Abstract
In addition to effecting the catalysis of sugar uptake, the bacterial phosphoenolpyruvate:sugar phosphotransferase system regulates a variety of physiological processes. Exposure of cells to glucose can result in repression or induction of gene expression. While the mechanism for carbon catabolite repression by glucose was well documented, that for glucose induction was not clearly understood in Escherichia coli. Recently, glucose induction of several E.coli genes has been shown to be mediated by the global repressor Mlc. Here, we elucidate a general mechanism for glucose induction of gene expression in E.coli, revealing a novel type of regulatory circuit for gene expression mediated by the phosphorylation state-dependent interaction of a membrane-bound protein with a repressor. The dephospho-form of enzyme IICBGlc, but not its phospho-form, interacts directly with Mlc and induces transcription of Mlc-regulated genes by displacing Mlc from its target sequences. Therefore, the glucose induction of Mlc-regulated genes is caused by dephosphorylation of the membrane-bound transporter enzyme IICBGlc, which directly recruits Mlc to derepress its regulon.
Keywords: enzyme IICBGlc/glucose induction/Mlc/protein–protein interaction/signal transduction
Introduction
Sensory transduction is an important device for monitoring the environment of all organisms. Bacteria sense continuous changes in their environment and adapt metabolically to compete effectively with other organisms for limiting nutrients. One system that plays an important part in this adaptation response is the phosphoenolpyruvate:sugar phosphotransferase system (PTS). The PTS is composed of two general cytoplasmic proteins, enzyme I (EI) and histidine phosphocarrier protein HPr, which are used for all sugars, and, in addition, some sugar-specific components collectively known as enzymes II (Postma et al., 1996). The primary function of this multifunctional system is the concomitant phosphorylation and translocation of its numerous sugar substrates across the cytoplasmic membrane. Glucose-specific enzyme II of Escherichia coli consists of two subunits: soluble enzyme IIAGlc (EIIAGlc) and membrane-bound enzyme IICBGlc (EIICBGlc). Thus, glucose transport in E.coli involves three soluble PTS components (EI, HPr and EIIAGlc, encoded by the ptsHIcrr operon) and one membrane-bound protein, enzyme IICBGlc (encoded by the ptsG gene). Glucose uptake entails sequential phosphoryl transfer via the PTS, as follows: phosphoenolpyruvate (PEP) ⇒ EI ⇒ HPr ⇒ EIIAGlc ⇒ EIICBGlc ⇒ glucose.
In addition to its sugar transport activities, the PTS takes part in a variety of physiological processes, including chemoreception (Lux et al., 1995), catabolite repression (Stülke and Hillen, 1999), carbohydrate transport and metabolism (Postma et al., 1996; Seok et al., 1997b), carbon storage (Seok et al., 1997a), and the coordination of carbon and nitrogen metabolism (Powell et al., 1995). The regulatory functions of the PTS depend on the phosphorylation state of the involved component, which increases in the absence and decreases in the presence of a PTS sugar substrate. The ratio of phosphorylated to dephosphorylated proteins in turn serves as signal input for the control of these physiological processes.
As expected for a system as central to bacterial metabolism as the PTS, synthesis of the PTS proteins is regulated in a highly sophisticated way. In enteric bacteria, expression of the pts operon increases during growth on glucose and requires an intact cAMP–cAMP receptor protein (CRP) system, although growth on glucose reduces the concentration of cAMP and CRP in the cell (Postma et al., 1993). While the mechanism for carbon catabolite repression by glucose is well understood (Postma et al., 1996; Stülke and Hillen, 1999), that for glucose induction was not clarified in E.coli. Glucose mediates transcriptional activation of several genes for PTS-related sugar transporters and some enzymes involved in glycolysis (Postma et al., 1996; Charpentier et al., 1998). Recently, glucose induction of several PTS operons and other genes was shown to be mediated by the Mlc (making large colonies) protein (Hosono et al., 1995). The mlc gene was originally found to cause the reduction of acetate accumulation when overexpressing cells grow in the presence of glucose. The mlc gene was shown to be identical to the previously characterized dgsA gene (Roehl and Vinopal, 1980; Morris et al., 1985; Plumbridge, 1998a). The Mlc protein (44 kDa) has been shown to act as a repressor for several catabolic operons whose products are linked to sugar metabolism. These Mlc-regulated genes include the manXYZ operon encoding the mannose PTS (Plumbridge, 1998a), the malT gene encoding the transcriptional activator of the maltose regulon (Decker et al., 1998), the ptsG gene (Kimata et al., 1998; Plumbridge, 1998b) and the ptsHIcrr operon (Kim et al., 1999; Plumbridge, 1999; Tanaka et al., 1999) as well as the mlc gene itself (Decker et al., 1998). A common feature of the five operons thus far identified as members of the Mlc regulon is that they possess at least one cAMP–CRP binding site as well as an Mlc binding site, and thus are under dual regulation. This seems to be necessary for the intricate regulation of expression of these genes in order to respond flexibly to various environmental conditions.
Although Mlc was proven to be a mediator of glucose induction of several genes, the mechanism of this mediation was not understood. In this report, we describe a general mechanism for glucose induction of gene expression in E.coli. We provide experimental evidence showing that glucose induction is mediated through the direct physical interaction between Mlc and EIICBGlc, dependent on the phosphorylation state of EIICBGlc.
Results
Overproduction of EIICBGlc induces transcription of Mlc-regulated genes in vivo
The activities of several transcription factors are regulated by interaction with other proteins, for example, the regulation of several sigma factors by their anti-sigma factors (Helman, 1999; Kang et al., 1999). Since it is also well established that PTS proteins interact with and regulate several proteins (Postma et al., 1996; Seok et al., 1997a), we addressed the possibility that glucose induction might be mediated by a protein–protein interaction involving the PTS, as proposed previously (Kim et al., 1999; Plumbridge, 1999). If this were the case, overexpression of the partner protein interacting with Mlc should induce expression of the Mlc regulon by sequestration of the repressor. We examined this question using E.coli strain GI698 (LaVallie et al., 1993; Seok et al., 1996) transformed with pRE1-based recombinant plasmids constructed for overexpression of the four PTS proteins necessary for glucose uptake. In plasmids pRE1-ptsG and pRE1-ptsHIcrr used in this work, genes are under the control of the strong λPL promoter–cII ribosome binding site combination (Reddy et al., 1989).
The level of transcription from the two Mlc-regulated operons on the chromosome, ptsHIcrr and ptsG, was examined in different cells (Figure 1). The transcripts from the plasmid genes were selectively excluded by using a probe specific for the untranslated region of the chromosomal gene absent in the recombinant plasmids. While overproduction of three soluble PTS proteins (EI, HPr and EIIAGlc) did not change the level of the endogenous ptsG transcript (lanes labeled pRE1-ptsHIcrr), it was increased by the overproduction of EIICBGlc (lanes labeled pRE1-ptsG) (Figure 1A). Thus, EIICBGlc exerts a strong stimulatory effect on ptsG expression that was not observed with EI, HPr and EIIAGlc.
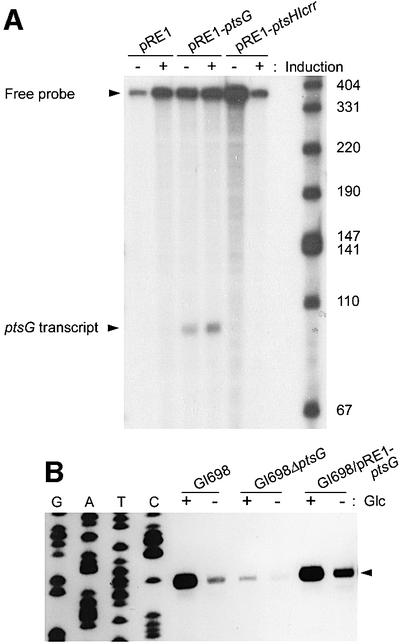
Fig. 1. Overproduced EIICBGlc induces the expression of genes encoding PTS proteins in vivo. (A) S1 nuclease protection analysis of ptsG transcripts. RNA samples prepared from GI698 cells harboring the indicated plasmids grown with and without induction of protein expression were hybridized with the 32P-labeled probe and digested with S1 nuclease as described in Materials and methods. S1 mapping analysis was accomplished four times and representative data are shown. The transcriptional level of Mlc-regulated ptsG increased in GI698 cells harboring pRE1-ptsG after induction of EIICBGlc production. (B) Effect of EIICBGlc on transcription of the ptsHIcrr operon analyzed by primer extension analysis. RNA samples were prepared from the indicated strains grown in tryptone broth with or without glucose (Glc). The arrowhead indicates the pts P0 transcripts.
The effect of EIICBGlc depletion or overproduction on another Mlc-regulated operon (ptsHIcrr) was also examined. Primer extension analysis of the ptsHIcrr transcripts demonstrated that the lack of EIICBGlc (compare lanes labeled GI698 with lanes labeled GI698ΔptsG) caused a reduction in the level of the pts P0 transcript (see arrowhead, Figure 1B) whereas overproduction of EIICBGlc (lanes labeled GI698/pRE1-ptsG) increased the level, even in the absence of glucose (Figure 1B). In all cases, the pts P0 transcript was increased by glucose. These results imply that dephosphorylated EIICBGlc in the membrane, but not the soluble PTS proteins, may act as a positive regulator of the Mlc regulon, possibly by neutralizing Mlc activity. A previous report arguing that the signal for the induction of pts gene expression might be an increase in the amount of unphosphorylated EIICBGlc is consistent with these data (De Reuse and Danchin, 1991).
Direct interaction between Mlc and EIICBGlc
We then examined whether purified Mlc interacts directly with the purified PTS components, EI, HPr and EIIAGlc. As expected from the data in Figure 1, real-time interaction analyses using BIAcore and electrophoretic mobility shift assays with purified proteins did not show any interaction between Mlc and the soluble PTS components (EI, HPr and EIIAGlc) regardless of their phosphorylation states (data not shown). Thus, we explored the interaction between Mlc and EIICBGlc in the form of membrane vesicles. EIICBGlc-depleted and -enriched membrane vesicles were prepared from E.coli GI698ΔptsG cells transformed with pRE1 and pRE1-ptsG, respectively. PEP phosphorylated PTS components including membrane-bound EIICBGlc, while glucose completely dephosphorylated those proteins. No Mlc was co-pelleted with the membrane vesicles prepared from strain GI698ΔptsG transformed with the control vector pRE1, regardless of the phosphorylation state of the added soluble PTS proteins (Figure 2, lanes labeled –EIICBGlc). However, when glucose was added to the reaction mixture containing EIICBGlc-enriched membrane vesicles, Mlc was completely co-precipitated with the membrane vesicles and no Mlc was detected in the supernatant (compare lanes for supernatants in Figure 2). When PEP was added to a reaction mixture containing EIICBGlc-enriched membrane vesicles and the three soluble PTS proteins, most of the added Mlc was partitioned into the soluble fraction. Therefore, the binding of Mlc to EIICBGlc occurred in a phosphorylation-dependent manner. These results provide strong evidence that there is a direct physical interaction between the membrane-bound glucose transporter EIICBGlc and the global repressor Mlc, and that this interaction is dependent on the availability of glucose and other PTS proteins that can dephosphorylate EIICBGlc concomitant with glucose transport. It should be noted, however, that EIICBGlc-enriched membrane vesicles prepared from GI698ΔptsHIcrr interacted with Mlc even in the absence of glucose, implying that the direct regulator for the Mlc–EIICBGlc interaction is not glucose but the phosphorylation state of EIICBGlc (data not shown). This also implies that the soluble PTS proteins are not involved in the interaction.
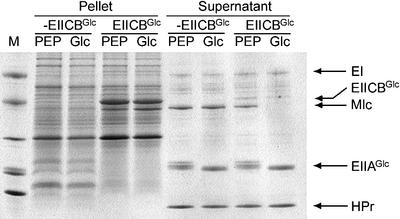
Fig. 2. Direct interaction between the dephospho-form of EIICBGlc and Mlc. Membrane vesicles (350 µg) in which EIICBGlc is enriched (lanes labeled EIICBGlc) or EIICBGlc is lacking (lanes labeled –EIICBGlc) were mixed with Mlc and three soluble PTS proteins (EI, HPr and EIIAGlc) in the presence of 2 mM PEP or 2 mM glucose (Glc), as indicated. After incubation for 5 min, EIICBGlc-bound proteins were separated by pelleting membrane vesicles using an Airfuge. The supernatants and pellets were run on SDS–PAGE gels (see Materials and methods). M, Mark 12™ protein standard (Novex).
The soluble EIIB domain is sufficient for the phosphorylation state-dependent interaction with Mlc
Since the interaction between Mlc and EIICBGlc was only dependent on the phosphorylation state of EIICBGlc, we tested the possibility that the isolated EIIB domain is sufficient for the interaction with Mlc in a phosphorylation state-dependent manner. Consequently, we cloned plasmid pJHK (see Materials and methods) to overexpress EIIB by removing the linker and the IIC domain from EIICBGlc. The purified EIIB domain could be phosphorylated by sequential phosphoryl transfer from PEP via EI, HPr and EIIAGlc (Figure 3A). Thus, we purified to homogeneity the phosphorylated EIIB by employing FPLC Mono Q 5/5 column chromatography, and mixed it with unphosphoryl ated EIIAGlc. The phosphoryl transfer reaction between EIIB and EIIAGlc was shown to be reversible (Figure 3B).
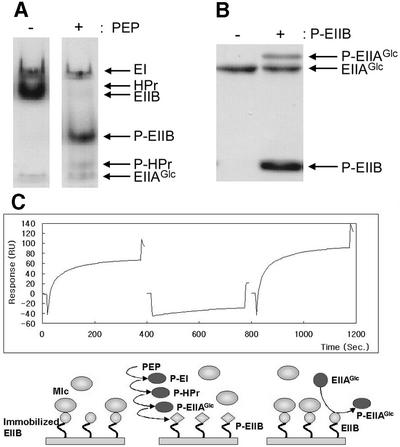
Fig. 3. The EIIB domain interacts with Mlc in a phosphorylation state-dependent manner. (A) The EIIB domain of EIICBGlc can be phosphorylated by EIIAGlc in the presence of PEP, EI and HPr. EIIB (3 µg) was incubated with EI, HPr and EIIAGlc (1 µg each) in the absence (–) or presence (+) of 1 mM PEP in the reaction buffer (10 mM Tris–HCl pH 7.5, 10 mM KCl, 1 mM MgCl2, 0.5 mM EDTA and 1 mM DTT) at room temperature for 5 min, and run on a 15% polyacrylamide gel under non-denaturing conditions. The gel was stained with Coomassie Blue. (B) Reversible phosphotransfer reaction between EIIB and EIIAGlc. Phosphorylated EIIB was purified and incubated with unphosphorylated EIIAGlc in the reaction buffer described in (A). After incubation at room temperature for 5 min, the mixture was examined by SDS–PAGE using a 15% polyacrylamide gel. The gel was stained with Coomassie Blue. (C) Phosphorylation state-dependent interaction between EIIB and Mlc measured by changes in SPR. Purified EIIB was immobilized on the carboxymethylated dextran surface of a CM5 sensor chip. Mlc (20 µg/ml) was allowed to flow over the EIIB surface for 6 min in each sensorgram. The phospho- and dephospho-EIIB surfaces were generated by the reversible phosphoryl transfer reactions between EIIB and EIIAGlc, as described in Materials and methods, and are shown schematically below each sensorgram. The first sensorgram shows Mlc binding to the immobilized EIIB surface without any treatment. In the second sensorgram, Mlc was injected after the immobilized EIIB surface had been phosphorylated by flowing the mixture of EI, HPr and EIIAGlc in the presence of PEP, and flushed with the running buffer to remove PEP and other PTS proteins. In the third sensorgram, dephosphorylated EIIAGlc was allowed to flow over the phospho-EIIB surface generated in the second sensorgram to dephosphorylate it before Mlc was injected.
The direct phosphorylation state-dependent interaction of EIIB and Mlc was demonstrated by surface plasmon resonance (SPR) using a BIAcore optical biosensor (Seok et al., 1997a; Kang et al., 1999). When purified Mlc was exposed to immobilized EIIB, high affinity interaction was detected (Figure 3C, left sensorgram). In contrast, after the immobilized EIIB domain was phosphorylated by flowing the mixture of EI, HPr and EIIAGlc in the presence of PEP, and subsequently washed with the running buffer, interaction of Mlc was hardly detectable (center sensorgram). Furthermore, the immobilized EIIB domain recovered Mlc binding activity after flowing dephosphorylated EIIAGlc through the flow cell and flushing with the running buffer (Figure 3C, right sensorgram). These results provide direct evidence for the interaction between EIICBGlc and Mlc, and indicate that the EIIB domain of EIICBGlc is sufficient for the phosphorylation state-dependent interaction with Mlc.
Binding affinity of Mlc with EIICBGlc and its target promoters
To determine the strength of the EIICBGlc–Mlc interaction, we incubated Mlc with various concentrations of EIICBGlc in the form of membrane vesicles. Then we estimated the amount of free Mlc, remaining soluble after pelleting membrane vesicles and proteins bound to them, by densitometric quantification of the Mlc bands in SDS–PAGE (Figure 4). As the amount of added EIICBGlc increased, that of Mlc in the soluble fractions decreased, while the level of the added soluble PTS proteins remained constant. This also supports the premise that Mlc interacts directly with EIICBGlc without involving any soluble PTS proteins. Recently, single particle analysis of EIICBGlc in proteoliposomes showed it to be a dimer (Zhuang et al., 1999), whereas gel filtration chromatography of the purified Mlc reveals its molecular weight to be ∼172 kDa, indicating that Mlc is a tetramer (T.-W.Nam and Y.-J.Seok, unpublished data). From the titration data in Figure 4A, the dissociation constant (Kd) was calculated to be ∼10–7 M, assuming interaction between a tetrameric form of Mlc and a dimeric form of EIICBGlc.
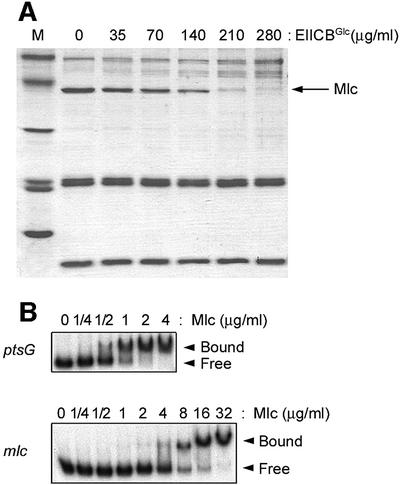
Fig. 4. Determination of the binding affinity of Mlc interaction with EIICBGlc and its target sequences of the Mlc regulon. (A) Interaction between dephosphorylated EIICBGlc and Mlc. Following incubation of Mlc (100 µg/ml) with the indicated concentrations of membrane-bound EIICBGlc in the presence of EI, HPr, EIIAGlc and 2 mM glucose, the reaction mixtures were centrifuged to separate Mlc bound to EIICBGlc in membrane vesicles. The supernatant fractions were electrophoresed by SDS–PAGE and stained with Coomassie Blue. The calculated Kd of the interaction between EIICBGlc and Mlc was ∼10–7 M. (B) Interaction between Mlc and its target sites on the ptsG and mlc promoters. 32P-labeled DNA probe (600 ng/ml) was mixed with the indicated concentrations of Mlc and then electrophoresed on 6% polyacrylamide gels. The calculated Kd of the interaction was ∼10–8 M between Mlc and the ptsG promoter, and ∼10–7 M between Mlc and the mlc promoter.
The strength of the interaction between the dephospho-form of EIICBGlc and Mlc was compared with that of the interaction between Mlc and its target DNA sites, in order to obtain a perspective on whether the dephospho-form of EIICBGlc could sequester Mlc from binding to the operator sites in the target promoters. The dissociation constants between Mlc and its target promoters were measured by gel shift assays (Figure 4B). The Kd of the interaction between the pts P0 promoter and Mlc (Figure 4B, upper panel) was ∼10–8 M, which is ∼10-fold lower than the Kd between Mlc and EIICBGlc. The Kd of the interaction between Mlc and the ptsG promoter was almost the same as that of the Mlc–pts P0 interaction (data not shown), while the strength of the interaction between Mlc and the mlc promoter was ∼10 times weaker (Kd of ∼10–7 M) (Figure 4B, lower panel). Since the intracellular level of EIICBGlc is at least 100 times higher than that of Mlc and its target promoters (Kimata et al., 1998; Kim et al., 1999; Rohwer et al., 2000), and the Kd of the interaction between Mlc and the pts P0 and ptsG promoters is only ∼10-fold lower than that between EIICBGlc and Mlc, it is reasonable to assume that Mlc could be completely displaced from its target DNA sites by interacting with EIICBGlc under physiological conditions in the presence of glucose in the medium.
The dephospho-form of EIICBGlc displaces Mlc from its target promoters to induce transcription in vitro
To support the idea that EIICBGlc induces the Mlc regulon by interacting with and displacing Mlc from its target DNA in the presence of glucose, the effect of EIICBGlc on the binding of Mlc to its target sites on the pts P0, ptsG and mlc promoters was examined by gel shift assays and in vitro transcription analysis (Figure 5). The addition of EIICBGlc-enriched membrane vesicles to the reaction mixture inhibited the binding of Mlc to its target sites on the pts P0, ptsG and mlc promoters in the presence of glucose, whereas EIICBGlc-deficient membrane vesicles did not inhibit the binding of Mlc (lanes 1–3 and 7–9, Figure 5A; data not shown for the mlc promoter). EIICBGlc-enriched membrane vesicles incubated with Mlc under phosphorylating conditions (in the presence of PEP and the three purified soluble PTS proteins) exerted little effect on the binding activity of Mlc to its target DNA (lanes 4–6). Displacement of Mlc by the dephospho-form of EIICBGlc from its target site was also verified by in vitro transcription assays. Using a supercoiled plasmid containing the ptsHIcrr promoter region, we recently showed that Mlc selectively repressed activity of the P0 promoter but not that of the P1 promoter (Kim et al., 1999). As shown in Figure 5B, EIICBGlc dephosphorylated by adding glucose could relieve repression of the pts P0 transcription by Mlc, whereas the phosphorylated form of EIICBGlc did not affect Mlc action. Neither EIICBGlc nor Mlc had any effect on transcription from the P1 promoter, indicating that EIICBGlc specifically relieves Mlc-dependent repression. These results indicate that glucose-dependent dephosphorylation enables EIICBGlc to interact tightly with Mlc, resulting in sequestration of the repressor from its binding sites in the target promoters, thereby derepressing gene expression in response to glucose.
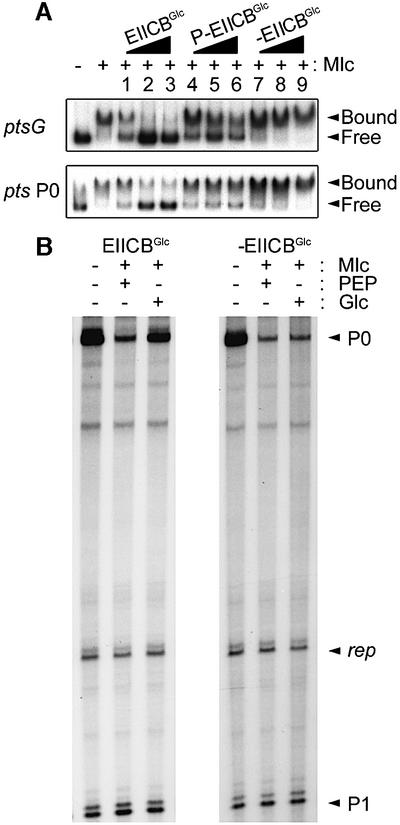
Fig. 5. The dephospho-form of EIICBGlc displaces Mlc from its target DNA to induce transcription. (A) Inhibition of Mlc binding to its target DNA by dephosphorylated EIICBGlc. 32P-labeled promoter DNA probes [ptsG (upper panel) and pts P0 (lower panel)] were mixed with 1 µg/ml Mlc and membrane vesicles in the binding buffer described in Figure 4. Membrane-bound EIICBGlc in the reaction mixture was phosphorylated with 2 mM PEP (lanes labeled P-EIICBGlc) and dephosphorylated with 2 mM glucose (lanes labeled EIICBGlc). Only dephosphorylated EIICBGlc efficiently displaced Mlc from its target DNA. Membrane vesicles lacking EIICBGlc were used as control (lanes labeled –EIICBGlc). Membrane vesicles were at 3.3 µg/ml (lanes 1, 4 and 7), 10 µg/ml (lanes 2, 5 and 8) and 30 µg/ml (lanes 3, 6 and 9). (B) Induction of pts P0 transcription by EIICBGlc in the presence of glucose in vitro. The ptsHIcrr promoter on the supercoiled plasmid pHX was used for in vitro transcription assays (Kim et al., 1999). A 185 nucleotide transcript from P0 and an 85 nucleotide transcript from P1 promoters are indicated by arrowheads. The transcripts from the plasmid origin of replication (106/107 nucleotides) are marked as rep.
Discussion
Glucose induces expression of several genes, many of which have been shown to be under the control of Mlc in E.coli. The inducing signal between glucose and Mlc, however, had not previously been identified. Several possibilities have been considered for the mechanism of glucose induction of the Mlc regulon. For many transcriptional repressors of specific operons for sugar transport, the inducer is a low-molecular-weight metabolite related to the transported substrate. Since the sequence of Mlc is ∼40% identical to NagC, a repressor and activator of the genes encoding enzymes for amino sugar degrada tion in response to N-acetylglucosamine-6-phosphate (Plumbridge, 1998a), the possibility that glucose or one of its metabolic intermediates may act as the inducer of Mlc-regulated genes has been considered. However, no simple sugars or their metabolic intermediates have been found to inactivate Mlc in vitro (Plumbridge, 1998b; Kim et al., 1999).
Regulation of the activities of several transcription factors has been reported to be mediated by direct phosphorylation. Many response regulators of the two-component signal transduction systems act as transcription factors whose activities are regulated by signal-dependent phosphorylation (Hoch, 2000). Transcriptional regulation of the bgl operon in E.coli was also shown to involve phosphotransferase system-mediated phosphorylation of BglG, a transcriptional antiterminator and the response regulator of the bgl sensory system (Amster-Choder and Wright, 1997; Chen et al., 1997). De Reuse and Danchin (1991) previously proposed a signal transduction mechanism to explain how EIICBGlc induced ptsHIcrr expression in response to glucose, based on the weak homology between the C-terminal EIIB domain of EIICBGlc and the sensor kinases of the two-component signal transduction systems. Although Mlc did not exhibit any homology to known response regulators, it might be imagined that PTS-mediated phosphorylation of Mlc would change its activity, since both the repression of several genes by Mlc and the phosphorylation of PTS proteins are dependent on the presence of its sugar substrate. This possibility, however, has been ruled out by our previous experiments (Kim et al., 1999).
Several other transcription factors are regulated by the interaction with their partner proteins, as demonstrated for the regulation of several sigma factors by their anti-sigma factors (Helman, 1999). In Bacillus subtilis, CcpA, the repressor/activator mediating carbon catabolite repression and glucose activation, was reported to form a complex with seryl-phosphorylated HPr for regulation of the pta gene (Presecan-Siedal et al., 1999). There are also a few reports indicating the involvement of membrane-bound or membrane-associated proteins in controlling the activity of transcription factors in E.coli. A recent report proposed that the RpoE heat-shock sigma factor is modulated by physical interaction with the membrane-bound anti-sigma factor RseA, although the mechanism for the regulation of the interaction was not clearly elucidated (Missiakas et al., 1997). It has also been demonstrated that MalK, a component of the ABC-type transporter for maltose, physically interacts with MalT to downregulate its transcriptional activity in response to the levels of ATP hydrolysis associated with maltose transport (Panagiotidis et al., 1998). Thus, we considered the protein–protein interaction as a reasonable mechanism for glucose induction of the Mlc regulon. If one of the PTS proteins interacted with Mlc to mediate signal transduction between glucose and Mlc, overproduction of the PTS proteins should increase both their phospho- and dephospho-forms, and result in induction of the expression of Mlc-regulated genes. We showed that overproduction of EIICBGlc resulted in induction of ptsHIcrr as well as ptsG in vivo, whereas a lack of EIICBGlc caused reduction of their expression. It was recently reported that ptsG–lacZ expression was enhanced only when ptsG was expressed from its own promoter on a multicopy plasmid, but not when the ptsG promoter was replaced by the lac promoter (Plumbridge, 1999). On the basis of this result, Plumbridge suggested that induction of ptsG–lacZ expression might not result from overproduction of EIICBGlc but from an operator titration effect displacing Mlc from its chromosomal locations; Plumbridge also suggested that EIICBGlc might possibly interact with Mlc. To clarify the mechanism for induction of Mlc-regulated genes by EIICBGlc overproduction, as shown in Figure 1 and in previous reports (De Reuse and Danchin, 1991; Plumbridge, 1999), direct interaction between Mlc and PTS proteins was measured using purified Mlc, EI, HPr, EIIAGlc and EIIBGlc. In this report, we demonstrated that glucose induction is mediated by the direct interaction between the membrane-bound glucose transporter EIICBGlc and Mlc, in a manner dependent on the phosphorylation state of EIICBGlc in vitro.
Every protein component of the E.coli PTS involved in glucose uptake has been shown to regulate the activity of other proteins by direct protein–protein interactions. The regulatory functions of the PTS depend on the phosphorylation state of its components. Unphosphorylated EI interacts with and regulates the autophosphorylation activity of CheA to trigger chemotaxis towards PTS carbohydrates (Lux et al., 1995). Unphosphorylated HPr interacts with and stimulates the activity of glycogen phosphorylase (Seok et al., 1997a). Unphosphorylated EIIAGlc inhibits glycerol kinase and several sugar permeases by a mechanism termed inducer exclusion (Hurley et al., 1993; Postma et al., 1993). Phosphorylated EIIAGlc stimulates cAMP synthesis (Peterkofsky et al., 1993). Here, we present the first evidence for the involvement of membrane-bound EIICBGlc in the regulation of expression of several genes, by direct interaction with the global repressor Mlc in a phosphorylation state-dependent manner in E.coli; this is illustrated by the model in Figure 6. When glucose is phosphorylated in the course of transport, PTS proteins are dephosphorylated. The resulting increase in unphosphorylated EIICBGlc causes the increase in the formation of the Mlc–EIICBGlc complex, derepressing expression of its target genes. Mlc controls the mlc gene itself, as well as the pts operon coding for EI, HPr and EIIAGlc, and the ptsG gene encoding EIICBGlc. Since the ptsG promoter is much stronger than that of mlc, and the concentration of Mlc in E.coli is limiting (Kimata et al., 1998; Kim et al., 1999), the increased level of EIICBGlc should completely sequester the induced level of Mlc. Thus, cells will make more PTS proteins necessary for the uptake of their sugar substrate, constituting a positive feedback loop. When glucose is depleted, the overproduced Mlc will rapidly dissociate from the phosphorylated EIICBGlc and shut down its target genes, forming a negative feedback loop (Figure 6).
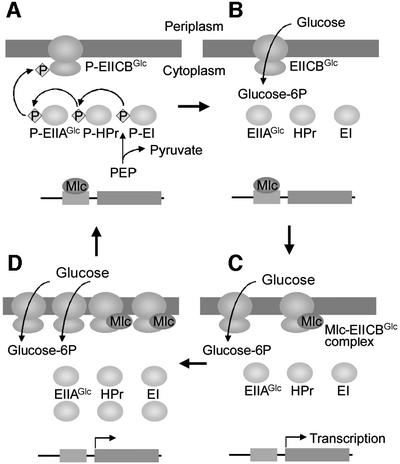
Fig. 6. A model showing that glucose induction is mediated by the direct interaction between Mlc and EIICBGlc in a phosphorylation state-dependent manner. In the absence of glucose, PTS proteins are phosphorylated by PEP, and the phospho-form of EIICBGlc does not interact with Mlc. The free Mlc binds to and represses its target genes, such as those for PTS proteins necessary for glucose transport (A). In the presence of glucose, PTS proteins are dephosphorylated, as glucose entering the bacteria is phosphorylated (B). The resulting unphosphorylated EIICBGlc binds Mlc and sequesters it from binding to its target promoters, thus derepressing gene expression to make more PTS proteins necessary for the efficient uptake of glucose (C). As EIICBGlc is dephosphorylated faster than it can be rephosphorylated by PEP in the presence of glucose, the induced EIICBGlc sequesters more Mlc, forming a positive feedback loop (D). When glucose is depleted, the overproduced Mlc dissociates from phosphorylated EIICBGlc and rapidly shuts down its target genes, forming a negative feedback loop (A).
It was shown previously that the levels of EI and HPr in E.coli and Salmonella typhimurium were higher in cells grown in media containing glucose, fructose or mannitol than in cells grown in glycerol or lactate (Mattoo and Waygood, 1983). Since all PTS and several non-PTS sugars can also lower the phosphorylation state of the PTS proteins (Postma et al., 1993; Hogema et al., 1998), these sugars might also enhance the interaction between EIICBGlc and Mlc to induce expression of the PTS proteins.
Materials and methods
Strains and plasmids
A ΔptsG mutant of E.coli strain GI698 (LaVallie et al., 1993) was constructed by P1 transduction of the CmR region from E.coli SR704, in which the ptsG gene is replaced by the chloramphenicol resistance gene, into E.coli GI698 (Seok et al., 1996), which encodes the gene for the λ cI repressor under control of the trp promoter. The DNA sequence from nucleotide 18 to 1505 of the E.coli ptsG gene (DDBJ/EMBL/GenBank accession No. J02618) was amplified by PCR, using mutagenic primers to create an NdeI site (underlined) at the ATG start codon (5′-TACTCAGGAGCACTCTCACATATGTTTAAG-3′) and a BamHI site 15 nucleotides downstream from the TAA stop codon (5′-CTGGCTGCCTTAGGATCCCCAACGTCTTAC-3′). The PCR product digested with NdeI and BamHI was cloned into vector pRE1 (Reddy et al., 1989), resulting in the recombinant plasmid pRE1-ptsG for EIICBGlc overproduction. pRE1-ptsHIcrr was prepared by employing similar procedures to overproduce EI, HPr and EIIAGlc simultaneously. The recombinant plasmid pJHK for overexpression of the EIIB domain of EIICBGlc was prepared similarly, using a forward primer possessing the synthetic NdeI site (underlined) at the new ATG start codon (5′-GCCGGGTCGTGAAGACCATATGGAAGATGC-3′) and the reverse primer used to make pRE1-ptsG. In pJHK, Thr390 of EIICBGlc was changed into a new N-terminal Met to make the EIIB protein with 88 amino acids.
Protein purification
To purify Mlc in a day, we devised a single column chromatography procedure. In a previous report (Kim et al., 1999), we showed that overproduced Mlc was insoluble at neutral pH but could be solubilized in glycine–NaOH buffer pH 9.5. Thus, the cell pellet containing overexpressed Mlc was resuspended in 10 mM Tris–HCl pH 7.5 containing 50 mM NaCl, disrupted by passing twice through a French press at 10 000 p.s.i., and centrifuged at 10 000 g for 5 min to precipitate Mlc. The Mlc pellet was solubilized using 10 mM glycine–NaOH pH 9.5 containing 50 mM NaCl. After removing insoluble cell debris by centrifugation at 10 000 g for 5 min, the Mlc was ∼90% pure. Solubilized Mlc was further chromatographed through an FPLC Mono Q 5/5 column (Pharmacia) using a gradient of 50–500 mM NaCl in 10 mM glycine–NaOH pH 9.5 (total volume 20 ml) to obtain homogeneous Mlc (>95% pure). Escherichia coli GI698 transformed with pRE1-ptsHIcrr was used for overproduction of the soluble PTS proteins (EI, HPr and EIIAGlc), and these proteins were purified as described previously (Seok et al., 1996). To overexpress the EIIB domain of EIICBGlc, E.coli GI698Δpts (Nosworthy et al., 1998) transformed with pJHK was used. The EIIB domain was purified to homogeneity according to the procedure previously described for the purification of the N-terminal domain of E.coli EI (Seok et al., 1996).
Preparation of membrane vesicles
EIICBGlc-enriched membrane vesicles were prepared from E.coli GI698ΔptsG transformed with the pRE1-ptsG plasmid after induction by adding tryptophan to a final concentration of 100 µg/ml (Seok et al., 1997a), whereas membrane vesicles lacking EIICBGlc were prepared from E.coli GI698ΔptsG cells transformed with pRE1 according to the method described previously, with slight modifications (Seok et al., 1997b). The prepared membranes were washed twice with buffer containing 1 M NaCl and 2 mM dithiothreitol (DTT), and pelleted again at 100 000 g for 90 min. The resulting membrane vesicles were resuspended in 100 mM Tris–HCl pH 7.5 containing 2 mM DTT (25 mg protein/ml), and stored frozen at –80°C until use. Protein was estimated by using bicinchoninic acid protein assay reagents (Pierce). Expression of EIICBGlc was estimated to constitute 15% of the total membrane protein.
Determination of the direct interaction between EIICBGlc and Mlc
Incubation mixtures contained 100 mM Tris–HCl pH 7.5, 2 mM DTT, 2 mM MgCl2, three soluble PTS proteins (5 µg of EI, 15 µg of HPr, 15 µg of EIIAGlc) and 10 µg of Mlc, with the indicated amounts of membrane vesicles in a total volume of 100 µl. Incubations were carried out in polyallomer tubes (5 × 20 mm) designed for use in the Beckman Airfuge (Seok et al., 1997b). After incubation for 5 min at room temperature, the membrane vesicles with bound proteins were separated by centrifugation at 100 000 g for 15 min in an Airfuge. The pellets were resuspended in 100 µl of SDS–PAGE loading buffer, and 5 µl aliquots were run on SDS–PAGE gels along with 5 µl of each supernatant. To measure the binding affinity of the interaction, Mlc was incubated with various concentrations of membrane-bound EIICBGlc with 2 mM glucose in the presence of EI, HPr and EIIAGlc. After centrifugation, the supernatants containing unbound Mlc were examined by SDS–PAGE using a 15% polyacrylamide gel. After the gel was stained with Coomassie Blue, the amounts of unbound Mlc were quantified by densitometric tracing of the stained gel. The dissociation constant (Kd) was calculated by plotting the amount of bound Mlc, obtained by subtracting unbound from added Mlc, versus added EIICBGlc.
Phosphorylation state-dependent interaction between EIIB and Mlc
Real-time interactions of Mlc with the EIIB domain and other soluble PTS proteins were monitored by SPR detection under different conditions using a BIAcore 2000 (Pharmacia Biosensor AB, Uppsala, Sweden). EI, HPr and EIIAGlc were separately immobilized on a CM5 sensor chip as described previously (Seok et al., 1997a). The EIIB domain (60 µl, 20 µg/ml) in coupling buffer (10 mM Na acetate pH 5.0) was allowed to flow over a CM5 sensor chip at 10 µl/min to couple the protein to the carboxymethylated dextran matrix by a NHS/EDC reaction (70 µl of mix). Unreacted N-hydroxysuccinimide was inactivated by injecting 70 µl of 1 M ethanolamine–HCl pH 8.0. The EIIB domain was immobilized to a surface concentration of 1.2 ng/mm2. The standard running buffer was 10 mM HEPES pH 7.2, 150 mM NaCl, 10 mM KCl, 1 mM MgCl2, 0.5 mM EDTA and 1 mM DTT, and all reagents were introduced at a flow rate of 10 µl/min. To phosphorylate the immobilized EIIB domain, a mixture (50 µl) of PEP (0.1 mM) and three soluble PTS proteins (10 µg/ml each) in the standard running buffer was allowed to flow into the flow cell for 5 min. To remove the phosphoryl group from the immobilized phospho-EIIB domain, EIIAGlc (0.1 mg/ml in the standard running buffer) was allowed to flow in the flow cell for 5 min. The sensor surface was regenerated between injections by flowing the standard running buffer at a flow rate of 100 µl/min for 10 min to remove bound analytes.
Gel shift assay
DNA fragments covering the promoter regions of ptsG, pts P0 and mlc (from –264 to +108, –249 to +211 and –146 to +183 with respect to the transcriptional start sites, respectively) were amplified by PCR and labeled with [γ-32P]ATP and T4 polynucleotide kinase. The binding buffer for the electrophoretic mobility shift assays contained 100 mM HEPES pH 8.0, 25 mM monosodium glutamate and 1 mg/ml bovine serum albumin, in a total volume of 10 µl. The binding mixtures were incubated at room temperature for 10 min, and analyzed by electrophoresis on 6% polyacrylamide gels in 0.5 × TBE at room temperature for 90 min.
In vivo transcript analysis
For S1 nuclease protection analysis, a DNA fragment covering the ptsG promoter region (from –363 to –1 with respect to the translational start codon) was amplified by PCR and used as the probe to detect mRNA originating from chromosomal ptsG but not from the plasmid pRE1-ptsG. RNA was prepared from GI698 cells harboring pRE1, pRE1-ptsG or pRE1-ptsHIcrr grown in synthetic medium containing 0.5% glycerol as carbon source (Seok et al., 1996). Cells were grown to A600 = 1.0 with or without induction at A600 = 0.5 by adding 100 µg/ml of tryptophan. For each sample, 150 µg of RNA were hybridized with 60 000 c.p.m. of the 32P-labeled probe, followed by S1 nuclease digestion as described previously (Smith, 1991). The protected fragments were electrophoresed on a 6% polyacrylamide gel containing 7 M urea. Primer extension analyses were carried out as described previously (Kim et al., 1999) using RNA samples (30 µg each) prepared from the indicated strains grown in tryptone broth (Kim et al., 1999) with or without glucose.
In vitro transcription assay
Reactions were carried out as described previously in a total volume of 25 µl, with slight modifications (Ryu and Garges, 1994; Kim et al., 1999). Mlc (100 ng), EI (200 ng), HPr (200 ng), EIIAGlc (200 ng) and membrane vesicles (500 ng of protein) were added as indicated. Membrane-bound EIICBGlc was phosphorylated by adding 2 mM PEP, and dephosphorylated by adding 2 mM glucose to the transcription buffer in the presence of EI, HPr and EIIAGlc, and incubating at room temperature for 5 min. The transcription mixture was incubated at 37°C for 10 min before adding nucleotides. Transcription was initiated by adding nucleotides and terminated after 10 min by adding 25 µl of formamide loading buffer. RNA was resolved by electrophoresis using 6% polyacrylamide gels with 8 M urea.
Note added in proof
Two papers were recently published presenting data documenting the interaction between EIICBGlc and Mlc: Lee,S.J., Boos,W., Bouche,J.P. and Plumbridge,J. (2000) Signal transduction between a membrane-bound transporter, PtsG, and a soluble transcription factor, Mlc, of Escherichia coli. EMBO J., 19, 5353--5361. Tanaka,Y., Kimata,K. and Aiba,H. (2000) A novel regulatory role of glucose transporter of Escherichia coli: membrane sequestration of a global repressor Mlc. EMBO J., 19, 5344–5352.
Acknowledgments
Acknowledgements
We are grateful to Byoung-Mo Koo and Chang-Ro Lee for their technical assistance. T.-W.N., J.-Y.J. and D.S. are recipients of the graduate fellowship provided by the Ministry of Education through the Brain Korea 21 Project. This work was supported by the Korea Research Foundation (to Y.-J.S.) made in the program year 1998.
References
- Amster-Choder O. and Wright,A. (1997) BglG, the response regulator of the Escherichia coli bgl operon, is phosphorylated on a histidine residue. J. Bacteriol., 179, 5621–5624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charpentier B., Bardey,V., Robas,N. and Branlant,C. (1998) The EIIGlc protein is involved in glucose mediated activation of Escherichia coli gapA and gapB pgk transcription. J. Bacteriol., 180, 6476–6483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q., Arents,J.C., Bader,R., Postma,P.W. and Amster-Choder,O. (1997) BglF, the sensor of the E.coli bgl system, uses the same site to phosphorylate both a sugar and a regulatory protein. EMBO J., 16, 4617–4627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker K., Plumbridge,J. and Boos,W. (1998) Negative transcriptional regulation of a positive regulator: the expression of malT, encoding the transcriptional activator of the maltose regulon of Escherichia coli, is negatively controlled by Mlc. Mol. Microbiol., 27, 381–390. [DOI] [PubMed] [Google Scholar]
- De Reuse H. and Danchin,A. (1991) Positive regulation of the pts operon of Escherichia coli: genetic evidence for a signal transduction mechanism. J. Bacteriol., 173, 727–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helman J.D. (1999) Anti-sigma factors. Curr. Opin. Microbiol., 2, 135–141. [DOI] [PubMed] [Google Scholar]
- Hoch J.A. (2000) Two-component and phosphorelay signal transduction. Curr. Opin. Microbiol., 3, 165–170. [DOI] [PubMed] [Google Scholar]
- Hogema B.M., Arents,J.C., Bader,R., Eijkemans,K., Yoshida,H., Takahashi,H., Aiba,H. and Postma,P.W. (1998) Inducer exclusion in Escherichia coli by non-PTS substrates: the role of the PEP to pyruvate ratio in determining the phosphorylation state of enzyme IIAGlc. Mol. Microbiol., 30, 487–498. [DOI] [PubMed] [Google Scholar]
- Hosono K., Kakuda,H. and Ichihara,S. (1995) Decreasing accumulation of acetate in rich medium by Escherichia coli on introduction of genes on a multicopy plasmid. Biosci. Biotechnol. Biochem., 59, 256–261. [DOI] [PubMed] [Google Scholar]
- Hurley J.H., Faber,H.R., Worthylake,D., Meadow,N.D., Roseman,S., Pettigrew,D.W. and Remington,S.J. (1993) Structure of the regulatory complex of Escherichia coli IIIGlc with glycerol kinase. Science, 259, 673–677. [PubMed] [Google Scholar]
- Kang J.-G., Paget,M.S., Seok,Y.-J., Hahn,M.-Y., Bae,J.-B., Hahn,J.-S., Kleanthous,C., Buttner,M.J. and Roe,J.-H. (1999) RsrA, an anti-sigma factor regulated by redox change. EMBO J., 18, 4292–4298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.-Y., Nam,T.-W., Shin,D., Koo,B.-M., Seok,Y.-J. and Ryu,S. (1999) Purification of Mlc and analysis of its effects on the pts expression in Escherichia coli.J. Biol. Chem., 274, 25398–25402. [DOI] [PubMed] [Google Scholar]
- Kimata K., Inada,T., Tagami,H. and Aiba,H. (1998) A global repressor (Mlc) is involved in glucose induction of the ptsG gene encoding major glucose transporter in Escherichia coli.Mol. Microbiol., 29, 1509–1519. [DOI] [PubMed] [Google Scholar]
- LaVallie E.R., DiBlasio,E.A., Kovacic,S., Grant,K.L., Schendel,P.F. and McCoy,J.M. (1993) A thioredoxin gene fusion expression system that circumvents inclusion body formation in the E.coli cytoplasm. Biotechnology (NY), 11, 187–193. [DOI] [PubMed] [Google Scholar]
- Lux R., Jahreis,K., Bettenbrock,K., Parkinson,J.S. and Lengeler,J.W. (1995) Coupling the phosphotransferase system and the methyl-accepting chemotaxis protein-dependent chemotaxis signaling pathways of Escherichia coli. Proc. Natl Acad. Sci. USA, 92, 11583–11587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattoo R.L. and Waygood,E.B. (1983) Determination of the levels of HPr and enzyme I of the phosphoenolpyruvate-sugar phosphotransferase system in Escherichia coli and Salmonella typhimurium. Can. J. Biochem. Cell Biol., 61, 29–37. [DOI] [PubMed] [Google Scholar]
- Missiakas D., Mayer,M.P., Lemaire,M., Georgopoulos,C. and Raina,S. (1997) Modulation of the Escherichia coliσE (RpoE) heat-shock transcription-factor activity by the RseA, RseB and RseC proteins. Mol. Microbiol., 24, 355–371. [DOI] [PubMed] [Google Scholar]
- Morris P.W., Binkley,J.P., Henson,J.M. and Kuempel,P.L. (1985) Cloning and location of the dgsA gene of Escherichia coli. J. Bacteriol., 163, 785–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nosworthy N.J., Peterkofsky,A., Konig,S., Seok,Y.-J., Szczepanowski, R.H. and Ginsburg,A. (1998) Phosphorylation destabilizes the amino-terminal domain of enzyme I of the Escherichia coli phosphoenolpyruvate:sugar phosphotransferase system. Biochemistry, 37, 6718–6726. [DOI] [PubMed] [Google Scholar]
- Panagiotidis C.H., Boos,W. and Shuman,H.A. (1998) The ATP-binding cassette subunit of the maltose transporter MalK antagonizes MalT, the activator of the Escherichia coli mal regulon. Mol. Microbiol., 30, 535–546. [DOI] [PubMed] [Google Scholar]
- Peterkofsky A., Reizer,A., Reizer,J., Gollop,N., Zhu,P.-P. and Amin,N. (1993) Bacterial adenylyl cyclases. Prog. Nucleic Acid Res. Mol. Biol., 44, 31–65. [DOI] [PubMed] [Google Scholar]
- Plumbridge J. (1998a) Control of the expression of the manXYZ operon in Escherichia coli: Mlc is a negative regulator of the mannose PTS. Mol. Microbiol., 27, 369–380. [DOI] [PubMed] [Google Scholar]
- Plumbridge J. (1998b) Expression of ptsG, the gene for the major PTS transporter in Escherichia coli, is repressed by Mlc and induced by growth on glucose. Mol. Microbiol., 29, 1053–1063. [DOI] [PubMed] [Google Scholar]
- Plumbridge J. (1999) Expression of the phosphotransferase system (PTS) both mediates and is mediated by Mlc regulation in Escherichia coli.Mol. Microbiol., 33, 260–273. [DOI] [PubMed] [Google Scholar]
- Postma P.W., Lengeler,J.W. and Jacobson,G.R. (1993) Phosphoenolpyruvate:carbohydrate phosphotransferase systems of bacteria. Microbiol. Rev., 57, 543–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postma P.W., Lengeler,J.W. and Jacobson,G.R. (1996) Phosphoenolpyruvate:carbohydrate phosphotransferase system. In Neidhardt,F.C. (ed.), Escherichia coli and Salmonella typhimurium: Cellular and Molecular Biology. ASM Press, Washington, DC, pp. 1149–1174.
- Powell B.S., Court,D.L., Inada,T., Nakamura,Y., Michotey,V., Cui,X., Reizer,A., Saier,M.H.,Jr and Reizer,J. (1995) Novel proteins of the phosphotransferase system encoded within the rpoN operon of Escherichia coli. Enzyme IIANtr affects growth on organic nitrogen and the conditional lethality of an erats mutant. J. Biol. Chem., 270, 4822–4839. [DOI] [PubMed] [Google Scholar]
- Presecan-Siedel E., Galinier,A., Longin,R., Deutscher,J., Danchin,A., Glaser,P. and Martin-Verstraete,I. (1999) Catabolite regulation of the pta gene as part of carbon flow pathways in Bacillus subtilis. J. Bacteriol., 181, 6889–6897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy P., Peterkofsky,A. and McKenney,K. (1989) Hyperexpression and purification of Escherichia coli adenylate cyclase using a vector designed for expression of lethal gene products. Nucleic Acids Res., 17, 10473–10488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roehl R.A. and Vinopal,R.T. (1980) Genetic locus, distant from ptsM, affecting enzyme IIA/IIB function in Escherichia coli K-12. J. Bacteriol., 142, 120–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohwer J.M., Meadow,N.D., Roseman,S., Westerhoff,H.V. and Postma,P.W. (2000) Understanding glucose transport by the bacterial phosphoenolpyruvate:glycose phosphotransferase system on the basis of kinetic measurements in vitro. J. Biol. Chem., 275, 34909–34921. [DOI] [PubMed] [Google Scholar]
- Ryu S. and Garges,S. (1994) Promoter switch in the Escherichia coli pts operon. J. Biol. Chem., 269, 4767–4772. [PubMed] [Google Scholar]
- Seok Y.-J., Lee,B.-R., Zhu,P.-P. and Peterkofsky,A. (1996) Importance of the carboxyl-terminal domain of enzyme I of the Escherichia coli phosphoenolpyruvate: sugar phosphotransferase system for phosphoryl donor specificity. Proc. Natl Acad. Sci. USA, 93, 347–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seok Y.-J., Sondej,M., Badawi,P., Lewis,M.S., Briggs,M.C., Jaffe,H. and Peterkofsky,A. (1997a) High affinity binding and allosteric regulation of Escherichia coli glycogen phosphorylase by the histidine phosphocarrier protein, HPr. J. Biol. Chem., 272, 26511–26521. [DOI] [PubMed] [Google Scholar]
- Seok Y.-J., Sun,J., Kaback,H.R. and Peterkofsky,A. (1997b) Topology of allosteric regulation of lactose permease. Proc. Natl Acad. Sci. USA, 94, 13515–13519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith C.P. (1991) Essential Molecular Biology, A Practical Approach. Oxford University Press, New York, NY.
- Stülke J. and Hillen,W. (1999) Carbon catabolite repression in bacteria. Curr. Opin. Microbiol., 2, 195–201. [DOI] [PubMed] [Google Scholar]
- Tanaka Y., Kimata,K., Inada,T., Tagami,H. and Aiba,H. (1999) Negative regulation of the pts operon by Mlc: mechanism underlying glucose induction in Escherichia coli.Genes Cells, 4, 391–399. [DOI] [PubMed] [Google Scholar]
- Zhuang J., Gutknecht,R., Flukiger,K., Hasler,L., Erni,B. and Engel,A. (1999) Purification and electron microscopic characterization of the membrane subunit (IICBglc) of the Escherichia coli glucose transporter. Arch. Biochem. Biophys., 372, 89–96. [DOI] [PubMed] [Google Scholar]


