Abstract
Activation of RAS proteins can lead to multiple outcomes by virtue of regulated signal traffic through alternate effector pathways. We demonstrate that the RAS effector protein RIN1 binds to activated RAS with an affinity (Kd, 22 nM) similar to that observed for RAF1. At concentrations close to their equilibrium dissociation constant values, RIN1 and RAF1 compete directly for RAS binding. RIN1 was also observed to inhibit cellular transformation by activated mutant RAS. This distinguishes RIN1 from other RAS effectors, which are transformation enhancing. Blockade of transformation was mediated by the RAS binding domain but required membrane localization. RIN1 recognizes endogenous RAS following transient activation by epidermal growth factor, and a portion of RIN1 fractionates to the cell membrane in a manner consistent with a reversible interaction. RIN1 also binds to 14-3-3 proteins through a sequence including serine 351. Mutation of this residue abolished the 14-3-3 binding capacity of RIN1 and led to more efficient blockade of RAS-mediated transformation. The mutant protein, RIN1S351A, showed a shift in localization to the plasma membrane. Serine 351 is a substrate for protein kinase D (PKD [also known as PKCμ]) in vitro and in vivo. These data suggest that the normal localization and function of RIN1, as well as its ability to compete with RAF, are regulated in part by 14-3-3 binding, which in turn is controlled by PKD phosphorylation.
Genetic and biochemical studies have demonstrated that RAS plays a pivotal role in the transduction of external signals that activate a variety of cellular processes, including proliferation, differentiation, and metabolism (63). RAS accomplishes its diversity of functions through a variety of mechanisms. These include expression of different RAS gene products (H-, K-, and N-RAS) in cell type and developmentally restricted manners (24, 30, 34, 44). In addition, RAS responds to regulatory factors that promote activation (guanine nucleotide exchange factors [49]) or stimulate the return to an inactive state (GTPase activating proteins [2]). The critical step in determining cell response is the physical interaction with downstream RAS interaction partners (effectors) that function to accept the activation message and dispatch it appropriately. Differences in availability (e.g., restricted expression and subcellular sequestration) and biochemical properties (e.g., binding affinities) of these effector proteins lead to signaling specificity. RAF proteins are the best characterized of the RAS effectors. The interaction of RAS(GTP) with RAF1 activates this proximal kinase of a MAP kinase cascade (reviewed in reference 31). Other RAS effectors that have been identified include PI3 kinase (58), RGF (also known as RalGDS) (reviewed in reference 10), RIN1 (16), AF6 (33, 71) and Nore1 (73).
RAS effectors do not share extensive primary sequence identity. They do, however, show significant similarities in their RAS binding structures (12, 46, 50). This similarity is reflected in the shared biochemical features of effector binding to RAS. These interactions are characterized by a strong preference for the GTP-dependent conformation of RAS. In addition, each effector so far identified interacts, at least in part, directly through a short amino acid region (effector domain) within RAS. Severe mutations within this sequence block all effector binding, while some single site alterations appear to block only selective effector interactions (27, 59).
RIN1 binds directly to RAS, as demonstrated by both in vitro and cell extract coimmunoprecipitation experiments (16, 17). RIN1 binding shows the RAS effector domain requirement and GTP dependency common among effectors. The RAS binding domain (RBD) of RIN1 is localized in the carboxyl-terminal region that has been shown to also interact with 14-3-3 proteins (17). Because 14-3-3 binding sites include an obligate phosphoserine residue, this suggests a possible mechanism for regulated RAS interactions. In addition, the amino terminus of RIN1 encodes a domain that promotes interaction with, and phosphorylation by, the ABL tyrosine kinase (1, 17) as well as the closely related protein ARG (H. Hu and J. Colicelli, unpublished data). RIN1 also enhances the transforming properties of BCR-ABL in vitro and in vivo (1).
The work reported here characterizes the high-affinity binding of RIN1 to RAS and demonstrates that this binding is competitive with that of RAF1 in vitro and in vivo. Unlike RAF and other RAS effectors, however, RIN1 is antagonistic to transformation. These findings are consistent with a dynamic signal flux that is coordinated through RAS proteins and modulated by the interplay among multiple RAS effectors. In addition, we present evidence that 14-3-3 proteins act as negative regulators of RIN1 membrane localization and RAS association. We also demonstrate that the critical serine of the 14-3-3 binding site in RIN1 is a substrate for protein kinase D (PKD [also known as PKCμ], providing another level of control for this pathway.
MATERIALS AND METHODS
Cell culture, transfection, and infection.
Mammalian cells were cultured in Dulbecco’s modified Eagle’s medium (Gibco BRL) supplemented with 10% fetal bovine serum (HyClone), penicillin (30 μg/ml), and streptomycin (60 μg/ml). For transfection and retrovirus production, helper-free retrovirus was produced by transient cotransfection of 293T cells (51) with retroviral vectors and an ecotropic packaging vector (43) using calcium phosphate. Supernatants from 293T cells were collected 24 to 48 h posttransfection. To generate stable transfectants, Rat1 and NIH 3T3 cells were infected with virus stocks that had been normalized to give equivalent protein expression for individual constructs. Forty-eight hours after infection, the cells were trypsinized and placed in medium containing 600 μg of G418 per ml to select for virus-infected cells for 2 weeks. Protein expression levels were assayed by immunoblotting. H-RASQ61L cells were generously provided by Adrienne Cox, University of North Carolina. These cells were grown in the absence of G418 selection prior to soft-agar colony assays. Lipofectin-mediated transfections of COS-7 cells were carried out as previously described (76).
For endogenous RAS activation assays, NIH 3T3 cells were serum starved for 24 h, stimulated with epidermal growth factor (EGF) (50 ng/ml) for various lengths of time, and then washed with cold phosphate-buffered saline (PBS). Cell extracts were prepared with lysis-immunoprecipitation (IP) buffer (77), incubated with immobilized RBD(His6) for 1 h at 4°C and washed four times with the same buffer. Bound material was released with sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer and analyzed by immunoblotting.
Protein purification.
RIN1(His6), RBD(His6), and RIN1S351A(His6) baculovirus constructs were used to infect Sf21 cells (infectivity ratio = 10) cultured in Grace’s medium supplemented with 10% fetal bovine serum (HyClone), penicillin, and streptomycin at 27°C. After 48 h, extracts were prepared by sonication of cells in lysis buffer (20 mM HEPES [pH 7.2], 2 mM MgCl2, 1 mM dithiothreitol [DTT], 160 mM NaCl, 0.1% Triton X-100, 5 mM imidazole) with a protease inhibitor cocktail and cleared at 16,000 × g in a microcentrifuge for 30 min at 4°C. Proteins were purified using Talon metal affinity resin (Clontech) under nondenaturing conditions, released with 150 mM imidazole, and dialyzed.
The RAS binding domain of RAF1 (RAF1[54–131]) was expressed as an MBP fusion, and RASG12V was expressed as a glutathione S-transferase (GST) fusion (74). Both were generously provided by A. Vojtek and J. Cooper. Proteins were expressed in Escherichia coli BL21 cells. Cultures (volume, 400 ml) were induced with 0.3 mM isopropyl-β-D-thiogalactopyranoside and disrupted by sonication in 10 ml of PBS plus 1% (vol/vol) Triton X-100 and a protease inhibitor cocktail (phenylmethylsulfonyl fluoride [0.1 mM], leupeptin [2 μg/ml], pepstatin [1 μg/ml]). Purifications of GST-RAS and MBP-RAF1[54–131] were performed using glutathione and maltose resins, respectively. Protein concentrations were quantified using Bradford and Lowry assays, and purity was determined by SDS-PAGE.
Surface plasmon resonance analysis.
The GST-H-RASG12V fusion protein was purified from bacteria, and guanine nucleotides (Sigma) were loaded onto the protein as previously described (77). RBD(His6) was purified from Sf21 insect cells using Talon affinity resin (Clontech). The BiaCore 2000 (Phamacia Biosensor, Uppsala, Sweden) was used to analyze the interaction between RIN1 and RAS in real time. Monoclonal anti-GST (BiaCore AB, Uppsala, Sweden) was first coupled to carboxy-methylated dextran on a CM5 sensor chip using standard 1-ethyl-3 (3 dimethylaminopropyl) carbodiimide-N-hydroxysuccinimide coupling chemistry (26). A 10-μl volume of GST (10 μg/ml) or GST-RASG12V (5 μg/ml) preloaded with either GDP or GTPγS was immobilized on the anti-GST-CM5 chip at a flow rate of 5 μl/min at 9°C. This yielded responsive units as follows: for GST, 400; for GST-RASG12V(GDP), 312; and for GST-RASG12V(GTPγS), 250. For kinetic measurements, RBD(His6) (32 to 255 nM) was injected at a flow rate of 15 μl/min. The binding surface was regenerated with 750 mM NaCl with no decrease in binding capacity, and all measurements were completed within 8 h. Data were collected after 10-min delays, and the Kd values were determined using BIA evaluation software (version 3.0).
Cell fractionation and subcellular localization.
Approximately 108 HBL100 cells were washed three times with ice-cold PBS containing phenylmethylsulfonyl fluoride (0.1 mM), pelleted at 1,000 × g for 10 min, resuspended in ice-cold hypotonic buffer (10 mM Tris-HCl [pH 7.4], 10 mM KCl, 1.5 mM MgCl2, and protease inhibitor cocktail) and incubated on ice for 45 min. The cells were then disrupted with a Dounce homogenizer, and the cell lysate was centrifuged at 4,000 × g for 30 min at 4°C. The supernatant was centrifuged at 100,000 × g for 1 h. The resulting pellet was resuspended in 5 mM Tris-HCl (pH 8.5) containing 8.6% sucrose; loaded onto a discontinuous sucrose gradient consisting of 16, 31, 45, and 60% sucrose in 5 mM Tris-HCl (pH 7.4); and then centrifuged for 4 h at 100,000 × g (57). Interface fractions were collected and diluted with 5 ml of 5 mM Tris-HCl. These samples were centrifuged at 100,000 × g for another hour. The protein from each fraction was quantified by Bradford assay, and equal amounts of protein from pellets and soluble fractions were subjected to SDS-PAGE. The results were further analyzed by immunoblotting. Pellet material from the 16/31 interface was treated with 1% SDS, 1.5 M NaCl, 6 M urea, or 10 mM EDTA at 4°C for 1 h and spun in a microcentrifuge for 30 min. The proteins from both supernatant and pellet were separated by SDS-PAGE, and the results were analyzed by immunoblotting.
Immunofluorescence.
NIH 3T3 or NIH 3T3 RASQ61L cells were infected with MSCV-RIN1 (wild type or mutant) and 2 days later were plated on coverslips coated with 0.01% poly-lysine−0.1% gelatin (50:50). After 24 h, the cells were fixed in PBS containing 3% paraformaldehyde for 15 min and then permeabilized with 0.2% TritonX-100 for 5 min. The cells were incubated with rabbit anti-RIN1 (Transduction Laboratories) for 1 h. After extensive washing, the cells were incubated with a 1:400 dilution of Cy3-conjugated affinity-purified sheep anti-rabbit immunoglobulin G (Sigma) for 1 h, washed and mounted onto slides with mounting solution (10 mM Tris-HCl [pH.7.4], 2% DABCO, 90% glycerol). A Leica TCS-SP microscope and 40× objective lens were used. The images were analyzed with Leica confocal software.
Phosphorylation and kinase assays.
For in vitro PKD assays, COS-7 cells were transfected with vector pcDNA3, an expression construct encoding wild-type PKD (pcDNA3-PKD), or a kinase-inactive mutant (pcDNA3-PKD-K618N) (23, 72, 82). After 72 h, indicated cultures were stimulated with phorbol 12,13 dibutyrate (PDB) for 10 min. Cells were lysed in buffer (1% Triton X-100, 2 mM EDTA, 2 mM EGTA, 2 mM DTT, 1 mM AEBSF [4-(2-aminoethyl)-benzenesulfonyl fluoride], aprotinin [100 μg/ml], and leupeptin [10 μg/ml] in 50 mM Tris-HCl, pH 7.4) and clarified by centrifugation at 16,000 × g at 4°C for 10 min. PKD was immunoprecipitated for 3 h at 4°C using PA-1 antiserum (72) together with protein A agarose. PKD immunocomplexes were washed twice with lysis buffer and then twice with kinase buffer (10 mM MgCl2 and 2 mM DTT in 30 mM Tris-HCl, pH 7.4). This material was incubated in kinase buffer with [γ-32P]ATP (5 μCi/reaction; final ATP concentration, 100 μM) at 30°C for 25 min in the presence of purified RIN1 protein (0.1 μg), RIN1S351A protein (0.1 μg), or elution buffer. The reactions were terminated by addition of sample buffer and analyzed by SDS-PAGE. The gels were dried and subjected to autoradiography.
For in vivo PKD assays, COS-7 cells were transfected with RIN1 or RIN1S351A, either alone or with PKD. After 72 h, the growth medium was replaced with fresh Dulbecco’s modified Eagle’s medium lacking phosphate and incubation was continued for 30 min. The medium was then replaced with 5 ml of the same medium containing 100 μCi of carrier-free 32PO4i per ml, and the cells were metabolically labeled at 37°C for 5 h. In the final 10 min, selected cultures (as indicated below) were stimulated with PDB (200 nM). The labeling medium was removed, and the cells were rinsed with cold PBS and then lysed as described above. RIN1 was immunoprecipitated using polyclonal anti-RIN1 (Transduction Laboratories) and protein A agarose. The immune complexes were analyzed by SDS-PAGE (8% acrylamide), with dried gels subjected to autoradiography.
In vitro kinase assays were also performed for JNK2 (UBI), p38 (UBI), and ERK2 (Uppsala Biotechnology, Inc.) using supplier-provided buffers.
RIN1-RAS pulldowns and RIN1/RAF competition assays.
For RIN1-RAS binding and RAF1 competition assays, 200 ng of GST-RASG12V(GTPγS) was immobilized on the glutathione beads and incubated at 4°C for 30 min. The beads were washed with 10 column volumes of binding buffer 2 (77), and mixtures of 50 nM RIN1(His6) with different concentrations of MBP-RAF were further incubated with GST-RAS-bound beads for 1 h at 4°C. The beads were then washed three times with washing buffer (77). The bound proteins were eluted with SDS-PAGE sample buffer and analyzed by immunoblotting.
For RAS pull-down experiments, RIN1(His6) proteins were expressed in Sf21 cells and immobilized onto Talon resin (Clontech). RASG12V wild-type and effector mutants (provided by Michael White, University of Texas Southwest Medical Center) were expressed in 293T cells. The cells were then sonicated in lysis/IP buffer (77). The clear lysates were incubated with RIN1-Talon resin for 1 h at 4°C. Protein-bound resin was further washed with lysis/IP buffer. Bound RAS was assayed by immunoblot with monoclonal anti-RAS (Transduction Laboratories).
Plasmid construction and two-hybrid assays.
Full-length RIN1 was cloned into pQE60 (Qiagen) as described previously (77). The 14-3-3 binding site mutation was engineered in this plasmid by using double-strand site-directed mutagenesis (Clontech). The primer used for Ser351Ala mutagenesis, 5′CTGCTTCGGTCCATGGCCGCCTTCTGCTC, also introduced an NcoI restriction site (underlined). A primer that eliminated the vector XmnI site was used to select for mutant plasmids. The EcoRI-BglII fragment from QE60-RIN1 (wild type or mutant) was ligated into EcoRI- and SalI-digested pBTM117 (16) in the presence of adapter oligonucleotides (BglII-EcoRI-SalI) to create LexA DNA binding domain fusions. RIN1 sequences were subcloned as EcoRI fragments into pMSCV (to generate retrovirus) and pcDNA3. pGAD425-14-3-3e constructs have been described previously (17). Two-hybrid assays were performed with yeast strain L40 (22) transformed with pBTM117-RIN1 and pGAD425-14-3-3e. Cells were selected on synthetic medium lacking tryptophan (pBTM117 marker), leucine (pGAD425 marker), and histidine (two-hybrid reporter).
RESULTS
RIN1 binds to RAS with high affinity.
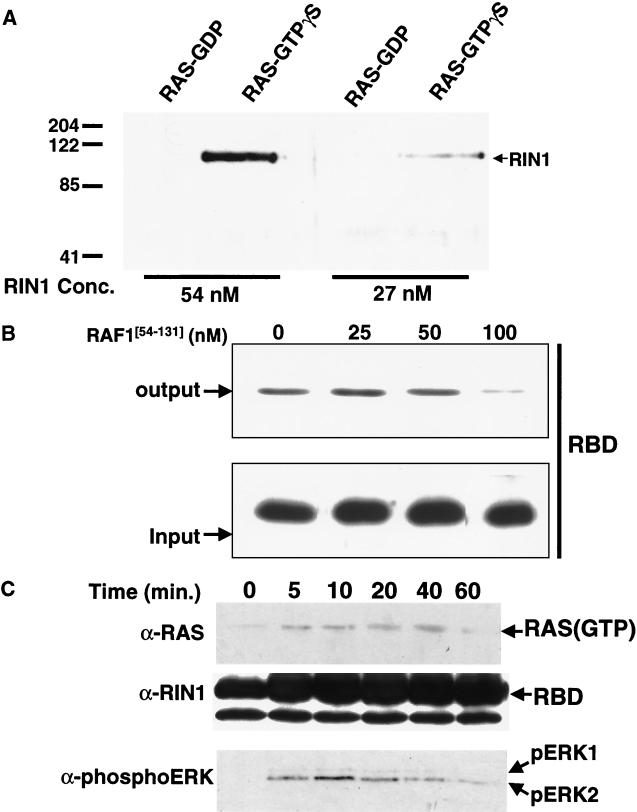
The affinity and specificity of RIN1 for RAS was examined using in vitro binding assays. Immobilized GST-H-RAS was loaded with either GTPγS or GDP and incubated with increasing concentrations of RIN1(His6). The bound protein was analyzed by immunoblotting. The results demonstrated that RIN1 bound preferentially to RAS loaded with the nonhydrolyzable GTP analog (Fig. 1A). The complex with RAS(GTPγS) was detectable at RIN1 concentrations as low as 27 nM, and the binding was concentration dependent. To confirm the specific association of RIN1 with RAS, we performed a binding assay in the presence of the RBD of RAF1 (74) at increasing concentrations as a competitor (Fig. 1B). We observed that RAF1 at concentrations in the range of 50 to 100 nM effectively blocked RIN1 (present at 50 nM) from binding to RAS. This result reflects the comparable high affinity binding of these two effectors.
FIG. 1.
RIN1 binds active RAS and competes with RAF1. (A) Binding of RIN1 to GST-RAS. Immobilized GST-RAS was loaded with guanine nucleotide (GDP or GTPγS) and incubated with RIN1(His6) (27 or 54 nM). The purified protein complex was analyzed by immunoblotting with anti-RIN1. The numbers at the left indicate molecular mass markers in kilodaltons. Conc., concentration. (B) Confirmation of specific association of RIN1 with RAS. Immobilized GST-RAS(GTPγS) was incubated with 50 nM RIN1-RBD and the indicated concentration of RAF1[54–131]. Bound RIN1-RBD was determined by immunoblotting. (C) Binding of RIN1 to RAS activated by EGF. Serum-starved NIH 3T3 cells were stimulated with EGF (time after stimulation is indicated in minutes [min.]), and endogenous RAS was pulled down by immobilized RBD(His6). The bound protein was examined by immunoblotting with anti-RAS. The activation of ERK1 and ERK2 was assessed using phosphospecific antibody. The total quantity of immobilized RBD(His6) for the zero time point was less than that for the other time points but was still in excess of the quantity of endogenous RAS protein in the cell extracts. Data gathered at the 60-min (signal attenuation) time point demonstrate that the concentration of activated RAS determines binding.
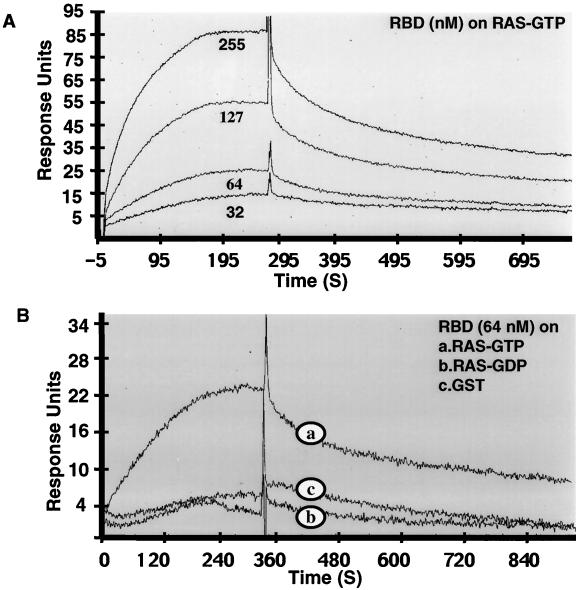
To directly measure the affinity of RIN1 for RAS, we performed surface plasmon resonance analysis. Recombinant GST, GST-RAS(GDP), and GST-RAS(GTPγS) proteins were loaded on sensor chips, and RIN1-RBD binding was examined. RBD formed complexes with RAS(GTPγS) selectively and in a concentration-dependent manner. In contrast, only background levels of association were seen with GST or GST-RAS(GDP) (Fig. 2). Kinetic measurements were used to calculate an equilibrium dissociation constant (Kd) of approximately 22 nM for RIN1-RBD-RAS binding.
FIG. 2.
RIN1 binds to RAS(GTP) with high affinity and specificity. (A) Affinity. The GST-RAS(GTPγS) fusion protein was immobilized onto a BiaCore CM5 sensor chip, and the surface plasmon resonance at increasing concentrations of RIN1-RBD was determined. (B) Specificity, as indicated by the interaction of RIN1-RBD with immobilized GST, GST-RAS(GTPγS), or GST-RAS(GDP). Response units are plotted as a function of time (in seconds [S]).
Mutations within the effector domain of H-RAS can influence binding to downstream effectors differentially (78), revealing that distinct effector pathways control different cellular processes and cell fates (25, 41, 55, 59, 67). We examined the ability of RIN1 to bind to RAS effector mutants using a pulldown protocol. 293T cells were transfected with wild-type H-RAS, H-RASG12V, H-RASG12V,T35S, H-RASG12V,E37G, or H-RASG12V,Y40C, and cell extracts were incubated with immobilized RIN1(His6). RAS proteins extracted by RIN1-coated resin were detected by immunoblotting. Position 35 (T/S) and 40 (Y/C) mutations respectively reduced and abolished RIN1 binding. In contrast, the position 37 (E/G) mutation had little effect on RIN1 binding (data not shown). This extends previous findings from two-hybrid studies (17) by showing that full-length and posttranslationally modified RIN1 (from insect cell culture) can quantifiably distinguish among RAS effector mutants produced in mammalian cells.
RIN1 binds endogenous, transiently activated RAS.
To determine whether RIN1 binds RAS activated in response to physiological stimulation signals, we employed a pulldown protocol (13). Quiescent NIH 3T3 cells were stimulated with EGF, and cell extracts were incubated with immobilized RIN1-RBD(His6). Within 5 min of growth factor treatment, there was a substantial increase in the level of RAS associated with RIN1-RBD (Fig. 1C). This signal persisted for close to 1 h. A parallel detection of phosphorylated ERK proteins demonstrated a close correlation between RAS binding to RIN1-RBD and activation of ERK proteins that are known downstream elements in the RAF1-initiated MAP kinase pathway (31). Binding to RIN1 follows the natural, transient course of RAS activation and inactivation by growth factor receptor stimulation. This reinforces the selectivity of RIN1 for the GTP-bound conformation of RAS. Also, a similar result has been reported for RAF, suggesting that RIN1 and RAF may bind to the same, transiently activated form of RAS, consistent with a competition model.
RIN1 is localized to both plasma membrane and cytoplasm.
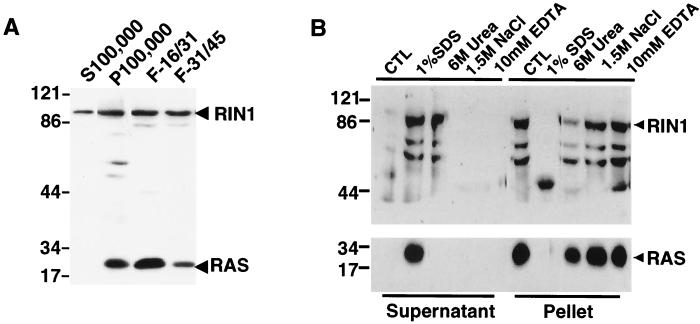
To characterize the localization of RIN1, we performed fractionation studies using HBL100 cells. This human breast tissue-derived epithelial cell line naturally expresses RIN1. Cell lysates were first separated into soluble cytoplasmic proteins and a pellet of membrane derived material. The pellet was further purified by discontinuous sucrose gradient centrifugation resulting in separation of plasma and microsomal membranes. RIN1 protein, detected by immunoblotting, was enriched in the plasma membrane fractions (Fig. 3A). As expected, RAS was also localized to the same fractions but was absent from the soluble material. The results demonstrate that some RIN1 is in close proximity to RAS and situated appropriately for regulated interactions.
FIG. 3.
Determination of RIN1 subcellular localization in HBL100 cells. HBL100 cell extracts were prepared under hypotonic conditions and separated into soluble cytosolic proteins (S100,000) and a pellet of membrane-derived material (P100,000). The pellet was further separated into plasma (F-16/31) and microsomal (F-31/45) membrane fractions. (A) Immunoblotting analysis of RIN1 with monoclonal anti-RIN1 or anti-RAS. (B) Immunoblotting analysis of supernatant and pellet fractions of plasma membrane fraction samples, treated as indicated in the text. CTL, control.
A variety of conditions were explored to characterize the nature of the plasma membrane association of RIN1. The plasma membrane fraction was treated with high salt (1.5 M NaCl), a denaturing agent (6 M urea), a cationic chelating agent (10 mM EDTA), or a detergent (1% SDS). While the high salt and chelating agent treatments did not disrupt the association of RIN1 with the plasma membrane, the presence of urea or 1% SDS resulted in significant release of RIN1 (Fig. 3B). In contrast, RAS protein could only be dislodged from the plasma membrane by detergent treatment. These results indicated that RIN1 is associated with the plasma membrane through a relatively weak interaction and probably not by a lipid modification.
RIN1 can block RAS-mediated cell transformation.
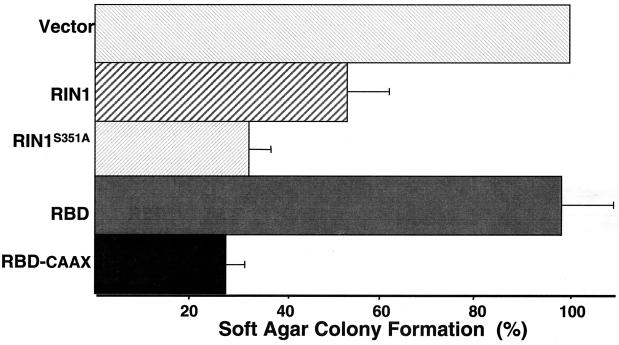
A soft-agar colony growth assay was used to determine the potential for RIN1 to interfere with RAS-mediated cell transformation. Consistent with its ability to interfere with RAS-RAF association in vitro, we observed that full-length RIN1 produced a twofold reduction in the number of soft-agar colonies when introduced into RASQ61L cells (Fig. 4). In a complementary experiment, RASG12V was introduced into Rat1 cells with or without stable expression of RIN1 and a similar block in transformation was detected (data not shown). Surprisingly, the RBD alone, which had been shown to bind tightly to RAS in vitro, had no suppression activity in this assay. We examined the effect of plasma membrane localization of RBD by appending a farnesylation signal (CAAX) onto the carboxy terminus. The resulting RBD-CAAX showed increased membrane localization, compared to that of full-length RIN1 or RBD, when expressed in NIH 3T3 cells with or without activated RAS (Fig. 5). This shift toward membrane association also correlated with potent suppression (three- to fourfold) of the soft-agar colony growth induced by RASQ61L (Fig. 4). These results are consistent with RIN1 acting as an endogenous RAS effector that, when localized to the plasma membrane, can directly and efficiently compete with RAF. Unlike other RAS effectors, however, full-length RIN1 does not promote fibroblast transformation.
FIG. 4.
RIN1 blocks RAS transformation. NIH 3T3 RASQ61L cells were infected with a retrovirus expressing the indicated construct and subjected to soft-agar growth assays (29). Colony formation is reported as a percentage of that seen for vector transduced cells. The results shown are the means of three experiments, each performed in duplicate.
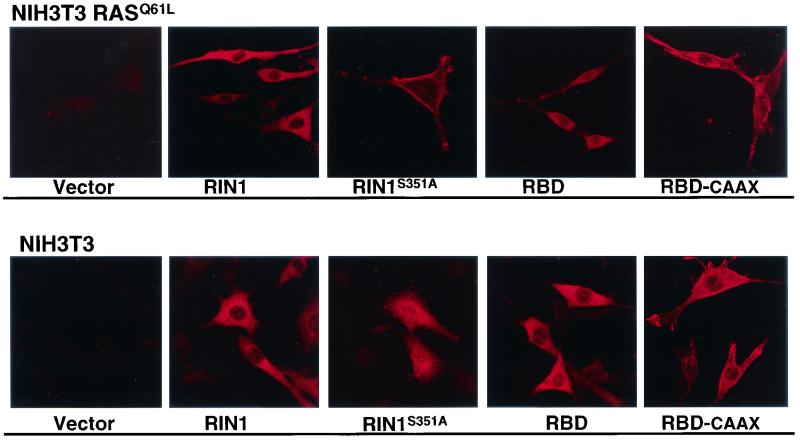
FIG. 5.
Mutation of 14-3-3 binding site changes RIN1 localization. NIH 3T3 or NIH 3T3 RASQ61L cells were infected with the indicated retroviral constructs. RIN1 proteins were visualized by anti-RIN1 immunofluorescence.
The regulatory role of 14-3-3 proteins in RIN1-RAS interactions.
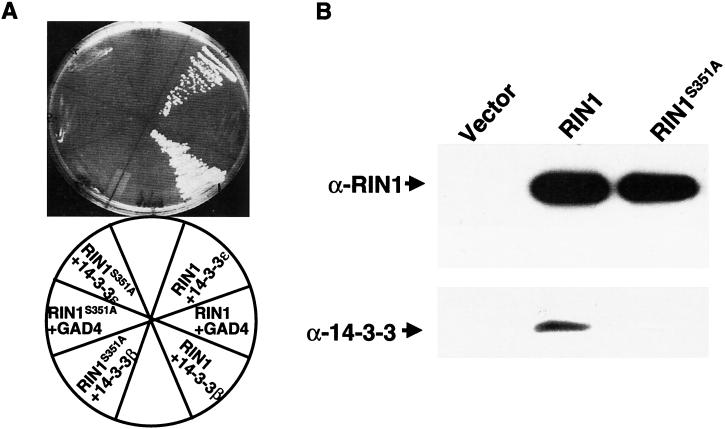
14-3-3 proteins can dramatically influence signaling by RAF (37, 54, 61, 68, 69, 80). RIN1 is also known to interact with multiple isoforms of 14-3-3 proteins, and the binding domain has been localized within the RBD region (17). Amino acid sequence analysis of RIN1 revealed a potential 14-3-3 binding site at residues 348 to 353 (RSMSAA) that matches the conserved 14-3-3 binding motif (RSXpSXP) (the lowercase p indicates a phosphate) except at position 6 which is known to show considerable degeneracy (reviewed in reference 11). Phosphorylation of the serine residue at position 4 is known to be critical for 14-3-3 binding (45). To determine the contribution of this potential 14-3-3 binding site to RIN1 function, we introduced a substitution (alanine for serine) at residue 351 of RIN1. Two-hybrid experiments indicated that this mutation blocked binding to 14-3-3 proteins (Fig. 6A). Similarly, in a coimmunoprecipitation assay the RIN1S351A mutant showed loss of binding to endogenous 14-3-3 (Fig. 6B). The mutant protein was expressed at levels comparable to those for wild-type RIN1, as determined by immunoblotting.
FIG. 6.
Binding of 14-3-3 is abolished in RIN1S351A. (A) Two-hybrid assay results. LexA fusions of RIN1 or RIN1S351A and GAL4 activation domain fusions of 14–3-3 (ε or β) were transformed into strain L40, and growth on histidine-deficient medium was used to select for activation of the HIS3 reporter gene. (B) Coimmunoprecipitation assay results. Cell lysates prepared from 293T cells transfected with the indicated RIN1 construct were immunoprecipitated with anti-RIN1 then subjected to immunoblot analysis for RIN1 and 14-3-3.
The reduction in 14-3-3 binding by RIN1S351A also correlated with an increased capacity for suppression of activated RAS (Fig. 4), suggesting that 14-3-3 proteins may function to inhibit RAS binding by RIN1. To determine if this involved regulation of access to the plasma membrane, we directly examined the subcellular localization of mutant RIN. In RASQ61L-expressing NIH 3T3 cells, RIN1S351A showed a marked shift to the membrane compared with wild-type RIN1 (Fig. 5). This was similar to what was seen for RBD-CAAX when compared with RBD. Both RIN1S351A and RBD-CAAX continued to show heightened plasma membrane localization in the absence of RASQ61L, demonstrating that this effect is not driven by RAS binding. Interestingly, however, NIH 3T3 cells expressing RIN1S351A (but not activated RAS) were rounded and showed a roughened membrane appearance, with the RIN1S351A protein distributed over the entire surface along with some enhanced staining at membrane edges.
PKD phosphorylates serine 351 of RIN1.
Based on the role of RIN1 serine 351 in binding to 14-3-3, as well as the consequences on subcellular localization and function, we sought to identify the kinase responsible for phosphorylation of this site. PKD is a protein serine kinase with a modular structure consisting of an N-terminal regulatory domain that includes a hydrophobic segment, two phorbol ester/diacylglycerol binding cysteine-rich motifs, a pleckstrin homology domain, and a C-terminal catalytic domain with a distinctive primary sequence and substrate specificity (70, 72, 75). The substrate specificity of PKD can be clearly distinguished from that of PKCs, indicating that this protein kinase selects a unique set of biological targets. Syntide-2 (PLARTLSVAGLPGKK) is phosphorylated by PKD with high efficiency (70, 72). In contrast, a PKCɛ substrate peptide (ERMRPRKRQGSVRRRV) is an excellent substrate for all PKC isoforms (28, 38) but not for PKD. A syntide-2 variant peptide (PLAATLSVAGLPGKK) with a single arginine-to-alanine change (syntide-2-R4A) was a poor substrate, however, indicating PKD’s preference for basic residues upstream of the targeted serine. In addition, PKD showed specificity for peptides containing arginine at position −3 and leucine at position −5, relative to the targeted serine (47). Finally, an optimized peptide incorporating preferred amino acids at positions −7 to +5 was efficiently phosphorylated by PKD but not by various PKCs (47).
A database search (BLAST) revealed close similarity between the putative optimal PKD substrate (L3VRQMSVAF14) (47) and a sequence in RIN1 (L346LRSMSAAF354). We examined RIN1 as a potential PKD substrate using in vitro kinase assays with PKD immunoprecipitated from transfected COS-7 cells. RIN1 was clearly phosphorylated by PKD (Fig. 7A). Prior stimulation of PKD by treatment of cells with the phorbol ester PDB was required, and RIN1 phosphorylation correlated with PKD autophosphorylation. A small amount of active PKD was isolated from PDB-stimulated control cells, and this endogenous PKD phosphorylated RIN1 to a minor extent (Fig. 7A). Assays using kinase-deficient enzyme (PKDK618N) demonstrated that PKD kinase activity, as opposed to that from a coprecipitating kinase, was responsible for RIN1 phosphorylation. Note that RIN1 phosphorylation in reactions using PKDK618N from PDB-stimulated cells was similar to that in lysates from PDB-stimulated control cells (Fig. 7A). The mutant RIN1S351A protein was not phosphorylated to a significant extent by activated PKD. In contrast, PKD autophosphorylation in these reactions was unchanged.
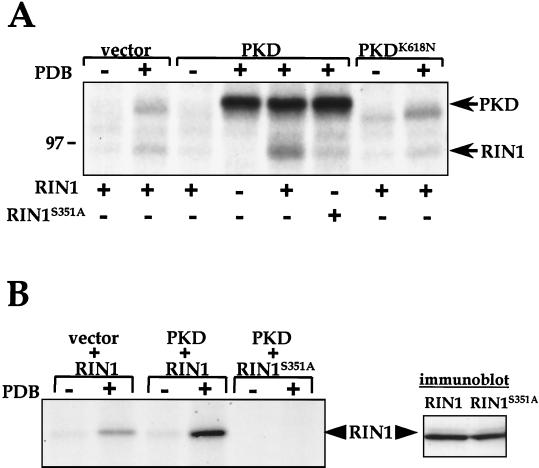
FIG. 7.
PKD phosphorylates Ser351 of RIN1. (A) Phosphorylation of RIN1 by PKD. PKD was immunoprecipitated from COS-7 cells transfected with vector (pcDNA3), wild-type PKD, or a kinase-inactive mutant (PKDK618N) and treated with phorbol 12,13 dibutyrate (PDB) where indicated. The resulting material was used for in vitro kinase assays with purified wild-type RIN1 or a phosphorylation site mutant (RIN1S351A) and analyzed by SDS-PAGE and autoradiography. A molecular marker (97 kDa) is shown at left. This experiment was performed three times with similar results. (B) Ability of PKD to use RIN1 as a cellular substrate. COS-7 cells transfected with RIN1 or RIN1S351A, either alone or with wild-type PKD, were metabolically labeled with 32P. Cells were then treated (or not) with PDB, as indicated, and lysed. The RIN1 immunoprecipitates were analyzed by SDS-PAGE and autoradiography. This experiment was performed three times with similar results. The results (shown at right) of immunoblotting analysis of COS-7 cells transfected with PKD, together with either RIN1 or RIN1S351A, and lysed directly in SDS-PAGE sample buffer, demonstrated comparable expression (immunoblotting carried out with anti-RIN1).
We next examined the ability of PKD to utilize RIN1 as a cellular substrate. RIN1 protein showed a low level of phosphorylation when immunoprecipitated from unstimulated, metabolically 32P-labeled COS-7 cells (Fig. 7B). In contrast, a marked increase in RIN1 phosphorylation was induced by PDB stimulation of cells, a treatment that activates PKD via a PKC-dependent signal transduction pathway involving PKD activation loop phosphorylation (76, 82). Cotransfection of RIN1 together with PKD resulted in strong, PDB stimulation-dependent RIN1 phosphorylation. In addition, RIN1 phosphorylated by PKD was predominantly at Ser351, since the RIN1S351A mutant cotransfected with PKD showed no phosphorylation, either with or without PDB stimulation (Fig. 7B). Taken together, these data indicate that Ser351, the 14-3-3 binding determinant, is an efficient and specific substrate for phosphorylation by PKD. They do not, however, demonstrate that Ser351 is phosphorylated exclusively by PKD in vivo.
There are likely to be phosphorylation events by other kinases acting at other sites on RIN1. Particularly noteworthy are two PXSP motifs that fit the reported optimum ERK substrate site (14, 35) and the observation that RIN1 was an in vitro substrate for the MAP kinases ERK2, JNK2 and p38 (data not shown). RIN1 was also phosphorylated by PKC (isoforms α, β, and γ), but not by PKA (data not shown). These data, although generated outside a cellular context, suggest additional levels of RIN1 regulation that will require further study.
DISCUSSION
RIN1 shows the established hallmarks of a RAS effector: binding is GTP dependent and requires an intact RAS effector domain (reference 17 and this work). The RBD of RIN1 has an affinity of approximately 22 nM for RAS(GTP). This is strikingly similar to the affinity of 18 nM measured for RAF1 and markedly stronger than the binding constants determined for other RAS effectors (20, 50). Notably, the full-length RIN1 protein has a somewhat lower RAS-binding affinity than does the RBD fragment. Although it is true that minimum RBDs of other effectors have shown binding affinities greater than those of the intact proteins, the RBD of RIN1 is unusually large in comparison to the minimal RBDs of other effectors (4, 21, 60, 62). This raises the possibility that the RAS-binding determinants of RIN1 are inextricably combined with structural features that are required for distinct functions such as RAB-directed nucleotide exchange (66) and 14-3-3 interactions (this work).
The observation that the RBDs of RIN1 and RAF1 directly compete for binding to activated RAS reflects the overlapping nature of these interactions. The approximately equal molar competition observed in vitro is consistent with the similarity in binding affinity for RAS(GTP). The RBD of RIN1 was also seen to bind transiently activated wild-type RAS from stimulated NIH 3T3 cells. This behavior has been reported for the RBD of RAF, and suggests that RIN1 may indeed compete with endogenous RAF. More importantly, while RAF activation is itself transforming in fibroblast cells (29, 64) and the effectors PI3K and RGF synergistically enhance transformation (59, 79), we report that full-length RIN1 blocked transformation by activated RAS. This result implies that RIN1 normally functions in a pathway either not required for or antagonistic to transformation of fibroblast cells. Possible antagonistic signals might be mediated by RIN1 effectors that include the ABL family tyrosine kinases (1) (H. Hu and J. Colicelli, unpublished) that regulate cytoskeleton remodeling and perhaps RAB proteins (66) that facilitate receptor downregulation.
The competition between RAF and RIN1 for activated RAS raises the possibility that the signal transmission output from RAS may be modulated physiologically through the regulated expression of these effectors. Although RAF expression appears to be ubiquitous (65), RIN1 is expressed at low or undetectable levels in most tissues except for a subset of brain neurons (17) (A. Dhaka and J. Colicelli, unpublished data) to concentrations that may support direct competition between these effectors. In this model, RAS occupation by RIN1 would result in a concerted blockade of RAF activation together with a redirection of RAS signaling through RIN1 to its downstream effectors. Indeed, recent findings support neuronal functions for RAS (3, 5, 8), for ABL family kinases (32), and for RAB proteins (53).
There are likely to be some differences in the specific contacts between RAS and the alternate effectors RAF and RIN1, as highlighted by distinctions in binding to RASG12V,T35S (binds RAF, not RIN1) and RASG12V,E37G (binds RIN1, not RAF) This may reflect subtle differences that could be exploited in cells to further discriminate among effectors and selectively shunt RAS signaling. It is of interest to note that RASG12V, E37G shows RGF binding capacity but that some of its biological effects appear to be independent of this effector (52, 55) and may be mediated by RIN1.
We have also characterized a specific 14-3-3 binding site within the RBD sequence of RIN1. Mutation of this site eliminated 14-3-3 binding and simultaneously produced a shift in localization to the plasma membrane. The membrane-staining pattern of RIN1S351A in NIH 3T3 cells was somewhat altered by RASQ61L. Although the RIN1 mutant appeared throughout a roughened membrane in wild-type NIH 3T3 cells, it was concentrated at membrane edges in cells with RASQ61L. Taken together, these observations raise the possibility that membrane compartmentalization (and proximity to RAS) of RIN1 may be reversible and regulated through phosphorylation and dephosphorylation of a core serine within the 14-3-3 recognition site. Engagement with 14-3-3 may reduce access to the plasma membrane through an allosteric change in RIN1 structure, by an induced covalent modification, and/or by simple sequestration. Indeed, 14-3-3 proteins can participate at multiple levels of signal regulation, as demonstrated by extensive studies with RAF proteins. There are at least two, characterized 14-3-3 binding sites in RAF1. While binding at one site has been demonstrated to stabilize the inactive kinase, binding at another site appears to facilitate RAF1 activation (7, 61, 68, 80). Further studies will be required to determine the full role of 14-3-3 in RIN1 function.
Both in vitro and in vivo experiments support a role for PKD-mediated phosphorylation of RIN1 at the site (serine 351) that controls 14-3-3 binding. Others have noted that the consensus PKD substrate site resembles some 14-3-3 binding motifs, and identified within the PKD regulatory domain a putative autophosphorylation site compatible with regulated 14-3-3 binding (19). Our findings provide evidence that PKD may indeed function as a regulator of 14-3-3 binding to at least some partner proteins. Because PKD is dynamically partitioned between the cytoplasm and plasma membrane (40, 56), it is well positioned to play a role in mediating RAS effector accessibility. PKD-mediated RIN1 Ser351 phosphorylation may, together with other events, serve as an attenuation mechanism to uncouple RIN1 from activated RAS through the promotion of 14-3-3 binding and cytoplasmic relocation. This might further result in the promotion of alternate (e.g., RAF) effector pathways and might in part explain the observation that PKD selectively stimulates the ERK1/2 mitogen-activated protein kinase cascade in some cells (18). Return of RIN1 to a signal-ready state (i.e., available for RAS interaction) would require the action of an as-yet-unidentified phosphatase.
RIN1 is subject to other modifications, including a previously characterized tyrosine phosphorylation by ABL (1, 36). We also report here on potential MAP kinase-mediated RIN1 phosphorylations. It should be noted that RAF proteins are regulated by multiple phosphorylations from distinct kinases, a complex system that has been only partly characterized (6, 9, 15, 39, 42, 81).
The propensity for RIN1 to associate with the plasma membrane, and perhaps to reside in particular subregions, is likely to be controlled at multiple levels. We have previously reported that in an epithelial cell line (HeLa) endogenous RIN1 showed primarily a punctate pattern of membrane staining. Here we demonstrate that in a fibroblast cell line (NIH 3T3) ectopically expressed RIN1 is somewhat more cytoplasmic. This may be in part due to the high level of expression. Also, fibroblast and epithelial cells differ substantially in the quality and quantity of cell-cell and cell-substrata adhesions, and they respond differently to activated RAS (48). These distinctions likely reflect additional levels of signal traffic regulation.
Taken together, these data suggest a multilevel regulatory system that controls RAS signaling output based on the affinity, cell-specific availability, and localization of downstream effectors.
Acknowledgments
We thank Pablo Rodriguez-Viciana, Frank McCormick, Michael White, and Adriana Cox for providing reagents. We also thank Elizabeth Williamson and Phillip Koeffler for guidance with kinase assays and Martin Phillips for assistance with BiaCore instrumentation. Walter Kolch and Fuyu Tamanoi provided critical evaluation during the preparation of the manuscript.
This work was supported by National Institutes of Health grants CA 56301 (to J.C.) and DK 55003 (to E.R.) and by Mentored Research Scientist Career Development Award KO1 DK 02834 (to R.T.W.).
REFERENCES
- 1.Afar, D. E., L. Han, J. McLaughlin, S. Wong, A. Dhaka, K. Parmar, N. Rosenberg, O. N. Witte, and J. Colicelli. 1997. Regulation of the oncogenic activity of BCR-ABL by a tightly bound substrate protein RIN1. Immunity 6:773–782. [DOI] [PubMed] [Google Scholar]
- 2.Boguski, M. S., and F. McMormick. 1993. Proteins regulating Ras and its relatives. Nature 366:643–654. [DOI] [PubMed] [Google Scholar]
- 3.Brambilla, R., L. Gnesutta, L. Minichiello, G. White, A. J. Roylance, C. E. Herron, W. Ramsey, D. P. Wolfer, V. Cestari, C. Rossi-Arnaud, S. G. Grant, P. F. Chapman, H. P. Lipp, E. Sturani, and R. Klein. 1997. A role for the Ras signalling pathway in synaptic transmission and long-term memory. Nature 390:281–286. [DOI] [PubMed] [Google Scholar]
- 4.Brtva, T. R., J. K. Drugan, S. Ghosh, R. S. Terrell, S. Campbell-Burk, R. M. Bell, and C. J. Der. 1995. Two distinct Raf domains mediate interaction with Ras. J. Biol. Chem. 270:9809–9812. [DOI] [PubMed] [Google Scholar]
- 5.Chen, H. J., M. Rojas-Soto, A. Oguni, and M. B. Kennedy. 1998. A synaptic Ras-GTPase activating protein (p135 SynGAP) inhibited by CaM kinase II. Neuron 20:895–904. [DOI] [PubMed] [Google Scholar]
- 6.Chong, H., J. Lee, and K. L. Guan. 2001. Positive and negative regulation of Raf kinase activity and function by phosphorylation. EMBO J. 20:3716–3727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clark, G. J., J. K. Drugan, K. L. Rossman, J. W. Carpenter, K. Rogers-Graham, H. Fu, C. J. Der, and S. L. Campbell. 1997. 14–3-3 ζ negatively regulates raf-1 activity by interactions with the Raf-1 cysteine-rich domain. J. Biol. Chem. 272:20990–20993. [DOI] [PubMed] [Google Scholar]
- 8.Costa, R. M., T. Yang, D. P. Huynh, S. M. Pulst, D. H. Viskochil, A. J. Silva, and C. I. Brannan. 2001. Learning deficits, but normal development and tumor predisposition, in mice lacking exon 23a of Nf1. Nat. Genet. 27:399–405. [DOI] [PubMed] [Google Scholar]
- 9.Diaz, B., D. Barnard, A. Filson, S. MacDonald, A. King, and M. Marshall. 1997. Phosphorylation of Raf-1 serine 338-serine 339 is an essential regulatory event for Ras-dependent activation and biological signaling. Mol. Cell. Biol. 17:4509–4516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Feig, L. A., T. Urano, and S. Cantor. 1996. Evidence for a Ras/Ral signaling cascade. Trends Biochem. Sci. 21:438–441. [DOI] [PubMed] [Google Scholar]
- 11.Fu, H., R. R. Subramanian, and S. C. Masters. 2000. 14–3-3 proteins: structure, function, and regulation. Annu. Rev. Pharmacol. Toxicol. 40:617–647. [DOI] [PubMed] [Google Scholar]
- 12.Geyer, M., C. Herrmann, S. Wohlgemuth, A. Wittinghofer, and H. R. Kalbitzer. 1997. Structure of the Ras-binding domain of RalGEF and implications for Ras binding and signalling. Nat. Struct. Biol. 4:694–699. [DOI] [PubMed] [Google Scholar]
- 13.Gille, H., and J. Downward. 1999. Multiple ras effector pathways contribute to G(1) cell cycle progression. J. Biol. Chem. 274:22033–22040. [DOI] [PubMed] [Google Scholar]
- 14.Gonzalez, F. A., D. L. Raden, and R. J. Davis. 1991. Identification of substrate recognition determinants for human ERK1 and ERK2 protein kinases. J. Biol. Chem. 266:22159–22163. [PubMed] [Google Scholar]
- 15.Guan, K. L., C. Figueroa, T. R. Brtva, T. Zhu, J. Taylor, T. D. Barber, and A. B. Vojtek. 2000. Negative regulation of the serine/threonine kinase B-Raf by Akt. J. Biol. Chem. 275:27354–27359. [DOI] [PubMed] [Google Scholar]
- 16.Han, L., and J. Colicelli. 1995. A human protein selected for interference with Ras function interacts directly with Ras and competes with Raf1. Mol. Cell. Biol. 15:1318–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Han, L., D. Wong, A. Dhaka, D. Afar, M. White, W. Xie, H. Herschman, O. Witte, and J. Colicelli. 1997. Protein binding and signaling properties of RIN1 suggest a unique effector function. Proc. Natl. Acad. Sci. USA 94:4954–4959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hausser, A., P. Storz, S. Hubner, I. Braendlin, M. Martinez-Moya, G. Link, and F. J. Johannes. 2001. Protein kinase C μ selectively activates the mitogen-activated protein kinase (MAPK) p42 pathway. FEBS Lett. 492:39–44. [DOI] [PubMed] [Google Scholar]
- 19.Hausser, A., P. Storz, G. Link, H. Stoll, Y. C. Liu, A. Altman, K. Pfizenmaier, and F. J. Johannes. 1999. Protein kinase C μ is negatively regulated by 14–3-3 signal transduction proteins. J. Biol. Chem. 274:9258–9264. [DOI] [PubMed] [Google Scholar]
- 20.Herrmann, C., G. Horn, M. Spaargaren, and A. Wittinghofer. 1996. Differential interaction of the ras family GTP-binding proteins H-Ras, Rap1A, and R-Ras with the putative effector molecules Raf kinase and Ral-guanine nucleotide exchange factor. J. Biol. Chem. 271:6794–6800. [DOI] [PubMed] [Google Scholar]
- 21.Hofer, F., S. Fields, C. Schneider, and G. S. Martin. 1994. Activated Ras interacts with the Ral guanine nucleotide dissociation stimulator. Proc. Natl. Acad. Sci. USA 91:11089–11093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hollenberg, S. M., R. Sternglanz, P. F. Cheng, and H. Weintraub. 1995. Identification of a new family of tissue-specific basic helix-loop-helix proteins with a two-hybrid system. Mol. Cell Biol. 15:3813–3822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Iglesias, T., N. Cabrera-Poch, M. P. Mitchell, T. J. Naven, E. Rozengurt, and G. Schiavo. 2000. Identification and cloning of Kidins220, a novel neuronal substrate of protein kinase D. J. Biol. Chem. 275:40048–40056. [DOI] [PubMed] [Google Scholar]
- 24.Johnson, L., D. Greenbaum, K. Cichowski, K. Mercer, E. Murphy, E. Schmitt, R. T. Bronson, H. Umanoff, W. Edelmann, R. Kucherlapati, and T. Jacks. 1997. K-ras is an essential gene in the mouse with partial functional overlap with N-ras. Genes Dev. 11:2468–2481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Joneson, T., M. A. White, M. H. Wigler, and D. Bar-Sagi. 1996. Stimulation of membrane ruffling and MAP kinase activation by distinct effectors of RAS. Science 271:810–812. [DOI] [PubMed] [Google Scholar]
- 26.Jonsson, U., L. Fagerstam, B. Ivarsson, B. Johnsson, R. Karlsson, K. Lundh, S. Lofas, B. Persson, H. Roos, I. Ronnberg, et al. 1991. Real-time biospecific interaction analysis using surface plasmon resonance and a sensor chip technology. BioTechniques 11:620–627. [PubMed] [Google Scholar]
- 27.Kaur, K. J., and M. A. White. 2001. Isolation of effector-selective Ras mutants by yeast two-hybrid screening. Methods Enzymol. 332:270–277. [DOI] [PubMed] [Google Scholar]
- 28.Kazanietz, M. G., L. B. Areces, A. Bahador, H. Mischak, J. Goodnight, J. F. Mushinski, and P. M. Blumberg. 1993. Characterization of ligand and substrate specificity for the calcium-dependent and calcium-independent protein kinase C isozymes. Mol. Pharmacol. 44:298–307. [PubMed] [Google Scholar]
- 29.Khosravi-Far, R., M. A. White, J. K. Westwick, P. A. Solski, M. Chrzanowska-Wodnicka, L. Van Aelst, M. H. Wigler, and C. J. Der. 1996. Oncogenic Ras activation of Raf/mitogen-activated protein kinase-independent pathways is sufficient to cause tumorigenic transformation. Mol. Cell. Biol. 16:3923–3933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Koera, K., K. Nakamura, K. Nakao, J. Miyoshi, K. Toyoshima, T. Hatta, H. Otani, A. Aiba, and M. Katsuki. 1997. K-ras is essential for the development of the mouse embryo. Oncogene 15:1151–1159. [DOI] [PubMed] [Google Scholar]
- 31.Kolch, W. 2000. Meaningful relationships: the regulation of the Ras/Raf/MEK/ERK pathway by protein interactions. Biochem. J. 351(Pt. 2):289–305. [PMC free article] [PubMed] [Google Scholar]
- 32.Koleske, A. J., A. M. Gifford, M. L. Scott, M. Nee, R. T. Bronson, K. A. Miczek, and D. Baltimore. 1998. Essential roles for the Abl and Arg tyrosine kinases in neurulation. Neuron 21:1259–1272. [DOI] [PubMed] [Google Scholar]
- 33.Kuriyama, M., N. Harada, S. Kuroda, T. Yamamoto, M. Nakafuku, A. Iwamatsu, D. Yamamoto, R. Prasad, C. Croce, E. Canaani, and K. Kaibuchi. 1996. Identification of AF-6 and canoe as putative targets for Ras. J. Biol. Chem. 271:607–610. [DOI] [PubMed] [Google Scholar]
- 34.Leon, J., I. Guerrero, and A. Pellicer. 1987. Differential expression of the ras gene family in mice. Mol. Cell. Biol. 7:1535–1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lewis, T. S., P. S. Shapiro, and N. G. Ahn. 1998. Signal transduction through MAP kinase cascades. Adv. Cancer Res. 74:49–139. [DOI] [PubMed] [Google Scholar]
- 36.Lim, Y. M., S. Wong, G. Lau, O. Witte, and J. Colicelli. 2000. BCR/ABL inhibition by an escort/phosphatase fusion protein. Proc. Natl. Acad. Sci. USA 97:12233–12238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.MacNicol, M. C., A. J. Muslin, and A. M. MacNicol. 2000. Disruption of the 14–3-3 binding site within the B-Raf kinase domain uncouples catalytic activity from PC12 cell differentiation. J. Biol. Chem. 275:3803–3809. [DOI] [PubMed] [Google Scholar]
- 38.Marais, R. M., and P. J. Parker. 1989. Purification and characterisation of bovine brain protein kinase C isotypes alpha, beta and gamma. Eur. J. Biochem. 182:129–137. [DOI] [PubMed] [Google Scholar]
- 39.Mason, C. S., C. J. Springer, R. G. Cooper, G. Superti-Furga, C. J. Marshall, and R. Marais. 1999. Serine and tyrosine phosphorylations cooperate in Raf-1, but not B-Raf activation. EMBO J. 18:2137–2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Matthews, S. A., T. Iglesias, E. Rozengurt, and D. Cantrell. 2000. Spatial and temporal regulation of protein kinase D (PKD). EMBO J. 19:2935–2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miller, M. J., L. Rioux, G. V. Prendergast, S. Cannon, M. A. White, and J. L. Meinkoth. 1998. Differential effects of protein kinase A on Ras effector pathways. Mol. Cell. Biol. 18:3718–3726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Morrison, D. K., G. Heidecker, U. R. Rapp, and T. D. Copeland. 1993. Identification of the major phosphorylation sites of the Raf-1 kinase. J. Biol. Chem. 268:17309–17316. [PubMed] [Google Scholar]
- 43.Muller, A. J., J. C. Young, A. M. Pendergast, M. Pondel, N. R. Landau, D. R. Littman, and O. N. Witte. 1991. BCR first exon sequences specifically activate the BCR/ABL tyrosine kinase oncogene of Philadelphia chromosome-positive human leukemias. Mol. Cell. Biol. 11:1785–1792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Muller, R., D. J. Slamon, E. D. Adamson, J. M. Tremblay, D. Muller, M. J. Cline, and I. M. Verma. 1983. Transcription of c-onc genes c-rasKi and c-fms during mouse development. Mol. Cell. Biol. 3:1062–1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Muslin, A. J., J. W. Tanner, P. M. Allen, and A. S. Shaw. 1996. Interaction of 14–3-3 with signaling proteins is mediated by the recognition of phosphoserine. Cell 84:889–897. [DOI] [PubMed] [Google Scholar]
- 46.Nassar, N., G. Horn, C. Herrmann, A. Scherer, F. McCormick, and A. Wittinghofer. 1995. The 2.2 A crystal structure of the Ras-binding domain of the serine/threonine kinase c-Raf1 in complex with Rap1A and a GTP analogue. Nature 375:554–560. [DOI] [PubMed] [Google Scholar]
- 47.Nishikawa, K., A. Toker, F. J. Johannes, Z. Songyang, and L. C. Cantley. 1997. Determination of the specific substrate sequence motifs of protein kinase C isozymes. J. Biol. Chem. 272:952–960. [DOI] [PubMed] [Google Scholar]
- 48.Oldham, S. M., G. J. Clark, L. M. Gangarosa, R. J. Coffey, Jr., and C. J. Der. 1996. Activation of the Raf-1/MAP kinase cascade is not sufficient for Ras transformation of RIE-1 epithelial cells. Proc. Natl. Acad. Sci. USA 93:6924–6928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Overbeck, A. F., T. R. Brtva, A. D. Cox, S. M. Graham, S. Y. Huff, R. Khosravi-Far, L. A. Quilliam, P. A. Solski, and C. J. Der. 1995. Guanine nucleotide exchange factors: activators of Ras superfamily proteins. Mol. Reprod. Dev. 42:468–476. [DOI] [PubMed] [Google Scholar]
- 50.Pacold, M. E., S. Suire, O. Perisic, S. Lara-Gonzalez, C. T. Davis, E. H. Walker, P. T. Hawkins, L. Stephens, J. F. Eccleston, and R. L. Williams. 2000. Crystal structure and functional analysis of Ras binding to its effector phosphoinositide 3-kinase gamma. Cell 103:931–943. [DOI] [PubMed] [Google Scholar]
- 51.Pear, W. S., G. P. Nolan, M. L. Scott, and D. Baltimore. 1993. Production of high-titer helper-free retroviruses by transient transfection. Proc. Natl. Acad. Sci. USA 90:8392–8396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Peyssonnaux, C., S. Provot, M. P. Felder-Schmittbuhl, G. Calothy, and A. Eychene. 2000. Induction of postmitotic neuroretina cell proliferation by distinct Ras downstream signaling pathways. Mol. Cell. Biol. 20:7068–7079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pierce, J. P., T. Mayer, and J. B. McCarthy. 2001. Evidence for a satellite secretory pathway in neuronal dendritic spines. Curr. Biol. 11:351–355. [DOI] [PubMed] [Google Scholar]
- 54.Qiu, W., S. Zhuang, F. C. von Lintig, G. R. Boss, and R. B. Pilz. 2000. Cell type-specific regulation of B-Raf kinase by cAMP and 14–3-3 proteins. J. Biol. Chem. 275:31921–31929. [DOI] [PubMed] [Google Scholar]
- 55.Ramocki, M. B., M. A. White, S. F. Konieczny, and E. J. Taparowsky. 1998. A role for RalGDS and a novel Ras effector in the Ras-mediated inhibition of skeletal myogenesis. J. Biol. Chem. 273:17696–17701. [DOI] [PubMed] [Google Scholar]
- 56.Rey, O., S. H. Young, D. Cantrell, and E. Rozengurt. 2001. Rapid protein kinase D translocation in response to G protein-coupled receptor activation: dependence on protein kinase C. J. Biol. Chem. 276:32616–32626. [DOI] [PubMed] [Google Scholar]
- 57.Riordan, J. R., and V. Ling. 1979. Purification of P-glycoprotein from plasma membrane vesicles of Chinese hamster ovary cell mutants with reduced colchicine permeability. J. Biol. Chem. 254:12701–12705. [PubMed] [Google Scholar]
- 58.Rodriguez-Viciana, P., P. H. Warne, R. Dhand, B. Vanhaesebroeck, I. Gout, M. J. Fry, M. D. Waterfield, and J. Downward. 1994. Phosphatidylinositol-3-OH kinase as a direct target of Ras. Nature 370:527–532. [DOI] [PubMed] [Google Scholar]
- 59.Rodriguez-Viciana, P., P. H. Warne, A. Khwaja, B. M. Marte, D. Pappin, P. Das, M. D. Waterfield, A. Ridley, and J. Downward. 1997. Role of phosphoinositide 3-OH kinase in cell transformation and control of the actin cytoskeleton by Ras. Cell 89:457–467. [DOI] [PubMed] [Google Scholar]
- 60.Rodriguez-Viciana, P., P. H. Warne, B. Vanhaesebroeck, M. D. Waterfield, and J. Downward. 1996. Activation of phosphoinositide 3-kinase by interaction with Ras and by point mutation. EMBO J. 15:2442–2451. [PMC free article] [PubMed] [Google Scholar]
- 61.Roy, S., R. A. McPherson, A. Apolloni, J. Yan, A. Lane, J. Clyde-Smith, and J. F. Hancock. 1998. 14–3-3 facilitates Ras-dependent Raf-1 activation in vitro and in vivo. Mol. Cell. Biol. 18:3947–3955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Scheffler, J. E., D. S. Waugh, E. Bekesi, S. E. Kiefer, J. E. LoSardo, A. Neri, K. M. Prinzo, K. L. Tsao, B. Wegrzynski, S. D. Emerson, et al. 1994. Characterization of a 78-residue fragment of c-Raf-1 that comprises a minimal binding domain for the interaction with Ras-GTP. J. Biol. Chem. 269:22340–22346. [PubMed] [Google Scholar]
- 63.Shields, J. M., K. Pruitt, A. McFall, A. Shaub, and C. J. Der. 2000. Understanding Ras: ‘it ain’t over ’til it’s over’. Trends Cell Biol. 10:147–154. [DOI] [PubMed] [Google Scholar]
- 64.Stang, S., D. Bottorff, and J. C. Stone. 1997. Interaction of activated Ras with Raf-1 alone may be sufficient for transformation of rat2 cells. Mol. Cell Biol. 17:3047–3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Storm, S. M., J. L. Cleveland, and U. R. Rapp. 1990. Expression of raf family proto-oncogenes in normal mouse tissues. Oncogene 5:345–351. [PubMed] [Google Scholar]
- 66.Tall, G. G., M. A. Barbieri, P. D. Stahl, and B. F. Horazdovsky. 2001. Ras-activated endocytosis is mediated by the Rab5 guanine nucleotide exchange activity of RIN1. Dev. Cell. 1:73–82. [DOI] [PubMed] [Google Scholar]
- 67.Therrien, M., A. M. Wong, E. Kwan, and G. M. Rubin. 1999. Functional analysis of CNK in RAS signaling. Proc. Natl. Acad. Sci. USA 96:13259–13263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Thorson, J. A., L. W. Yu, A. L. Hsu, N. Y. Shih, P. R. Graves, J. W. Tanner, P. M. Allen, H. Piwnica-Worms, and A. S. Shaw. 1998. 14–3-3 proteins are required for maintenance of Raf-1 phosphorylation and kinase activity. Mol. Cell. Biol. 18:5229–5238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tzivion, G., Z. Luo, and J. Avruch. 1998. A dimeric 14–3-3 protein is an essential cofactor for Raf kinase activity. Nature 394:88–92. [DOI] [PubMed] [Google Scholar]
- 70.Valverde, A. M., J. Sinnett-Smith, J. Van Lint, and E. Rozengurt. 1994. Molecular cloning and characterization of protein kinase D: a target for diacylglycerol and phorbol esters with a distinctive catalytic domain. Proc. Natl. Acad. Sci. USA 91:8572–8576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Van Aelst, L., M. A. White, and M. H. Wigler. 1994. Ras partners. Cold Spring Harb. Symp. Quant. Biol. 59:181–186. [DOI] [PubMed] [Google Scholar]
- 72.Van Lint, J. V., J. Sinnett-Smith, and E. Rozengurt. 1995. Expression and characterization of PKD, a phorbol ester and diacylglycerol-stimulated serine protein kinase. J. Biol. Chem. 270:1455–1461. [DOI] [PubMed] [Google Scholar]
- 73.Vavvas, D., X. Li, J. Avruch, and X. F. Zhang. 1998. Identification of Nore1 as a potential Ras effector. J. Biol. Chem. 273:5439–5442. [DOI] [PubMed] [Google Scholar]
- 74.Vojtek, A. B., S. M. Hollenberg, and J. A. Cooper. 1993. Mammalian Ras interacts directly with the serine/threonine kinase Raf. Cell 74:205–214. [DOI] [PubMed] [Google Scholar]
- 75.Waldron, R. T., T. Iglesias, and E. Rozengurt. 1999. The pleckstrin homology domain of protein kinase D interacts preferentially with the eta isoform of protein kinase C. J. Biol. Chem. 274:9224–9230. [DOI] [PubMed] [Google Scholar]
- 76.Waldron, R. T., O. Rey, T. Iglesias, T. Tugal, D. Cantrell, and E. Rozengurt. 2001. Activation loop Ser744 and Ser748 in protein kinase D are transphosphorylated in vivo. J. Biol. Chem. 276:32606–32615. [DOI] [PubMed] [Google Scholar]
- 77.Wang, Y., and J. Colicelli. 2001. RAS interaction with effector target RIN1. Methods Enzymol. 332:139–151. [DOI] [PubMed] [Google Scholar]
- 78.White, M. A., C. Nicolette, A. Minden, A. Polverino, L. Van Aelst, M. Karin, and M. H. Wigler. 1995. Multiple Ras functions can contribute to mammalian cell transformation. Cell 80:533–541. [DOI] [PubMed] [Google Scholar]
- 79.White, M. A., T. Vale, J. H. Camonis, E. Schaefer, and M. H. Wigler. 1996. A role for the Ral guanine nucleotide dissociation stimulator in mediating Ras-induced transformation. J. Biol. Chem. 271:16439–16442. [DOI] [PubMed] [Google Scholar]
- 80.Yip-Schneider, M. T., W. Miao, A. Lin, D. S. Barnard, G. Tzivion, and M. S. Marshall. 2000. Regulation of the Raf-1 kinase domain by phosphorylation and 14–3-3 association. Biochem. J. 351:151–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zimmermann, S., and K. Moelling. 1999. Phosphorylation and regulation of Raf by Akt (protein kinase B). Science 286:1741–1744. [DOI] [PubMed] [Google Scholar]
- 82.Zugaza, J. L., J. Sinnett-Smith, J. Van Lint, and E. Rozengurt. 1996. Protein kinase D (PKD) activation in intact cells through a protein kinase C-dependent signal transduction pathway. EMBO J. 15:6220–6230. [PMC free article] [PubMed] [Google Scholar]