Abstract
Transcription is a complex process, the regulation of which is crucial for cellular and organismic growth and development. Deciphering the molecular mechanisms that define transcription is essential to understanding the regulation of RNA synthesis. Here we describe the molecular mechanism of escape commitment, a critical step in early RNA polymerase II transcription. During escape commitment ternary transcribing complexes become stable and committed to proceeding forward through promoter escape and the remainder of the transcription reaction. We found that the point in the transcription reaction at which escape commitment occurs depends on the length of the transcript RNA (4 nucleotides [nt]) as opposed to the position of the active site of the polymerase with respect to promoter DNA elements. We found that single-stranded nucleic acids can inhibit escape commitment, and we identified oligonucleotides that are potent inhibitors of this specific step. These inhibitors bind RNA polymerase II with low nanomolar affinity and sequence specificity, and they block both promoter-dependent and promoter-independent transcription, the latter occurring in the absence of general transcription factors. We demonstrate that escape commitment involves translocation of the RNA polymerase II active site between synthesis of the third and fourth phosphodiester bonds. We propose that a conformational change in ternary transcription complexes occurs during translocation after synthesis of a 4-nt RNA to render complexes escape committed.
Transcription is a critical first step in gene expression that is catalyzed by DNA-dependent RNA polymerases, which synthesize RNA transcripts using a single strand of DNA as a template. RNA polymerases range in size and complexity from the single-subunit bacteriophage T7 RNA polymerase through the multisubunit Escherichia coli RNA polymerase to eukaryotic RNA polymerase II, which requires a minimum of 14 different protein subunits for promoter-specific transcription (23, 24, 30, 32). Despite the differences in the complexity and organization of these diverse RNA polymerases, they each catalyze synthesis of RNA, and the essential elements of the transcription reaction appear to be conserved.
The transcription reaction is a multistep process that minimally involves the RNA polymerase and any associated factors first binding to the promoter DNA. After this point, transcription initiates in the presence of nucleoside triphosphates (NTPs). Initiated complexes of all RNA polymerases are capable of abortive initiation, the steady-state production of very short RNA products over time (11, 12, 21, 25). These initiated complexes then undergo a transition referred to as promoter escape in which they transform into elongation complexes. Ternary elongation complexes are quite stable and proceed through the remainder of the transcription reaction to complete RNA synthesis.
Numerous studies have characterized stable elongation complexes in transcription by bacteriophage T7, E. coli, and eukaryotic RNA polymerases (10, 14, 16, 17, 29, 33). While these studies reveal the nature of elongation complexes, little is known about the transitions that occur during early transcription to form stable elongation complexes. For example, initiated complexes undergo an intricate metamorphosis that simultaneously involves the release of accessory factors, extension of the melted region of the DNA, formation of an RNA-DNA duplex, and direction of the newly transcribed RNA out of the polymerase, all with the goal of forming a stable elongation complex that will complete synthesis of the transcript (17, 18, 35). Although we know that such events occur during early transcription, a unified view of the specific transitions and conformational changes that ternary transcription complexes undergo during this critical point in the transcription reaction has not yet emerged.
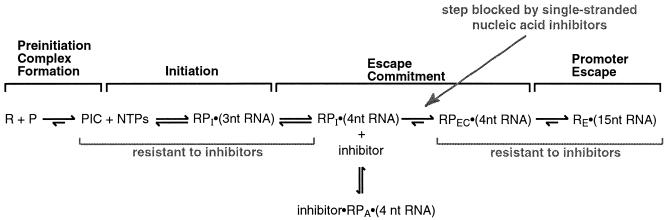
We are interested in understanding the transitions that occur during the early steps of mRNA transcription. Toward this goal, we previously studied the kinetics of several steps in the human RNA polymerase II transcription reaction (19, 20). We arrived at a model for eukaryotic transcription by experimentally isolating five distinct steps (see Fig. 1A): preinitiation complex formation, initiation, escape commitment, promoter escape, and transcript elongation. These studies revealed a crucial transition, termed escape commitment, which occurs during early transcription after initiation and prior to promoter escape (19). Escape commitment is complete after a 4-nucleotide (nt) RNA is synthesized, which occurs within 10 s after addition of NTPs to assembled preinitiation complexes. Notably, after this transition is complete, complexes are stable and committed to proceeding forward through promoter escape and the remainder of the transcription reaction. Escape-committed complexes [RPEC · (4nt RNA)] decay quite slowly compared to the rate at which they form (19). Other biochemical studies of early RNA polymerase II transcription have also characterized transitions that occur upon synthesis of a 4-nt RNA. Timmers and colleagues observed a distinct change in the melted region of the DNA when a 4-nt RNA was synthesized (13). In addition, Luse and colleagues found that stable ternary complexes formed when an RNA of 4 nt was transcribed (3, 22).
FIG. 1.
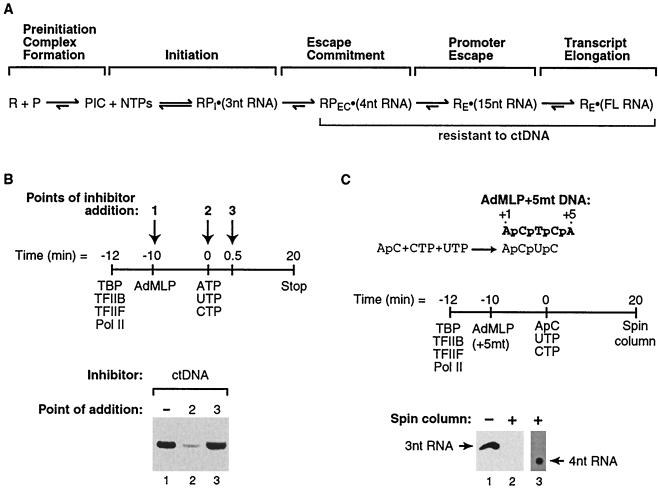
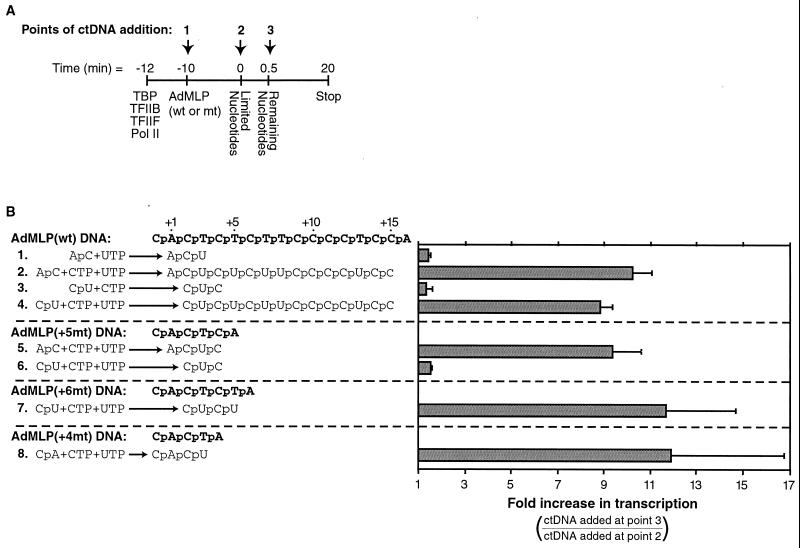
Escape commitment is a step in early transcription that occurs after initiation and prior to promoter escape. (A) Model depicting the steps in the RNA polymerase II transcription reaction. See the text for a description. Abbreviations: R, general transcription factors (TBP, TFIIB, TFIIF, and RNA polymerase II); P, promoter DNA (AdMLP); PIC, preinitiation complex; RPI · (3nt RNA), initiated complex containing a 3-nt RNA; RPEC · (4nt RNA), escape-committed complex containing a 4-nt RNA; RE · (15nt RNA), elongation complex containing a 15-nt RNA; RE · (FL RNA), elongation complex containing full-length RNA. (B) ctDNA inhibits escape commitment when added to reaction mixtures with the nucleotides. The method used to monitor escape commitment is depicted. ctDNA can be added at three different points in the transcription reaction: either with promoter DNA (point 1), with nucleotides (point 2), or 30 s after nucleotides (point 3). ctDNA (275 μg/ml) was added to reaction mixtures at points 2 and 3. The 390-nt G-less product is shown. (C) Escape-committed complexes contain 4-nt RNAs stably bound. The sequence of the nontemplate strand of the +5mt AdMLP is shown in boldface, with the sequence of the 4-nt RNA produced from it shown below. The schematic shows the method used to isolate ternary complexes. Transcription reactions were performed under conditions where a 4-nt RNA product is the longest that can be made at the AdMLP. Reaction mixtures were passed over G25 spin columns and phosphatase treated, and RNA products were resolved by 20% denaturing PAGE. Lane 3 is a longer exposure of lane 2. Positions of 3- and 4-nt RNA products are indicated.
To better understand the nature of transitions during the early steps of transcription, we investigated here the mechanism of escape commitment by human RNA polymerase II. We first asked which components of the transcription reaction are important for escape commitment to occur. Specifically, does the point at which escape commitment occurs depend on the length of the transcribed RNA or the position of the polymerase with respect to promoter DNA elements? Is a transcription factor released from the promoter and rebound during escape commitment? Is escape commitment inherent to RNA polymerase II, or are the general transcription factors involved? In addition, we asked which step in the nucleotide addition cycle (i.e., nucleotide binding, phosphodiester bond synthesis, or translocation) triggers escape commitment. Our results are consistent with a model in which translocation of the RNA polymerase II active site after synthesis of a 4-nt RNA induces a conformational change that renders transcription complexes stable and committed to proceeding forward through promoter escape and the remainder of the transcription reaction.
MATERIALS AND METHODS
Transcription factors and other reagents.
Recombinant (TATA-binding protein [TBP], transcription factor IIB [TFIIB], and TFIIF) and native (core RNA polymerase II) human transcription factors were prepared as described previously (reference 20 and references therein). The DNA template was negatively supercoiled plasmid DNA containing the adenovirus major late (AdMLP) core promoter (−53 to +10) fused to a 380-bp G-less cassette (11). The +4mt, +5mt, and +6mt AdMLP constructs were constructed via site-directed mutagenesis (20). Calf thymus DNA (ctDNA; Sigma) was subjected to extensive sonication, phenol-chloroform extraction, and ethanol precipitation. For the experiment for which results are shown in Fig. 3C, ctDNA was treated with Klenow fragment (Promega) at a final concentration of 1 U/μg in a buffer consisting of 50 mM Tris · HCl (pH 7.2), 10 mM MgSO4, 0.1 mM dithiothreitol (DTT), 20 μg of bovine serum albumin/ml, and 40 μM each dATP, dCTP, dGTP, and dTTP. After heat inactivation of the Klenow fragment, the DNA was ethanol precipitated. DNA oligonucleotide inhibitors were purchased from Life Technologies, and RNA oligonucleotide inhibitors were purchased from Dharmacon Research.
FIG. 3.
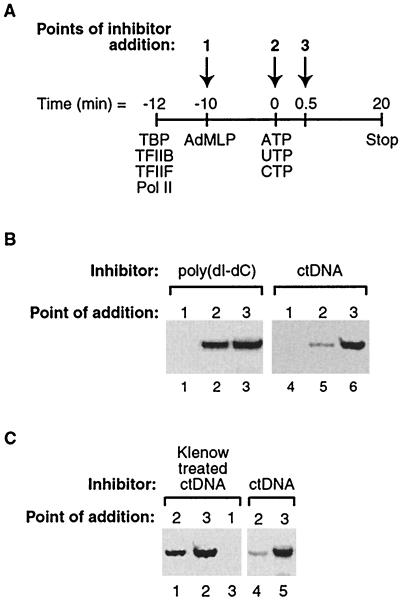
Single-stranded nucleic acid ends are required to inhibit escape commitment. (A) Method used to monitor escape commitment. Nucleic acid inhibitors were added to transcription reaction mixtures at points 1, 2, and 3. (B) Poly(dI-dC) does not inhibit escape commitment. Poly(dI-dC) and ctDNA were added to transcription reactions at the points indicated (see panel A for the method). The 390-nt G-less transcript is shown. (C) The single-stranded ends of ctDNA inhibit escape commitment. ctDNA was treated with Klenow fragment in the presence of dNTPs to remove 5′ and 3′ single-stranded overhangs and was subsequently added to assays at the points indicated. The 390-nt G-less transcript is shown.
In vitro transcription assays with human RNA polymerase II.
Transcription reactions were performed in buffer A containing 10 mM Tris · HCl (pH 7.9), 10 mM HEPES (pH 8.0), 10% glycerol, 1 mM DTT, 4 mM MgCl2, 50 mM KCl, 50 μg of bovine serum albumin/ml, and 15 U of RNA Guard (Amersham Pharmacia). Reaction mixtures contained 2 ng of TBP, 10 ng of TFIIB, 6 ng of TFIIF, 25 ng of RNA polymerase II, and 0.8 to 1.2 nM DNA template. Nucleotides were added in the combinations indicated in the figures and figure legends at final concentrations of 625 μM ATP, 625 μM CTP, 25 μM [α-32P]UTP (5 μCi per reaction), 1 mM ApC, 1 mM CpU, and 1 mM CpA. A general outline of the transcription reaction follows, with details and exceptions given in the figures and figure legends. The general transcription factors and RNA polymerase II were preincubated in buffer A for 2 min at 30°C (10 μl per reaction), after which promoter DNA in buffer A at 30°C (10 μl per reaction) was added. Proteins and DNA were incubated together for 10 min, at which point nucleotides were added. Transcription proceeded for 20 min at 30°C. Reactions were stopped with 100 μl of a stop solution containing 3.1 M ammonium acetate, 10 μg of carrier yeast RNA, and 15 μg of proteinase K. The samples were ethanol precipitated and resolved by 6% denaturing polyacrylamide gel electrophoresis (PAGE). For the experiments for which results are shown in Fig. 1C, 8B, and 8C, there were the following exceptions. Nucleotide concentrations were 1 mM ApC, 100 μM CTP, and 0.5 μM [α-32P]UTP (5 μCi per reaction). Before the reactions were stopped, reaction mixtures were passed through G25 spin columns (Amersham Pharmacia) preequilibrated in buffer A. Eluates were heated for 3 min at 70°C and then treated with calf intestinal alkaline phosphatase (10 U) at 37°C for 20 min. Three microliters of stop mix containing 200 mM EDTA, 20% glycerol, and 0.025% bromophenol blue was added. Products were resolved by 20% denaturing PAGE and sized by UV shadowing 2-, 3-, 4-, and 5-nt RNA standards of the appropriate sequences. For the promoter-independent transcription assays for which results are shown in Fig. 6B and C, poly(dA-dT) · poly(dA-dT) (0.5 mg/ml; Sigma) was added to reaction mixtures in place of promoter DNA. The general transcription factors were omitted, and final nucleotide concentrations were 625 μM ATP and 25 μM [α-32P]UTP (5 μCi per reaction).
FIG. 8.
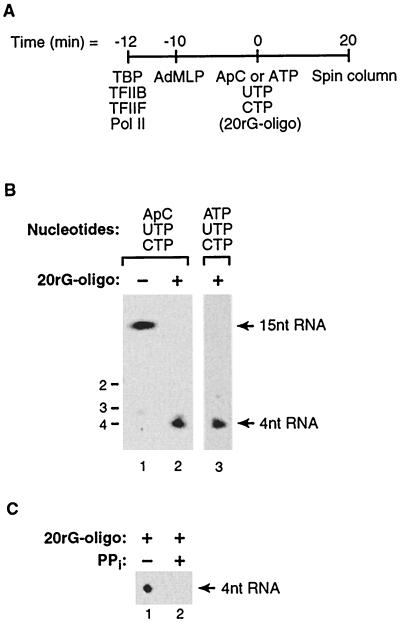
Escape commitment involves translocation of the RNA polymerase II active site between synthesis of the third and fourth phosphodiester bonds. (A) Method used to isolate ternary complexes. Transcription reactions were performed in the absence and presence of the 20rG oligonucleotide (250 nM) under conditions where a 15-nt RNA product is the longest that can be made at the AdMLP. Reaction mixtures were passed over G25 spin columns and then phosphatase treated, and RNA products were resolved by 20% denaturing PAGE. (B) The 4-nt RNA product is part of a stable inhibited complex at the AdMLP. Reactions were performed as described in the text. Note that the reactions for lanes 1 and 2 were initiated with ApC while the reaction for lane 3 was initiated with ATP. Size markers and positions of 4- and 15-nt RNA products are indicated. (C) Escape-committed complexes with the 20rG oligonucleotide bound can undergo pyrophosphorolysis. Ternary complexes containing the 20rG oligonucleotide (250 nM) were isolated as described for panel A and incubated in the presence or absence of pyrophosphate (1 mM) for 30 min at 30°C. The 4-nt RNA product is shown.
FIG. 6.
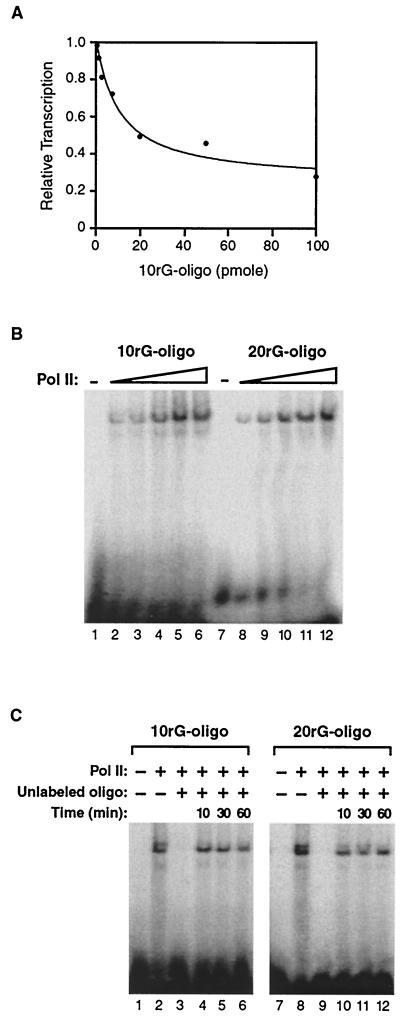
The site on RNA polymerase II to which the inhibitor oligonucleotides bind is removed from the site at which inhibition occurs. (A) An RNA oligonucleotide consisting of 10 guanosine residues (the 10rG oligonucleotide) does not inhibit escape commitment to the same extent as the 20rG oligonucleotide. The 10rG oligonucleotide was titrated into transcription reaction mixtures at point 2 (see Fig. 3A). The 390-nt RNA was quantitated and plotted. The IC50 is 0.5 μM. (B) The 10rG oligonucleotide and the 20rG oligonucleotide bind to RNA polymerase II with similar affinities. Electrophoretic mobility shift assays were performed with 32P-labeled 10rG and 20rG oligonucleotides (0.05 pmol) and increasing amounts of RNA polymerase II (3.5, 7, 14, 28, and 56 ng). (C) The 10rG oligonucleotide and the 20rG oligonucleotide form kinetically stable complexes with RNA polymerase II. RNA polymerase II (14 ng) was prebound to the 32P-labeled oligonucleotide indicated (0.05 pmol), and a 500-fold excess (25 pmol) of unlabeled oligonucleotide was subsequently added for the times indicated above lanes 4 to 6 and 10 to 12. In lanes 3 and 9, the unlabeled oligonucleotide was added prior to RNA polymerase II.
Abortive initiation assays.
Abortive initiation reactions were performed as described for the in vitro transcription reactions with the following exceptions. Final nucleotide concentrations in the reactions were 1 mM ApC and 0.5 μM [α-32P]UTP (5 μCi per reaction). Reactions were stopped at varying time points (up to 10 min) with heating at 70°C for 3 min, and then reaction products were treated with calf intestinal alkaline phosphatase (10 U) at 37°C for 20 min. Three microliters of a stop solution containing 200 mM EDTA, 20% glycerol, and 0.025% bromophenol blue was added. A 6-μl volume of each reaction product was loaded on a 20% denaturing gel. The amount of product produced at each time point was quantitated using a Molecular Dynamics PhosphorImager and normalized with respect to time. For each condition tested, a parallel reaction was performed with a mutant AdMLP in which the start site position was changed from an A to a G (nontemplate strand) (11). The product produced from the mutant template originated from nonspecific start sites on the plasmid DNA and was subtracted as background from the product produced from the wild-type template. The sizes of the RNA products were confirmed by UV shadowing 3-nt RNA standards of the appropriate sequences.
Electrophoretic mobility shift assays.
Reactions were performed in the buffers described for in vitro transcription assays. RNA polymerase II was added to 32P-labeled oligonucleotides, as indicated in the figure legends. Binding occurred at 30°C for 10 min. Complexes were resolved on native 4% polyacrylamide gels (37.5:1 arylamide-bis-acrylamide, 5% glycerol, and 0.5× Tris-borate-EDTA) at 150 V for 3 h.
RESULTS
We previously studied the kinetics of the eukaryotic transcription reaction using a minimal in vitro transcription system consisting of TBP, TFIIB, TFIIF, RNA polymerase II, and the AdMLP contained on a negatively supercoiled DNA template (19, 20). Using this system, we identified escape commitment as a distinct step in early transcription, because escape-committed complexes [RPEC · (4nt RNA)] (Fig. 1A) were stable to ctDNA added to transcription reaction mixtures as an inhibitor. In contrast, prior to the completion of escape commitment, ctDNA inhibited the majority of the transcription complexes; thus, they did not produce a full-length transcript. Figure 1B illustrates the method we developed to monitor escape commitment and shows representative results. ctDNA was added to transcription reaction mixtures at point 1, 2, or 3. When added at point 1, with the promoter DNA, ctDNA inhibited preinitiation complex formation by sequestering the general transcription factors before they bound to the promoter; thus, transcripts were not produced (data not shown). When ctDNA was added with the nucleotides (point 2), escape commitment had not yet occurred, the majority of complexes were sensitive to the inhibitor, and a low level of transcript was observed (Fig. 1B, lane 2). When ctDNA was added 30 s after the addition of nucleotides (point 3), escape commitment was already complete. As a result, complexes were stable to the inhibitor and a high level of transcript was observed (Fig. 1B, lane 3). The amount of transcript produced in the absence of ctDNA was similar to that produced when ctDNA was added at point 3 (Fig. 1B, lane 1).
Stable ternary complexes form only after synthesis of a 4-nt RNA.
Although the experiment for which results are shown in Fig. 1B was performed under conditions in which the full-length RNA transcript was observed, it should be noted that stability to ctDNA occurs once a 4-nt RNA is synthesized (19). If escape commitment is the distinct point in transcription at which stable ternary complexes form, transcribing complexes prior to this point should not be stable. To directly address ternary complex stability both before and after synthesis of a 4-nt RNA, we performed the experiment for which results are shown in Fig. 1C. As diagrammed in the schematic, we utilized a mutant of the AdMLP (+5mt) in which the T at position +5 was changed to an A. By use of this mutant, a 4-nt RNA was the longest product that could be made when transcription was initiated with the dinucleotide ApC, as well as UTP and CTP. To assess ternary complex stability, reaction mixtures were passed through gel filtration spin columns so that only those RNAs stably bound in ternary complexes would be present in the eluate (22). As shown in Fig. 1C, an abundance of 3-nt RNA was detected in the reaction product that had not been passed through a spin column (lane 1). As shown in lane 2, however, 3-nt RNA was no longer detected after ternary complexes were passed through a spin column. This indicates that the 3-nt RNA we observed was produced abortively and released; thus, it is not part of stable complexes. Strikingly, a longer exposure of lane 2 revealed 4-nt RNA that survived the spin column as part of ternary complexes (Fig. 1C, lane 3). This confirms that stable ternary complexes form only after synthesis of a 4-nt RNA, concomitant with the completion of escape commitment.
The point at which escape commitment occurs depends on the length of the transcribed RNA.
The point at which escape commitment is complete could be dictated by either the length of the transcribed RNA or the position of the polymerase active site with respect to the promoter DNA elements. To distinguish between these possibilities, we used the method shown in Fig. 2A, in which nucleotides were added to transcription reaction mixtures in two steps: limited nucleotides were added first, followed by remaining nucleotides 30 s later. Together, these nucleotides allowed synthesis of a full-length transcript; however, by altering the composition of the limited nucleotides, we controlled both the length of the RNA produced and the position of the polymerase on the template DNA during early transcription. We determined whether the limited nucleotides were sufficient for escape commitment to occur. As shown in Fig. 2B, transcription was initiated at either the −1, +1, or +2 position of the AdMLP and the polymerase was paused at different positions on the template DNA depending on which limited nucleotides were added. ctDNA was added either with the limited nucleotides (point 2) or with the remaining nucleotides (point 3). If the limited nucleotides were sufficient for escape commitment to occur, there was a ca. 10-fold increase in the level of transcript produced when ctDNA was added at point 3 compared to point 2, as reflected in the bar plot on the right. We found that irrespective of where transcription initiated and where the polymerase paused on the promoter DNA, escape commitment always occurred when an RNA of 4 nt or longer was made (reactions 2, 4, 5, 7, and 8) and did not occur when a 3-nt RNA was made (reactions 1, 3, and 6). These experiments showed that the length of the RNA transcript determines the point at which escape commitment is complete. Therefore, escape commitment depends on the distance between the active site of the polymerase and the 5′ end of the RNA, not on the distance between the active site and promoter DNA elements such as the TATA box. This indicates that the growing transcript itself participates in a transition that occurs during early transcription.
FIG. 2.
Escape commitment depends on mRNA length and not on the position of the polymerase along the DNA template. (A) The schematic shows the method used to monitor escape commitment and is described in the text. (B) Escape commitment is complete after synthesis of a 4-nt RNA, regardless of where transcription initiates on the template DNA. The sequences of the nontemplate strand of the AdMLP and three mutant versions of the AdMLP are shown in boldface. The +4mt, +5mt, and +6mt promoters alter the position of the first adenosine on the nontemplate strand. At each promoter, transcription pauses at the first adenosine on the nontemplate strand in the absence of ATP. Shown on the left for each reaction are the limited nucleotides added, and the longest products formed before addition of the remaining nucleotides. The remaining nucleotides consisted of ATP, with CTP or UTP as needed to produce a full-length transcript. For each reaction ctDNA (275 μg/ml) was added either at point 2 or at point 3. The 390-nt G-less RNA product was quantitated, and the ratio of product produced when ctDNA was added at point 3 to that produced when ctDNA was added at point 2 was plotted on the graph on the right. If escape commitment occurred, the increase in transcription was ninefold or greater. Each bar represents the average of at least three measurements, and each error bar represents 1 standard deviation.
In interpreting these experiments, we considered the possibility that if readthrough of any of the pause sites occurred, then we could not conclude with certainty that escape commitment depends on the transcript length. Experimental evidence indicates, however, that readthrough does not occur under our conditions. First, with multiple different combinations of nucleotides, as well as with the same nucleotides on different DNA templates, the results were consistent. This indicates that the nucleotide stocks were not contaminated with low levels of other nucleotides and that the polymerase did not misincorporate nucleotides. For example, CTP and UTP must not have been contaminated with ATP because reaction 6, utilizing the +5mt promoter, did not result in escape commitment; however, in reaction 7, the same set of nucleotides was sufficient for escape commitment on the +6mt promoter. Second, escape commitment was complete within 30 s after addition of the limited nucleotides; thus, any readthrough would have had to occur in less than 30 s, which seems unlikely. Third, the sizes of the short products synthesized from the +5mt template were directly monitored (Fig. 1C). In addition, when transcription was initiated with AMP as opposed to ApC, similar results were obtained (data not shown). Together, our data allow us to conclude with certainty that escape commitment depends on the length of the RNA; thus, a 4-nt transcript triggers a transition that occurs during early transcription.
Single-stranded ends of ctDNA directly attack transcription complexes to block escape commitment.
We reasoned that determining the property (or properties) of ctDNA responsible for inhibiting escape commitment would provide insight into the mechanism by which this step occurs. There are at least two models to explain the observation that ctDNA inhibits escape commitment. First, one of the general transcription factors might be released from the promoter during early transcription and would have to rebind for productive transcription to continue. ctDNA could sequester the released general transcription factor and prevent it from rebinding. Second, ctDNA might directly attack ternary complexes that contain all of the general transcription factors and inhibit a transition that occurs during escape commitment. For example, if one of the DNA or RNA binding grooves on RNA polymerase II is exposed, ctDNA could enter this site on the polymerase and block the transition to escape-committed complexes. To begin to distinguish between these possibilities, we tested whether a different nucleic acid inhibitor, poly(dI-dC) · poly(dI-dC), could inhibit escape commitment. Experiments were performed as diagrammed in Fig. 3A. As shown in Fig. 3B, poly(dI-dC), as opposed to ctDNA, was not able to inhibit escape commitment (compare lanes 2 and 3 with lanes 5 and 6). Poly(dI-dC) did, however, inhibit preinitiation complex formation completely when added with the AdMLP prior to addition of the general transcription machinery (Fig. 3B, lane 1). Furthermore, it inhibited each individual transcription factor from binding to the promoter after the others were prebound (data not shown). The observation that poly(dI-dC) cannot inhibit escape commitment but can prevent each protein from binding the promoter argues against protein release and rebinding as the mechanism by which ctDNA inhibits escape commitment. Instead we favor a model in which RNA polymerase II is directly attacked by ctDNA, and a change in ternary complexes during escape commitment renders the polymerase resistant to this attack.
We hypothesized that single-stranded ends generated during the preparation of sonicated ctDNA might be required for inhibition of escape commitment. To test this, we treated ctDNA with Klenow fragment (in the presence of dNTPs) to eliminate single-stranded 5′ and 3′ overhangs. The Klenow fragment-treated ctDNA did not inhibit escape commitment to the same extent as untreated ctDNA (Fig. 3C); hence, single-stranded ends are involved in inhibiting this step.
Identification of oligonucleotides that are potent inhibitors of escape commitment.
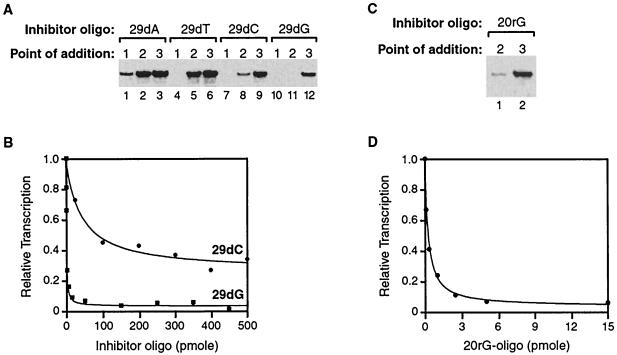
Because the single-stranded ends of ctDNA were required to inhibit escape commitment, we sought to identify one or more single-stranded oligonucleotides that could inhibit escape commitment and mimic the effects of ctDNA. Doing so would provide an essential experimental tool to further probe the mechanism of escape commitment. We tested several oligonucleotides with both random and homopolymeric sequences by adding them to transcription reaction mixtures at points 1, 2, and 3 (see Fig. 3A). Figure 4A shows the effects of four different homopolymeric oligonucleotides 29 residues in length composed of either deoxyadenosines, deoxythymidines, deoxycytidines, or deoxyguanosines. We found that the 29-dC and 29-dG oligonucleotides inhibited escape commitment (Fig. 4A, lanes 8 and 9 and lanes 11 and 12, respectively), while the 29-dA and 29-dT oligonucleotides did not (lanes 2 and 3 and lanes 5 and 6, respectively). Moreover, the 29-dG oligonucleotide is a rather potent inhibitor of escape commitment: 95% inhibition was obtained with only 5 to 10 pmol of this oligonucleotide added to reaction mixtures at point 2 (Fig. 4B). The 50% inhibitory concentration (IC50) for the 29-dG oligonucleotide is 45 nM, while that for the 29-dC oligonucleotide is 6.7 μM.
FIG. 4.
Poly(dG) and poly(G) are potent inhibitors of escape commitment. (A) Single-stranded oligonucleotides consisting of deoxyguanosines and deoxycytidines inhibit escape commitment. The 29-dA, 29-dT, 29-dC, and 29-dG oligonucleotides (34 μM) were added to reaction mixtures at points 1, 2, and 3 (see Fig. 3A). The 390-nt G-less product is shown. (B) The IC50s for the 29-dG and 29-dC oligonucleotides are 45 nM and 6.7 μM, respectively. Increasing amounts of the 29-dG and 29-dC oligonucleotides were added to transcription reaction mixtures at point 2. The 390-nt RNA was quantitated and plotted. (C) Poly(G) inhibits escape commitment. The 20rG oligonucleotide (250 nM) was added to transcription reaction mixtures at points 1 and 2. The 390-nt RNA is shown. (D) The IC50 for the 20rG oligonucleotide is 12 nM. Increasing amounts of the 20rG oligonucleotide were added to transcription reaction mixtures at point 2. The 390-nt RNA was quantitated and plotted.
Because the point at which escape commitment occurs depends on the length of the transcribed RNA (Fig. 2), we reasoned that the inhibitory nucleic acids might bind in the RNA groove of the polymerase. Accordingly, we asked whether an RNA oligonucleotide could inhibit escape commitment. As shown in Fig. 4C and D, an RNA oligonucleotide consisting of 20 guanosines (20rG) was a fourfold more potent inhibitor of escape commitment (IC50, 12 nM) than the 29-dG oligonucleotide. Hence, we identified an exceptional inhibitor of a step during early transcription and used this inhibitor as an experimental tool to probe the mechanism of escape commitment.
The oligonucleotide inhibitors target core RNA polymerase II.
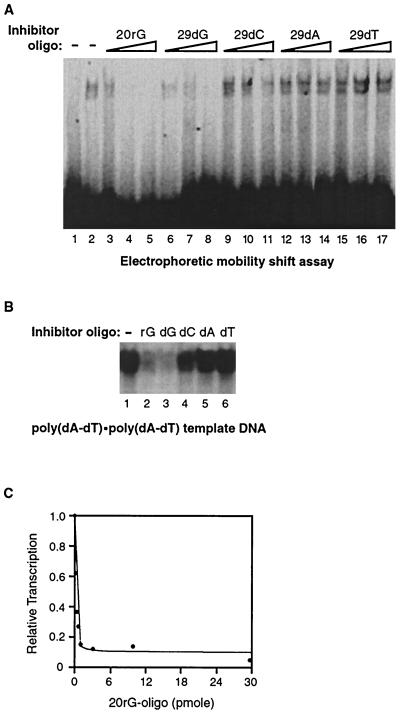
We hypothesized that single-stranded nucleic acids inhibit escape commitment by targeting RNA polymerase II and blocking the transition to escape-committed complexes. If so, the oligonucleotides that inhibit escape commitment should bind directly to RNA polymerase II. To test this, we performed an electrophoretic mobility shift assay (Fig. 5A). The 32P-labeled 20rG oligonucleotide formed a complex with RNA polymerase II (Fig. 5A, lane 2). When additional unlabeled oligonucleotide inhibitors were titrated into reaction mixtures, the complex was efficiently competed away only by the 20rG and 29-dG oligonucleotides. Hence, the oligonucleotides bind to RNA polymerase II with the same sequence specificity with which they inhibit escape commitment. Therefore, the sequence specificity with which the oligonucleotides inhibit transcription is likely dictated by their ability to bind to the polymerase.
FIG. 5.
Single-stranded 20rG and 29-dG oligonucleotides bind RNA polymerase II and inhibit transcription in the absence of other general transcription factors. (A) The 20rG and 29-dG oligonucleotides bind directly to RNA polymerase II. A 32P-labeled 20rG oligonucleotide (0.5 pmol) and, where indicated, unlabeled oligonucleotides (1, 5, and 50 pmol) were incubated with RNA polymerase II (35 ng). The reaction in lane 1 lacked RNA polymerase. (B) The 20rG and 29-dG oligonucleotides inhibit transcription from a poly(dA-dT) · poly(dA-dT) template DNA. RNA polymerase II was added to reaction mixtures containing poly(dA-dT), ATP, UTP, and inhibitor oligonucleotides (250 nM). Transcription proceeded for 20 min. The nonspecific transcript is shown. (C) The 20rG oligonucleotide inhibits promoter-independent transcription with an IC50 of 2.3 nM. Reactions were performed as described above. The nonspecific product was quantitated and plotted as shown.
We next asked whether the oligonucleotide inhibitors could block transcription from a promoter-independent template where general transcription factors are not required. To answer this question we assessed transcription by RNA polymerase II in the absence of accessory factors by using the promoter-independent template poly(dA-dT) · poly(dA-dT). Figure 5B shows that the 20rG and 29-dG oligonucleotides and, to a lesser extent, the 29-dC oligonucleotide inhibited transcription from the poly(dA-dT) template, while the 29-dA and 29-dT oligonucleotides did not. Hence, the sequence specificity observed for binding to RNA polymerase II and for inhibition of promoter-specific transcription from the AdMLP was mimicked in the promoter-independent assays in the absence of general transcription factors. We next titrated the 20rG oligonucleotide into transcription reaction mixtures on the poly(dA-dT) template with RNA polymerase II alone. As shown in Fig. 5C, the 20rG oligonucleotide inhibits promoter-independent transcription with an IC50 of 2.3 nM, even lower than that observed for promoter-specific transcription. This suggests that the mechanism of inhibition of transcription on the poly(dA-dT) template is similar to that on the AdMLP. Therefore, we propose that the conformational change blocked by the oligonucleotide inhibitors is inherent to transcription by core RNA polymerase II.
Poly(G) oligonucleotide inhibitors must be of sufficient length to inhibit escape commitment.
We next asked whether the ability of poly(G) RNA to inhibit escape commitment depends on its length. We titrated an RNA oligonucleotide consisting of 10 guanosine residues (10rG) into transcription reactions prior to escape commitment (point 2 in Fig. 3A). As shown in Fig. 6A, the 10rG oligonucleotide was able to inhibit escape commitment only when added at levels much higher than those observed for the 20rG oligonucleotide (see Fig. 4D). The IC50 for the 10rG oligonucleotide is 0.5 μM, which is at least 40-fold greater than that observed for the 20rG oligonucleotide. Moreover, complete inhibition was never observed with the 10rG oligonucleotide. This finding indicates that inhibitor oligonucleotides must be of a certain length to effectively block escape commitment.
We questioned if the difference in the abilities of the 10rG and 20rG oligonucleotides to inhibit escape commitment reflected their relative affinities for binding RNA polymerase II. To directly test this, we performed electrophoretic mobility shift assays with RNA polymerase II and either the 32P-labeled 10rG oligonucleotide or the 32P-labeled 20rG oligonucleotide. RNA polymerase II bound each oligonucleotide with an approximate Kd of ≤1.5 nM (Fig. 6B). Therefore, the >40-fold decrease in the ability of the 10rG oligonucleotide to inhibit escape commitment is not due to an impaired ability to bind to RNA polymerase II. We also asked whether the 10rG and 20rG oligonucleotides bind RNA polymerase II with similar kinetic stabilities. To test this, we monitored the rates at which the 10rG and 20rG oligonucleotides dissociated once bound to the polymerase. The respective oligonucleotides were prebound to RNA polymerase II; then a 500-fold excess of unlabeled oligonucleotide was added for various times. As shown in Fig. 6C, with both the 10rG and 20rG oligonucleotides, there was little reduction in the amount of 32P-labeled oligonucleotide bound to RNA polymerase II over the course of 1 h. As controls, lanes 2 and 8 show the amount of bound polymerase in the absence of unlabeled oligonucleotide, and lanes 3 and 9 show that the unlabeled oligonucleotides fully compete binding when added prior to RNA polymerase II. These data indicate that both oligonucleotides bind quite stably to RNA polymerase II.
The observation that the 10rG oligonucleotide cannot inhibit escape commitment but can bind tightly and stably to RNA polymerase II indicates that binding and inhibition are separable. This implies that the high-affinity site on the polymerase to which these oligonucleotides bind is removed from the site at which inhibition occurs. Because the inhibition of escape commitment is lost as the length of the oligonucleotide inhibitor is shortened, we favor a model in which the inhibitory oligonucleotides bind a distinct site on RNA polymerase II and then extend through the RNA exit groove toward the active site of the polymerase. The 20rG oligonucleotide reaches far enough through the exit groove to effectively inhibit escape commitment, whereas the 10rG oligonucleotide does not.
The 29-dG oligonucleotide does not block abortive synthesis of 3-nt RNA transcripts.
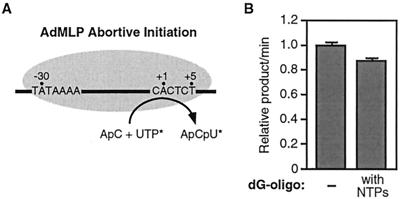
To better understand the mechanism of escape commitment, we needed to determine the exact point in the transcription reaction at which the inhibitors act, and therefore at which escape commitment occurs. We have shown that all ternary complexes after synthesis of a 4-nt RNA were both stable and resistant to ctDNA (Fig. 1C and 2) (19). We demonstrated that initiated complexes producing 3-nt RNAs were unstable (Fig. 1C), but we were not certain whether the inhibitors could target these complexes. We therefore asked whether the 29-dG oligonucleotide inhibited abortive initiation, using a steady-state assay that monitors the synthesis of 3-nt transcripts produced specifically from the start site of the AdMLP (diagrammed in Fig. 7A). As shown in Fig. 7B, there was little difference in the amounts of trinucleotide product made over time in the presence or absence of the inhibitor. We also observed no inhibition by the 20rG oligonucleotide during the production of 3-nt RNA by core RNA polymerase II on poly(dA-dT) templates (data not shown). From these data several important conclusions can be drawn. First, the inhibitors do not block phosphodiester bond synthesis per se. Second, the inhibitors do not block nucleotide binding by the polymerase. Third, the inhibitors act after synthesis of a 3-nt RNA; thus, escape commitment does not begin until after this point.
FIG. 7.
The 29-dG oligonucleotide does not inhibit abortive initiation. (A) Schematic of the abortive-initiation assay, a steady-state assay monitoring production of 3-nt RNAs. (B) Abortive initiation was performed on the AdMLP in the absence and presence of the 29-dG oligonucleotide (250 nM). When present, the 29-dG oligonucleotide was added at point 2 (see Fig. 3A). ApCpU product was quantitated at different time points after addition of nucleotides. Each error bar represents 1 standard deviation.
The 20rG oligonucleotide inhibits translocation of the RNA polymerase II active site after synthesis of the third phosphodiester bond.
The observation that escape commitment occurs after synthesis of a 3-nt RNA but is complete after synthesis of a 4-nt RNA indicates that a unique event in transcription takes place during or immediately after synthesis of the third phosphodiester bond. At least three events must repeatedly occur to add each nucleotide to a growing transcript: (i) binding of the incoming nucleotide in the active site of the polymerase, (ii) synthesis of the phosphodiester bond (together with pyrophosphate release), and (iii) translocation of the active site of the polymerase. In considering the mechanism of escape commitment, we demonstrated that the inhibitors cannot block nucleotide binding or actual phosphodiester bond synthesis (Fig. 2 and 7). Moreover, it seemed unlikely that synthesis of the third phosphodiester bond would be unique or different from synthesis of either of the two previous bonds or of all subsequent bonds. Similarly, it was doubtful that there would be anything distinct about binding of the fourth or fifth nucleotide as opposed to any other. Consequently, we predicted that translocation of the active site of the polymerase after synthesis of the third phosphodiester bond would involve a unique conformational change essential to completing escape commitment. Note that we use the term “translocation” in the manner of Kornberg and colleagues in describing the structure of an RNA polymerase II elongation complex (10); translocation involves movement of the active site of the polymerase with respect to the RNA-DNA hybrid. This translocation event positions the active site of the polymerase such that it is capable of synthesizing the subsequent phosphodiester bond. This is distinct from movement of the front end of the polymerase along the DNA template, and translocation of the active site does not necessarily require forward movement of the entire polymerase (28, 29).
If escape commitment involves translocation after synthesis of the third phosphodiester bond, then a 4-nt RNA and nothing longer would be produced in the presence of the inhibitor. To test this, we determined the sizes of RNA products stably bound in transcribing complexes in the absence and presence of inhibitor. As shown in Fig. 8A, nucleotides sufficient to allow synthesis of a 15-nt RNA were added to preinitiation complexes with or without the 20rG oligonucleotide. Reaction mixtures were then passed through gel filtration spin columns to separate RNA bound in stable complexes from RNA produced abortively and released (22). As shown in Fig. 8B, in the absence of the inhibitor a 15-nt RNA was present in ternary complexes (lane 1). In the presence of the inhibitor, however, only a 4-nt product and nothing longer was observed (lane 2). This clearly demonstrates that the third phosphodiester bond can be synthesized in the presence of the inhibitor but subsequent phosphodiester bonds cannot. In these reactions transcription was initiated with the dinucleotide ApC, which lacks 5′ phosphates. To determine if the presence of a 5′ triphosphate would alter the point at which the inhibitor acted, we performed a similar reaction in which the initiating nucleotide was ATP rather than ApC. Again, only a 4-nt RNA and nothing longer was observed (lane 3). Under these conditions, in the absence of an inhibitor a 390-nt RNA was produced (data not shown). We conclude that the inhibitors block translocation after synthesis of a 4-nt RNA irrespective of the number of phosphates on the 5′ end of the RNA transcript.
Lastly, we wanted to determine whether inhibited ternary complexes containing 4-nt RNAs were still catalytically active. This is important because it would demonstrate that the 20rG oligonucleotide does not cause a conformational change resulting in nonfunctional ternary complexes. Isolated escape-committed complexes cannot proceed forward to lengthen the 4-nt RNA because the 20rG oligonucleotide is still bound, given the slow off-rate (Fig. 6). Therefore, to assess catalytic activity, we asked whether the RNA polymerase II in the inhibited ternary complexes was capable of carrying out pyrophosphorolysis. It had been shown previously that stable RNA polymerase II ternary complexes can undergo pyrophosphorolysis in the presence of pyrophosphate (34). We used spin columns to isolate oligonucleotide-inhibited ternary complexes containing 4-nt RNAs. These were then incubated in the presence or absence of pyrophosphate. As shown in Fig. 8C, in the presence of pyrophosphate the 4-nt RNA disappeared, indicating that these complexes can carry out pyrophosphorolysis. Therefore, we can conclude that in blocking escape commitment the 20rG oligonucleotide does not inhibit catalysis per se.
DISCUSSION
In these studies we investigated the molecular mechanism of escape commitment, a crucial transition that occurs during early transcription. We identified potent oligonucleotide inhibitors of escape commitment that bind to RNA polymerase II with low nanomolar affinity and sequence specificity and that inhibit only this specific step. We demonstrated that escape commitment requires core RNA polymerase II and depends on the length of the RNA transcript rather than the position of the polymerase active site with respect to promoter elements. We found that the binding of poly(G) oligonucleotides to RNA polymerase II is not adequate to inhibit escape commitment; rather, the oligonucleotides must be of sufficient length for inhibition to occur. Lastly, we showed that escape commitment involves translocation of the RNA polymerase II active site between synthesis of the third and fourth phosphodiester bonds. We propose that a conformational change in ternary transcription complexes occurs during translocation after synthesis of a 4-nt RNA to render complexes escape committed.
Model for the molecular mechanism of escape commitment by RNA polymerase II.
The exact point at which the inhibitor oligonucleotides act, as well as where escape commitment occurs with respect to the remainder of the transcription reaction, is shown in Fig. 9. When preinitiation complexes are provided with nucleotides, transcription initiates. Initiation encompasses production of 2- and 3-nt RNA products, which can be abortively synthesized and released in the presence of the oligonucleotide inhibitors. After synthesis of a 4-nt RNA, but prior to translocation of the polymerase active site, the ternary complexes are susceptible to attack by the oligonucleotide inhibitors. Once the polymerase active site translocates to the position required for fourth phosphodiester bond synthesis, ternary complexes are escape committed and resistant to the addition of inhibitors. At this point, stable ternary complexes proceed forward through promoter escape, which occurs during synthesis of the 4th through the 14th phosphodiester bonds. We have previously reported that in the presence of ctDNA, the RNA polymerase II transcription reaction branches prior to completion of escape commitment (19). Here we developed new inhibitors that allowed us to determine that the transcription reaction branches when the polymerase active site attempts to translocate after synthesis of the third phosphodiester bond. We have also characterized the kinetic mechanism of the transcription reaction at the human interleukin-2 promoter. In these studies we found that escape commitment also occurs on the interleukin-2 promoter upon synthesis of a 4-nt RNA (9a). This suggests that escape commitment could be a generalizable step in early transcription by RNA polymerase II.
FIG. 9.
Model depicting escape commitment within the context of the transcription reaction. The oligonucleotide inhibitors block translocation of the polymerase active site immediately after a 4-nt RNA is produced. All other events in transcription are resistant to the inhibitors. A detailed explanation of the model is provided in the text.
The observation that the 20rG and 29-dG oligonucleotides inhibit only one translocation event suggests that there is something unique about translocation of the active site after synthesis of the third phosphodiester bond. We propose that this translocation is accompanied by a conformational change that renders ternary transcription complexes stable and resistant to inhibition. The presence of oligonucleotide inhibitors, likely bound in the RNA groove of the polymerase, prevents this conformational change from occurring. Because the point at which escape commitment occurs depends on the length of the RNA transcript, it is likely that the conformational change intimately involves both the polymerase and the RNA. It is formally possible that the inhibitors drive an unspecified conformational change that prevents the formation of a 5-nt RNA, although we believe our data do not support this model. The observations that inhibited complexes can carry out pyrophosphorolysis and that ternary complex stability occurs in the absence of the inhibitor are not consistent with the inhibitors driving an unfavorable conformational change resulting in inactive complexes.
The results we present here are significant because they demonstrate that translocation of the polymerase active site, and not phosphodiester bond synthesis, results in escape-committed complexes. Escape commitment also occurs independently of where the polymerase active site is located with respect to the promoter DNA. Our data show that RNA polymerase II senses the actual length of the transcript RNA during escape commitment as opposed to the number of phosphodiester bonds it has synthesized. Finally, the point of escape commitment is unaffected by the presence of phosphates on the 5′ ends of the transcript. The proposal that escape commitment is accompanied by a conformational change agrees with previous biochemical studies of RNA polymerase II transcription. We and others have observed an increase in ternary complex stability after the synthesis of a 4-nt RNA (3, 22), and Timmers and colleagues observed a change in the melted region of the DNA at the same juncture in the reaction (13).
Recently, Kornberg and colleagues solved a crystal structure of a yeast RNA polymerase II elongation complex (10). The structure contains the multisubunit RNA polymerase II bound to a tailed DNA template from which the polymerase transcribed a 9-nt RNA. Within the ternary elongation complex, the position of the RNA transcript with respect to the active site of the polymerase reveals that the RNA polymerase has completed phosphodiester bond synthesis, but the active site has not yet translocated with respect to the RNA-DNA hybrid. Additionally, Kornberg and colleagues solved the crystal structure of free RNA polymerase II in the absence of any nucleic acids (7, 8). Comparing the structure of the elongation complex to that of the free polymerase suggests that conformational changes occur during early transcription. For example, a series of switch regions in the polymerase undergo conformational changes and folding transitions that are likely induced by binding to the downstream DNA and/or the DNA-RNA hybrid (10). The 4 nt on the 3′ end of the RNA are buried in the transcribing complex and are in direct contact with the RNA polymerase (10). In considering the structure of the elongation complex, it was predicted that once translocation after production of a 4-nt RNA occurred, the 5′ end of the RNA would become exposed for the first time. At this point one additional long-range interaction between R497 of Rpb2 and the 5′ end of the RNA would take place (10). This is the exact point in the transcription reaction at which escape commitment occurs and stable ternary complexes form, and at which we predict a conformational change happens. The crystal structures suggest that changes in the polymerase might occur at this juncture; however, a detailed understanding of early transcription and how the conformations of ternary complexes change will require further biochemical studies and additional structural studies of ternary complexes trapped during early transcript synthesis.
A crystal structure by Cheetham and Steitz of a transcribing bacteriophage T7 RNA polymerase provided the first structural view of an initiated RNA polymerase and interestingly indicated that a transition at +4 occurs (5, 6). The conformations of the single-stranded promoter DNA, 3 nt of transcribed RNA in a heteroduplex with the DNA, and an incoming ribonucleoside triphosphate analog are clearly shown within the active-site pocket of the T7 RNA polymerase structure. The authors predict that extending the RNA-DNA duplex by even one additional base pair (i.e., to a total of 5 bp) would result in steric clashes between the 5′ end of the RNA and the N-terminal domain of the polymerase. Thus, it appears that a critical conformational change during early transcription must also occur at register +4 in bacteriophage T7 RNA polymerase for transcription to continue. Several studies of early transcription by E. coli RNA polymerase have also observed distinct transitions after synthesis of a 4-nt RNA (2, 9, 31). This juncture is the precise point in the eukaryotic RNA polymerase II transcription reaction where we found escape commitment to occur. Determining whether these transitions are related to escape commitment by RNA polymerase II will require future studies.
Mechanism by which inhibitor oligonucleotides block escape commitment.
We identified two oligonucleotides, the 20rG oligonucleotide and the 29-dG oligonucleotide, that potently inhibit escape commitment by binding directly to RNA polymerase II. It is not yet clear why a strong preference for guanosine residues exists. We predict that the 20rG and 29-dG oligonucleotides bind to a distinct site on the polymerase and extend through the RNA groove that exits the active-site pocket of the polymerase. This is based on (i) the dependence on RNA transcript length for completing escape commitment (Fig. 2), (ii) the potency with which the RNA oligonucleotide inhibits (Fig. 4), and (iii) the observation that inhibitor oligonucleotides must be of sufficient length to block escape commitment (Fig. 6). Our findings show that 10 guanosine residues are sufficient for high-affinity binding to the polymerase but that to block escape commitment the oligonucleotide must be of sufficient length to extend into the region of the ternary complex at which inhibition occurs (perhaps by channeling through the RNA exit groove). The inhibitor could then block a conformational change during escape commitment, thereby preventing productive transcription from continuing. We know that the conformation of the inhibited complex is quite stable, because it retains the 4-nt RNA for long periods of time and survives passage through a spin column. We also know that the inhibited complex is still active, because it is able to carry out pyrophosphorolysis. We are not yet certain, however, whether these inhibited complexes could recover and begin productive transcription again if the inhibitor were removed. Johnson and Chamberlin previously observed that RNA oligonucleotides could bind to yeast RNA polymerase II in a manner that mimicked a ternary complex (15). These interactions did not inhibit transcription, did not exhibit sequence specificity, and did not exhibit low nanomolar Kds; thus, they are likely distinct from the interactions reported here. Chamberlin and colleagues also found that E. coli RNA polymerase has two RNA binding sites that are thought to bind the elongating transcript (1, 27). If analogous sites exist on RNA polymerase II, one or both could be responsible for binding the oligonucleotide inhibitors.
The oligonucleotide inhibitors we described here inhibit a specific translocation event during early transcription. At a glance, this appears similar to the mechanism by which the antibiotic rifampin (RIF) inhibits transcription by bacterial RNA polymerases; however, the inhibition we observe is quite different from that observed for RIF. Most significantly, the step blocked by the inhibitors characterized here (i.e., escape commitment) is a critical transition that occurs naturally during early transcription by RNA polymerase II. Escape commitment results in stable ternary complexes and is a step that can be controlled by the helicase activity of TFIIH (3, 13, 19, 22). In contrast, the point at which RIF blocks transcription from continuing, as described in more detail below, is not considered to be a unique transition during early transcript synthesis by bacterial RNA polymerases. RIF blocks translocation during early transcription at a point dependent on the nature of the phosphate group(s) on the 5′ end of the transcript RNA. Transcripts initiated with NTPs are blocked after formation of a 2-nt RNA, whereas transcripts initiated with dinucleotides or nucleoside di- or monophosphates are blocked after synthesis of a 3-nt RNA (4, 26). Our data, however, demonstrate that inhibition of escape commitment by oligonucleotides does not depend on the number of phosphates on the 5′ end of the RNA transcript (Fig. 8). Moreover, when RIF blocks translocation during early transcription by bacterial RNA polymerases, the short RNA products that form are produced and released. In contrast, when the 20rG oligonucleotide inhibits escape commitment, the 4-nt RNA remains stably bound in ternary complexes. Finally, inhibition of escape commitment depends on the length of the single-stranded oligonucleotide inhibitor (Fig. 6). This supports a model in which the inhibitor oligonucleotides bind to RNA polymerase II at a site removed from the region at which inhibition occurs, as opposed to binding a RIF-like pocket near the region of inhibition. Although RIF and the oligonucleotide inhibitors both block translocation events, the mechanisms by which they do so differ.
Biochemical and structural data from several different RNA polymerases indicate that critical conformational changes likely occur during early transcription in order to facilitate the transition from an unstable initiation complex to a stable elongation complex. Our data suggest that one such change occurs during escape commitment as the RNA polymerase II active site translocates after synthesis of the third phosphodiester bond.
Acknowledgments
We thank Ray Fall, Heather Ferguson, Rob Kuchta, Tin Tin Su, and Olke Uhlenbeck for helpful discussions.
This research was supported by Research Project Grant RPG-00-271-01-MGO from the American Cancer Society. J.A.G. is a Pew Scholar in the Biomedical Sciences.
REFERENCES
- 1.Altmann, C. R., D. E. Solow-Cordero, and M. J. Chamberlin. 1994. RNA cleavage and chain elongation by Escherichia coli DNA-dependent RNA polymerase in a binary enzyme · RNA complex. Proc. Natl. Acad. Sci. USA 91:3784–3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bowser, C., and M. Hanna. 1991. Sigma subunit of Escherichia coli RNA polymerase loses contacts with the 3′ end of the nascent RNA after synthesis of a tetranucleotide. J. Mol. Biol. 220:227–239. [DOI] [PubMed] [Google Scholar]
- 3.Cai, H., and D. S. Luse. 1987. Transcription initiation by RNA polymerase II in vitro: properties of preinitiation, initiation, and elongation complexes. J. Biol. Chem. 262:298–304. [PubMed] [Google Scholar]
- 4.Campbell, E. A., N. Korzheva, A. Mustaev, K. Murakami, S. Nair, A. Goldfarb, and S. A. Darst. 2001. Structural mechanism for rifampicin inhibition of bacterial RNA polymerase. Cell 104:901–912. [DOI] [PubMed] [Google Scholar]
- 5.Cheetham, G. M., and T. A. Steitz. 2000. Insights into transcription: structure and function of single-subunit DNA-dependent RNA polymerases. Curr. Opin. Struct. Biol. 10:117–123. [DOI] [PubMed] [Google Scholar]
- 6.Cheetham, G. M. T., and T. A. Steitz. 1999. Structure of a transcribing T7 RNA polymerase initiation complex. Science 286:2305–2309. [DOI] [PubMed] [Google Scholar]
- 7.Cramer, P., D. Bushnell, J. Fu, A. Gnatt, B. Maier-Davis, N. Thompson, R. Burgess, A. Edwards, P. David, and R. Kornberg. 2000. Architecture of RNA polymerase II and implications for the transcription mechanism. Science 288:640–649. [DOI] [PubMed] [Google Scholar]
- 8.Cramer, P., D. A. Bushnell, and R. D. Kornberg. 2001. Structural basis of transcription: RNA polymerase II at 2.8 Å resolution. Science 292:1863–1876. [DOI] [PubMed] [Google Scholar]
- 9.DeRiemer, L. H., and C. F. Meares. 1981. Early steps in the path of nascent ribonucleic acid across the surface of ribonucleic acid polymerase, determined by photoaffinity labeling. Biochemistry 20:1612–1617. [DOI] [PubMed] [Google Scholar]
- 9a.Ferguson, H. A., J. F. Kugel, and J. A. Goodrich. J. Mol. Biol., in press. [DOI] [PubMed]
- 10.Gnatt, A. L., P. Cramer, J. Fu, D. A. Bushnell, and R. D. Kornberg. 2001. Structural basis of transcription: an RNA polymerase II elongation complex at 3.3 Å resolution. Science 292:1876–1882. [DOI] [PubMed] [Google Scholar]
- 11.Goodrich, J. A., and R. Tjian. 1994. Transcription factors IIE and IIH and ATP hydrolysis direct promoter clearance by RNA polymerase II. Cell 77:145–156. [DOI] [PubMed] [Google Scholar]
- 12.Gralla, J. D., A. J. Carpousis, and J. E. Stefano. 1980. Productive and abortive initiation of transcription in vitro at the lac UV5 promoter. Biochemistry 19:5864–5869. [DOI] [PubMed] [Google Scholar]
- 13.Holstege, F. C. P., U. Fiedler, and H. T. M. Timmers. 1997. Three transitions in the RNA polymerase II transcription complex during initiation. EMBO J. 16:7468–7480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang, J., and R. Sousa. 2000. T7 RNA polymerase elongation complex structure and movement. J. Mol. Biol. 303:347–358. [DOI] [PubMed] [Google Scholar]
- 15.Johnson, T. L., and M. J. Chamberlin. 1994. Complexes of yeast RNA polymerase II and RNA are substrates for TFIIS-induced RNA cleavage. Cell 77:217–224. [DOI] [PubMed] [Google Scholar]
- 16.Kireeva, M. L., N. Komissarova, D. S. Waugh, and M. Kashlev. 2000. The 8-nucleotide-long RNA:DNA hybrid is a primary stability determinant of the RNA polymerase II elongation complex. J. Biol. Chem. 275:6530–6536. [DOI] [PubMed] [Google Scholar]
- 17.Korzheva, N., A. Mustaev, E. Nudler, V. Nikiforov, and A. Goldfarb. 1998. Mechanistic model of the elongation complex of Escherichia coli RNA polymerase. Cold Spring Harbor Symp. Quant. Biol. 63:337–345. [DOI] [PubMed] [Google Scholar]
- 18.Krummel, B., and M. J. Chamberlin. 1989. RNA chain initiation by Escherichia coli RNA polymerase: structural transitions of the enzyme in early ternary complexes. Biochemistry 28:7829–7842. [DOI] [PubMed] [Google Scholar]
- 19.Kugel, J. F., and J. A. Goodrich. 2000. A kinetic model for the early steps of RNA synthesis by human RNA polymerase II. J. Biol. Chem. 275:40483–40491. [DOI] [PubMed] [Google Scholar]
- 20.Kugel, J. F., and J. A. Goodrich. 1998. Promoter escape limits the rate of transcription from the adenovirus major late promoter on negatively supercoiled templates. Proc. Natl. Acad. Sci. USA 95:9232–9237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Luse, D. S., and G. A. Jacob. 1987. Abortive initiation by RNA polymerase II in vitro at the adenovirus major late promoter. J. Biol. Chem. 262:14990–14997. [PubMed] [Google Scholar]
- 22.Luse, D. S., T. Kochel, E. D. Kuempel, J. A. Coppola, and H. Cai. 1987. Transcription initiation by RNA polymerase II in vitro. At least two nucleotides must be added to form a stable ternary complex. J. Biol. Chem. 262:289–297. [PubMed] [Google Scholar]
- 23.McAllister, W. T. 1993. Structure and function of the bacteriophage T7 RNA polymerase (or, the virtues of simplicity). Cell. Mol. Biol. Res. 39:385–391. [PubMed] [Google Scholar]
- 24.McClure, W. R. 1985. Mechanism and control of transcription initiation in prokaryotes. Annu. Rev. Biochem. 54:171–204. [DOI] [PubMed] [Google Scholar]
- 25.McClure, W. R. 1980. Rate-limiting steps in RNA chain initiation. Proc. Natl. Acad. Sci. USA 77:5634–5638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McClure, W. R., and C. L. Cech. 1978. On the mechanism of rifampicin inhibition of RNA synthesis. J. Biol. Chem. 253:8949–8956. [PubMed] [Google Scholar]
- 27.Milan, S., L. D’Ari, and M. J. Chamberlin. 1999. Structural analysis of ternary complexes of Escherichia coli RNA polymerase: ribonuclease footprinting of the nascent RNA in complexes. Biochemistry 38:218–225. [DOI] [PubMed] [Google Scholar]
- 28.Nudler, E., A. Goldfarb, and M. Kashlev. 1994. Discontinuous mechanism of transcription elongation. Science 265:793–796. [DOI] [PubMed] [Google Scholar]
- 29.Nudler, E., A. Mustaev, E. Lukhtanov, and A. Goldfarb. 1997. The RNA-DNA hybrid maintains the register of transcription by preventing backtracking of RNA polymerase. Cell 89:33–41. [DOI] [PubMed] [Google Scholar]
- 30.Orphanides, G., T. Lagrange, and D. Reinberg. 1996. The general transcription factors of RNA polymerase II. Genes Dev. 10:2657–2683. [DOI] [PubMed] [Google Scholar]
- 31.Ruetsch, N., and D. Dennis. 1987. RNA polymerase. Limit cognate primer for initiation and stable ternary complex formation. J. Biol. Chem. 262:1674–1679. [PubMed] [Google Scholar]
- 32.Sousa, R. 1996. Structural and mechanistic relationships between nucleic acid polymerases. Trends Biochem. Sci. 21:186–190. [PubMed] [Google Scholar]
- 33.Temiakov, D., P. E. Mentesana, K. Ma, A. Mustaev, S. Borukhov, and W. T. McAllister. 2000. The specificity loop of T7 RNA polymerase interacts first with the promoter and then with the elongating transcript, suggesting a mechanism for promoter clearance. Proc. Natl. Acad. Sci. USA 97:14109–14114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang, D., and D. K. Hawley. 1993. Identification of a 3′→5′ exonuclease activity associated with human RNA polymerase II. Proc. Natl. Acad. Sci. USA 90:843–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zawel, L., K. P. Kumar, and D. Reinberg. 1995. Recycling of the general transcription factors during RNA polymerase II transcription. Genes Dev. 9:1479–1490. [DOI] [PubMed] [Google Scholar]