Abstract
Class Ia phosphoinositide (PI) 3-kinase is a central component in growth factor signaling and is comprised of a p110 catalytic subunit and a regulatory subunit, the most common family of which is derived from the p85α gene (Pik3r1). Optimal signaling through the PI 3-kinase pathway depends on a critical molecular balance between the regulatory and catalytic subunits. In wild-type cells, the p85 subunit is more abundant than p110, leading to competition between the p85 monomer and the p85-p110 dimer and ineffective signaling. Heterozygous disruption of Pik3r1 results in increased Akt activity and decreased apoptosis by insulin-like growth factor 1 (IGF-1) through up-regulated phosphatidylinositol (3,4,5)-triphosphate production. Complete depletion of p85α, on the other hand, results in significantly increased apoptosis due to reduced PI 3-kinase-dependent signaling. Thus, a reduction in p85α represents a novel therapeutic target for enhancing IGF-1/insulin signaling, prolongation of cell survival, and protection against apoptosis.
Phosphoinositide (PI) 3-kinase plays a pivotal role in the metabolic and mitogenic actions of insulin and insulin-like growth factor 1 (IGF-1) (9, 43). Following IGF-1 and insulin stimulation, the tyrosine-phosphorylated pYMXM and pYXXM motifs in the insulin receptor substrate (IRS) proteins bind to class Ia PI 3-kinase, thereby increasing its activity (2, 43). The class Ia PI 3-kinases are dimers composed of a p110 catalytic subunit and a regulatory subunit with SH2 domains which can interact with IRS proteins (17, 52). At least eight isoforms of the regulatory subunits derived from three distinct genes have been identified. p85α and p85β represent the full-length versions of the regulatory subunits and contain an SH3 domain, a bcr homology (BH) domain flanked by two proline-rich domains, two SH2 domains (referred to as the amino-terminal and carboxy-terminal SH2 domains), and an inter SH2 domain containing the p110 binding region (35). The shorter versions of the regulatory subunits, AS53 (also known as p55α) (1, 23) and p50α (15, 24), are splicing variants derived from the same gene encoding p85α (Pik3r1) (15). They share the common amino-terminal SH2-inter SH2-carboxy-terminal SH2 structure with p85α but lack the amino-terminal half containing the SH3 domain, amino-terminal proline-rich domain, and BH domain, and in its place they have unique amino-terminal sequences consisting of 34 and 6 amino acids, respectively. Another small version of the regulatory subunit, p55PIK, has a homologous structure with AS53/p55α but is encoded by a different gene (36). Of these isoforms, p85α is predominantly and ubiquitously expressed in most tissues and is thought to be the major response pathway for most stimuli (35, 43). The spliced variants, AS53 and p50α, may have differing levels of potency for PI 3-kinase signaling (1, 24, 50) and appear to play specific roles in some selected tissues (1, 15, 24) or in particular states of insulin resistance (26).
To elucidate the physiological roles of the regulatory subunits encoded by the Pik3r1 gene, we and others have generated knockout (KO) mice with a null mutation of this gene. Terauchi et al. have found that the mice lacking only the full-length version of p85α can grow to adulthood and exhibit improved insulin sensitivity, presumably through up-regulation of p50α (48). On the other hand, KO mice with a disruption of all three isoforms of the Pik3r1 gene die within a few weeks of birth, indicating the importance of p85α and its spliced variants in normal growth and normal metabolism (16, 18). Interestingly, Pik3r1 gene heterozygous KO mice exhibit improved sensitivity to insulin and IGF-1 and help protect mice carrying heterozygous null mutations of insulin receptor and IRS-1 (6) from the development of overt diabetes (32). These data suggest the possibility that changes in the molecular balance between the regulatory subunit and the catalytic subunit may affect the PI 3-kinase-dependent signaling and its biological effects in response to growth factor stimuli.
In this study, we have used fibroblastic cell lines derived from Pik3r1 gene KO embryos to elucidate the functions of the regulatory subunits of PI 3-kinase in IGF-1 signaling and the molecular mechanisms of these complicated phenotypes in the KO mice. We find that normal cells exhibit an imbalance between the catalytic and regulatory subunits of PI 3-kinase and that modification of the molecular balance between the subunits of PI 3-kinase can play a major role in insulin/IGF-1 signaling, cellular metabolism, and cell survival.
MATERIALS AND METHODS
Cells and cell culture.
Mouse embryonic fibroblasts were isolated from Pik3r1+/+, Pik3r1+/− (heterozygous KO), and Pik3r1−/− (null) embryonic day 16.5 embryos. Established cell lines were produced by infection with a simian virus 40 large T antigen in a retroviral vector as described previously (18). Cells were maintained in Dulbecco’s modified Eagle medium supplemented with 10% fetal bovine serum. Three independent cell lines from different animals of each genotype were used and gave similar results. Cells were subjected to assays after they were serum starved for 24 h.
Antibodies.
Rabbit polyclonal anti-p85α (αp85pan) antibodies and mouse monoclonal anti-p85α (αp85α) antibodies were purchased from Upstate Biotechnology, Inc. Rabbit polyclonal anti-IRS-1 antibodies and anti-IRS-2 antibodies were generated as described previously (6), and rabbit polyclonal anti-IGF-1 receptor (αIGF-1R) antibodies and anti-Gab-1 antibodies were purchased from Santa Cruz Biotechnology. Rabbit polyclonal anti-p85β (αp85β) antibodies were generated as described previously (18). Rabbit polyclonal anti-p110α (αp110α) antibodies, anti-p110β antibodies, anti-p110 (αp110pan) antibodies, mouse monoclonal anti-PTEN antibodies, and rabbit polyclonal anti-SHIP antibodies were purchased from Santa Cruz Biotechnology. Goat polyclonal anti-Akt (αAkt) antibodies, rabbit polyclonal anti-p70 S6 kinase (αp70S6K) antibodies, and anti-p90 ribosomal 6S kinase (αp90RSK) antibodies for the kinase assays were purchased from Santa Cruz Biotechnology, and rabbit polyclonal anti-phospho-Akt antibodies recognizing phosphorylated Ser-473 of Akt1 and anti-phospho-p70S6K antibodies were purchased from New England BioLabs, Inc. Rabbit polyclonal anti-Bad (αBad) antibodies and anti-14-3-3β recognizing all isoforms of 14-3-3 were purchased from Santa Cruz Biotechnology; rabbit polyclonal anti-phospho-Bad antibodies recognizing phosphorylated Ser-112 of Bad were purchased from New England BioLabs, Inc. Rabbit polyclonal antibodies for FKHR, phospho-FKHR (Ser-256), CREB, and phospho-CREB (Ser-133) were purchased from New England BioLabs, Inc. Mouse monoclonal anti-phosphotyrosine (4G10) antibodies were purchased from Upstate Biotechnology, Inc.
Immunoprecipitation and immunoblotting.
After starvation, cells were treated with IGF-1 for the indicated period and then lysed with buffer A containing 25 mM Tris-HCl (pH 7.4), 2 mM Na3VO4, 10 mM NaF, 10 mM Na4P2O7, 1 mM EGTA, 1 mM EDTA, 10 nM okadaic acid, 5 μg of leupeptin/ml, 5 μg of aprotinin/ml, 1 mM phenylmethylsulfonyl fluoride, and 1% Nonidet P-40. The lysates were subjected to immunoprecipitation with the appropriate antibodies described above and immobilized on protein A- or G-Sepharose beads. The lysates or the immunoprecipitates were subjected to immunoblotting and visualized by an enhanced chemiluminescence system (Boehringer Mannheim Corp.). Developed films were scanned and quantitated by using NIH Image software (National Institutes of Health).
Affinity purification of regulatory subunits of PI 3-kinase using a pYMXM column.
Three milligrams of a 16-mer peptide (Lys-Lys-His-Thr-Asp-Asp-Gly-Tyr-Met-Pro-Met-Ser-Pro-Gly-Val-Ala) surrounding Tyr-608 of the rat IRS-1 protein (Biomol) was phosphorylated by the purified cytoplasmic domain of the β-subunit of human insulin receptor (Biomol) using γ-S-labeled ATP. The phosphorylated peptide was immobilized on Affi-gel 10 beads (Bio-Rad) and packed in a column. Thirty milligrams of the lysates of cells of each genotype was applied to the column and extensively washed with buffer A with 500 mM NaCl. The proteins bound to the pYMXM peptide were eluted with the elution buffer composed of 2.5 M glycine (pH 4.5) and 2 M NaCl and dialyzed with phosphate-buffered saline containing 1% glycerol. The purified proteins were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis and visualized by silver staining. The stained gels were scanned and quantitated by using NIH Image software (National Institutes of Health).
In vitro kinase assays.
For the PI 3-kinase assay, the immunoprecipitates with αp85pan, αp85α, αp85β, 4G10, or αp110α were washed three times with buffer A and twice with PI 3-kinase reaction buffer (20 mM Tris-HCl [pH 7.4], 100 mM NaCl, 0.5 mM EGTA) and suspended in 50 μl of PI 3-kinase reaction buffer containing 0.1 mg of PI (Avanti Polar Lipids)/ml. The reactions were performed, and the phosphorylated lipids were separated by thin-layer chromatography (TLC) as described previously (50). For the Akt kinase assay, cells were lysed with buffer A, and the lysates were subjected to immunoprecipitation with αAkt followed by an Akt kinase activity assay with Crosstide (51). For the p70S6K and p90RSK kinase assays, cells were lysed with buffer A and immunoprecipitated with αp70S6K or αp90RSK. The immunoprecipitates were washed and resuspended in 50 mM Tris-HCl, pH 7.5, 10 mM MgCl2, and 1 mM dithiothreitol to which 20 μM ATP, 5 μCi of [γ-32P]ATP, and 1 μg of S6 peptide (octomer peptide from the C-terminal sequence of ribosomal S6 protein; Santa Cruz Biotechnology) had been added. After 20 min at 30°C, the reaction was stopped, the aliquots were spotted on squares of P-81 paper and washed with 0.5% of phosphoric acid, and the radioactivity was counted (50).
In vivo generation of phosphatidylinositol (3,4,5)-triphosphate (PIP3).
Cells were washed with phosphate-free RPMI 1640 and incubated in phosphate-free RPMI 1640 containing 25 mM HEPES, pH 7.4, and [32P]orthophosphate (1 mCi/ml) for 3 h. Cells were then stimulated with 100 nM IGF-1 for the indicated periods, and the reaction was stopped by the addition of methanol-1 N HCl (1:1). The phosphorylated lipids were separated by TLC as described previously (27).
Apoptosis assay.
Cells were incubated in Dulbecco’s modified Eagle medium containing the indicated concentration of IGF-1 or serum for 5 h, and the lysates were applied to the enzyme-linked immunosorbent assay kit (Boehringer Mannheim) to determine the amount of nucleosomes that were present as a marker of apoptosis. An equal number of cells were plated in 96-well culture plates in serum-supplemented medium and grown to confluence for 72 h. At that time, the confluent cells were washed with phosphate-buffered saline and treated with or without IGF-1 or serum for 5 h. The cells (both attached and floating) were collected to prepare the cytosol fractions containing the nucleosomes. Equal volumes of these cytosolic fractions were incubated in antihistone antibody-coated wells (96-well plates), and the histones of the DNA fragments were allowed to bind to the antihistone antibodies. Peroxidase-labeled mouse monoclonal DNA antibodies were used to localize and detect the bound fragmented DNA using photometric detection with 2,2′-azino-di-[3-ethylbenzathiazoline sulfonate] (ABTS) as the substrate. Calcium ionophore-treated cells were used as positive controls. Cells cultured in serum-supplemented medium were used as negative controls. The reaction products in each 96-well plate were read using a microplate reader.
RESULTS
Effect of Pik3r1 gene disruption on signaling in the IGF-1-dependent PI 3-kinase pathway.
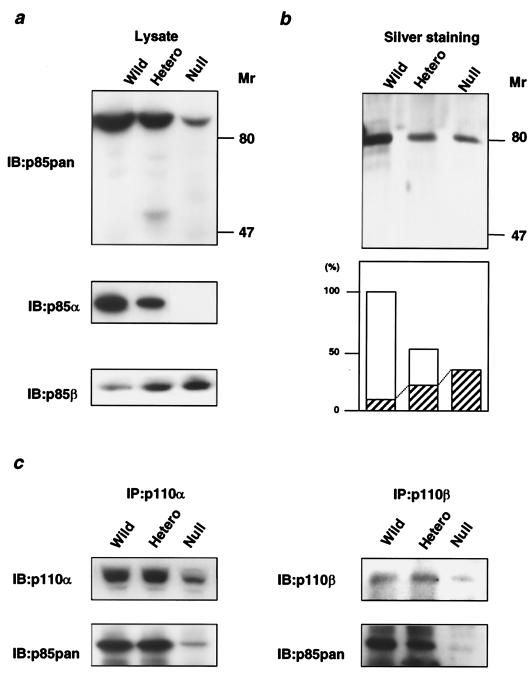
To clarify the molecular mechanisms involved in control of PI 3-kinase activity by the regulatory subunit, we established fibroblastic cell lines from null, heterozygous KO, and Pik3r1+/+ embryos and studied the effects of this gene disruption on growth factor signaling and cell survival. The expression and complex formation of the molecules involved in the IGF-1-dependent PI 3-kinase pathway in wild-type, heterozygous KO, and null cells were assessed by immunoblotting. In cells of all three genotypes, the full-length p85 proteins were the major component detected by the αp85pan antibody, which recognizes equally all isoforms derived from the Pik3r1 gene and also recognizes p85β and p55PIK, but to a lesser extent. In heterozygous and homozygous KO cells, the total p85 protein level detected by the αp85pan antibody was decreased by ∼40% (38.6% ± 3.4%, n = 4) and ∼90% (86% ± 1.3%, n = 4), with a small amount of p85β still detectable, respectively (Fig. 1a, top panel), while it was hard to detect p55PIK assessed by the specific antibody (data not shown). As expected, using a p85α-specific antibody, p85α protein was decreased by 50% in heterozygous KO cells and was undetectable in null cells (Fig. 1a, middle panel), whereas with a specific p85β antibody, p85β was up-regulated twofold in heterozygous KO cells and threefold in null cells (Fig. 1a, bottom panel).
FIG. 1.
Effect of disruption of Pik3r1 on class Ia PI 3-kinase complexes. (a) Expression levels of the regulatory subunits of PI 3-kinase in cells of each genotype. Cell lysates were subjected to immunoblotting with αp85pan (top panel), αp85α (middle panel), or αp85β (bottom panel). (b) Affinity purification of the regulatory subunits of PI 3-kinase from cells of each genotype using a phosphopeptide column. The cell lysates were applied to the column coupled with the phosphorylated p85-binding domain peptide of IRS-1 as described in Materials and Methods. The collected proteins were visualized by silver staining (top panel). In the bottom panel, each bar represents the mean level of eluted protein from the results of two independent experiments, and the shaded area represents the theoretical level of p85β estimated by the results shown in panel a. The value is expressed as a ratio to the total p85 protein level in wild-type cells. (c) Interaction of the regulatory subunit with p110α and p110β in cells of each genotype. The immunoprecipitates with αp110α (left panels) or anti-p110β (αp110β; right panels) antibody were subjected to immunoblotting with the same antibody (top panels) or the αp85pan antibody (bottom panels). Wild, wild type; hetero, heterozygous KO; IP, immunoprecipitate; IB, immunoblot.
To directly assess the levels of p85α and p85β and determine if there might be other regulatory subunits of PI 3-kinase not detected by αp85pan antibody, we purified all of the SH2 domain-containing proteins that can bind to the consensus binding motif for PI 3-kinase in each cell line using an affinity column coupled with a phospho-YMPM peptide corresponding to a region around Tyr-608 of IRS-1 (44, 45). As shown in Fig. 1b, in all three cell types the only detectable regulatory subunits bound to pYMXM motif were the p85 proteins, and the amount of p85 protein bound to pYMPM was decreased in heterozygous KO and null cells corresponding to the disruption of p85α. The p85 protein remaining in null cells, which corresponds to p85β, represented ∼30% (29.8%, n = 2) of the level of the total p85 observed in wild-type cells. Since p85β is up-regulated threefold in null cells compared to wild-type cells (as determined using a p85β specific antibody, results shown in Fig. 1a), we estimate that p85β represents ∼10% of the total p85 protein in wild-type cells. In heterozygous KO cells, total p85 protein is reduced to 60% (60.1%, n = 2) of its level in wild-type cells and p85β is up-regulated twofold, suggesting that p85β would represent ∼30% of the total p85 in the cells (Fig. 1b, bottom panel).
To effectively transmit the insulin or IGF-1 signal, the PI 3-kinase heterodimer composed of one regulatory subunit and one catalytic subunit of PI 3-kinase must bind to one of the tyrosine-phosphorylated IRS proteins using both SH2 domains (20, 43). In heterozygous KO cells, the amount of p85 regulatory subunit bound to p110α was almost equal to that in wild-type cells in culture (Fig. 1c) despite the 40% decrease in p85 protein, suggesting that under normal circumstances p85 exists in excess of the amount of p110α. A similar result was observed in the livers of heterozygous KO mice in vivo (32). By contrast, in null cells, the amount of p85 protein bound to p110α estimated by the αp85pan antibody was markedly decreased (88.3% ± 3.5%, n = 4) (Fig. 1c, left panel). This was due to a decrease in p85 protein as well as a significant decrease in p110α protein (76.5% ± 4.2%, n = 4) (Fig. 1c, left panel). This decrease in p110 is compatible with the hypothesis that interaction between the catalytic and regulatory subunits is important for the stability of the p110 catalytic subunit (56). Similar results were obtained in p110β immunoprecipitation (Fig. 1c, right panel).
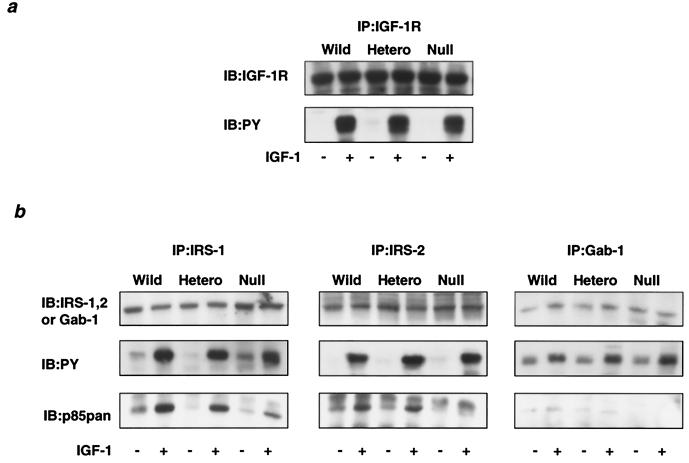
Disruption of p85α did not affect either the levels of the IGF-1 receptor protein or its phosphorylation in response to IGF-1 (Fig. 2a). In mouse embryonic fibroblasts, IRS-1 and IRS-2 are known to be major substrates for the IGF-1 receptor tyrosine kinase that directly interacts with PI 3-kinase (7), and Gab-1 is also phosphorylated and can bind to PI 3-kinase by IGF-1 stimulation but to a much lesser extent (54). As shown in Fig. 2b, there was no obvious change in the protein levels of IRS-1, IRS-2, or Gab-1 by the deletion of p85α. Of three substrates assessed, IRS-1 was most prominently phosphorylated, and interestingly, in heterozygous KO cells the amount of p85 protein bound to phosphorylated IRS-1 was decreased only ∼15% compared to that in the wild type (Fig. 2b, left panel), consistent with the hypothesis that p85α is also more abundant than phosphorylated IRS proteins. In null cells, the amount of p85 bound to IRS-1 was decreased ∼70% due to the absence of p85α and an only modest increase in p85β (Fig. 2b, left panel). Protein and phosphorylation levels of IRS-2 were not altered by the deletion of p85α either. In heterozygous KO cells, the p85 protein bound to IRS-2 was also slightly decreased, whereas in null cells, the amount of p85 bound to IRS-2 was decreased ∼90%. On the other hand, phosphorylation levels of Gab-1 were much smaller than those of IRS-1 and IRS-2. In parallel with phosphorylation levels in wild-type and heterozygous KO cells, the amount of p85 bound to Gab-1 appears to be very small, and in null cells, p85 in Gab-1 immunoprecipitation was almost undetectable (Fig. 2b, right panel).
FIG. 2.
Effect of disruption of Pik3r1 on the interaction between the regulatory subunit and phosphorylated IRS proteins in response to IGF-1. (a) Protein and phosphorylation levels of the IGF-1 receptor in cells of each genotype. The cells were starved for 24 h and then stimulated with 10 nM IGF-1 for 5 min. Cell lysates were subjected to immunoprecipitation with αIGF-1R followed by immunoblotting with αIGF-1R (top panel) or 4G10 (bottom panel). (b) Interaction between IRS proteins and the regulatory subunit. Cell lysates were subjected to immunoprecipitation with anti-IRS-1 (αIRS-1; left panels), anti-IRS-2 (αIRS-2; middle panels), or anti-Gab-1 (αGab-1; right panels) antibody followed by immunoblotting with the same antibody (top panels), 4G10 antibody (middle panels), or αp85pan antibody (bottom panels). Wild, wild type; hetero, heterozygous KO.
Effect of Pik3r1 gene disruption on PI 3-kinase activity.
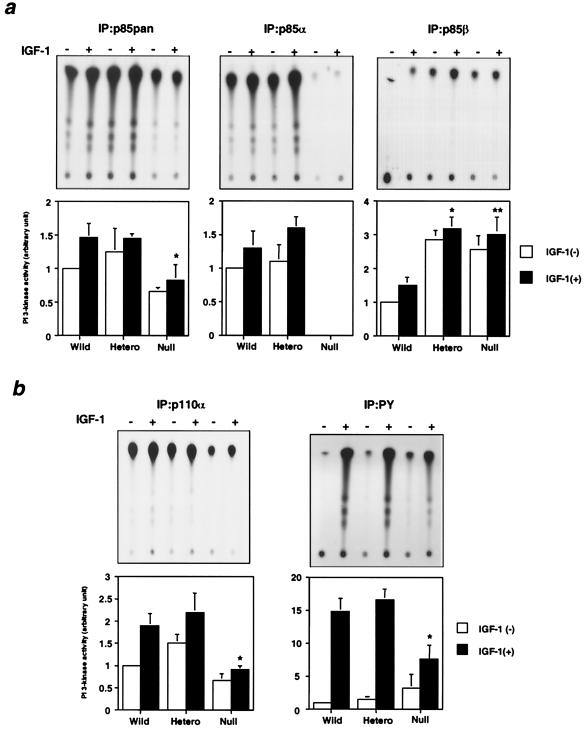
To understand the molecular mechanisms involved in PI 3-kinase-mediated signaling in each cell line, it is necessary to assess the PI 3-kinase activity associated with each regulatory isoform as well as that associated with tyrosine-phosphorylated proteins. PI 3-kinase activity in p85pan antibody precipitates, which reflects the total amount of the heterodimers composed of p85α or p85β and p110α or p110β, was similar in heterozygous KO cells and wild-type cells, and it was decreased ∼50% in null cells (Fig. 3a, left panel). As expected, PI 3-kinase activity associated with p85α was completely abolished in null cells but did not change in heterozygous KO cells compared to that in wild-type cells (Fig. 3a, middle panel).
FIG. 3.
Effect of disruption of Pik3r1 on the PI 3-kinase activity associated with each signaling molecule. (a) PI 3-kinase activities associated with the regulatory subunits. The cells were starved for 24 h and then stimulated with 10 nM IGF-1 for 5 min. Cell lysates were subjected to immunoprecipitation with αp85pan (left panels), αp85α (middle panels), or αp85β (right panels) antibody followed by the PI 3-kinase assay. The top panels show representative results, and in the bottom panels each bar represents the mean ± standard deviation of the relative PI-3 kinase activity calculated from the results of three independent experiments. In the αp85pan precipitation: *, P value of <0.05 for wild-type (Wild) versus null cells. In the p85β precipitation: *, P value of <0.01 for wild versus heterozygous KO (Hetero) cells; **, P value of <0.01 for wild versus null cells. (b) PI 3-kinase activities associated with the catalytic subunit and tyrosine-phosphorylated proteins. Cell lysates were subjected to immunoprecipitation with αp110α (left panels) or 4G10 (right panels) antibody followed by the PI 3-kinase assay. Top panels show representative results, and in the bottom panels each bar represents the mean ± standard deviation of the relative PI-3 kinase activity calculated from the results of three independent experiments. *, P value of <0.01 for wild versus null cells.
PI 3-kinase activities associated with p85β in both heterozygous KO cells and null cells were up-regulated two- to threefold, consistent with the increase in p85β protein (Fig. 3a, right panel). As a result, PI 3-kinase activity associated with p110α in heterozygous KO cells, which reflects the amount of the heterodimers composed of both p85α and p85β with p110α, was almost equal to that in wild-type cells, whereas the activity was significantly decreased in null cells (Fig. 2b, left panel). Similarly, there was no significant difference in IGF-1-induced PI 3-kinase activity associated with tyrosine-phosphorylated proteins, which reflects the total amount of the p85-p110 heterodimer bound to all IRS proteins, between the heterozygous KO cells and wild-type cells, whereas it was decreased ∼50% in null cells (Fig. 3b, right panel). On the other hand, basal PI 3-kinase activity in null cells was increased ∼3-fold and tended to be increased in heterozygous KO cells (Fig. 3b, right panel). This probably reflects the increase in the heterodimers composed of p85β-p110α or -p110β, since p85β associated with p110 has been reported to produce a higher basal activity than that associated with p85α (3).
Molecular balance between regulatory subunits, catalytic subunits, and IRS proteins.
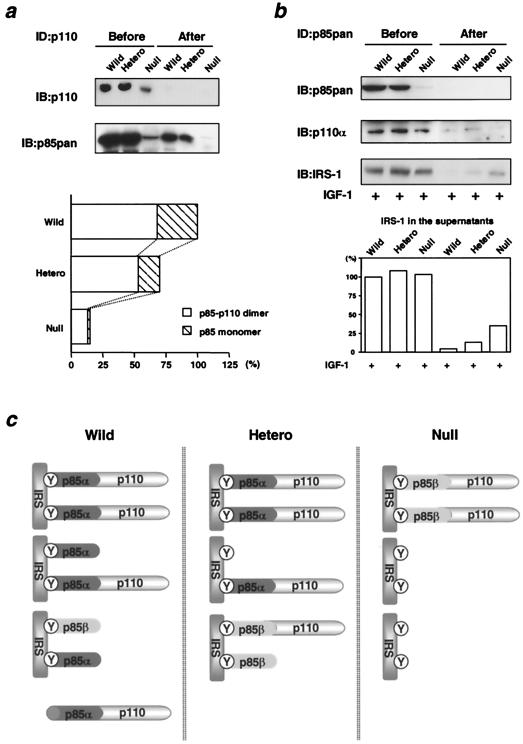
How can heterozygous KO cells maintain PI 3-kinase activities comparable to those in wild-type cells in spite of a 40% decrease in total p85 protein? As noted above, one possible explanation is that in wild-type cells the regulatory subunits are more abundant than the catalytic subunits, such that a reduction of p85α in heterozygous KO cells does not proportionally affect the amount of the p85-p110 heterodimer. To assess this possibility directly, we performed three rounds of sequential immunoprecipitation using αp110pan antibody to completely deplete p110 protein from the cell lysates (Fig. 4a, top panel). The p85 protein remaining in the lysates after immunodepletion represents the p85 monomer, and the amount of protein depleted should correspond to the p85-p110 dimer. Using this approach, we found that the ratio of p85-p110 dimer to p85 monomer was about 2:1 in wild-type cells (Fig. 4a, bottom panel). In the heterozygous KO cells, the ratio of the p85-p110 dimer to the p85 monomer was increased to 3:1 due to a reduction of p85 monomer that was greater than the reduction of p85-p110 dimer. In p85α null cells, the ratio of the p85-p110 dimer to p85 monomer was further increased to 7:1, although, in these cells, the absolute amount of the dimer was also dramatically decreased. (The decrease is somehow overestimated because all of the p85 protein in the null cells is p85β, which is less effectively recognized by the p85pan antibody, although the ratio of p85-p110 to p85 is not altered by this.) Thus, p85 is more abundant than p110 under normal conditions. Heterozygous disruption of p85α results in a large decrease in the p85α monomer but only in a small decrease in the p85-p110 dimer, whereas homozygous disruption of p85α results in a decrease in both the p85 monomer and p85-p110 dimer.
FIG. 4.
Molecular balance among p85 regulatory subunits, p110 catalytic subunits, and phosphorylated IRS proteins. (a) Excess of p85 regulatory subunits in relation to p110 catalytic subunits. Cell lysates were subjected to three rounds of immunodepletion using αp110pan antibody followed by immunoblotting with αp110pan (top panel) or αp85pan (bottom panel) antibody. The amount of the p85-p110 dimer and the p85 monomer was expressed as a ratio to the amount of total p85 in the wild-type cells. In the bottom graph, each bar represents the ratio normalized to the total p85 in wild-type (Wild) cells. (b) Molecular balance between p85 regulatory subunits and phosphorylated IRS proteins. Cell lysates were subjected to three rounds of immunodepletion using αp85pan antibody followed by immunoblotting with αp85pan (top panel), αp110α (middle panel), or 4G10 (bottom panel) antibody. In the bottom graph, each bar represents phosphorylated IRS proteins detected by 4G10 in the lysates before or after immunodepletion, expressed as a ratio to the amount in wild-type cells before immunodepletion. (c) A hypothetical model of the molecular balance between p85 regulatory subunits, p110 catalytic subunits, and phosphorylated IRS proteins in cells of each genotype. Hetero, heterozygous KO; Y, phosphorylated tyrosine; ID, immunodepletion.
To assess the molecular balance between p85 and phosphorylated IRS proteins, we performed immunodepletion using αp85pan antibody (Fig. 4b). In wild-type cells, no phosphorylated IRS proteins were detected in the lysates after immunodepletion; in heterozygous KO cells, 10% of phosphorylated IRS proteins remained in the supernatant; and in null cells, 25% of phosphorylated IRS proteins were detectable (Fig. 4b, bottom panel). These data indicate that in wild-type cells, p85 is more abundant than phosphorylated IRS proteins, while in heterozygous KO cells and null cells, phosphorylated IRS proteins are in excess of the remaining p85 protein. The changing patterns in the molecular balance among regulatory subunits, catalytic subunits, and phosphorylated IRS proteins in the three cell types are schematically represented in Fig. 4c.
Effect of Pik3r1 gene disruption on PIP3 level and downstream signaling events.
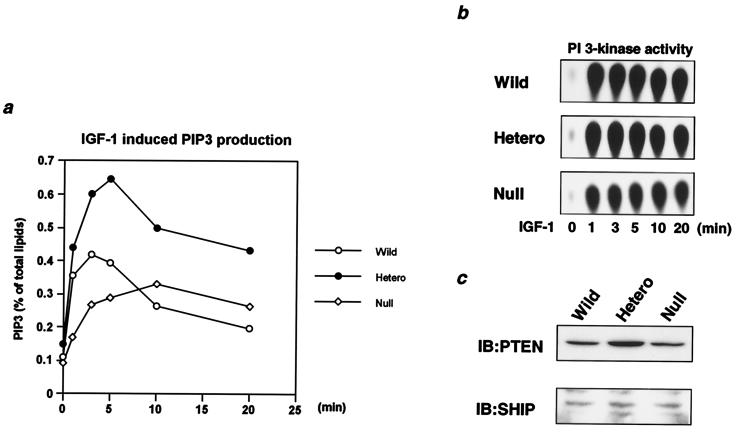
PIP3 is produced by PI 3-kinase and acts as a pivotal second messenger in the metabolic and mitogenic events regulated by growth factors, including insulin and IGF-1 (10, 49). The level of PIP3 is also regulated by lipid phosphatases, such as PTEN and SHIP (8, 42). In wild-type cells, the PIP3 level assessed by 32P labeling and TLC was transiently increased following IGF-1 stimulation, reaching a maximum at 2.5 min and then rapidly decreasing to the basal level within 20 min of stimulation (Fig. 5a). Interestingly, in heterozygous KO cells the PIP3 level was stimulated to a much greater extent than in wild-type cells, and a submaximal level was sustained for 20 min of stimulation (Fig. 5a). This occurred despite the fact that heterozygous KO and wild-type cells exhibited almost equal PI 3-kinase activities associated with tyrosine-phosphorylated proteins at all time points during this period (Fig. 5b). Furthermore, the level of PTEN protein in heterozygous KO cells was actually up-regulated, by twofold, and the level of SHIP protein was not altered (Fig. 5c). In null cells, although the maximal stimulated level of PIP3 was markedly decreased due to the decrease in PI 3-kinase activity associated with tyrosine-phosphorylated proteins during the period (Fig. 5b), the submaximal level of PIP3 was maintained for 20 min of stimulation (Fig. 5a), suggesting decreased PIP3 degradation. In these cells, no change was detectable in the level of either PTEN or SHIP compared to that of wild-type cells (Fig. 5c). Taken together, these data indicate that while there is enhanced functional PI 3-kinase activity in heterozygous KO cells, both heterozygous KO cells and null cells appear to exhibit a reduced clearance rate of PIP3 compared to that in wild-type cells.
FIG. 5.
Effect of disruption of Pik3r1 on production of PIP3 in response to IGF-1 in vivo. (a) IGF-1-induced PIP3 production in cells of each genotype. Cells were labeled with [32P]orthophosphate as described in Materials and Methods and stimulated with 10 nM IGF-1 for the indicated period. 32P-labeled phospholipids were extracted and separated by TLC. In the graph, the mean levels of PIP3 normalized to the total labeled phospholipids from two independent experiments are shown. (b) Time course of PI 3-kinase associated with phosphotyrosine complex in cells of each genotype. Cells were stimulated with 10 nM IGF-1 for the indicated period and subjected to immunoprecipitation with 4G10 followed by the PI 3-kinase assay. (c) Expression levels of PTEN and SHIP in cells of each genotype. Cell lysates were subjected to immunoblotting with anti-PTEN (αPTEN; top panel) or anti-SHIP (αSHIP; bottom panel) antibody. Wild, wild type; Hetero, heterozygous KO.
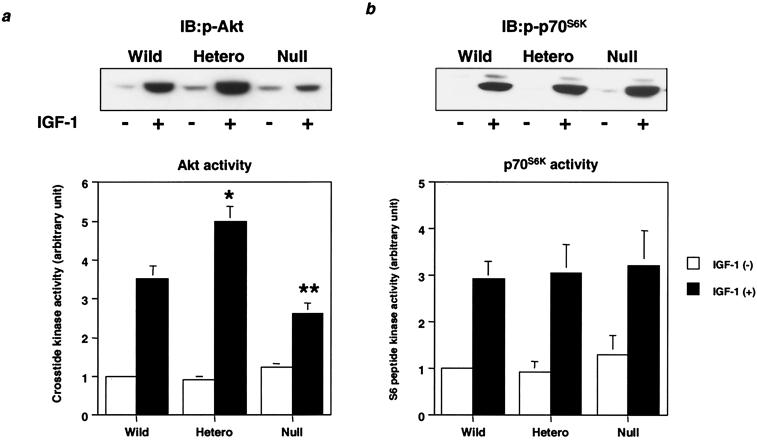
Corresponding to the increased PIP3 levels in the heterozygous KO cells, Akt phosphorylation and activity were up-regulated by 150% compared to those in wild-type cells (Fig. 6a), whereas in null cells, Akt phosphorylation and activity were decreased ∼30% (Fig. 6b). On the other hand, although p70S6K also lies downstream of PI 3-kinase and 3-phosphoinositide-dependent protein kinase 1 (PDK1) (9, 38), p70S6K phosphorylation and activity were almost equal among all cell lines (Fig. 6b).
FIG. 6.
Effect of disruption of Pik3r1 on downstream kinases from PI 3-kinase. (a) IGF-1-induced Akt activity in cells of each genotype. Cells were starved for 24 h and then stimulated with 10 nM IGF-1 for 5 min. Cell lysates were subjected to immunoblotting with anti-phospho-Akt (αphospho-Akt; top panel) antibody or immunoprecipitation with αAkt antibody. The immunoprecipitates were subjected to an immune complex kinase assay. In the bottom panel, each bar represents the mean ± standard deviation of the relative Akt kinase activity calculated from the results of three independent experiments. *, P value of <0.01 for wild-type (Wild) versus heterozygous KO (Hetero) cells; **, P value of <0.05 for wild versus null cells. (b) IGF-1-induced p70S6K activity in cells of each genotype. After 20 min of stimulation with 10 nM IGF-1, cell lysates were subjected to immunoblotting with anti-phospho-p70S6K (αphospho-p70S6K; top panel) antibody or immunoprecipitation with αp70S6K antibody. The immunoprecipitates were subjected to an immune complex kinase assay. In the bottom panel, each bar represents the mean ± standard deviation of the relative p70S6K kinase activity calculated from the results of three independent experiments.
Effect of Pik3r1 gene disruption on IGF-1-dependent antiapoptosis.
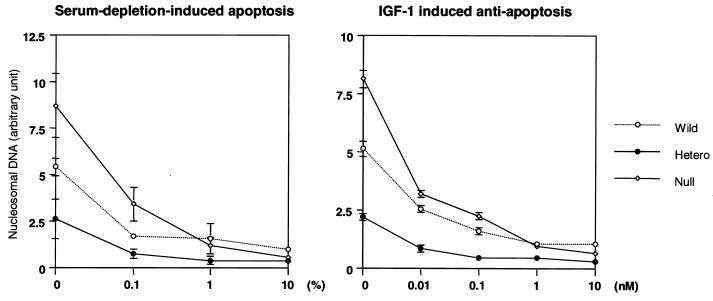
Since one of the important biological responses regulated via the PI 3-kinase/Akt pathway is its effect on cell survival (11, 14, 22), we measured serum depletion-induced apoptosis and the antiapoptotic effect of IGF-1 in each cell line. Consistent with the increase in Akt activity, p85α heterozygous KO cells were more resistant to serum depletion-induced apoptosis than were wild-type cells and more sensitive to the antiapoptotic effect of IGF-1 (Fig. 7). Null cells, on the other hand, were more prone to apoptosis and more resistant to rescue by IGF-1, consistent with the reduced PI 3-kinase and Akt activities (Fig. 7).
FIG. 7.
Effect of disruption of Pik3r1 on serum deprivation-induced apoptosis and IGF-1-dependent antiapoptosis. Cells were cultured in the indicated concentration of serum (left panel) or IGF-1 without serum (right panel) for 5 h. The level of apoptosis was assessed using an enzyme-linked immunosorbent assay for nucleosomal DNA as described in Materials and Methods. Each value is expressed as the ratio of the value of the wild-type cells treated with 10% serum and represents the mean ± standard deviation of three independent experiments. Wild, wild type; Hetero, heterozygous KO.
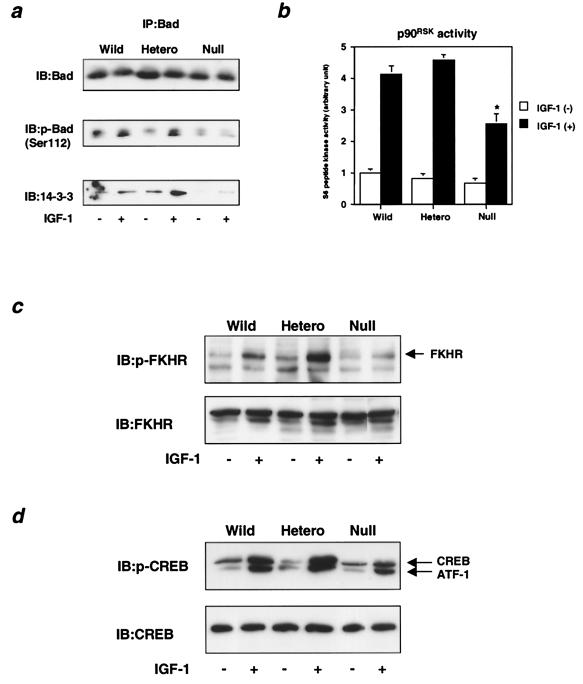
Three pathways downstream of PI 3-kinase are thought to be involved in antiapoptosis regulated by IGF-1. Survival factors, such as IGF-1, stimulate phosphorylation of the proapoptotic protein, Bad, on its two serine residues (Ser-112 and Ser-136) and promote its association with 14-3-3, leading to antiapoptosis (30). In heterozygous KO cells, Bad phosphorylation of Ser-112 was slightly increased compared to that in wild-type cells, whereas in null cells, Ser-112 phosphorylation was prominently decreased (Fig. 8a, middle panel), consistent with the changes in apoptosis in these cells. Since p90RSK has been shown to phosphorylate Ser-112 in Bad in response to survival factors (4) and Ser-136 has been shown to be phosphorylated by Akt (12), we assessed the kinase activity of p90RSK in all three cell lines. In null cells, the kinase activity of p90RSK was significantly decreased compared to that in wild-type cells (Fig. 8b), whereas mitogen-activated protein (MAP) kinase activity was almost comparable to that in wild-type cells (data not shown). In heterozygous KO cells, p90RSK activity tended to be increased (Fig. 8b). The amount of 14-3-3 bound to either of the Bad phosphorylation sites (Ser-112 and Ser-136) in heterozygous KO cells was increased by 140% compared to that in wild-type cells, whereas in null cells it was decreased by 70% (Fig. 8a, bottom panel).
FIG. 8.
Effect of disruption of Pik3r1 on IGF-1-dependent antiapoptotic signaling. (a) IGF-1-induced Bad phosphorylation and the interaction between Bad and 14-3-3 in cells of each genotype. Cells were starved for 24 h and then stimulated with 10 nM IGF-1 for 20 min. Cell lysates were subjected to immunoprecipitation with αBad antibody followed by immunoblotting. Immunoblots were probed with αBad (top panel), anti-phospho-Bad (αphospho-Bad; middle panel), or anti-14-3-3 (α14-3-3; bottom panel) antibody and visualized by enhanced chemiluminescence with protein A-conjugated peroxidase. (b) IGF-1-induced p90RSK activity in cells of each genotype. After 20 min of stimulation with 10 nM IGF-1, cell lysates were subjected to immunoprecipitation with αp90RSK antibody followed by an immune complex kinase assay. Each bar represents the mean ± standard deviation of the p90RSK activity calculated from the results of three independent experiments. *, P value of <0.01 for wild-type (Wild) versus null cells. (c) IGF-1-induced FKHR phosphorylation. After 20 min of stimulation with 10 nM IGF-1, cell lysates were subjected to immunoblotting with anti-phospho-FKHR (αphospho-FKHR; top panel) or anti-FKHR (αFKHR; bottom panel) antibody. (d) IGF-1-induced CREB phosphorylation in cells of each genotype. After 20 min of stimulation with 10 nM IGF-1, cell lysates were subjected to immunoblotting with anti-phospho-CREB (αphospho-CREB; top panel) or anti-CREB (αCREB; bottom panel) antibody. Hetero, heterozygous KO.
Finally, forkhead transcription factors, such as FKHR, and cyclic AMP-responsive element binding (CREB) protein have also been shown to be involved in IGF-1-dependent antiapoptosis. FKHR is phosphorylated and negatively regulated in response to survival factors. FKHR phosphorylation on Ser-256 in response to IGF-1 through a PI 3-kinase/Akt pathway (5, 47), reflecting a deactivation of transcriptional activity, was clearly increased in heterozygous KO cells compared to that in wild-type cells, whereas FKHR phosphorylation was decreased in null cells (Fig. 8c). Phosphorylation of CREB (Ser-133) in heterozygous KO cells was slightly up-regulated, suggesting increased transcriptional activity (19), whereas CREB phosphorylation was down-regulated in null cells (Fig. 8d). These changes could contribute to an induction or a decrease in the antiapoptosis protein Bcl-2 (53) and thereby also contribute to the increased and decreased effects of IGF-1 on apoptosis in these cells which were observed in heterozygous KO and null cells, respectively.
DISCUSSION
PI 3-kinase activity is required for a wide variety of IGF-1, insulin, growth factor, and cytokine signaling events, including stimulation of glucose transport and metabolism and antiapoptosis (9, 20, 34, 43). For insulin and IGF-1, an interaction between tyrosine-phosphorylated IRS proteins and class Ia PI 3-kinase initiates these various biological responses (2, 20, 43). The alternative spliced products of the Pik3r1 gene, p85α, AS53/p55α, and p50α, represent three of the regulatory subunits that are involved in PI 3-kinase signaling, and each may have a specific physiological role (1, 24, 50). KO mice lacking only the full-length form of p85α grow to adulthood with a moderate immunodeficiency syndrome (46), whereas disruption of all the spliced isoforms of the p85α gene (Pik3r1) results in perinatal lethality with abnormalities in multiple tissues (16, 18). Heterozygous disruption of Pik3r1, which reduces all isoforms of p85α by 50%, results in improved sensitivity to insulin and IGF-1 and decreases the incidence of diabetes in insulin receptor/IRS-1 double heterozygous KO mice (32). To clarify the molecular mechanism of PI 3-kinase-mediated signaling, we decided to establish cell lines from heterozygous and homozygous Pik3r1 KO mice to investigate the roles of the products of this gene in insulin/IGF-1 signaling and their role in cell survival (22).
We find that in wild-type cells, p85 is much more abundant than p110, such that normally at least 30% of p85 exits as a monomer that is not only unable to transmit a signal, but it actually acts to inhibit signaling via the p85-p110 dimer by competing for binding to phosphorylated IRS proteins. This natural inhibition is similar to that observed following overexpression of the wild-type regulatory subunit (39, 50) or the mutant p85 lacking the p110-binding site (20). In p85α heterozygous KO cells, there is a 50% reduction in p85α. Most of the decrease occurs in the p85 monomer with little change in the amount of the p85-p110 dimer. Thus, the level of p85 bound to p110 does not change, and the amount of p85 interacting with tyrosine-phosphorylated proteins is only slightly decreased. As a result, the activity of PI 3-kinase associated with p85α and the level of PI 3-kinase activity associated with p110 or tyrosine-phosphorylated proteins are normal or even tend to be increased.
In null cells, on the other hand, the amount of p85-p110 dimer is markedly diminished. This is due to a complete absence of p85α coupled with a secondary reduction of p110, probably due to a lack of the regulatory subunit to stabilize p110 (56). As a result, even though there is some up-regulation of p85β, PI 3-kinase activity induced by IGF-1 is significantly decreased.
Thus, the improvement of sensitivity of cells to IGF-1 or insulin following reduction of the p85 protein depends on the balance in the p85, p110, and phosphorylated IRS proteins. This may vary from tissue to tissue and with the intensity of stimulation. Thus, if the ratio of p85 to p110 is extremely high in a particular tissue, an increase in IGF-1-dependent PI 3-kinase activation may occur even with homozygous KO Pik3r1 cells because a sufficient amount of p85β-p110 dimer is preserved. On the other hand, if phosphorylated IRS proteins are much more abundant after ligand stimulation than p85 protein, the increase in the ratio of p85-p110 dimer to p85 monomer in the heterozygous KO may not affect PI 3-kinase-dependent signaling, since most of the p85-p110 dimer already binds IRS proteins even in the wild-type.
These changes in the molecular balance in the PI 3-kinase signaling complex can explain why Pik3r1 heterozygous KO cells and heterozygous KO mice exhibit preserved insulin- or IGF-1-induced PI 3-kinase activity; however, this may not totally account for the up-regulation of some downstream signals, such as Akt activity (32). The latter reflects PIP3 levels rather than PI 3-kinase activity (10), and PIP3 levels are regulated by both PI 3-kinase and lipid phosphatases, such as PTEN and SHIP (8, 42). Interestingly, although PI 3-kinase activities in wild-type and heterozygous KO cells are almost equal during the period of stimulation, the maximal PIP3 level is highly up-regulated and the submaximal level is more sustained in heterozygous KO cells than in wild-type cells. Furthermore, in null cells, although the maximal level of PIP3 is decreased due to the decrease in PI 3-kinase activity, the submaximal level is sustained. Since p21ras has been reported to directly bind and activate PI 3-kinase in a GTP-dependent manner (40, 41), it is possible that deletion of p85α may cause up-regulation of IGF-1-induced p21ras activity, leading to an increase in PI 3-kinase activity bound to p21ras. However, p21ras activity does not appear to be altered by the Pik3r1 KO, because MAP kinase activity in null cells is almost comparable to that in wild-type cells. Thus, these findings rather suggest that clearance of PIP3 in heterozygous KO and null cells is attenuated and occurs with no change in SHIP and even an up-regulation of PTEN in heterozygous KO cells, although we cannot completely rule out the possibility that unknown pathways up-regulate PI 3-kinase activation by a reduction in p85 protein in a phosphotyrosine-independent manner. Similar up-regulation of PIP3, in spite of a decrease in PI 3-kinase activity, is observed in mice lacking only the full-length version of p85α (48). Taken together, these data suggest that PTEN and/or SHIP activity or some other factor(s) contributing to PIP3 clearance may be positively regulated by the p85α regulatory subunit in a manner independent of actual PI 3-kinase activity. Corresponding to the PIP3 level (but not to the PI 3-kinase activity), Akt activity in p85α heterozygous KO cells is significantly up-regulated, whereas the activity in null cells is decreased compared to that in wild-type cells. On the other hand, there is no significant difference in p70S6K activity among cells of all genotypes, although p70S6K is known to be regulated by PI 3-kinase and PDK1 (9, 38). It is unclear whether this is due to the fact that only a small amount of PIP3 is required for full activation of p70S6K or that some alternative pathway of regulation takes over in the face of the reduced PI 3-kinase activity.
One of the important biological responses induced by IGF-1 through PI 3-kinase and Akt is antiapoptosis, which has been shown to play a pivotal role in regulating life span, carcinogenesis, and normal development (10, 11, 14). We find that p85α heterozygous KO cells are very resistant to apoptosis and sensitive to the antiapoptotic effects of IGF-1, whereas null cells are more prone to apoptosis and resistant to IGF-1 compared to wild-type cells. To date, several signaling cascades have been implicated in antiapoptosis by survival factors such as IGF-1. One of the most intensively investigated pathways is the phosphorylation-mediated regulation of the pro-apoptotic protein Bad, a member of the Bcl-2 family (30). In the absence of survival signals or in the presence of death signals, Bad binds antiapoptotic protein Bcl-2 or Bcl-XL and suppresses its activity. Survival factors promote phosphorylation of two serine residues of Bad (Ser-112 and Ser-136), leading to the dissociation of Bcl-2 and association with 14-3-3. This interaction prevents Bad from translocating to the mitochondrial membrane, thereby inhibiting apoptosis. Recently, Akt has been shown to phosphorylate Ser-136 on Bad (12), whereas p90RSK (4) and cyclic AMP-dependent kinase (21) have been demonstrated to phosphorylate Ser-112. As noted above, Akt activity regulated by PDK1 is up-regulated in heterozygous KO cells and significantly decreased in null cells, while in cells of all genotypes, IGF-1 induces MAP kinase activation to almost the same level (data not shown). p90RSK, on the other hand, is subject to phosphorylation in the amino-terminal kinase domain by PDK1 and the carboxy-terminal domain by MAP kinase (25). As a result of these two influences, in null cells p90RSK activity is markedly decreased, whereas in p85α heterozygous KO cells p90RSK activity tends to be increased. In parallel with the p90RSK activity, phosphorylation of Ser-112 in Bad is markedly decreased in null cells, and phosphorylation of Ser-112 is slightly increased in heterozygous KO cells. Finally, 14-3-3 bound to either Ser-112 or Ser-136 is significantly increased in heterozygous KO cells, whereas it is markedly decreased in null cells. Thus, the amount of 14-3-3 bound to Bad seems to correlate with the combined activities of Akt and in p90RSK and could account for why p85α heterozygous KO cells are resistant to apoptosis while null cells are prone to apoptosis.
Another pathway involved in apoptosis is mediated via the forkhead transcription factor family. Genetic studies using the nematode Caenorhabditis elegans have revealed that a forkhead transcription factor, DAF-16, is negatively regulated by AKT-1/2 (homologues of Akt) through AGE-1 (homologue of PI 3-kinase) and DAF-2 (homologue of insulin/IGF-1 receptor) (28, 31, 33). Recently, it has been shown that in mammalian cells, forkhead transcription factors (FKHR, FKHRL1, and AFX1) are negatively regulated by Akt in a phosphorylation-dependent manner (5, 29, 47). It has also been suggested that the phosphorylated forms of forkhead transcription factors bind 14-3-3 and cannot translocate to nuclei, thereby inhibiting transcription of apoptotic proteins such as the Fas ligand (5). Corresponding to the Akt activity, FKHR phosphorylation is up-regulated in heterozygous KO cells compared to wild-type cells, whereas in null cells FKHR phosphorylation is decreased. This may also contribute to the phenotype in apoptosis in each cell line.
Finally, the transcription factor CREB protein is also known to regulate IGF-1-dependent antiapoptosis in a phosphorylation-dependent manner (Ser-133), presumably through increasing the transcription of Bcl-2 (4, 53). Although the kinase responsible for IGF-1-induced phosphorylation of CREB (13, 37, 55) is still unclear, in both heterozygous KO and null cells, CREB phosphorylation on Ser-133 correlates with the activity of Akt or p90RSK, as previously shown, rather than that of p38 MAP kinase as suggested by others (37), since p38 MAP kinase activity is down-regulated (data not shown). Thus, all three pathways investigated are up-regulated in heterozygous KO cells, while they are down-regulated in null cells. This increased susceptibility to apoptosis in null cells may contribute to the shortened life span of Pik3r1−/− mice through intolerance for the environmental stresses and/or abnormal development of organs.
In summary, in normal cells, the regulatory subunit of PI 3-kinase (p85) is more abundant than the p110 catalytic subunits, and monomeric p85 inhibits the IRS protein-mediated signal by competing with the p85-p110 dimer. The 50% reduction in p85α in heterozygous KO cells results in improvement of some of the PI 3-kinase-mediated biological responses by IGF-1, such as Akt activity and antiapoptosis, through the decrease in the p85 monomer and the attenuation of PIP3 clearance. The latter effect appears to be regulated by p85 independent of its regulation of PI 3-kinase activity. Complete depletion of p85α, on the other hand, results in a significant decrease in the PI 3-kinase-mediated biological responses, such as antiapoptosis, by a marked reduction of PI 3-kinase activity, owing to a decrease in both p85 and p110. These data suggest that the appropriate amount of reduction of p85 could improve IGF-1 and insulin signaling, such as antiapoptosis, and possibly glucose metabolism (32). Thus, p85 may be a therapeutic target for prolongation of a life span as well as treatment of diabetes.
Acknowledgments
This work was supported by NIH grants DK33201 and DK55545 to C.R.K. and GM41890 to L.C.C. and by Joslin DERC grant DK34834 to C.R.K. D.A.F. was supported by fellowships from the Damon Runyon-Walter Winchell Cancer Research Fund and the Leukemia Society of America. S.M.B. was supported by a fellowship from Boehringer Ingelheim Funds.
We thank S. Paqutte for excellent secretarial assistance.
REFERENCES
- 1.Antonetti, D. A., P. Algenstaedt, and C. R. Kahn. 1996. Insulin receptor substrate 1 binds two novel splice variants of the regulatory subunit of phosphatidylinositol 3-kinase in muscle and brain. Mol. Cell. Biol. 16:2195–2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Backer, J. M., M. G. Myers, Jr., S. E. Shoelson, D. J. Chin, X. J. Sun, M. Miralpeix, P. Hu, B. Margolis, E. Y. Skolnik, J. Schlessinger, and M. F. White. 1992. Phosphatidylinositol 3′-kinase is activated by association with IRS-1 during insulin stimulation. EMBO J. 11:3469–3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baltensperger, K., L. M. Kozma, S. R. Jaspers, and M. P. Czech. 1994. Regulation by insulin of phosphatidylinositol 3′-kinase bound to alpha- and beta-isoforms of p85 regulatory subunit. J. Biol. Chem. 269:28937–28946. [PubMed] [Google Scholar]
- 4.Bonni, A., A. Brunet, A. E. West, S. R. Datta, M. A. Takasu, and M. E. Greenberg. 1999. Cell survival promoted by the Ras-MAPK signaling pathway by transcription-dependent and -independent mechanisms. Science 286:1358–1362. [DOI] [PubMed] [Google Scholar]
- 5.Brunet, A., A. Bonni, M. J. Zigmond, M. Z. Lin, P. Juo, L. S. Hu, M. J. Anderson, K. C. Arden, J. Blenis, and M. E. Greenberg. 1999. Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell 96:857–868. [DOI] [PubMed] [Google Scholar]
- 6.Bruning, J. C., J. Winnay, S. Bonner-Weir, S. I. Taylor, D. Accili, and C. R. Kahn. 1997. Development of a novel polygenic model of NIDDM in mice heterozygous for IR and IRS-1 null alleles. Cell 88:561–572. [DOI] [PubMed] [Google Scholar]
- 7.Bruning, J. C., J. Winnay, B. Cheatham, and C. R. Kahn. 1997. Differential signaling by insulin receptor substrate 1 (IRS-1) and IRS-2 in IRS-1-deficient cells. Mol. Cell. Biol. 17:1513–1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cantley, L. C., and B. G. Neel. 1999. New insights into tumor suppression: PTEN suppresses tumor formation by restraining the phosphoinositide 3-kinase/AKT pathway. Proc. Natl. Acad. Sci. USA 96:4240–4245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheatham, B., C. J. Vlahos, L. Cheatham, L. Wang, J. Blenis, and C. R. Kahn. 1994. Phosphatidylinositol 3-kinase activation is required for insulin stimulation of pp70 S6 kinase, DNA synthesis, and glucose transporter translocation. Mol. Cell. Biol. 14:4902–4911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coffer, P. J., J. Jin, and J. R. Woodgett. 1998. Protein kinase B (c-Akt): a multifunctional mediator of phosphatidylinositol 3-kinase activation. Biochem. J. 335:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Datta, S. R., A. Brunet, and M. E. Greenberg. 1999. Cellular survival: a play in three Akts. Genes Dev. 13:2905–2927. [DOI] [PubMed] [Google Scholar]
- 12.Datta, S. R., H. Dudek, X. Tao, S. Masters, H. Fu, Y. Gotoh, and M. E. Greenberg. 1997. Akt phosphorylation of BAD couples survival signals to the cell-intrinsic death machinery. Cell 91:231–241. [DOI] [PubMed] [Google Scholar]
- 13.Du, K., and M. Montminy. 1998. CREB is a regulatory target for the protein kinase Akt/PKB. J. Biol. Chem. 273:32377–32379. [DOI] [PubMed] [Google Scholar]
- 14.Franke, T. F., D. R. Kaplan, and L. C. Cantley. 1997. PI3K: downstream AKTion blocks apoptosis. Cell 88:435–437. [DOI] [PubMed] [Google Scholar]
- 15.Fruman, D. A., L. C. Cantley, and C. L. Carpenter. 1996. Structural organization and alternative splicing of the murine phosphoinositide 3-kinase p85α gene. Genomics 37:113–121. [DOI] [PubMed] [Google Scholar]
- 16.Fruman, D. A., F. Mauvais-Jarvis, D. A. Pollard, C. M. Yballe, D. Brazil, R. T. Bronson, C. R. Kahn, and L. C. Cantley. 2000. Hypoglycaemia, liver necrosis and perinatal death in mice lacking all isoforms of phosphoinositide 3-kinase p85α. Nat. Genet. 26:379–382. [DOI] [PubMed] [Google Scholar]
- 17.Fruman, D. A., R. E. Meyers, and L. C. Cantley. 1998. Phosphoinositide kinases. Annu. Rev. Biochem. 67:481–507. [DOI] [PubMed] [Google Scholar]
- 18.Fruman, D. A., S. B. Snapper, C. M. Yballe, L. Davidson, J. Y. Yu, F. W. Alt, and L. C. Cantley. 1999. Impaired B cell development and proliferation in absence of phosphoinositide 3-kinase p85α. Science 283:393–397. [DOI] [PubMed] [Google Scholar]
- 19.Gonzalez, G. A., and M. R. Montminy. 1989. Cyclic AMP stimulates somatostatin gene transcription by phosphorylation of CREB at serine 133. Cell 59:675–680. [DOI] [PubMed] [Google Scholar]
- 20.Hara, K., K. Yonezawa, H. Sakaue, A. Ando, K. Kotani, T. Kitamura, Y. Kitamura, H. Ueda, L. Stephens, T. R. Jackson, P. T. Hawkins, R. Dahnd, A. E. Clark, G. D. Holman, M. D. Waterfield, and M. Kasuga. 1994. 1-Phosphatidylinositol 3-kinase activity is required for insulin-stimulated glucose transport but not for RAS activation in CHO cells. Proc. Natl. Acad. Sci. USA 91:7415–7419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harada, H., B. Becknell, M. Wilm, M. Mann, L. J. Huang, S. S. Taylor, J. D. Scott, and S. J. Korsmeyer. 1999. Phosphorylation and inactivation of BAD by mitochondria-anchored protein kinase A. Mol. Cell 3:413–422. [DOI] [PubMed] [Google Scholar]
- 22.Hemmings, B. A. 1997. Akt signaling: linking membrane events to life and death decisions. Science 275:628–630. [DOI] [PubMed] [Google Scholar]
- 23.Inukai, K., M. Anai, E. Van Breda, T. Hosaka, H. Katagiri, M. Funaki, Y. Fukushima, T. Ogihara, Y. Yazaki, M. Kikuchi, Y. Oka, and T. Asano. 1996. A novel 55-kDa regulatory subunit for phosphatidylinositol 3-kinase structurally similar to p55PIK is generated by alternative splicing of the p85α gene. J. Biol. Chem. 271:5317–5320. [DOI] [PubMed] [Google Scholar]
- 24.Inukai, K., M. Funaki, T. Ogihara, H. Katagiri, A. Kanda, M. Anai, Y. Fukushima, T. Hosaka, M. Suzuki, B. C. Shin, K. Takata, Y. Yazaki, M. Kikuchi, Y. Oka, and T. Asano. 1997. p85α gene generates three isoforms of regulatory subunit for phosphatidylinositol 3-kinase (PI 3-kinase), p50α, p55α, and p85α, with different PI 3-kinase activity elevating responses to insulin. J. Biol. Chem. 272:7873–7882. [DOI] [PubMed] [Google Scholar]
- 25.Jensen, C. J., M. B. Buch, T. O. Krag, B. A. Hemmings, S. Gammeltoft, and M. Frodin. 1999. 90-kDa ribosomal S6 kinase is phosphorylated and activated by 3-phosphoinositide-dependent protein kinase-1. J. Biol. Chem. 274:27168–27176. [DOI] [PubMed] [Google Scholar]
- 26.Kerouz, N. J., D. Horsch, S. Pons, and C. R. Kahn. 1997. Differential regulation of insulin receptor substrates-1 and -2 (IRS-1 and IRS-2) and phosphatidylinositol 3-kinase isoforms in liver and muscle of the obese diabetic (ob/ob) mouse. J. Clin. Investig. 100:3164–3172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kimura, K., S. Hattori, Y. Kabuyama, Y. Shizawa, J. Takayanagi, S. Nakamura, S. Toki, Y. Matsuda, K. Onodera, and Y. Fukui. 1994. Neurite outgrowth of PC12 cells is suppressed by wortmannin, a specific inhibitor of phosphatidylinositol 3-kinase. J. Biol. Chem. 269:18961–18967. [PubMed] [Google Scholar]
- 28.Kimura, K. D., H. A. Tissenbaum, Y. Liu, and G. Ruvkun. 1997. daf-2, an insulin receptor-like gene that regulates longevity and diapause in Caenorhabditis elegans. Science 277:942–946. [DOI] [PubMed] [Google Scholar]
- 29.Kops, G. J., N. D. de Ruiter, A. M. De Vries-Smits, D. R. Powell, J. L. Bos, and B. M. Burgering. 1999. Direct control of the Forkhead transcription factor AFX by protein kinase B. Nature 398:630–634. [DOI] [PubMed] [Google Scholar]
- 30.Korsmeyer, S. J. 1999. BCL-2 gene family and the regulation of programmed cell death. Cancer Res. 59:1693s–1700s. [PubMed] [Google Scholar]
- 31.Lin, K., J. B. Dorman, A. Rodan, and C. Kenyon. 1997. daf-16: an HNF-3/forkhead family member that can function to double the life-span of Caenorhabditis elegans. Science 278:1319–1322. [DOI] [PubMed] [Google Scholar]
- 32.Mauvais-Jarvis, F., K. Ueki, D. A. Fruman, M. F. Hirshman, K. Sakamoto, L. J. Goodyear, M. Iannacone, D. Accili, L. C. Cantley, and C. R. Kahn. Reduction in the p85α subunit of phosphoinositide 3-kinase improves insulin signaling and ameliorates diabetes in mice. J Clin. Investig., in press. [DOI] [PMC free article] [PubMed]
- 33.Ogg, S., S. Paradis, S. Gottlieb, G. I. Patterson, L. Lee, H. A. Tissenbaum, and G. Ruvkun. 1997. The Forkhead transcription factor DAF-16 transduces insulin-like metabolic and longevity signals in C. elegans. Nature 389:994–999. [DOI] [PubMed] [Google Scholar]
- 34.Okada, T., Y. Kawano, T. Sakakibara, O. Hazeki, and M. Ui. 1994. Essential role of phosphatidylinositol 3-kinase in insulin-induced glucose transport and antilipolysis in rat adipocytes. Studies with a selective inhibitor wortmannin. J. Biol. Chem. 269:3568–3573. [PubMed] [Google Scholar]
- 35.Otsu, M., I. Hiles, I. Gout, M. J. Fry, F. Ruiz-Larrea, G. Panayotou, A. Thompson, R. Dhand, J. Hsuan, and N. Totty. 1991. Characterization of two 85 kd proteins that associate with receptor tyrosine kinases, middle-T/pp60c-src complexes, and PI3-kinase. Cell 65:91–104. [DOI] [PubMed] [Google Scholar]
- 36.Pons, S., T. Asano, E. Glasheen, M. Miralpeix, Y. Zhang, T. L. Fisher, M. G. Myers, Jr., X. J. Sun, and M. F. White. 1995. The structure and function of p55PIK reveal a new regulatory subunit for phosphatidylinositol 3-kinase. Mol. Cell. Biol. 15:4453–4465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pugazhenthi, S., T. Boras, D. O’Connor, M. K. Meintzer, K. A. Heidenreich, and J. E. Reusch. 1999. Insulin-like growth factor I-mediated activation of the transcription factor cAMP response element-binding protein in PC12 cells. Involvement of p38 mitogen-activated protein kinase-mediated pathway. J. Biol. Chem. 274:2829–2837. [DOI] [PubMed] [Google Scholar]
- 38.Pullen, N., P. B. Dennis, M. Andjelkovic, A. Dufner, S. C. Kozma, B. A. Hemmings, and G. Thomas. 1998. Phosphorylation and activation of p70s6k by PDK1. Science 279:707–710. [DOI] [PubMed] [Google Scholar]
- 39.Rameh, L. E., C. S. Chen, and L. C. Cantley. 1995. Phosphatidylinositol (3,4,5)P3 interacts with SH2 domains and modulates PI 3-kinase association with tyrosine-phosphorylated proteins. Cell 83:821–830. [DOI] [PubMed] [Google Scholar]
- 40.Rodriguez-Viciana, P., P. H. Warne, R. Dhand, B. Vanhaesebroeck, I. Gout, M. J. Fry, M. D. Waterfield, and J. Downward. 1994. Phosphatidylinositol-3-OH kinase as a direct target of Ras. Nature 370:527–532. [DOI] [PubMed] [Google Scholar]
- 41.Rodriguez-Viciana, P., P. H. Warne, B. Vanhaesebroeck, M. D. Waterfield, and J. Downward. 1996. Activation of phosphoinositide 3-kinase by interaction with Ras and by point mutation. EMBO J. 15:2442–2451. [PMC free article] [PubMed] [Google Scholar]
- 42.Rohrschneider, L. R., J. F. Fuller, I. Wolf, Y. Liu, and D. M. Lucas. 2000. Structure, function, and biology of SHIP proteins. Genes Dev. 14:505–520. [PubMed] [Google Scholar]
- 43.Shepherd, P. R., D. J. Withers, and K. Siddle. 1998. Phosphoinositide 3-kinase: the key switch mechanism in insulin signalling. Biochem. J. 333:471–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Songyang, Z., S. E. Shoelson, M. Chaudhuri, G. Gish, T. Pawson, W. G. Haser, F. King, T. Roberts, S. Ratnofsky, R. J. Lechleider, B. G. Neel, R. B. Birge, J. E. Fajardo, M. M. Chou, H. Hanafusa, B. Schaffhausen, and L. C. Cantley. 1993. SH2 domains recognize specific phosphopeptide sequences. Cell 72:767–778. [DOI] [PubMed] [Google Scholar]
- 45.Sun, X. J., P. Rothenberg, C. R. Kahn, J. M. Backer, E. Araki, P. A. Wilden, D. A. Cahill, B. J. Goldstein, and M. F. White. 1991. Structure of the insulin receptor substrate IRS-1 defines a unique signal transduction protein. Nature 352:73–77. [DOI] [PubMed] [Google Scholar]
- 46.Suzuki, H., Y. Terauchi, M. Fujiwara, S. Aizawa, Y. Yazaki, T. Kadowaki, and S. Koyasu. 1999. Xid-like immunodeficiency in mice with disruption of the p85α subunit of phosphoinositide 3-kinase. Science 283:390–392. [DOI] [PubMed] [Google Scholar]
- 47.Tang, E. D., G. Nunez, F. G. Barr, and K. L. Guan. 1999. Negative regulation of the forkhead transcription factor FKHR by Akt. J. Biol. Chem. 274:16741–16746. [DOI] [PubMed] [Google Scholar]
- 48.Terauchi, Y., Y. Tsuji, S. Satoh, H. Minoura, K. Murakami, A. Okuno, K. Inukai, T. Asano, Y. Kaburagi, K. Ueki, H. Nakajima, T. Hanafusa, Y. Matsuzawa, H. Sekihara, Y. Yin, J. C. Barrett, H. Oda, T. Ishikawa, Y. Akanuma, I. Komuro, M. Suzuki, K. Yamamura, T. Kodama, H. Suzuki, and T. Kadowaki. 1999. Increased insulin sensitivity and hypoglycaemia in mice lacking the p85α subunit of phosphoinositide 3-kinase. Nat. Genet. 21:230–235. [DOI] [PubMed] [Google Scholar]
- 49.Toker, A., and L. C. Cantley. 1997. Signalling through the lipid products of phosphoinositide-3-OH kinase. Nature 387:673–676. [DOI] [PubMed] [Google Scholar]
- 50.Ueki, K., P. Algenstaedt, F. Mauvais-Jarvis, and C. R. Kahn. 2000. Positive and negative regulation of phosphoinositide 3-kinase-dependent signaling pathways by three different gene products of the p85α regulatory subunit. Mol. Cell. Biol. 20:8035–8046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ueki, K., R. Yamamoto-Honda, Y. Kaburagi, T. Yamauchi, K. Tobe, B. M. Burgering, P. J. Coffer, I. Komuro, Y. Akanuma, Y. Yazaki, and T. Kadowaki. 1998. Potential role of protein kinase B in insulin-induced glucose transport, glycogen synthesis, and protein synthesis. J. Biol. Chem. 273:5315–5322. [DOI] [PubMed] [Google Scholar]
- 52.White, M. F., and C. R. Kahn. 1994. The insulin signaling system. J. Biol. Chem. 269:1–4. [PubMed] [Google Scholar]
- 53.Wilson, B. E., E. Mochon, and L. M. Boxer. 1996. Induction of bcl-2 expression by phosphorylated CREB proteins during B-cell activation and rescue from apoptosis. Mol. Cell. Biol. 16:5546–5556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Winnay, J. N., J. C. Bruning, D. J. Burks, and C. R. Kahn. 2000. Gab-1-mediated IGF-1 signaling in IRS-1-deficient 3T3 fibroblasts. J. Biol. Chem. 275:10545–10550. [DOI] [PubMed] [Google Scholar]
- 55.Xing, J., D. D. Ginty, and M. E. Greenberg. 1996. Coupling of the RAS-MAPK pathway to gene activation by RSK2, a growth factor-regulated CREB kinase. Science 273:959–963. [DOI] [PubMed] [Google Scholar]
- 56.Yu, J., Y. Zhang, J. McIlroy, T. Rordorf-Nikolic, G. A. Orr, and J. M. Backer. 1998. Regulation of the p85/p110 phosphatidylinositol 3′-kinase: stabilization and inhibition of the p110α catalytic subunit by the p85 regulatory subunit. Mol. Cell. Biol. 18:1379–1387. [DOI] [PMC free article] [PubMed] [Google Scholar]