Abstract
Mammalian telomeres form large duplex loops (t-loops) that may sequester chromosome ends by invasion of the 3′ TTAGGG overhang into the duplex TTAGGG repeat array. Here we document t-loops in Trypanosoma brucei, a kinetoplastid protozoan with abundant telomeres due to the presence of many minichromosomes. These telomeres contained 10–20 kb duplex TTAGGG repeats and a 3′ TTAGGG overhang. Electron microscopy of psoralen/UV cross-linked DNA revealed t-loops in enriched telomeric restriction fragments and at the ends of isolated minichromosomes. In mammals, t-loops are large (up to 25 kb), often comprising most of the telomere. Despite similar telomere lengths, trypanosome t-loops were much smaller (∼1 kb), indicating that t-loop sizes are regulated. Coating of non-cross-linked minichromosomes with Escherichia coli single-strand binding protein (SSB) often revealed 3′ overhangs at both telomeres and several cross-linked minichromosomes had t-loops at both ends. These results suggest that t-loops and their prerequisite 3′ tails can be formed on the products of both leading and lagging strand synthesis. We conclude that t-loops are a conserved feature of eukaryotic telomeres.
Keywords: telomere/t-loop/trypanosome
Introduction
The advent of linear chromosomes in eukaryotes was accompanied by the acquisition of specialized terminal structures that preserve chromosome ends. Most eukaryotic telomeres feature a tandem array of short repeats and a 3′ overhang (Wellinger and Sen, 1997). These telomeres are maintained by telomerase, which adds telomeric repeats to the 3′ end of the chromosome (Greider and Blackburn, 1985; Nugent and Lundblad, 1998). Telomerase-mediated telomere elongation is the predominant mechanism by which eukaryotes compensate for the failure of lagging strand synthesis to complete the replication of terminal sequences.
Telomeres protect chromosome ends against degradation and end-to-end fusion, and they prevent inappropriate activation of checkpoint pathways that respond to chromosome breaks (Muller, 1938; McClintock, 1941; Sandell and Zakian, 1993; van Steensel et al., 1998; Karlseder et al., 1999). This capping function is mediated by telomere-associated proteins. The stability of Saccharomyces cerevisiae telomeres depends on Cdc13p, a single-stranded telomeric DNA binding protein that protects telomeres from degradation and prevents activation of the RAD9 checkpoint pathway (Garvik et al., 1995; Lin and Zakian, 1996; Nugent et al., 1996). Similarly, hypotrichous ciliates have short [<20 nucleotides (nt)] telomeric overhangs that are bound by a single-strand binding protein, but the in vivo function of this complex has not been established (Gottschling and Cech, 1984; Gottschling and Zakian, 1986; Price, 1999). Mammalian telomeres are protected by TRF2, which binds along the duplex array of telomeric TTAGGG repeats (Bilaud et al., 1997; Broccoli et al., 1997; Smogorzewska et al., 2000). Interference with TRF2 function results in immediate deprotection of telomeres, as evidenced by loss of the telomeric 3′ overhang, formation of end-to-end chromosome fusions, activation of an ATM/p53-dependent DNA damage checkpoint, cell cycle arrest and apoptosis (van Steensel et al., 1998; Karlseder et al., 1999).
Electron microscopy (EM) of mammalian telomeric DNA has revealed large duplex loops (t-loops) at chromosome ends in vivo (Griffith et al., 1999). t-loops were stabilized by cross-linking the DNA in chromatin with the psoralen 4′ aminomethyltrioxalen (AMT) and UV light. Telomeric DNA was enriched by gel-filtration chromatography after digestion of the bulk chromosomal DNA with frequently cutting restriction enzymes that do not cleave TTAGGG repeats. t-loops are composed of duplex TTAGGG repeats and vary with the length of the telomeric repeat array from 1 to 25 kb. Single-stranded DNA binding protein (SSB) from Escherichia coli bound to the tail–loop junction, indicating that there is a segment of single-stranded DNA at this site, most likely formed by strand displacement upon invasion of the 3′ overhang into the duplex telomeric repeat tract. t-loops were observed frequently in a variety of mammalian DNA sources, including HeLa cells, mouse liver, HT1080 cells and primary peripheral blood leukocytes. A recent report showed loops at the ends of amplified micronuclear chromosomes in Oxytricha fallax (Murti and Prescott, 1999). These loops were not found in DNA from macronuclei, in which telomeres are extremely short and have a protein complex bound to the 3′ overhang.
Although the mechanism by which t-loops are formed in vivo has not been established, both TRF2 and the related telomeric protein TRF1 have biochemical features suggestive of a role in loop formation and/or maintenance. TRF1 can loop and pair telomeric DNA, and TRF2 promotes the formation of t-loops from a model telomere substrate (Bianchi et al., 1997, 1999; Griffith et al., 1998, 1999). In addition, proteins involved in DNA recombination and repair have been proposed to contribute to t-loop dynamics. An example is the Rad50–Mre11–Nbs1 recombinational repair complex that binds to TRF2 and interacts with mammalian telomeres (de Lange and Petrini, 2000; Zhu et al., 2000).
t-loops could provide an architectural solution to the problems posed by telomeres by hiding the telomere terminus in the duplex part of the telomere, preventing activation of DNA damage checkpoints and protecting chromosome ends from inappropriate DNA repair (Griffith et al., 1999). In addition, insertion of the 3′ terminus would presumably block telomerase from adding repeats to the chromosome end. Therefore, the rate at which t-loops are refolded after DNA replication might contribute to the regulation of telomere length.
To determine whether t-loops are conserved, we employed the advantages of trypanosome genomes. The Trypanosomatidae are protozoan parasites, of which Trypanosoma brucei is best known for the phenomenon of antigenic variation, allowing extracellular survival in the bloodstream of its mammalian host. Each bloodstream form (BF) cell expresses 10 million copies of a single species of variant surface glycoprotein (VSG) (Cross, 1975). The expressed VSG gene (VSG) is invariably positioned at a telomere (de Lange and Borst, 1982), at the end of a polycistronic transcription unit called an expression site (ES). Only one ES is transcriptionally active at any time, and antigenic variation is achieved either by gene conversion of the transcribed VSG by one of the several hundred VSGs dispersed around the genome, or by coordinated activation/inactivation of different telomeric ESs (Cross, 1996; Rudenko et al., 1998). ESs are not transcribed in the non-infectious insect stage (procyclic form, PF) of the life cycle.
Trypanosome telomeres contain long TTAGGG repeat arrays, which grow by 9–12 bp per cell division (Bernards et al., 1983; Blackburn and Challoner, 1984; van der Ploeg et al., 1984), presumably through the action of telomerase (Cano et al., 1999). Transcriptionally active telomeres grow slightly faster and are more susceptible to truncations than inactive ones (Pays et al., 1983; Horn and Cross, 1997), suggesting complex mechanisms of telomere maintenance and protection. Position effects within the ES (Rudenko et al., 1995; Horn and Cross, 1997), chromatin remodeling and developmentally regulated repression effects close to telomeres suggest a role for telomeres in regulating antigenic variation (Navarro et al., 1999).
Trypanosoma brucei contains 11 large chromosome pairs (Melville et al., 1998, 2000), ranging from 1 to >5 Mbp, which carry the essential genes, and ∼100 minichromosomes, of 25–150 kb, which are predominantly composed of tandem 177 bp repeats (Weiden et al., 1991). Minichromosomes have canonical telomeres at both ends and some telomeres carry silent VSG genes that can contribute to the expressed VSG repertoire through transposition to ESs in the large chromosomes (van der Ploeg et al., 1984; Weiden et al., 1991). Minichromosomes represent an abundant source of telomeres, which were used in this study to address the evolutionary conservation of t-loops.
Results
Trypanosoma brucei telomeres carry ∼10–20 kb of TTAGGG repeats and have a 3′ overhang
It was previously shown that T.brucei telomeres are composed of TTAGGG repeats and that most TTAGGG repeats in the trypanosome genome occupy terminal sites, based on their sensitivity to exonuclease treatment of intact genomic DNA (Blackburn and Challoner, 1984; van der Ploeg et al., 1984). In order to determine the median length of the TTAGGG repeat arrays in the T.brucei line used in this study, we applied a technique previously used to measure the length of human telomeres (Saltman et al., 1993). The rate at which the exonuclease Bal31 removes TTAGGG repeat hybridization signal is compared with the rate at which the enzyme shortens terminal DNA fragments. In T.brucei, this approach is facilitated by the availability of probes for subtelomeric VSG genes and the detailed knowledge of the restriction maps of these loci, allowing precise measurements of Bal31 digestion rates on well defined terminal restriction fragments.
Digestion of DNA from BF and PF T.brucei with frequently cutting enzymes yielded telomeric fragments in the 10–20 kb range (Figure 1A), suggesting that the telomeres contain long arrays of TTAGGG repeats. To measure the length of the telomeric repeat array directly, intact PF DNA was treated with Bal31 exonuclease for increasing times, digested with HinfI–AluI–RsaI and hybridized to a TTAGGG repeat probe (Figure 1B). Quantification of the TTAGGG repeat signal at each time point indicated that the exonuclease removed ∼2.6% of the TTAGGG repeat signal per minute. The rate at which Bal31 shortened the telomeric fragment that carries VSG 221 was determined in parallel (Figure 1B; see Figure 1C for restriction map). The same shortening rate was found for the telomeric restriction fragment carrying VSG 121 (Figure 1B; see Figure 1C for restriction map). As expected, Bal31 did not affect chromosome-internal restriction fragments, such as those carrying non-telomeric copies of VSG 121 (Figure 1B). Comparison of the two rates (Figure 1D) showed that Bal31 removed ∼10% of the TTAGGG repeat signal in the time needed to shorten the telomere by 1.5 kb, implying that the average length of the TTAGGG repeat array was ∼15 kb. This value for the length of the TTAGGG repeats array was in agreement with the median length for the telomeric fragments observed in DNA digested with HinfI and RsaI (Figure 1A), which are expected to remove most of the subtelomeric sequences from the terminal fragments.
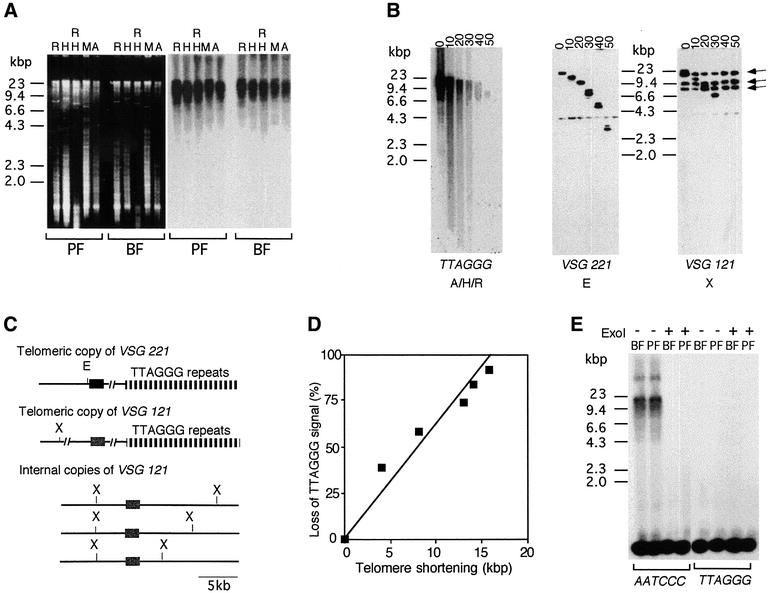
Fig. 1. Analysis of T.brucei telomeric DNA. (A) Southern blotting analysis of telomeric restriction fragments in DNA from PF and BF trypanosomes. The DNAs were digested with RsaI (R), HinfI (H), RsaI–HinfI (RH), MboI (M) or AluI (A). The left panel shows the ethidium bromide; the right panel shows a Southern blot probed with a (TTAGGG)27 probe. (B) Bal31 digestion of trypanosome telomeric DNA. Intact genomic DNA from BF trypanosomes was treated with Bal31 for the indicated times (in minutes) and digested with AluI–HinfI–RsaI (left panel), EcoRI (middle) or XmnI (right panel) and probed as indicated below the panels. The arrows indicate three non-telomeric VSG 121 fragments. (C) Restriction maps of the telomeric VSG 221 and 121 loci and the chromosome internal 121 genes. E, EcoRI; X, XmnI. (D) Graph of the rate at which Bal31 removed the TTAGGG repeat signal in (B) plotted against the rate at which the exonuclease shortened the VSG 221 telomeric EcoRI fragment in (B). (E) Overhang assay. DNA from BF and PF was digested with AluI–HinfI–RsaI after treatment with (+) or without (–) E.coli exonuclease I. DNA was incubated with radiolabeled single-stranded [TTAGGG]4 or [AATCCC]4 probes as indicated and separated by agarose gel electrophoresis (van Steensel et al., 1998). The gel was dried and exposed on a PhosphorImager (Molecular Dynamics). The signal at the front represents free probe.
We next determined whether T.brucei chromosome ends carry an overhang of the TTAGGG repeat strand. Mammalian chromosomes have up to 200 nt of single-stranded TTAGGG repeats at their 3′ termini (Makarov et al., 1997; McElligott and Wellinger, 1997; Wright et al., 1997; van Steensel et al., 1998; Huffman et al., 2000) and this overhang is presumed to be important for the formation of t-loops. The presence of single-stranded TTAGGG repeats can be assessed by annealing labeled C-strand-specific oligonucleotides to genomic DNA. Using this approach, we found that native DNA from T.brucei contained single-stranded TTAGGG repeats (Figure 1E). The signal was not detected in DNA digested with E.coli exonuclease I, which is specific for 3′ single-stranded tails, or in a control hybridization with an oligonucleotide representing the G-rich telomeric strand, consistent with the signal being derived from 3′ single-stranded TTAGGG tails. Furthermore, the signals were present on large fragments (>10 kb) in DNA that was digested with AluI–HinfI–RsaI, as would be expected if the G-tails are present at the ends of the trypanosome telomeres. The presence of single-stranded telomeric tails was corroborated by EM analysis of minichromosomes coated with E.coli SSB (see below). These data indicate that T.brucei telomeres resemble human telomeres in both the length of the TTAGGG repeat array and the presence of a 3′ [TTAGGG]n overhang.
t-loops in T.brucei telomeric DNA from procyclic and bloodstream forms
The telomeric repeat arrays of T.brucei are sufficiently long to allow their isolation by differential size fractionation after digestion of genomic DNA with frequently cutting restriction endonucleases (see Figure 1A). We previously employed this approach to isolate telomeric DNA from the human genome, for cloning and for EM visualization (de Lange et al., 1990; Griffith et al., 1999). Furthermore, the TTAGGG sequence of trypanosome telomeres, like their human counterparts, lends itself to cross-linking with psoralen (AMT) and UV light, which cross-links T residues on opposite strands at AT steps, potentially stabilizing t-loops during their isolation. Accordingly, PF or BF T.brucei were permeabilized with digitonin and treated with AMT and UV light. Following deproteinization, the DNA was cleaved with AluI– HinfI–RsaI and size fractionated on a Bio-Gel A-15m matrix. Fractions containing large DNA fragments were then prepared for EM by spreading on a denatured film of cytochrome c protein, followed by rotary shadowcasting.
EM examination revealed the presence of long DNA molecules is the early eluting (high molecular weight) fractions of the Bio-Gel column, and many of these molecules contained loops at one end. In five experiments, the fraction of long linear DNAs (≥10 kb) containing a loop at one end varied from 8% to as much as 25% (n ≥100 for each experiment), which is a frequency of t-loops very similar to that observed in mammalian telomeric restriction fragments. The structure of trypanosome t-loops appeared to be similar to that of t-loops from mammalian cells, containing a single terminal loop of variable size and a variably sized unforked tail. Examples of trypanosome t-loops are shown in Figure 2, where the loops vary in size from 6.3 (A) to 0.63 kb (F). No difference was observed in t-loop frequency or structure in DNA from BF and PF trypanosomes.
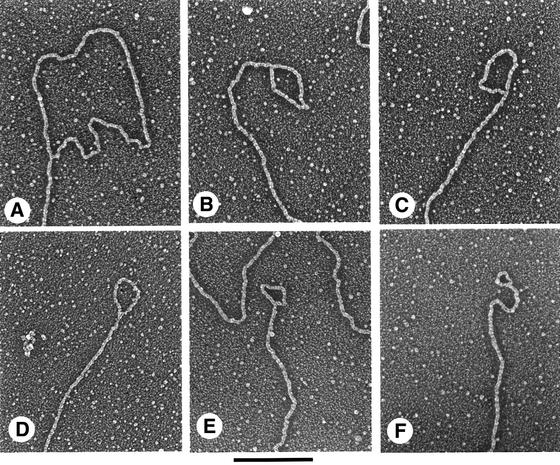
Fig. 2. Visualization of t-loops from T.brucei DNA photo-cross-linked with AMT and UV light. Trypanosomes were permeabilized with digitonin and treated with AMT and UV light, followed by endonuclease cleavage of purified DNA and isolation of the telomeric restriction fragments by gel filtration. DNA fragments were prepared for EM by spreading on a denatured film of cytochrome c protein and rotary shadowcasting with platinum:paladium. Shown in reverse contrast. t-loops shown in (A–F) measured 6.3, 1.75, 1.5, 1.2, 0.99 and 0.63 kb, respectively. Bar is equivalent to 1 kb.
Trypanosome t-loops are small
Measurement of loop contour lengths of t-loops in enriched telomeric restriction fragments showed a wide variety of sizes ranging from as small as 0.3 kb to as large as 8 kb. However, >65% of the loops were quite small (<1.5 kb) and the median length of the 48 t-loops from telomeric restriction fragments was ∼1.1 kb. Similarly, t-loops at the ends of isolated minichromosomes (see below) showed a range in loop sizes from as small as 570 bp to as large as 8.4 kb, with a median value of 1.0 kb for 21 loops analyzed. The combined data on the size range of the t-loops in both types of DNA preparations are given in Figure 3. Overall, the median size of the loops was 1.1 kb, and 42 out of 69 t-loops analyzed were very small, ranging between 0.5 and 1.0 kb. Furthermore, a number of trypanosome t-loops measured <500 bp. Large t-loops (>3 kb) were rare in both the enriched telomeric restriction fragments and in isolated minichromosomes.
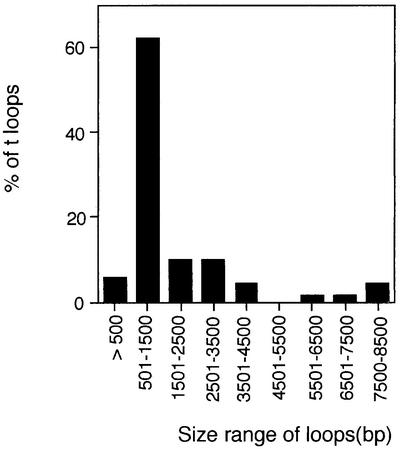
Fig. 3. Size distribution of trypanosome t-loops. Bar graph depicting the size distribution of T.brucei t-loops. The data were obtained from measurements of 48 t-loops in enriched telomeric restriction fragments (examples shown in Figure 2) and from measurements of 21 t-loops at the ends of minichromosomes (examples shown in Figure 5).
The level of resolution of the surface spreading method employed here is such that circles of <150–200 bp would frequently appear as balls rather than small loops or donuts. Examination of the minichromosomes by directly adsorbing the samples to carbon supports and rotary shadowcasting in the absence of denatured protein allowed a higher resolution inspection of the DNA ends. However, even using this technique, no examples of circles smaller than those detected by surface spreading were observed (data not shown). Nonetheless, it remains possible that some very small loops were present and not scored in these experiments.
t-loops and single-stranded tails at both ends of T.brucei minichromosomes
We next asked whether t-loops can occur at both ends of a chromosome. Trypanosome minichromosomes are sufficiently small to allow their visualization as intact molecules, allowing inspection of both ends of each chromosome (Weiden et al., 1991). To isolate minichromosomes for this purpose, permeabilized trypanosomes were treated with psoralen and UV, gently lysed, deproteinized, and sedimented through a 5–20% sucrose gradient. Gradient fractions containing minichromosomes were identified by gel electrophoresis, under conditions that separate minichromosomes from larger chromosomes, followed by detection of telomeric DNA with a TTAGGG repeat probe. Using this approach, fractions were identified that were highly enriched for minichromosomal DNA (Figure 4A and B). These fractions appeared to lack DNA derived from the larger chromosomes because they did not contain detectable amounts of an abundant 50 bp repeat element that is present upstream of ESs on the larger chromosomes (Melville et al., 1998) (compare fractions to total DNA in Figure 4C).
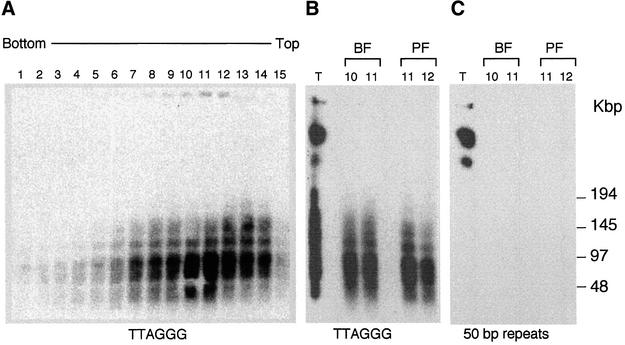
Fig. 4. Isolation and analysis of minichromosomes. (A) Southern blot of fractions from sedimentation of BF trypanosome chromosomal DNA through a linear 5–20% sucrose gradient. Fractions were analyzed by rotating agarose gel electrophoresis (RAGE) and probed with [TTAGGG]27. Two fractions from BF chromosomes (10 and 11) and two fractions from a parallel sedimentation of PF chromosomes (11 and 12) are compared alongside input PF DNA prior to sedimentation (T). Blots were probed with TTAGGG repeats (B) or a 50 bp repeat probe (C).
The enriched minichromosomal fractions were analyzed by EM and found to contain linear DNA molecules ranging from 20 to 50 kb. This size range is about half that expected from measurements by EM and gel electrophoresis (Figure 4), suggesting that some of the minichromosomal DNAs were broken during isolation. However, EM analysis showed that 14 out of 144 molecules contained a small t-loop at one end. In a second experiment, 23 out of 115 large molecules showed a loop at one end. Thus, overall, ∼15% of the ends had a t-loop. In these experiments we found four minichromosomal DNAs that carried t-loops at both ends (Figure 5) and a fifth double-looped minichromosome was found in a third experiment. If the t-loop frequencies in our preparations were primarily determined by the extent to which t-loops were lost during DNA isolation due to incomplete cross-linking or breakage, we would expect that the frequency of double-looped molecules would be ∼2.25% (15% of 15%), predicting approximately six double-looped molecules in the 259 DNAs that were examined. This number is in reasonable agreement with the four double-looped molecules that were observed, suggesting that t-loops often occur at both ends of minichromosomes.
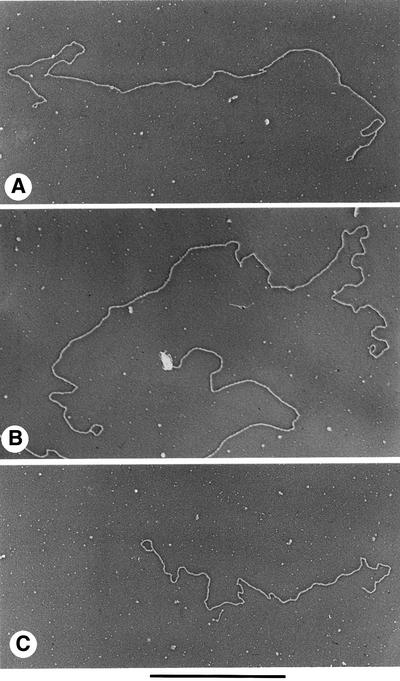
Fig. 5. t-loops in T.brucei minichromosomes. Minichromosomes enriched by sucrose gradient sedimentation were prepared for EM as described in Materials and methods. The minichromosome in (A) measures 28.7 kb, and the loops at the left and right ends measure 650 and 710 bp, respectively. The minichromosome in (B) is 30.5 kb, with loops of 1310 and 790 bp at the left and right ends, respectively. The molecule in (C) is 21.1 kb (most likely a broken minichromosome) and the loop at the left end is 1470 bp. Shown in reverse contrast. Bar is equivalent to 5 kb.
A 3′ overhang of single-stranded telomeric repeats is likely to be a prerequisite for t-loop formation. However, based on the mechanism of DNA replication, 3′ overhangs are not expected to occur at chromosome ends formed by leading strand DNA synthesis and there are conflicting reports on whether both ends of human chromosomes have a single-stranded tail (Makarov et al., 1997; Wright et al., 1997).
To address this issue we used E.coli SSB to query the status of the DNA at the ends of trypanosome minichromosomes (Figure 6). Trypanosoma brucei minichromosomes were prepared by lysis of PF cells and sucrose gradient sedimentation in the presence of sarcosyl without AMT and UV treatment. Aliquots were then chromatographed over Bio-Gel A-15m to remove the detergent and the minichromosomes incubated with E.coli SSB protein to bind any single-stranded DNA. Single tetramers or octamers of SSB bound along the length or at the ends of otherwise duplex DNA can be distinguished by EM, and represent the presence of ∼75 (single tetramer) or 150 nt (octamer) of single-stranded DNA (Chrysogelos and Griffith, 1982). The minimum length of a single-stranded DNA overhang that will allow binding of an SSB tetramer has not been established. Thus, overhangs less than ∼75 nt may be missed using this approach. Following preparation of the complexes for EM, examination of 138 minichromosomes judged to be ≥50 kb revealed that 70% showed no SSB on either end, 23% showed SSB binding at one end and 7% had SSB at both ends (Figure 6).
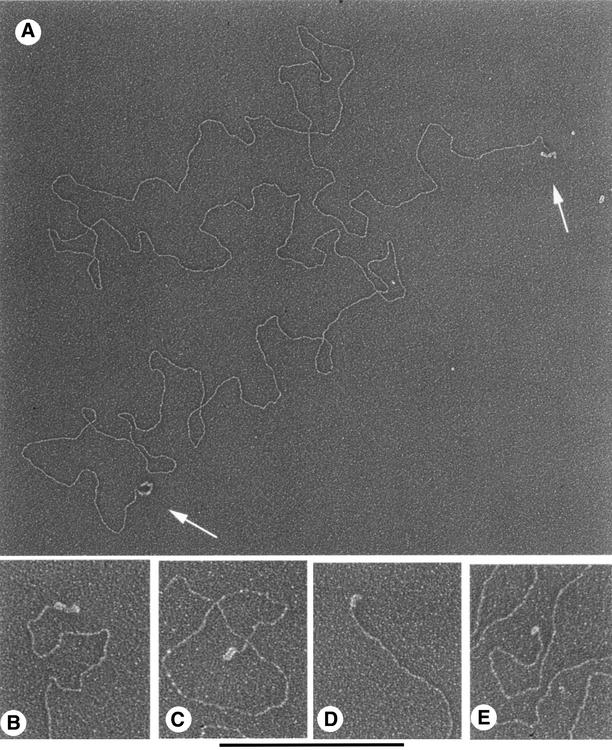
Fig. 6. Trypanosoma brucei minichromosomes with single-strand overhangs stained with SSB. Non-cross-linked minichromosomes enriched by sucrose gradient sedimentation were incubated with E.coli SSB and prepared for EM as described in Materials and methods, including adsorption to thin carbon foils, dehydration and rotary shadowcasting with tungsten. (A) A 31 kb minichromosome with single-stranded overhangs at both ends. The overhangs on this molecule are longer than most and were selected for greater visibility at low magnification. (B–E) Individual minichromosome ends with SSB bound. The size of the particle in (E) corresponds to a single SSB tetramer. Shown in reverse contrast. Bar equals 0.5 µm (A) and 0.27 µm (B–E).
To evaluate the length of the overhang, the number of SSB tetramers bound at an end was counted. For the minichromosomes with SSB bound at just one end or at both ends, 71 and 70%, respectively, of the ends showed from one to three tetramers bound, suggestive of overhangs in the range of 75–225 nt. The remaining 30% of the ends showed longer SSB-bound tracts ranging up to ∼500 nt. Thus, consistent with the annealing data in Figure 1, trypanosome chromosome ends contain substantial regions of single-stranded DNA and these overhangs can occur at both ends of the same chromosome.
Discussion
This report documents the presence of t-loops at chromosome ends in trypanosomes. Although they have the same sequence and overall length, trypanosome telomeres had loops that were significantly smaller than those of mammalian telomeres, indicating that t-loop sizes are determined by a specific mechanism. A significant fraction of isolated intact trypanosome minichromosomes had two t-loops and carried single-stranded overhangs at both ends, showing that telomeres generated by both leading and lagging strand synthesis can be remodeled into t-loops. Generation of the 3′ overhang for t-loop formation at the end duplicated by leading strand synthesis must involve post-replicative modification since the replication product is predicted to be blunt. Our findings, together with the demonstration of t-loops in mammals and ciliates, indicate that they are a conserved feature of eukaryotic telomeres and their presence at both ends of a chromosome is consistent with a requirement for t-loops in the function of all telomeres.
Trypanosome t-loops are relatively small
The trypanosome telomeres analyzed in this study are composed of 10–20 kb of TTAGGG repeats. Human telomeres have a very similar structure, containing a duplex TTAGGG repeat array in the 5–20 kb range. Despite these similarities, the size distribution of the t-loops observed in these two species was significantly different. Trypanosome telomeres often had small t-loops (median size 1.1 kb), whereas human telomeres very rarely showed t-loops in that size range. For instance, HeLa cells with telomeres in the 20 kb range had t-loops with a median size of 14 kb and <2% of the t-loops were ≤1 kb. The smallest human t-loops were observed in cells with telomeres composed of ∼5 kb TTAGGG repeats, but these t-loops were still significantly larger (median 3 kb) than those of trypanosomes. Similarly, the loops observed at the ends of micronuclear chromosomes of O.fallax were much larger [5–10 kb loops (Murti and Prescott, 1999)] than trypanosome t-loops and more comparable to those of mammalian cells. Although it is not clear what determines the size of the t-loops, the data suggest that there is an active process involved in establishing or maintaining t-loops of a specific size.
Conservation of t-loops
This study focused on telomeres in trypanosomes because of their experimental advantages and because they represent a very ancient lineage. Trypanosomes probably branched off >500 million years ago, long before the origin of their metazoan hosts (Stevens and Gibson, 1999). The molecular biology of these highly diverged protozoa is quite distinct from the perceived norm as represented by yeast, plants and mammals. For instance, trypanosomes have a specialized organelle for glycolysis, their mitochondria contain an unusual network of small circular DNAs, mitochondrial RNAs are edited, and nuclear mRNAs are formed by trans-splicing. Within this context, the conservation of t-loops in trypanosomes stands out as highly significant and predicts that t-loops play an essential role at telomeres in many eukaryotes.
The previous demonstration of looped structures at the ends of O.fallax micronuclear chromosomes (Murti and Prescott, 1999) is in agreement with the proposal that t-loops are highly conserved . Interestingly, this organism also provides an example of functional telomeres that lack t-loops. The macronuclear DNA of Oxytricha and other hypotrichous ciliates is formed by extensive fragmentation and processing of the micronuclear genome, resulting in amplified small DNA fragments each carrying one gene. These gene-sized molecules are all endowed with short telomeres that contain <50 bp of duplex telomeric DNA and a short single-stranded overhang (Price, 1999). Given their extreme short size, it was anticipated that these telomeres would lack t-loops, (Griffith et al., 1999) a prediction consistent with the EM analysis. Instead, the ends of the macronuclear DNAs may be protected by the tenaciously bound protein complex (Gottschling and Zakian, 1986; Horvath et al., 1998; Price, 1999). Collectively, the presence of t-loops in organisms as diverged as mammals, ciliates and Kinetoplastidae indicates that this aspect of telomere structure is highly conserved.
t-loops at yeast telomeres?
The finding of t-loops in diverged eukaryotes has raised the question of whether they occur in budding yeast, where telomeres have been characterized extensively. A t-loop-like structure was proposed by Li and Lustig (1996) as an intermediate in the rapid deletions that can occur when yeast telomeres are excessively long. Telomere folding (without strand invasion) was also proposed by McEachern and Blackburn (1995) to explain the mechanism of telomere length regulation in Kluyveromyces lactis, and Zakian and Ptashne and their colleagues proposed a fold-back structure for telomeres in S.cerevisiae based on studies of transcriptional regulation of subtelomeric genes (de Bruin et al., 2000, 2001). Similarly, based on the ability to cross-link the telomeric DNA binding protein Rap1p to subtelomeric Y′ elements, Grunstein, (1997) proposed that yeast telomeres form a higher order structure in which the telomere is folded back along the subtelomeric DNA. Technical limitations of the current t-loop assays have hindered direct examination of yeast telomere structure.
A significant difference between yeast telomeres and those of trypanosomes and mammals is that yeast telomeres appear to lack long single-stranded protrusions [except for a short window late in S phase (Wellinger et al., 1993)]. Such telomere tails are presumed to be required for the strand invasion that creates the t-loop, although different scenarios can be envisioned. Furthermore, t-loop formation in mammals has been proposed to depend on the telomeric protein TRF2, and a recent comparison of the mammalian and yeast telomeric complexes has suggested that budding yeast has lost the genes encoding both TRFs (Li et al., 2000). Interestingly, the major yeast telomeric DNA binding protein Rap1p has the ability to promote the pairing of single-stranded telomeric DNA with duplex repeat tracts in vitro (Gilson et al., 1994), an activity that could be indicative of a role in higher order remodeling of telomeric DNA .
So far, the protein components of trypanosome telomeres have not been identified. Specifically, it will be of interest to determine whether trypanosomes have TRF and Rap1p orthologs. Given the ease of gene targeting in trypanosomes and the abundance of their telomeres, trypanosomes could become a fruitful system for telomere biology once telomeric proteins are in hand.
t-loops and telomeric tails at both ends of each chromosome
Several trypanosome minichromosomes showed t-loops at both ends. The frequency of double-looped molecules was high enough to suggest that t-loops are formed at DNA ends created by both lagging and leading strand synthesis. The two modes of DNA synthesis are predicted to generate different ends. Lagging strand synthesis generates a 3′ overhang with a length that depends primarily on the site where primase synthesized the last RNA primer; removal of the RNA primer could contribute an additional 8–12 nt to the protrusion. By contrast, leading strand synthesis should result in a blunt end and formation of a 3′ overhang was therefore suggested to require a nuclease. This dilemma was previously recognized in the context of tests for the presence of 3′ overhangs at both ends of each chromosome and there are conflicting reports on the terminal structure of human chromosome ends (Makarov et al., 1997; Wright et al., 1997). Our data are compatible with the view that all chromosome ends carry a 3′ overhang, as an overhang is likely to be required for the maintenance of t-loops. Indeed, EM analysis of minichromosomes with bound SSB showed frequent occurrence of single-stranded DNA (presumably the G-strand overhang) at both chromosome ends. There are several mechanisms by which the end created by leading strand synthesis could acquire a 3′ overhang. Telomerase could synthesize the overhangs, they could be generated by an (unknown) 5′→3′ exonuclease, or the newly generated blunt end could invade the duplex part of the telomere and the 3′ end could then be extended by the replication machinery. Regardless of the mechanism of their formation, the presence of overhangs and t-loops at both ends of trypanosome chromosomes further corroborates the idea that t-loops are required for the protection of all chromosome ends.
Materials and methods
Trypanosomes
Molteno Institute Trypanozoon antigenic type 1.2 (MITat 1.2), clone 221a, derived from strain 427 was used. Mice were infected by intraperitoneal injection and observed until the parasitemia reached ∼5 × 108 trypanosomes/ml. A total of 109 trypanosomes were purified through DEAE–cellulose as described (Cross, 1975), centrifuged gently and suspended in ice-cold trypanosome dilution buffer, TDB (5 mM KCl, 80 mM NaCl, 1 mM MgSO4⋅7H2O, 20 mM Na2HPO4, 2 mM NaH2PO4⋅2H2O, 20 mM glucose). Procyclic forms of the same strain were cultured at 27°C in SDM-79 supplemented with fetal bovine serum, to a concentration of 107 trypanosomes/ml. A total of 109 trypanosomes were collected, washed and resuspended in ice-cold phosphate- buffered saline (PBS) pH 7.3 (137 mM NaCl, 2.7 mM KCl, 4.3 mM Na2HPO4⋅7H2O, 1.4 mM KH2PO4).
Genomic blotting
To isolate genomic DNA, ∼5 × 108 trypanosomes were resuspended in TNE (10 mM Tris pH 7.4, 10 mM EDTA, 100 mM NaCl) and lysed in TNES (10 mM Tris pH 7.4, 100 mM NaCl, 10 mM EDTA, 1% SDS) in the presence of 100 µg/ml proteinase K. After overnight incubation with proteinase K at 37°C, and phenol/chloroform extractions, DNA was precipitated with isopropanol and resuspended in TE (10 mM Tris pH 7.5/1 mM EDTA). RNase A treatment, phenol/chloroform extractions and isopropanol precipitation followed. Bal31 digestions were performed as described elsewhere (de Lange and Borst, 1982). At each time point, the reaction was stopped by increasing the temperature to 65°C for 10 min and DNA was purified using Sephacryl MicroSpin columns (Amersham). For telomere blots, DNA was digested overnight with the restriction enzymes AluI, HinfI, RsaI, MboI or XmnI, and size fractionated and blotted as described (de Lange and Borst, 1982). Telomeric restriction fragments were detected using a probe containing TTAGGG repeats and labeled as previously described (de Lange, 1992). The sequence of the 50 bp repeat was obtained from a plasmid provided by P.Borst (pBL-50), and a probe consisting of one repeat (GTGTACTTGCCTGTACTAAAAGTATTCTTACAGGGGTTGCAGTATACTGT) was synthesized and end labeled. Probes for VSGs 221 and 121 were labeled by standard methods. Signals were quantified using ImageQuant software and PhosphorImager data.
Permeabilization, cross-linking and telomeric DNA preparation for EM
BF or PF trypanosomes (5 × 108) washed in TDB or PBS, respectively, were resuspended in resuspension buffer (15 mM Tris–HCl pH 7.4, 15 mM NaCl, 60 mM KCl, 1 mM EDTA, 0.25 mM sucrose) and incubated on ice in the presence of 40 µM digitonin (Sigma) for 5 min. Trypanosomes were collected by spinning at 3000 r.p.m. for 30 s in a microcentrifuge, and treated for cross-linking and DNA extraction following scaled-down adaptations of previously published protocols. Specifically, 50 µl of a solution of AMT (Sigma; 5 µg/ml in H2O) were added to 1 ml of permeabilized cells. Trypanosomes were stirred slowly and exposed to UV light (350 nm) for 30 min. An equal volume of TNES was added and supplemented with 200 µg of proteinase K. The samples were incubated at 55°C for 2 h. After two extractions with phenol/chloroform and precipitation of the DNA with isopropanol, the DNA was resuspended in TNE and treated with RNase A (20 µg/ml) for 1 h at 37°C. DNA was extracted twice with phenol/chloroform, collected by precipitation with isopropanol, dissolved and digested with RsaI–HinfI–AluI. The digest was phenol extracted, ethanol precipitated and resuspended in TE.
Sucrose gradients and pulsed-field rotating gel electrophoresis of minichromosomal DNA
The procedure was adapted from the protocol of Weiden et al. (1991). Briefly, digitonin-permeabilized trypanosomes cross-linked with psoralen/UV as above were resuspended in 100 µl of TNE and immediately lysed by adding 1 ml of lysis solution [200 mM EDTA, 1% sodium lauryl sarcosinate (SLS), 0.5 mg/ml proteinase K] and incubating for 2–3 h at room temperature. The lysate was loaded onto a 35 ml linear 5–20% sucrose gradient in 100 mM EDTA, 25 mM Tris pH 7.5, 1% SLS. Gradients were centrifuged at 25°C for 16 h at 10 000 r.p.m. in an SW28 ultracentrifuge rotor. Fractions of 2 ml were collected from the bottom of the gradient. Aliquots of 30 µl from each fraction were mixed with 30 µl of 1.6% low melting point agarose at 65°C and loaded onto a 0.8% agarose gel. Electrophoresis was carried at an angle of 120°, 1–12 s linear ramp, and a constant voltage of 180 V for 15 h at 13°C in a rotating agarose gel electrophoresis (RAGE) apparatus (Stratagene).
Gel chromatography of T.brucei telomeric restriction fragments
Following restriction of total cross-linked T.brucei DNA, the DNA was precipitated with ethanol and resuspended in 10 mM Tris pH 7.5, 1 mM EDTA at a concentration of ∼200 µg/ml and applied to a 20 ml column of Bio-Gel A-5m equilibrated in the same buffer. The chromatography was controlled with a Pharmacia Gradifract apparatus. The DNA profile was determined by absorbance readings at 260 nm and fractions containing the telomeric restriction fragments were prepared for EM.
Staining of minichromosomes with SSB
Aliquots of T.brucei minichromosomes in sucrose–sarcosyl were chromatographed over 2 ml columns of Bio-Gel A-5m equilibrated in 20 mM HEPES pH 7.5/1 mM EDTA. Escherichia coli SSB was added to 1 µg/ml and the sample incubated on ice for 10 min. Glutaraldehyde was then added to 0.6% for 5 min at room temperature and the sample chromatographed over a second Bio-Gel column to remove the free protein and fixative. Minichromosomes in the excluded fractions were prepared for EM by direct adsorption onto glow-charged carbon films in the presence of spermidine, washed, air dried and rotary shadowcast with tungsten (Griffith and Christiansen, 1978).
EM methods
To examine cross-linked T.brucei minichromosomes separated by sucrose sedimentation, the pooled DNA fractions were chromatographed through 2 ml columns of Bio-Gel A-5m (Bio-Rad Inc.) equilibrated in TE to remove the sucrose and detergent. Following cross-linking and processing, DNA was precipitated with ethanol and dissolved in TE. Cross-linked DNA samples in TE were prepared for EM by spreading on a denatured film of cytochrome c using the droplet variation of the method of Kleinschmidt as described (Griffith et al., 1999). The grids were air dried and rotary shadowcast with platinum:paladium (20:1), and examined in a Philips CM12 instrument at 40 kV. DNA lengths were measured by projecting images on EM sheet film onto a Summagraphics digitizing tablet attached to a Macintosh computer programmed with software developed in the Griffith laboratory. Images for publication were scanned using a Nikon LS4500 film scanner, and the contrast adjusted and images arranged into figures using Adobe Photoshop software.
Acknowledgments
Acknowledgements
We thank Piet Borst for the pBL-50bp clone and Matthew Berriman for preparing the probe. Agata Smogorzewska and Giulia Celli are thanked for their assistance with telomere blots and sucrose gradients, and additional members of the de Lange laboratory are acknowledged for their helpful suggestions during the course of this work and their comments on this manuscript. This work was funded by grants for the National Institutes of Health AI21729 (G.A.M.C.), 1-F31-AI09893 (J.L.M.-J.), GM31819 (J.D.G.), CA70343 (J.D.G.), GM49046 (T.d.L.) and AG16642 (T.d.L.). T.d.L. is supported by an Ellison Medical Foundation Senior Scholar Award.
References
- Bernards A., Michels,P.A.M., Lincke,C.R. and Borst,P. (1983) Growth of chromosome ends in multiplying trypanosomes. Nature, 303, 592–597. [DOI] [PubMed] [Google Scholar]
- Bianchi A., Smith,S., Chong,L., Elias,P. and de Lange,T. (1997) TRF1 is a dimer and bends telomeric DNA. EMBO J., 16, 1785–1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchi A., Stansel,R.M., Fairall,L., Griffith,J.D., Rhodes,D. and de Lange,T. (1999) TRF1 binds a bipartite telomeric site with extreme spatial flexibility. EMBO J., 18, 5735–5744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilaud T., Brun,C., Ancelin,K., Koering,C.E., Laroche,T. and Gilson,E. (1997) Telomeric localization of TRF2, a novel human telobox protein. Nature Genet., 17, 236–239. [DOI] [PubMed] [Google Scholar]
- Blackburn E.H. and Challoner,P.B. (1984) Identification of a telomeric DNA sequence in Trypanosoma brucei. Cell, 36, 447–457. [DOI] [PubMed] [Google Scholar]
- Broccoli D., Smogorzewska,A., Chong,L. and de Lange,T. (1997) Human telomeres contain two distinct Myb-related proteins, TRF1 and TRF2. Nature Genet., 17, 231–235. [DOI] [PubMed] [Google Scholar]
- Cano M.I.N., Dungan,J.M., Agabian,N. and Blackburn,E.H. (1999) Telomerase in kinetoplastid parasitic protozoa. Proc. Natl Acad. Sci. USA, 96, 3616–3621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chrysogelos S. and Griffith,J. (1982) E.coli single strand DNA binding protein organizes single stranded DNA in nucleosome-like units. Proc. Natl Acad. Sci. USA, 79, 5803–5807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross G.A.M. (1975) Identification, purification and properties of clone-specific glycoprotein antigens constituting the surface coat of Trypanosoma brucei. Parasitology, 71, 393–417. [DOI] [PubMed] [Google Scholar]
- Cross G.A.M. (1996) Antigenic variation in trypanosomes: secrets surface slowly. BioEssays, 18, 283–291. [DOI] [PubMed] [Google Scholar]
- de Bruin D., Kantrow,S.M., Liberatore,R.A. and Zakian,V.A. (2000) Telomere folding is required for the stable maintenance of telomere position effects in yeast. Mol. Cell. Biol., 20, 7991–8000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bruin D., Zaman,Z. Liberatore,R.A. and Ptashne,M. (2001) Telomere looping permits gene activation by a downstream UAS in yeast. Nature, 209, 109–113. [DOI] [PubMed] [Google Scholar]
- de Lange T. (1992) Human telomeres are attached to the nuclear matrix. EMBO J., 11, 717–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lange T. and Borst,P. (1982) Genomic environment of the expression-linked extra copies of genes for surface antigens of Trypanosoma brucei resembles the end of a chromosome. Nature, 299, 451–453. [DOI] [PubMed] [Google Scholar]
- de Lange T. and Petrini,J.H.J. (2000) A new connection at human telomeres: association of the Mre11 complex with TRF2. Cold Spring Harb. Symp. Quant. Biol., in press. [DOI] [PubMed] [Google Scholar]
- de Lange T., Shiue,L., Myers,R.M., Cox,D.R., Naylor,S.L., Killery,A.M. and Varmus,H.E. (1990) Structure and variability of human chromosome ends. Mol. Cell. Biol., 10, 518–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garvik B., Carson,M. and Hartwell,L. (1995) Single-stranded DNA arising at telomeres in cdc13 mutants may constitute a specific signal for the RAD9 checkpoint. Mol. Cell. Biol., 15, 6128–6138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilson E., Muller,T., Sogo,J., Laroche,T. and Gasser,S.M. (1994) RAP1 stimulates single- to double-strand association of yeast telomeric DNA: implications for telomere–telomere interactions. Nucleic Acids Res., 22, 5310–5320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottschling D.E. and Cech,T.R. (1984) Chromatin structure of the molecular ends of Oxytricha macronuclear DNA: phased nucleosomes and a telomeric complex. Cell, 38, 501–510. [DOI] [PubMed] [Google Scholar]
- Gottschling D.E. and Zakian,V.A. (1986) Telomere proteins: specific recognition and protection of the natural termini of Oxytricha macronuclear DNA. Cell, 47, 195–205. [DOI] [PubMed] [Google Scholar]
- Greider C.W. and Blackburn,E.H. (1985) Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell, 43, 405–413. [DOI] [PubMed] [Google Scholar]
- Griffith J. and Christiansen,G. (1978) Electron microscopic visualization of chromatin and other DNA–protein complexes. Annu. Rev. Biophys. Bioeng., 7, 19–35. [DOI] [PubMed] [Google Scholar]
- Griffith J., Bianchi,A. and deLange,T. (1998) TRF1 promotes parallel pairing of telomeric tracts in vitro. J. Mol. Biol., 278, 79–88. [DOI] [PubMed] [Google Scholar]
- Griffith J.D., Comeau,L., Rosenfield,S., Stansel,R.M., Bianchi,A., Moss,H. and de Lange,T. (1999) Mammalian telomeres end in a large duplex loop. Cell, 97, 503–514. [DOI] [PubMed] [Google Scholar]
- Grunstein M. (1997) Molecular model for telomeric heterochromatin in yeast. Curr. Opin. Cell Biol., 9, 383–387. [DOI] [PubMed] [Google Scholar]
- Horn D. and Cross,G.A.M. (1997) Analysis of Trypanosoma brucei vsg expression site switching in vitro. Mol. Biochem. Parasitol., 84, 189–201. [DOI] [PubMed] [Google Scholar]
- Horvath M.P., Schweiker,V.L., Bevilacqua,J.M., Ruggles,J.A. and Schultz,S.C. (1998) Crystal structure of the Oxytricha nova telomere end binding protein complexed with single strand DNA. Cell, 95, 963–974. [DOI] [PubMed] [Google Scholar]
- Huffman K.E., Levene,S.D., Tesmer,V.M., Shay,J.W. and Wright,W.E. (2000) Telomere shortening is proportional to the size of the G-rich telomeric 3′-overhang. J. Biol. Chem., 275, 19719–19722. [DOI] [PubMed] [Google Scholar]
- Karlseder J., Broccoli,D., Dai,Y., Hardy,S. and de Lange,T. (1999) p53- and ATM-dependent apoptosis induced by telomeres lacking TRF2. Science, 283, 1321–1325. [DOI] [PubMed] [Google Scholar]
- Li B. and Lustig,A.J. (1996) A novel mechanism for telomere size control in Saccharomyces cerevisiae. Genes Dev., 10, 1310–1326. [DOI] [PubMed] [Google Scholar]
- Li B., Oestreich,S. and de Lange,T. (2000) Identification of human Rap1: implications for telomere evolution. Cell, 101, 471–483. [DOI] [PubMed] [Google Scholar]
- Lin J.J. and Zakian,V.A. (1996) The Saccharomyces CDC13 protein is a single-strand TG1-3 telomeric DNA-binding protein in vitro that affects telomere behavior in vivo. Proc. Natl Acad. Sci. USA, 93, 13760–13765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C. and Lustig,A.J. (1996) Genetic analysis of Rap1p/Sir3p interactions in telomeric and HML silencing in Saccharomyces cerevisiae. Genetics, 143, 81–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarov V.L., Hirose,Y. and Langmore,J.P. (1997) Long G tails at both ends of human chromosomes suggest a C strand degradation mechanism for telomere shortening. Cell, 88, 657–666. [DOI] [PubMed] [Google Scholar]
- McClintock B. (1941) The stability of broken ends of chromosomes in Zea mays. Genetics, 26, 234–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEachern M.J. and Blackburn,E.H. (1995) Runaway telomere elongation caused by telomerase RNA gene mutations. Nature, 376, 403–409. [DOI] [PubMed] [Google Scholar]
- McElligott R. and Wellinger,R.J. (1997) The terminal DNA structure of mammalian chromosomes. EMBO J., 16, 3705–3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melville S.E., Leech,V., Gerrard,C.S., Tait,A. and Blackwell,J.M. (1998) The molecular karyotype of the megabase chromosomes of Trypanosoma brucei and the assignment of chromosome markers. Mol. Biochem. Parasitol., 94, 155–173. [DOI] [PubMed] [Google Scholar]
- Melville S.E., Leech,V., Navarro,M. and Cross,G.A.M. (2000) The molecular karyotype of the megabase chromosomes of Trypanosoma brucei stock 427. Mol. Biochem. Parasitol., 111, 261–273. [DOI] [PubMed] [Google Scholar]
- Muller H.J. (1938) The remaking of chromosomes. The Collecting Net, Woods Hole, 8, 182–195. [Google Scholar]
- Murti K.G. and Prescott,D.M. (1999) Telomeres of polytene chromosomes in a ciliated protozoan terminate in duplex DNA loops. Proc. Natl Acad. Sci. USA, 96, 14436–14439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro M., Cross,G.A.M. and Wirtz,E. (1999) Trypanosoma brucei variant surface glycoprotein regulation involves coupled activation/inactivation and chromatin remodeling of expression sites. EMBO J., 18, 2265–2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nugent C.I. and Lundblad,V. (1998) The telomerase reverse transcriptase: components and regulation. Genes Dev., 12, 1073–1085. [DOI] [PubMed] [Google Scholar]
- Nugent C.I., Hughes,T.R., Lue,N.F. and Lundblad,V. (1996) Cdc13p: a single-strand telomeric DNA-binding protein with a dual role in yeast telomere maintenance. Science, 274, 249–252. [DOI] [PubMed] [Google Scholar]
- Pays E., Laurent,M., Delinte,K., van Meirvenne,N. and Steinert,M. (1983) Differential size variations between transcriptionally active and inactive telomeres of Trypanosoma brucei. Nucleic Acids Res., 11, 8137–8147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price C. (1999) Telomeres. Capping off the ends. Nature, 397, 213–214. [DOI] [PubMed] [Google Scholar]
- Rudenko G., Blundell,P.A., Dirks-Mulder,A., Kieft,R. and Borst,P. (1995) A ribosomal DNA promoter replacing the promoter of a telomeric VSG gene expression site can be efficiently switched on and off in T. brucei. Cell, 83, 547–553. [DOI] [PubMed] [Google Scholar]
- Rudenko G., Cross,M. and Borst,P. (1998) Changing the end: antigenic variation orchestrated at the telomeres of African trypanosomes. Trends Microbiol., 6, 113–116. [DOI] [PubMed] [Google Scholar]
- Saltman D., Morgan,R., Cleary,M.L. and de Lange,T. (1993) Telomeric structure in cells with chromosome end associations. Chromosoma, 102, 121–128. [DOI] [PubMed] [Google Scholar]
- Sandell L.L. and Zakian,V.A. (1993) Loss of a yeast telomere: arrest, recovery and chromosome loss. Cell, 75, 729–739. [DOI] [PubMed] [Google Scholar]
- Smogorzewska A., van Steensel,B., Bianchi,A., Oelmann,S., Schaefer,M.R., Schnapp,G. and de Lange,T. (2000) Control of human telomere length by TRF1 and TRF2. Mol. Cell. Biol., 20, 1659–1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens J.R. and Gibson,W. (1999) The molecular evolution of trypanosomes. Parasitol. Today, 15, 432–437. [DOI] [PubMed] [Google Scholar]
- van der Ploeg L.H.T., Liu,A.Y.C. and Borst,P. (1984) Structure of the growing telomeres of trypanosomes. Cell, 36, 459–468. [DOI] [PubMed] [Google Scholar]
- van Steensel B., Smogorzewska,A. and de Lange,T. (1998) TRF2 protects human telomeres from end-to-end fusions. Cell, 92, 401–413. [DOI] [PubMed] [Google Scholar]
- Weiden M., Osheim,Y.N., Beyer,A.L. and Van der Ploeg,L.H. (1991) Chromosome structure: DNA nucleotide sequence elements of a subset of the minichromosomes of the protozoan Trypanosoma brucei. Mol. Cell. Biol., 11, 3823–3834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellinger R.J. and Sen,D. (1997) The DNA structures at the ends of eukaryotic chromosomes. Eur. J. Cancer, 33, 735–749. [DOI] [PubMed] [Google Scholar]
- Wellinger R.J., Wolf,A.J. and Zakian,V.A. (1993) Saccharomyces telomeres acquire single-strand TG1-3 tails late in S phase. Cell, 72, 51–60. [DOI] [PubMed] [Google Scholar]
- Wright W.E., Tesmer,V.M., Huffman,K.E., Levene,S.D. and Shay,J.W. (1997) Normal human chromosomes have long G-rich telomeric overhangs at one end. Genes Dev., 11, 2801–2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X.D., Kuster,B., Mann,M., Petrini,J.H. and Lange,T. (2000) Cell-cycle-regulated association of RAD50/MRE11/NBS1 with TRF2 and human telomeres. Nature Genet., 25, 347–352. [DOI] [PubMed] [Google Scholar]


