overexpressed as described (27). All purification steps were performed at 0–4°C. Potassium phosphate buffer (pH 7.0) was used throughout the purification. Centrifugation was carried out for 30 min at 15,000 × g.
The cells were harvested by centrifugation, washed twice with 100 mM buffer, and then disrupted by sonication (Insonator Model 201M; Kubota, Tokyo) to prepare a cell-free extract. Cell debris was removed by centrifugation. The resulting supernatant was fractionated with ammonium sulfate (35–60% saturation), followed by dialysis against 10 mM buffer. The dialyzed solution was applied to a DEAE-Sephacel column (5 × 25 cm) (Amersham Pharmacia Biosciences) equilibrated with 10 mM buffer. The column was washed with 2.0 liters of 10 mM buffer containing 0.1 M KCl, and then the protein was eluted from the column with 2.0 liters of 10 mM buffer, with the concentration of KCl increased linearly from 0.1 to 0.5 M. The active fractions were collected, and then ammonium sulfate was added to give 20% saturation. The enzyme solution was placed on a TSK gel Butyl-Toyopearl 650M column (4.2 × 35 cm) (Tosoh, Tokyo) equilibrated with 10 mM buffer 20% saturated with ammonium sulfate. The column was washed with 1.0 liter of the same buffer, and then the enzyme was eluted by lowering the concentration of ammonium sulfate (20% to 0%) in 2.0 liters of the buffer. The active fractions were combined and then precipitated with ammonium sulfate at 70% saturation. The precipitate was collected by centrifugation, dissolved in 0.1 M buffer, and then dialyzed against three changes of 5.0 liters of 1 mM buffer (pH 6.8). After centrifugation, the enzyme solution was loaded onto a Cellulofine HAp column (4.2 × 35 cm) (Seikagaku Kogyo, Tokyo) equilibrated with 1 mM buffer (pH 6.8). The column was washed with 2.0 liters of the same buffer, and then the protein was eluted with a linear gradient, 1–100 mM, of the buffer (pH 6.8). The active fractions were collected and then dialyzed against 10 mM buffer. The homogeneity of the purified protein was confirmed by SDS/PAGE.
Analytical Methods. Ferrous OxdA, which is an active form of OxdA, was prepared by adding sodium dithionite to purified OxdA under anaerobic conditions. The reaction buffer for observation of OS-II comprised 10 mM sodium dithionite and 100 mM potassium phosphate (pH 7.0). The reaction was started by the addition of excess butyraldoxime to ferrous OxdA under anaerobic conditions. The reaction product (butyronitrile) was determined with a gas chromatograph (GC-14BPF; Shimadzu) equipped with a flame ionization detector and a glass column (3.2 mm × 2.1 m) packed with Gaskuropack 56 (80/100% mesh; GL-Science, Tokyo).
The absorption spectrum was recorded with a Shimadzu UV-1700 PharmaSpec spectrophotometer.
RR Spectroscopy. RR spectra were obtained with excitation at 413.1 nm with a Kr+ ion laser (Spectra-Physics, model 2060). The excitation light was focused into the cell; the laser power was 2 mW at the cell for OxdA samples. The sample solutions for the Raman measurements were sealed in quartz cells, which were rotated at 5 × g at room temperature. Typically, 50-μl aliquots of 5 μM protein in 10 mM sodium dithionite and 100 mM potassium phosphate buffer, pH 7.0, were put into the cell. The scattered light at the right angle was dispersed with a single polychromator (DG-1000; Ritsu Oyo Kogaku, Saitama, Japan) equipped with a liquid nitrogen-cooled charge-coupled device camera. Raman shifts were calibrated by using indene and CCl4 as frequency standards, providing an accuracy of ±1 cm–1 for intense isolated lines.
Synthesis of 15N-Butyraldoxime. 15N-labeled butyraldoxime (C3H7CH=15NOH) was basically synthesized by using 15N-labeled hydroxylamine and butyraldehyde, according to the method of Tselinskii et al. (51). To a solution of 3.0 g (0.04 mol) of [15N]hydroxylamine hydrochloride in 4.0 ml of water, we first added, with stirring below 10°C, 3.1 g (0.04 mol) of freshly distilled butyraldehyde and then a solution of 1.7 g (0.04 mol) of NaOH in 2.3 ml of water. The mixture was kept for 20 h, and then [15N]butyraldoxime was recrystallized and confirmed by means of IR and 1H NMR spectra. Yield was 1.5 g (41.3%), mp 50–52°C (from hexane). IR spectrum, ν, cm–1: 3,270, 2,960, 2,940, 2,450, 1,600, 1,460, 1,380, 1,280, 1,080, 930, and 860. 1H NMR spectrum (chloroform-d3), δ, ppm: 0.96 t (3H, CH3), 1.53 m (2H, CH2), 2.25 m (2H, CH2), 7.07 m (1H, HC=N), 9.10 d (1H, NOH). Furthermore, the UV-vis spectrum of OS-II formed by using [15N]butyraldoxime was identical to that of OS-II formed by using [14N]butyraldoxime.
Acknowledgments
This work was supported in part by the 21st Century Centers of Excellence Program of the Ministry of Education, Culture, Sports, Science, and Technology (MEXT); by a Grant-in-Aid for Scientific Research from MEXT; and by the National Project on Protein Structural and Functional Analyses. This work was also supported by Japan Society for the Promotion of Science Research Fellowships for Young Scientists (to T.O. and K.-I.O.) and by a Grant-in-Aid for Specifically Promoted Research (14001004, to T.K.).
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: OxdA, aliphatic aldoxime dehydratase; RR, resonance Raman; OS-I, OxdA–substrate complex I; OS-II, OxdA–substrate complex II.
References
- 1.Kobayashi, M. & Shimizu, S. (1998) Nat. Biotechnol. 16, 733–736. [DOI] [PubMed] [Google Scholar]
- 2.Komeda, H., Kobayashi, M. & Shimizu, S. (1996) J. Biol. Chem. 271, 15796–15802. [DOI] [PubMed] [Google Scholar]
- 3.Komeda, H., Kobayashi, M. & Shimizu, S. (1996) Proc. Natl. Acad. Sci. USA 93, 4267–4272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kobayashi, M., Komeda, H., Yanaka, N., Nagasawa, T. & Yamada, H. (1992) J. Biol. Chem. 267, 20746–20751. [PubMed] [Google Scholar]
- 5.Goda, M., Hashimoto, Y., Shimizu, S. & Kobayashi, M. (2001) J. Biol. Chem. 276, 23480–23485. [DOI] [PubMed] [Google Scholar]
- 6.Goda, M., Hashimoto, Y., Takase, M., Herai, S., Iwahara, Y., Higashibata, H. & Kobayashi, M. (2002) J. Biol. Chem. 277, 45860–45865. [DOI] [PubMed] [Google Scholar]
- 7.Fukatsu, H., Hashimoto, Y., Goda, M., Higashibata, H. & Kobayashi, M. (2004) Proc. Natl. Acad. Sci. USA 101, 13726–13731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kobayashi, M., Nagasawa, T. & Yamada, H. (1992) Trends Biotechnol. 10, 402–408. [DOI] [PubMed] [Google Scholar]
- 9.Yamada, H. & Kobayashi, M. (1996) Biosci. Biotechnol. Biochem. 60, 1391–1400. [DOI] [PubMed] [Google Scholar]
- 10.Kobayashi, M., Yanaka, N., Nagasawa, T. & Yamada, H. (1992) Biochemistry 31, 9000–9007. [DOI] [PubMed] [Google Scholar]
- 11.Komeda, H., Hori, Y., Kobayashi, M. & Shimizu, S. (1996) Proc. Natl. Acad. Sci. USA 93, 10572–10577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kobayashi, M., Izui, H., Nagasawa, T. & Yamada, H. (1993) Proc. Natl. Acad. Sci. USA 90, 247–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kobayashi, M., Suzuki, T., Fujita, T., Masuda, M. & Shimizu, S. (1995) Proc. Natl. Acad. Sci. USA 92, 714–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kobayashi, M. & Shimizu, S. (1999) Eur. J. Biochem. 261, 1–9. [DOI] [PubMed] [Google Scholar]
- 15.Asano, Y., Tani, Y. & Yamada, H. (1980) Agric. Biol. Chem. 44, 2251–2252. [Google Scholar]
- 16.Popescu, V. C., Munck, E., Fox, B. G., Sanakis, Y., Cummings, J. G., Turner, I. M., Jr., & Nelson, M. J. (2001) Biochemistry 40, 7984–7991. [DOI] [PubMed] [Google Scholar]
- 17.Kobayashi, M., Fujiwara, Y., Goda, M., Komeda, H. & Shimizu, S. (1997) Proc. Natl. Acad. Sci. USA 94, 11986–11991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kobayashi, M., Goda, M. & Shimizu, S. (1998) FEBS Lett. 439, 325–328. [DOI] [PubMed] [Google Scholar]
- 19.Kobayashi, M., Komeda, H., Nagasawa, T., Nishiyama, M., Horinouchi, S., Beppu, T., Yamada, H. & Shimizu, S. (1993) Eur. J. Biochem. 217, 327–336. [DOI] [PubMed] [Google Scholar]
- 20.Yamada, H., Shimizu, S. & Kobayashi, M. (2001) Chem. Rec. 1, 152–161. [DOI] [PubMed] [Google Scholar]
- 21.Endo, I., Odaka, M. & Yohda, M. (1999) Trends Biotechnol. 17, 244–248. [DOI] [PubMed] [Google Scholar]
- 22.Nagasawa, T., Nanba, H., Ryuno, K., Takeuchi, K. & Yamada, H. (1987) Eur. J. Biochem. 162, 691–698. [DOI] [PubMed] [Google Scholar]
- 23.Nagasawa, T., Ryuno, K. & Yamada, H. (1989) Experientia 45, 1066–1070. [Google Scholar]
- 24.Hann, E. C., Eisenberg, A., Fager, S. K., Perkins, N. E., Gallagher, F. G., Cooper, S. M., Gavagan, J. E., Stieglitz, B., Hennessey, S. M. & DiCosimo, R. (1999) Bioorg. Med. Chem. 7, 2239–2245. [DOI] [PubMed] [Google Scholar]
- 25.Oinuma, K-I., Hashimoto, Y., Konishi, K., Goda, M., Noguchi, T., Higashibata, H. & Kobayashi, M. (2003) J. Biol. Chem. 278, 29600–29608. [DOI] [PubMed] [Google Scholar]
- 26.Xie, S. X., Kato, Y. & Asano, Y. (2001) Biosci. Biotechnol. Biochem. 65, 2666–2672. [DOI] [PubMed] [Google Scholar]
- 27.Oinuma, K.-I., Ohta, T., Konishi, K., Hashimoto, Y., Higashibata, H., Kitagawa, T. & Kobayashi, M. (2004) FEBS Lett. 568, 44–48. [DOI] [PubMed] [Google Scholar]
- 28.Konishi, K., Ishida, K., Oinuma, K.-I., Ohta, T., Hashimoto, Y., Higashibata, H., Kitagawa, T. & Kobayashi, M. (2004) J. Biol. Chem. 279, 47619–47625. [DOI] [PubMed] [Google Scholar]
- 29.Oinuma, K.-I., Kumita, H., Ohta, T., Konishi, K., Hashimoto, Y., Higashibata, H., Kitagawa, T., Shiro, Y. & Kobayashi, M. (2005) FEBS Lett. 579, 1394–1398. [DOI] [PubMed] [Google Scholar]
- 30.Green, M. T., Dawson, J. H. & Gray, H. B. (2004) Science 304, 1653–1656. [DOI] [PubMed] [Google Scholar]
- 31.Spolitak, T., Dawson, J. H. & Ballou, D. P. (2005) J. Biol. Chem. 280, 20300–20309. [DOI] [PubMed] [Google Scholar]
- 32.Rohde, J. U., In, J. H., Lim, M. H., Brennessel, W. W., Bukowski, M. R., Stubna, A., Munck, E., Nam, W. & Que, L., Jr. (2003) Science 299, 1037–1039. [DOI] [PubMed] [Google Scholar]
- 33.Oshima, R., Fushinobu, S., Su, F., Zhang, L., Takaya, N. & Shoun, H. (2004) J. Mol. Biol. 342, 207–217. [DOI] [PubMed] [Google Scholar]
- 34.Morimoto, A., Tanaka, M., Takahashi, S., Ishimori, K., Hori, H. & Morishima, I. (1998) J. Biol. Chem. 273, 14753–14760. [DOI] [PubMed] [Google Scholar]
- 35.Kitagawa, T., Kyogoku, Y., Iizuka, T. & Saito, M. I. (1976) J. Am. Chem. Soc. 98, 5169–5173. [DOI] [PubMed] [Google Scholar]
- 36.Hashimoto, S., Tatsuno Y. & Kitagawa, T. (1984) Proc. Japan Acad. 60, 345–348. [Google Scholar]
- 37.Kitagawa, T. (1988) Biological Applications of Raman Spectroscopy, ed. Spiro, T. G. (Wiley, New York), Vol. III, pp. 97–131. [Google Scholar]
- 38.Aono, S., Kato, T., Matsuki, M., Nakajima, H., Ohta, T., Uchida, T. & Kitagawa, T. (2002) J. Biol. Chem. 277, 13528–13538. [DOI] [PubMed] [Google Scholar]
- 39.Ogura, T., Takahashi, S., Shinzawa-Itoh, K., Yoshikawa, S. & Kitagawa, T. (1990) J. Biol. Chem. 265, 14721–14723. [PubMed] [Google Scholar]
- 40.Egawa, T., Ogura, T., Makino, R., Ishimura, Y. & Kitagawa, T. (1991) J. Biol. Chem. 266, 10246–10248. [PubMed] [Google Scholar]
- 41.Lukat-Rodgers, G. S. & Rodgers, K. R. (1997) Biochemistry 36, 4178–4187. [DOI] [PubMed] [Google Scholar]
- 42.Coyle, C. M., Vogel, K. M., Rush, T. S., III, Kozlowski, P. M., Williams, R., Spiro, T. G., Dou, Y., Ikeda-Saito, M., Olson, J. S. & Zgierski, M. Z. (2003) Biochemistry 42, 4896–4903. [DOI] [PubMed] [Google Scholar]
- 43.Wagner, W. D. & Nakamoto, K. (1988) J. Am. Chem. Soc. 110, 4044–4045. [Google Scholar]
- 44.Wagner, W. D. & Nakamoto, K. (1989) J. Am. Chem. Soc. 111, 1590–1598. [Google Scholar]
- 45.Dey, A. & Ghosh, A. (2002) J. Am. Chem. Soc. 124, 3206–3207. [DOI] [PubMed] [Google Scholar]
- 46.Dolphin, D., Forman, A., Borg, D. C., Fajer, J. & Felton, R. H. (1971) Proc. Natl. Acad. Sci. USA 68, 614–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Oertling, W. A., Salehi, A., Chung, Y. C., Leroi, G. E., Chang, C. K. & Babcock, G. T. (1987) J. Phys. Chem. 91, 5887–5898. [Google Scholar]
- 48.Hart-Davis, J., Battioni, P., Boucher, J.-L. & Mansuy, D. (1998) J. Am. Chem. Soc. 120, 12524–12530. [Google Scholar]
- 49.Boucher, J.-L., Delaforge, M. & Mansuy, D. (1994) Biochemistry 33, 7811–7818. [DOI] [PubMed] [Google Scholar]
- 50.Paddock, M. L., Graige, M. S., Feher, G. & Okamura, M. Y. (1999) Proc. Natl. Acad. Sci. USA 96, 6183–6188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tselinskii, I. V., Mel'nikova, S. F. & Romanova, T. V. (2001) Russ. J. Org. Chem. 37, 430–436. [Google Scholar]