Abstract
The complete set of nuclear genes that encode proteins targeted to mitochondria in plants is currently undefined and thus the full range of mitochondrial functions in plants is unknown. Analysis of two-dimensional gel separations of Arabidopsis cell culture mitochondrial protein revealed approximately 100 abundant proteins and 250 low-abundance proteins. Comparison of subfractions of mitochondrial protein on two-dimensional gels provided information on the soluble, membrane, or integral membrane locations of this protein set. A total of 170 protein spots were excised, trypsin-digested, and matrix-assisted laser desorption ionization/time of flight mass spectrometry spectra obtained. Using this dataset, 91 of the proteins were identified by searching translated Arabidopsis genomic databases. Of this set, 81 have defined functions based on sequence comparison. These functions include respiratory electron transport, tricarboxylic acid cycle metabolism, amino acid metabolism, protein import, processing, and assembly, transcription, membrane transport, and antioxidant defense. A total of 10 spectra were matched to Arabidopsis putative open reading frames for which no specific function has been determined. A total of 64 spectra did not match to an identified open reading frame. Analysis of full-length putative protein sequences using bioinformatic tools to predict subcellular targeting (TargetP, Psort, and MitoProt) revealed significant variation in predictions, and also a lack of mitochondrial targeting prediction for several characterized mitochondrial proteins.
The mitochondrion is the organelle within the eukaryotic cell that is primarily concerned with the synthesis of ATP in the fundamental process known as respiration. The origins of this organelle can be traced back to an event in which a prokaryotic cell was engulfed by another prokaryote to form a cell lineage containing two independent genomes. Over time, these genomes became codependent and mitochondria lost the ability to be viable organisms outside the host cell. A substantial transfer of genetic information occurred from the mitochondrial to the nuclear genome during this time. Today, it is predicted that mitochondria synthesize 2% to 5% of the proteins required for their function, with the remaining 95% to 98% of proteins required encoded by the nuclear genome and targeted back to the mitochondria as protein precursors using encrypted targeting information in the protein sequence (Gray et al., 1999). The detection of these encryptions and thus identification of the full set of these genes within the nuclear genome is a major challenge for biologists.
The recent sequencing of whole genomes has encouraged an increasing effort to develop bioinformatic tools to predict the cellular localization of putative protein sequences. Psort was developed as an expert system that uses a set of 100 “if-then”-type of rules based on analysis of characterized protein sequences from a variety of subcellular locations (Nakai and Kanehisa, 1992). MitoProt was developed to predict mitochondrial targeting and presequence cleavage sites based on a set of 47 known characteristics of presequences and cleavage sites (Claros and Vincens, 1996). More recently, TargetP, based on neural network programming, was developed to predict targeting of protein sequences to chloroplasts, mitochondria, and the secretory system using a knowledge based derived from Swiss-Prot sequence entries (Emanuelsson et al., 2000).
The full sequences of the five chromosomes from the model plant Arabidopsis were recently published (The Arabidopsis Genome Initiative, 2000). The analysis of this data included a full genome analysis using TargetP that predicted the number of nuclear-encoded, mitochondrial-targeted gene products. This number could be as high as 2,897, or, using a >0.95 specificity cut-off in TargetP, it could be as low at 349. The breadth of this prediction range (8.1-fold) was in stark contrast to the range for chloroplast targeting (0.7-fold) and secretory pathway targeting (0.07-fold) when using the same high- and low-stringency settings in TargetP.
Analysis of the presequences that direct proteins to mitochondria in plants shows substantial differences to the consensus sequences present in yeast (Saccharomyces cerevisiae) and mammals. This means that identifications in plants cannot simply rely on existing bioinformatic tools (Sjoling and Glaser, 1998). In addition, a range of proteins that are known to be targeted to plant mitochondria do not utilize cleavable N-terminal presequences and thus remain undetected by algorithms or training regimes based on identification of these signal peptides. Examples include a range of outer membrane proteins and also the inner membrane superfamily of carrier proteins (Laloi, 1999). All the prediction programs available have been largely established and/or trained on non-plant sequences, and thus the robustness of these tools for prediction in plants is still uncertain.
Density gradient purification of plant mitochondria has been optimized from a range of plant tissues and allows the rapid and high-purity recovery of these organelles (Day et al., 1985; Neuberger, 1985). Biochemical investigations have revealed that isolated plant mitochondria share many similarities to those from animals and fungi. However, plant mitochondria also contain additional features such as non-phosphorylating bypasses of the electron transport chain, specialized metabolite carriers, and enzymes involved in the synthesis of folate, lipoic acid, and vitamin C (Rebeille et al., 1997; Bartoli et al., 2000; Gueguen et al., 2000). Only a few of the nuclear genes encoding proteins that maintain these unique functions have been identified. In addition, the predictions by TargetP suggest that an array of other proteins whose functions have yet to be characterized are present in the nuclear genome of plants.
Several analyses of subcellular proteomes from Arabidopsis have been reported for the plasma membrane and endoplasmic reticulum systems (Santoni et al., 1998; Prime et al., 2000) and the chloroplast (Peltier et al., 2000). These studies have used a combination of Edman degradation and mass spectrometry (MS)/MS-based sequencing to identify proteins from two-dimensional gels. To determine which putative proteins from the Arabidopsis genome sequence represent constitutively expressed proteins for mitochondrial function, we have used two-dimensional analysis of purified Arabidopsis mitochondria coupled with peptide mass fingerprinting identification. This approach aims to identify proteins localized in plant mitochondria, thus providing new insights into the function of plant mitochondria and also providing a larger set of experimentally characterized mitochondrial proteins. This set of experimental knowledge could be used to improve neural network and/or rule based programming for targeting prediction of products from plant nuclear genomes.
RESULTS
Isolation of High-Purity Mitochondria from Arabidopsis Cell Culture
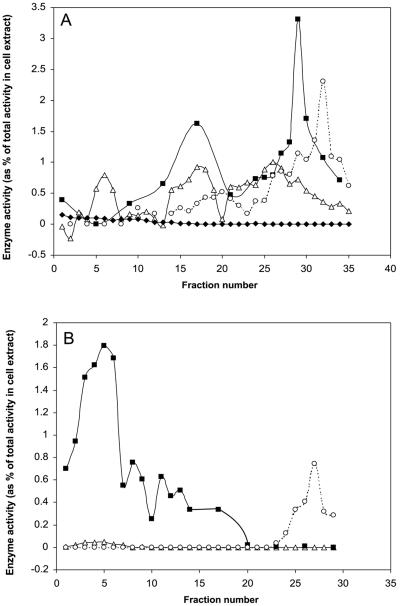
The integrity of a subcellular proteome, such as that of mitochondria, is largely dependent on the purification of the isolated compartment away from other cellular contaminants. We have used two Percoll gradient density separations that yield mitochondria that are essentially free of contamination by cytosol, peroxisomes, plastids, and other membranes (Fig. 1). On the first step-gradient, mitochondria band at the interface of the 23% and 40% Percoll steps (corresponding to the peak of cytochrome c oxidase activity in fractions 26–35; Fig. 1A). These mitochondria were contaminated by peroxisomes and plastids as revealed by the activity of catalase and alkaline pyrophosphatase, respectively. However, alcohol dehydrogenase could not be detected in these fractions, indicating the absence of contamination by cytosol. Interestingly, plastids resolved into three separate populations, and the presence of carotenoids (a commonly used marker for plastids) only correlated with one of the plastid peaks (fractions 13–20; data not shown). Thus, for Arabidopsis cells, the presence of carotenoids is not a reliable marker for the presence of all types of plastids. To further purify the mitochondria, a second self-forming gradient consisting of 28% Percoll was employed (Fig. 1B). Mitochondria formed a broad band in the upper part of the gradient, corresponding to a peak of cytochrome c oxidase activity at fraction 5. The mitochondrial band was free of contamination by peroxisomes (catalase activity was not coincident with cytochrome c oxidase activity, instead forming a peak lower down the gradient between fractions 23–29). A small peak of alkaline pyrophosphatase activity did coincide with the mitochondrial peak, indicating a slight contamination by plastids. However, based on the activity of the marker enzymes in fractions 1 through 14 in comparison with that present in the initial cell extract, the yield of mitochondria was 12.4%, whereas the yield of plastids in the same fractions was only 0.2%. This level of contamination is sufficiently minor that it is unlikely that we would detect plastid proteins in two-dimensional gels of the mitochondrial fraction.
Figure 1.
Isolation of Arabidopsis mitochondria by density centrifugation. The organelle pellet from homogenized Arabidopsis cell culture was loaded onto a Percoll step gradient consisting of steps of 40% (fractions 31–35), 23% (fractions 11–30), and 18% Percoll (fractions 1–10; A). After centrifugation, mitochondria were recovered from the 40%:23% interface (fractions 26–35) and were loaded onto a self-forming Percoll gradient containing 28% Percoll (B). One-milliliter fractions were collected from both gradients (from top to bottom) and the activities of cytochrome c oxidase (▪), catalase (○), alkaline pyrophosphatase (▵), and alcohol dehydrogenase (♦) were assayed in each fraction. Values are expressed as a percentage of the total activity in the initial cell extract.
Integrity and Function of Isolated Arabidopsis Mitochondria
We assayed the integrity and function of the purified mitochondria to ensure that proteins were not being lost by rupture during isolation and that key functions were maintained. The outer membrane of mitochondrial samples was found to be 97% intact based on the latency of cytochrome c oxidase activity initiated by exogenously added cytochrome c (Table I). Whole electron transport chain activities showed that oxygen consumption by the mitochondria could be supported by succinate, external NADH, or a combination of malate and pyruvate. Oxygen consumption supported by each of these substrates was significantly and transiently stimulated by addition of ADP. These data suggested the presence and function of the electron transport chain, the F1F0-ATP synthase, the tricarboxylic acid (TCA) cycle in the mitochondrial matrix, and the integrity of the inner membrane. Oxygen consumption by mitochondrial samples was largely inhibited by addition of the cytochrome oxidase inhibitor, KCN, and the small remaining rate was inhibited by n-propylgallate (nPG), an alternative oxidase inhibitor (Table I).
Table I.
Oxygen consumption by isolated Arabidopsis mitochondria following addition of different substrates, effectors, and inhibitors
| Substrate | +ADP | −ADP | +KCN | +nPG |
|---|---|---|---|---|
| nmol O2 min−1 mg−1 protein | ||||
| NADH | 233 ± 32 | 123 ± 13 | 10 ± 5 | 2 ± 2 |
| Succinate | 250 ± 34 | 122 ± 19 | 15 ± 5 | 1 ± 2 |
| NADH and succinate | 390 ± 52 | 166 ± 24 | 25 ± 7 | 1 ± 2 |
| Malate and pyruvate | 172 ± 18 | 66 ± 11 | 7 ± 3 | 1 ± 1 |
| Cytochrome c | 640 ± 86 | – | 3 ± 2 | – |
| Outer membrane Integrity (%) | 97 ± 1 | – | – | – |
Respiratory assays were performed according to “Materials and Methods.” Means ± sd (n = 3) for oxygen consumption rates are presented and outer membrane integrity is presented as a percentage of intact mitochondria.
Fractionation of Mitochondrial Proteins
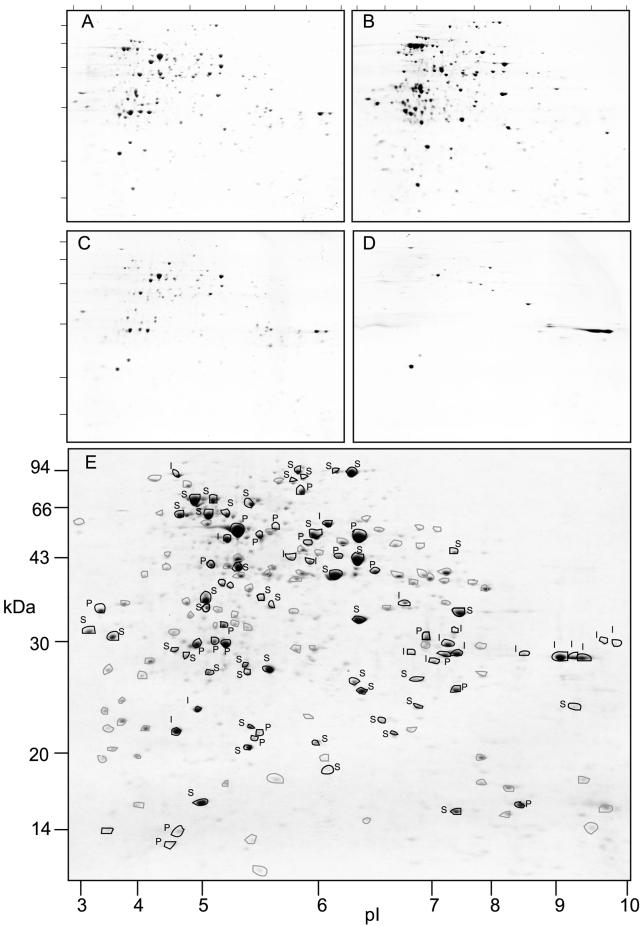
Mitochondrial protein samples were further fractionated based on the degree of membrane association (DMA). Total protein (A), soluble protein (B), membrane protein (C), and integral membrane protein (D) samples were separated by two-dimensional gel electrophoresis (Fig. 2, A–D). This analysis revealed varying patterns of protein spots in each gel corresponding to the relative abundance of each protein in the different compartments. Based on these gels, an overlay of the whole mitochondrial protein gel was made with protein spots highlighted that could be clearly identified by spot abundance changes to be peaking in amount in a particular fraction. In total, 163 protein spots were tracked for location in this manner. A set of 43 protein spots were identified as soluble proteins (S; present in A, peaked in B, and largely absent in C and D). A total of 21 protein spots were identified as peripheral membrane proteins (P; present in A, absent in B, peaking in abundance in C, but absent in D). Only 18 protein spots were deemed integral membrane (I; present in A, absent in B, and present in C and D in similar abundance). An additional 81 spots could not be easily assigned to any group as they appeared in abundance in several fractions (circled, unmarked). Allocation of protein spots to DMA groupings was based on densitometry analysis of gels by ImageMaster two-dimensional analysis software (Amersham Pharmacia Biotech, Sydney, Australia). Allocations to S, P, and I were only made if a protein spot abundance was more than 5-fold in the “present” versus “absent” gels for each class. For example, the abundance of an S protein in A and B was more than 5-fold that in C and D. The S, P, and I classes and no class (N) are also annotated on Table II in the DMA column.
Figure 2.
Two-dimensional separations of Arabidopsis mitochondrial proteins (pI 3–10) fractionated on the degree of membrane association. A, Total mitochondrial proteins; B, soluble proteins; C, total membrane proteins; D, integral membrane proteins based on Na2CO3 treatment; and E, predicted localization of 165 protein spots on the basis of distribution in A through D. Circled, black spots are annotated as I, S, or P. Circled, unannotated gray spots could not be assigned as I, S, or P based on their distribution between the gels. Numbers on the x axis are pI and numbers on the y axis are apparent molecular mass (kilodaltons) in E. A through D, Ticks marks show the position of pI 3 to 10 and the six molecular mass markers presented in E.
Table II.
Identification of two-dimensional separated protein spots from purified Arabidopsis mitochondria using matrix-assisted laser desorption ionization-time of flight (MALDI-ToF) spectra of trypsinated peptides matched against TrEMBL (TE), Genbank (GB), and the Arabidopsis Information Resource (AT) database (DB) entries
| Sample | DB | Entry | Details of Match | No. MP | Percentage Covered | MM (Da) Match | pl Match | Molecular Mass (Da) Gel | pl Gel | DMA | TargetP | MitoProt | Psort | GRAVY |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Tricarboxylic acid cycle | ||||||||||||||
| 3 | TE | O82661 | Succinyl-CoA-ligase α subunit | 7 | 31 | 36,152 | 8.7 | 36,000 | 7.6 | S | C5 | 0.9709 | Mma (0.898) | 0.000 |
| 100 | TE | O82662 | Succinyl-CoA-ligase β subunit | 9 | 27 | 45,346 | 6.7 | 44,000 | 5.4 | S | M2 | 0.9344 | Mma (0.508) | −0.053 |
| 101, 116, 165 | TE | Q9SIB9 | Aconitate hydratase | 10, 17, 18 | 15, 22, 33 | 98,155 | 6.1 | 93,000–98,000 | 5.9–6.3 | S | 2 | 0.0215 | C (0.450) | −0.184 |
| 21 | TE | Q9SZ36 | Aconitase hydratase | 9 | 15 | 98,953 | 6.2 | 97,000 | 5.8 | S | 2 | 0.0479 | C (0.450) | −0.161 |
| 15, 108 | TE | Q9M5K2 | Lipoamide dehydrogenase | 10, 14 | 36, 43 | 53,986 | 6.6 | 53,000–57,000 | 6.3–6.7 | PM | M1 | 0.9858 | Mma (0.845) | −0.023 |
| 86 | GB | AAF34795 | Lipoamide dehydrogenase | 12 | 34 | 52,872 | 6.5 | 52,000 | 6.7 | N | M3 | 0.9678 | Mma (0.845) | −0.041 |
| 44 | TE | O48685 | Pyruvate dehydrogenase E1 α subunit | 11 | 31 | 43,359 | 8.3 | 42,000 | 7.1 | N | M5 | 0.9982 | Mma (0.810) | −0.317 |
| 132 | TE | Q9SXC2 | Pyruvate dehydrogenase E1 α subunit | 12 | 23 | 43,059 | 7.6 | 41,000 | 6.8 | N | C5 | 0.9995 | Mma (0.903) | −0.330 |
| 18 | SP | ODPB_ARATH | Pyruvate dehydrogenase E1 β subunit | 6 | 21 | 39,188 | 5.8 | 37,000 | 5.2 | S | M1 | 0.9103 | Mma (0.788) | −0.002 |
| 56, 34 | TE | Q9ZP06 | Malate dehydrogenase | 5, 9 | 25, 37 | 35,805 | 8.7 | 35,000–37,000 | 5.7–6.5 | S | M4 | 0.9980 | Mma (0.920) | 0.165 |
| 87 | TE | P93033 | Fumarase | 9 | 34 | 52,830 | 8.2 | 50,000 | 7.5 | S | M4 | 0.6453 | Cls (0.826) | −0.168 |
| 104, 33 | TE | O64869 | Citrate synthase | 15, 13 | 39, 37 | 52,783 | 6.8 | 48,000 | 6.2–6.4 | S | M2 | 0.9710 | Mma (0.883) | −0.211 |
| 112 | TE | O65501 | NAD-isocitrate dehydrogenase subunit 1 | 11 | 28 | 39,627 | 8.5 | 41,000 | 7.2 | N | M2 | 0.9855 | Mma (0.576) | −0.050 |
| Electron transport chain | ||||||||||||||
| 46 | TE | O82663 | Succinate dehydrogenase α subunit | 15 | 32 | 69,657 | 6.2 | 69,000 | 5.5 | PM | M2 | 0.9416 | Mma (0.810) | −0.355 |
| 72 | SP | NUIM_ARATH | NADH-ubiquinone reductase 23-kDa subunit | 9 | 31 | 25,503 | 5.4 | 29,000 | 4.3 | N | M1 | 0.9680 | Mma (0.920) | −0.559 |
| 80 | SP | NUHM_ARATH | NADH-ubiquinone reductase 24-kDa subunit | 6 | 29 | 27,182 | 8 | 31,000 | 6 | N | M2 | 0.8675 | Mma (0.717) | −0.381 |
| 20 | GB | BAB10432 | NADH-ubiquinone reductase 13-kDa subunit | 6 | 35 | 19,179 | 4.7 | 22,000 | 4.3 | N | M2 | 0.8923 | Mma (0.470) | −0.478 |
| 22 | GB | BAB10668 | NADH-ubiquinone reductase 75-kDa subunit | 5 | 16 | 81,183 | 6.2 | 80,000 | 5.8 | PM | M3 | 0.9890 | Mma (0.648) | −0.129 |
| 91 | GB | AAG51074 | NADH-ubiquinone reductase B14 subunit | 7 | 58 | 15,082 | 9.1 | 16,000 | 9.5 | N | M5 | 0.2240 | C (0.450) | −0.238 |
| 4 | SP | ATPO_ARATH | ATP synthase Δ chain | 9 | 54 | 26,207 | 9.7 | 26,000 | 7.5 | PM | M4 | 0.8111 | Mma (0.683) | −0.106 |
| 24 | SP | ATP0_ARATH | ATP synthase α chain | 13 | 36 | 54,971 | 6.5 | 56,000 | 6.4 | PM | 4 | 0.2910 | Mma (0.479) | −0.070 |
| 23 | TE | O24345 | ATP synthase, β subunit | 10 | 36 | 49,135 | 5.3 | 57,000 | 5.4 | PM | – | – | – | – |
| 164 | TE | Q9SJ12 | ATP synthase Fad subunit | 8 | 38 | 27,597 | 6.6 | 31,000 | 5.3 | PM | M1 | 0.9000 | N (0.760) | −0.656 |
| 1 | GB | T46100 | ATP synthase D subunit | 8 | 48 | 19,586 | 5.1 | 23,000 | 4.8 | PM | 3 | 0.5303 | C (0.450) | −0.896 |
| 170 | TE | Q9SUU5 | Ubiquinol-cytochrome c reductase | 7 | 40 | 14,527 | 9.9 | 15,000 | 9.9 | N | M4 | 0.8264 | C (0.450) | −0.633 |
| 151 | TE | Q9SSB8 | Cytochrome c oxidase VB subunit | 6 | 29 | 18,583 | 5.7 | 15,000 | 4 | N | C4 | 0.9175 | Mma (0.759) | −0.321 |
| 82 | TE | Q9SRR8 | Adrenodoxin/mitochondrial ferridoxin | 5 | 30 | 17,602 | 8.3 | 17,000 | 5.7 | N | C5 | 0.9430 | Cls (0.623) | −0.275 |
| 10 | TE | Q9SRK3 | MN superoxide dismutase | 8 | 55 | 25,444 | 8.7 | 26,000 | 6.5 | S | M2 | 0.9609 | Mma (0.699) | −0.338 |
| 79 | GB | BAA96953 | UQ biosynthesis methyltransferase | 5 | 25 | 32,290 | 6.7 | 31,000 | 5.9 | N | M2 | 0.9970 | Mma (0.920) | −0.166 |
| Membrane carriers | ||||||||||||||
| 57, 96 | TE | Q9SRH5 | V-DEP anion-selective channel protein | 8, 8 | 45, 47 | 29,425 | 9 | 30,000–31,000 | 9.3–9.5 | IM | 2 | 0.1328 | N (0.760) | −0.114 |
| 14, 128 | TE | Q9SMX3 | V-DEP anion-selective channel protein | 7, 8 | 43, 51 | 29,211 | 8.4 | 30,000 | 8.1–9.2 | IM | 3 | 0.1576 | P (0.560) | −0.183 |
| RNA metabolism and translational apparatus | ||||||||||||||
| 47, 94, 168 | TE | Q9ZT91 | Mitochondrial elongation factor TU | 12, 12, 22 | 41, 39, 52 | 49,410 | 6.6 | 43,000 | 5.4–6.2 | N | C5 | 0.8743 | Cls (0.570) | −0.117 |
| 102, 103 | TE | Q9SHD6 | Mitochondrial elongation factor G | 9, 20 | 16, 31 | 83,112 | 6.3 | 86,000 | 6.7–6.8 | S | M4 | 0.8737 | ERm (0.600) | −0.259 |
| 145 | TE | O82505 | Translation elongation factor TS | 10 | 27 | 43,566 | 6 | 40,000 | 5.3 | N | M4 | 0.9538 | Mma (0.704) | −0.129 |
| 2 | TE | Q39105 | Gly-rich RNA-binding protein | 4 | 41 | 15,702 | 6.7 | 15,000 | 5.1 | S | M4 | 0.7432 | P (0.640) | −0.294 |
| 163 | GB | BAB01770 | Dead-box RNA helicase | 17 | 32 | 65,359 | 9.5 | 61,000 | 9.3 | N | M3 | 0.9822 | Mma (0.867) | −0.315 |
| Heat shock protein, chaperonin, and protein processing | ||||||||||||||
| 97 | GB | 116229 | Chaperonin 60 | 4 | 11 | 61,351 | 5.7 | 66,000 | 5.2 | S | M3 | 0.9898 | Mma (0.685) | −0.069 |
| 45 | TE | O49314 | Chaperonin 60 | 11 | 35 | 55,254 | 5.3 | 66,000 | 5.3 | S | 1 | 0.0495 | C (0.450) | −0.023 |
| 32, 99 | TE | Q9SZJ3 | Heat shock protein 70 | 10 | 18, 22 | 71,174 | 5.3 | 67,000, 74,000 | 4.8, 5.1 | S | C5 | 0.8846 | Cls (0.520) | −0.298 |
| 28 | GB | T49939 | Heat shock protein 70 | 13 | 35 | 72,991 | 5.6 | 74,000 | 5.3 | S | M4 | 0.9879 | Mma (0.907) | −0.306 |
| 30 | TE | Q9S7E7 | Putative heat shock protein 90 | 18 | 33 | 90,956 | 5.3 | 94,000 | 4.8 | IM | M4 | 0.9919 | Mma (0.793) | −0.600 |
| 31 | TE | Q9SIF2 | Putative heat shock protein 90 | 9 | 16 | 88,663 | 5 | 90,000 | 4.5 | N | C3 | 0.7324 | N (0.600) | −0.552 |
| 65 | GB | BAB09831 | ATP-DEP protease (clpP-like) | 8 | 30 | 26,283 | 6.7 | 27,000 | 6.4 | S | M3 | 0.7529 | P (0.640) | 0.031 |
| 25, 167 | TE | Q9SGA7 | Processing peptidase β subunit | 12, 18 | 34, 39 | 59,160 | 6.7 | 56,000, 61,000 | 5.6, 6.2 | IM | M2 | 0.9910 | Mma (0.920) | −0.342 |
| 17 | TE | Q9ZU25 | Processing peptidase α subunit | 12 | 32 | 54,402 | 6.2 | 54,000 | 5.3 | IM | M3 | 0.9556 | Mma (0.898) | −0.126 |
| 13 | TE | O04331 | Prohibitin | 9 | 51 | 30,400 | 7.7 | 31,000 | 7.5 | IM | S2 | 0.5098 | C (0.450) | −0.013 |
| 58 | TE | O49460 | Prohibitin | 6 | 33 | 31,707 | 9.5 | 32,000 | 9.7 | IM | 3 | 0.5580 | ERm (0.550) | −0.128 |
| 156 | TE | Q9ZNT7 | Prohibitin 2 | 10 | 50 | 31,811 | 9.6 | 31,000 | 9.8 | IM | S2 | 0.5916 | C (0.650) | −0.155 |
| 166 | TE | Q9SIL6 | Prohibitin | 12 | 58 | 31,637 | 9.9 | 31,000 | 9.8 | IM | 4 | 0.3587 | ERm (0.550) | −0.140 |
| 169 | GB | BAB02123 | Prohibitin | 10 | 44 | 30,638 | 6.9 | 31,000 | 6.7 | IM | S1 | 0.3396 | Cls (0.471) | −0.083 |
| 89 | GB | BAB03165 | TOM40 protein receptor | 11 | 39 | 32,316 | 5.9 | 37,000 | 6.8 | IM | 2 | 0.0737 | P (0.397) | −0.224 |
| Carbon metabolism | ||||||||||||||
| 16 | TE | Q9SU63 | NAD+ aldehyde dehydrogenase | 12 | 35 | 58,589 | 7.6 | 56,000 | 6 | S | M3 | 0.9988 | Mma (0.920) | −0.072 |
| 158 | GB | BAB11503 | Proline/aldehyde dehydrogenase | 7 | 20 | 61,742 | 6.3 | 62,000 | 6.5 | N | M3 | 0.9841 | Mma (0.808) | −0.159 |
| 40, 43 | TE | Q43314 | Glutamate dehydrogenase 1 | 5, 8 | 15, 25 | 44,524 | 6.8 | 43,000 | 7–7.4 | S | M4 | 0.6226 | Mma (0.665) | −0.174 |
| 41 | TE | Q38946 | Glutamate dehydrogenase 2 | 10 | 34 | 44,699 | 6.4 | 44,000 | 6.5 | PM | M3 | 0.7886 | Mma (0.659) | −0.148 |
| 26 | TE | Q9SEK4 | Succinic semialdehyde dehydrogenase | 17 | 48 | 56,559 | 6.9 | 57,000 | 5.6 | PM | M3 | 0.9856 | Mma (0.601) | 0.036 |
| 54 | SP | CYSM_ARATH | Cys synthase | 8 | 26 | 45,149 | 8.8 | 39,000 | 5.5 | N | C4 | 0.9698 | Mma (0.741) | −0.086 |
| 76 | TE | O64530 | 3-Mercapopyruvate sulphurtransferase | 9 | 28 | 41,893 | 6.3 | 37,000 | 5 | N | C4 | 0.9860 | Mma (0.814) | −0.292 |
| 107 | TE | Q9SWG0 | Isovaleryl-CoA-dehydrogenase | 7 | 18 | 44,772 | 7.9 | 43,000 | 5.9 | N | M3 | 0.9893 | Mma (0.884) | −0.181 |
| 27 | TE | Q9SI43 | Methylmalonate semi-aldehyde DH | 6 | 11 | 65,927 | 9 | 59,000 | 5.7 | PM | M4 | 0.7168 | P (0.300) | −0.176 |
| 111 | GB | U15026 | Asp aminotransferase | 12 | 32 | 47,758 | 8.5 | 41,000 | 7.8 | N | M4 | 0.9264 | Mma (0.674) | −0.303 |
| 154 | GB | AAF827821 | Ala aminotransferase | 8 | 15 | 59,807 | 5.9 | 60,000 | 5.2 | N | M5 | 0.8319 | Mma (0.609) | −0.254 |
| 141 | TE | Q9SZ48 | Enoyl-CoA hydratase-like protein | 6 | 24 | 46,063 | 5.1 | 48,000 | 4.6 | N | M1 | 0.8055 | Mma (0.647) | −0.171 |
| 130 | TE | Q9SMN1 | Acetyltransferase-like protein | 4 | 26 | 27,956 | 9 | 28,000 | 7.9 | N | M5 | 0.9958 | Mma (0.908) | 0.041 |
| 11 | GB | AAG52641 | Acetyltransferase-like protein | 12 | 59 | 30,065 | 6.7 | 32,000 | 7.3 | IM | M2 | 0.7411 | C (0.650) | −0.157 |
| 131 | GB | BAB10927 | Ferripyrochelin-binding protein-like | 6 | 50 | 22,601 | 5.6 | 33,000 | 7.5 | IM | 2 | 0.0480 | C (0.650) | −0.223 |
| 50 | TE | O82514 | Adenylate kinase | 7 | 41 | 26,934 | 8.5 | 32,000 | 7 | PM | 2 | 0.0150 | C (0.450) | −0.400 |
| 53 | TE | O23443 | Isomerase like protein | 5 | 33 | 24,154 | 7.6 | 27,000 | 6.9 | S | M5 | 0.9880 | Mma (0.464) | −0.023 |
| Unknown function | ||||||||||||||
| 19 | TE | Q9SX77 | Unknown function | 6 | 36 | 28,106 | 6.6 | 20,000 | 4.2 | N | M2 | 0.3842 | C (0.650) | −0.323 |
| 105 | TE | Q9SZT3 | Unknown function | 13 | 30 | 54,217 | 7.9 | 48,000 | 6 | N | M3 | 0.0709 | Mma (0.681) | −0.363 |
| 106, 148 | TE | Q9T082 | Stomatin like protein | 11, 11 | 23, 24 | 55,923 | 5.2 | 47,000, 49,000 | 5.8, 5.9 | IM | M3 | 0.9912 | Mma (0.497) | −0.375 |
| 48 | GB | T50795 | Bacterial inosine-5′-MP dehydrogenase-like | 8 | 38 | 22,729 | 9.1 | 19,000 | 7.8 | N | M5 | 0.8549 | C (0.650) | −0.224 |
| 125 | GB | AAF75065 | Unknown function | 5 | 23 | 21,410 | 5.5 | 22,000 | 5.6 | PM | C2 | 0.0148 | Cls (0.871) | −0.339 |
| 143 | TE | Q9SHJ6 | Unknown function | 7 | 27 | 36,119 | 4.8 | 42,000 | 4.3 | N | 1 | 0.0048 | PM (0.700) | −0.844 |
| 144 | AT | AT2G28000 | Unknown function | 12 | 25 | 62,072 | 5.1 | 65,000 | 4.5 | N | C2 | 0.9984 | Cls (0.927) | 0.002 |
| 78 | TE | Q9ZVC3 | Unknown function | 9 | 42 | 30,558 | 5.4 | 34,000 | 5.3 | PM | 5 | 0.1412 | P (0.559) | −0.409 |
| 124 | TE | O25390 | Bacterial oxidoreductase-like | 5 | 28 | 32,941 | 6 | 35,000 | 5.8 | N | 4 | 0.2212 | P (0.640) | −0.159 |
No. MP, No. of peptides matching ± 50 ppm to predicted protein sequence; percentage covered, percentage of predicted protein sequence covered by matched peptides; predicted molecular mass and pI of matched sequence and observed MM and pI of the sample is from the gel in Figure 3. TargetP, Predicted localization of sequence by TargetP; M1–5, mitochondrial; C1–5, chloroplast; S1–5, secretory pathway; 1–5, other (1, high probability; 5, low probability); MitoProt, probability of mitochondrial targeting (P = 0–1); Psort, predicted localization; Mma, mitochondrial matrix; Cls, chloroplast; C, cytosol; N, nucleus; ER, endoplasmic reticulum; PM, plasma membrane; P, peroxisome (P = 0–1). DMA, Degree of membrane association as determined from Figure 2; GRAVY, grand average of hydropathicity index calculated using the matched protein sequence.
MALDI-ToF Peptide Mass Fingerprinting of Arabidopsis Mitochondrial Proteome
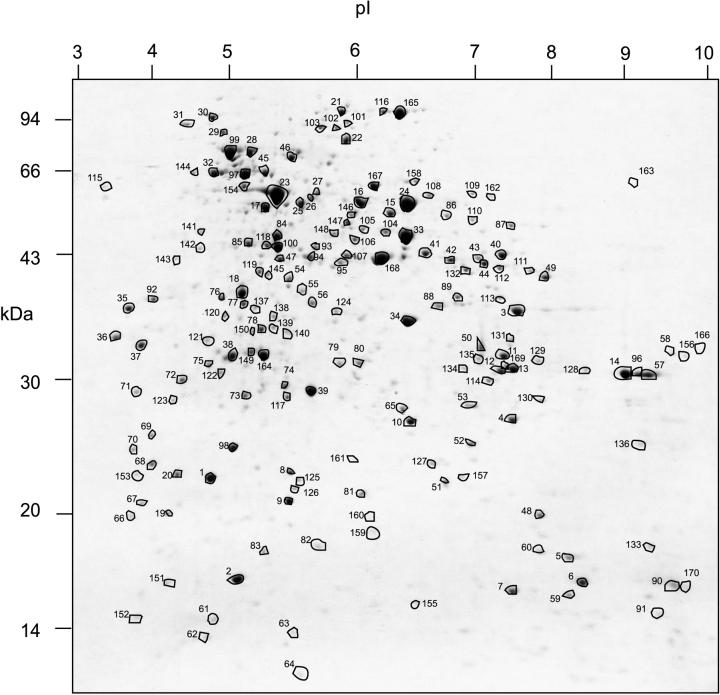
A set of 170 protein spots, including those identified in Figure 2 and several other less-abundant proteins, were chosen because they were reproducibly observed in the mitochondrial proteome throughout the different experiments undertaken. These protein spots were excised from whole mitochondria profile gels and were in-gel digested for further MS analysis of the resulting peptide fragments (Fig. 3). MALDI-ToF provided a peptide mass spectrum for each protein spot. From this total of 170 proteins, 15 samples provided poor spectra that could not be used for further analysis (6, 29, 36, 42, 52, 63, 64, 70, 71, 75, 113, 114, 123, 134, and 135), and the remaining 155 were used for database (DB) searches to identify genes encoding proteins with similar peptide mass fingerprints. A total of 81 of the spectra were matched to 68 predicted Arabidopsis protein sequences in the DB with known function based on functional studies or sequence comparison with proteins of known function (Table II). A further 10 spectra matched to nine predicted Arabidopsis protein sequences, largely from genome sequencing for which no function has been identified by functional studies or comparative genomics. The apparent and predicted molecular mass and pI of the matched samples are consistent with expectation. In cases where a mitochondrial targeting presequence is present, we expect a decrease of 1 to 4 kD between predicted and apparent molecular mass, and we expect a more major shift in the pI of the protein in the acidic direction following the removal of the typically basic presequence. In cases where no presequence is present (such as the porins, TOM40, and aconitases), the apparent and predicted molecular mass and pI values are much closer. Overall, the sequences, excluding those known not to contain presequences, apparent molecular masses were 1.5 ± 0.5 kD smaller than precursor proteins predicted masses. The pI values of protein spots observed on gels were also shifted 0.85 ± 0.15 units in the acidic direction compared with predicted precursor protein pI values (Table II).
Figure 3.
Two-dimensional separation of Arabidopsis total mitochondrial proteins (pI 3–10). A total of 170 protein spots from this map were excised, digested, and analyzed by MALDI-ToF to yield a peptide mass fingerprint for DB searching. Numbers indicate spot number for comparison with text and Table II. Numbers on the x axis are pI and numbers on the y axis are apparent molecular mass (kilodaltons).
Membrane Channels/Carriers
The voltage-dependent anion-selective channel proteins, or porins, allow channel transport of small molecules (<8 kD) across the outer mitochondrial membrane. Four proteins in the Arabidopsis mitochondrial profile (14, 57, 96, and 128) were matched to two separate Arabidopsis genes encoding this class of channel protein. All four of these proteins were identified in the integral membrane protein set based on the DMA analysis of Figure 2.
Protein Import, Processing, Chaperonins, and Degradation
Three types of the classical heat shock/chaperonin proteins were identified, those in the 60-, 70-, and 90-kD classes (45, 97, 32, 99, 28, 30, and 31). The partner protein of HSP60, known as HSP10, was not identified; it would have been too small (approximately 10 kD) to be observed on the SDS-PAGE gels in these experiments. The new class of molecular chaperonins recently characterized in mammalian mitochondria, known as prohibitins (Nijtmans et al., 2000), were also identified in our mitochondrial samples. A total of five prohibitin-like proteins were identified (13, 58, 156, 166, and 169) and matched as the products of five separate genes in Arabidopsis. In addition, the TOM40 receptor for protein import on the outer mitochondrial membrane was identified (98), along with both subunits of the mitochondrial-processing peptidase responsible for removal of N-terminal targeting presequences from mitochondrial proteins (17, 25, and 168). A clpP-like ATP-dependent protease probably involved in proteolysis in mitochondria (65) was also identified. Recently, antibodies raised to a recombinant Arabidopsis protein, designated by homology to be a putative plant mitochondrial clpP, have been used to localize a cross-reacting protein in mitochondria from pea (Pisum sativum; Halperin et al., 2001). In yeast, clpP-like proteases are known to be associated in large supercomplexes with prohibitins in mitochondria, and genetic evidence suggests prohibitins have a negative regulatory effect on protease function (Steglich et al., 1999). Further investigation of a prohibitin/ATP-dependent protease complex in plant mitochondria is warranted.
TCA Cycle Enzymes
Proteins from all of the TCA cycle enzyme complexes were identified, and these included subunits of pyruvate dehydrogenase complex (44, 132, 18, 15, 108, and 86), citrate synthase (104 and 33), isocitrate dehydrogenase (112), 2-oxoglutarate dehydrogenase (15, 108, and 86), succinyl coenzyme A (CoA) ligase (3 and 100), succinate dehydrogenase (46), fumarase (87), and malate dehydrogenase (56 and 34). The two pyruvate dehydrogenase E1α subunits (44 and 132) share 87% sequence identity and are only 50% to 52% identical to the plastidic E1α-subunit sequence (GB U80185), suggesting they are mitochondrial E1α subunits. Notable absences from the list include NAD-malic enzyme that catalyzes the conversion of malate to pyruvate in the plant TCA cycle. The activity of this enzyme was present in Arabidopsis mitochondria (data not shown). Previous studies by several of the authors in potato mitochondria showed that the α- and β-subunits of NAD-malic enzyme enter two-dimensional gels and cross-react with antibodies raised to NAD-malic enzyme (Jenner et al., 2001). None of the E2 subunits of pyruvate or 2-oxoglutarate dehydrogenase complexes were identified, although these proteins are present in two-dimensional gels in mitochondrial samples from other plant species (data not shown). The modification of these proteins by lipoic acid moieties might explain why they are not identified by mass fingerprinting, as these modifications will alter the apparent molecular masses of peptides derived from these protein sequences.
Aconitase
In yeast, a single nuclear gene encodes aconitase, and “inefficient import” of this protein is believed to explain the cytosolic and mitochondrial localization of this protein (Gangloff et al., 1990). It had been suggested that a single gene also encoded aconitase in Arabidopsis (Peyret et al., 1995); however, subsequent genome sequencing has shown there are four aconitase genes in this plant species. We have identified four aconitase proteins (101, 116, 165, and 21) as the products of two aconitase genes (Q9SZ36 and Q9SIB9) in the mitochondrial profile. The predicted protein sequences from these genes are only 75% and 84% identical to the original aconitase cloned and entered into SWISS-PROT (Q42560) as the cytoplasmic aconitase hydratase by Peyret et al. (1995). Neither of the two predicted protein products has putative mitochondrial targeting sequences. The mechanism of import of these large proteins into the mitochondrial matrix in the absence of a presequence is not known.
Lipoamide Dehydrogenase
Two distinct lipoamide dehydrogenase proteins were identified as the products of Q9M5K2 and AAF34795. These predicted proteins are identical in length (507 amino acids) and share 94% identity at the amino acid level. The power of peptide mass fingerprinting is shown by the ease of differentiation of these two proteins by peptide matches. Only four peptides in both spectra of identical size were matched to identical sequence in both predicted protein sequences, an additional six to eight peptides in each case matched to regions with one or two amino acid substitutions altering peptides mass in each predicted protein sequence (data not shown). The presence of these two mitochondrial proteins is in contrast to a long-standing debate in the literature about the number of lipoamide genes and protein products that serve as the dehydrogenases for the 2-oxo acid multienzyme complexes and Gly decarboxylase complex in mitochondria and chloroplasts. It has been held that a single product of a single gene encoding lipoamide dehydrogenase was targeted to both organelles and partnered all these multienzyme complexes. Recently, two genes encoding plastid specific lipoamide dehydrogenases have been identified (Lutziger and Oliver, 2000). The same authors cloned and entered both of these mitochondrial lipoamide dehydrogenases sequences (Q9M5K2 and AAF34795) into public DBs in 2000. This is the first evidence to our knowledge that these two genes are translated to form lipoamide dehydrogenase proteins that accumulate in plant mitochondria. Further, it appears there is significantly more Q9M5K2 than AAF34795 protein in our mitochondrial samples (Fig. 3).
Electron Transport Chain
Representative subunits from each of the classical electron transport chain complexes were identified. These included five subunits of complex 1 (NADH-ubiquinone [UQ] oxidoreductase 72, 80 20, 22, and 91), three subunits of complex III (UQ-cytc oxidoreductase 171, 17, 168, and 25), five subunits of complex V (ATP synthetase complex, 1, 4, 24, 23, and 164), and one subunit each from complex II (succinate dehydrogenase, 46) and complex IV (cytochrome c oxidase, 151). No sequence entry for the β-subunit of the mitochondrial ATP synthase from Arabidopsis could be found by the authors. Matches to the spectrum from spot 23 were found to the β-subunit from a wide range of other plant species; in Table II, the β-subunit from Sorghum bicolor was matched to 205. The reason for the absence of an entry for this major mitochondrial protein from National Center for Biotechnology Information, Translated European Molecular Biology Laboratory Nucleotide Sequence Database, and the The Arabidopsis Information Resource DBs is unclear. No peptide fingerprint spectra matched to protein sequences for the non-phosphorylating bypasses of the plant electron transport chains, notably the alternative oxidase protein sequences (Saisho et al., 1997) or the recently identified putative rotonone-insensitive NADH-dehydrogenase protein sequences (Rasmusson et al., 1999). This absence was despite the evidence from Table I of the operation of both of these bypasses in intact Arabidopsis mitochondria. A protein with significant homology with the mitochondrial class of ferrodoxins in mammals known as adrenodoxins was also found (82). In mammals, this class of ferrodoxin specifically function in a redox chain with an adrenodoxin reductase to reduce cytochrome P450s in mitochondria (Solish et al., 1988). Some evidence for a mitochondrial cytochrome P450 in plants has been presented (Lindemann and Luickner, 1997), but none of the large number of P450 genes in Arabidopsis have been characterized as mitochondrial, and a redox partner for this adrenodoxin in plant mitochondria remains elusive. The mitochondrial matrix isoform of superoxide dismutase with Mn at its active site was also identified (10), as was a methyltransferase involved in the final step of UQ synthesis (79).
Amino Acid Metabolism
A variety of enzymes associated with amino acid metabolism were also identified. Ala and Asp aminotransferases (111 and 154) allow the interconversion of TCA cycle intermediates with amino acid pools. Several broad range enzymes were also identified that have the potential to be involved in a variety of amino acid degradation pathways with an array of different substrates; these include the two aldehyde dehydrogenases (158 and 16) and enoyl CoA hydratase (141). The aldehyde dehydrogenases function broadly in amino acid and fatty acid metabolism to catalyze the oxidation of aldehyde or oxo groups to form carboxylates through the reduction of NAD(P) to NAD(P) H. Interestingly, it was a putative aldehyde dehydrogenase that was identified as the rf2 nuclear restorer of the T-cytoplasm maize (Zea mays) cytoplasmic male sterility line (Cui et al., 1996). Enoyl CoA hydratase catalyzes the removal of water from a variety of unsaturated acyl-CoAs in the degradation pathways of Val, Leu, iso-Leu, Lys, Trp, and β-Ala. Methylmalonate semi-aldehyde dehydrogenase (27) acts downstream of enoyl coA hydratase in the catabolism of branched chain amino acids.
Pyruvate is converted to Cys via the intermediate, 3-mercaptopyruvate, and Ser is converted to Cys via an acetyltransferase and Cys synthase. The presence of 3-mercaptopyruvate sulfur transferase (76), Cys synthase (54), and acetyltransferase-like proteins (11 and 130) suggests that such metabolic pathways may be in action in plant mitochondria. One of the substrates for 3-mercaptopyruvate sulfur transferase is hydrogen cyanide, which is also a potent inhibitor of respiration by the cytochrome pathway. It would be interesting to consider if the operation of this metabolic pathway may be correlated with the expression of the alternative oxidase, which is insensitive to cyanide and thus able to maintain electron transport. Recently, a β-cyano-Ala synthase, which also uses hydrogen cyanide as a substrate and is structurally related to the Cys synthases, was identified as a mitochondrial protein in plants (Hatzfeld et al., 2000).
γ-Aminobutyric Acid (GABA) Shunt Metabolism
Several enzymes potentially involved in a GABA shunt of carbon intermediates between 2-oxoglutarate and succinate were also identified. Glu dehydrogenase (40 and 43) forms Glu from 2-oxoglutarate to begin the shunt and succinic semialdehyde dehydrogenase (26) concludes the shunt by the formation of succinate. Recently, work by Busch and Fromm (1999) has identified that an Arabidopsis succinic semialdehyde dehydrogenase was localized in mitochondria. Previously, they had identified a Glu decarboxylase in the cytosol (Snedden et al., 1996), but have not found evidence of GABA aminotransferase gene(s), which are likely to code for mitochondrial protein(s), for the completion of this putative shunt (Busch and Fromm, 1999). Several aminotransferases were identified in this study (154 and 111) and have been annotated based on average homology to transferases with known substrates. A range of other aminotransferases with some homology to GABA and β-Ala aminotransferases are also present in Arabidopsis protein DBs, and several have predicted mitochondrial presequences. Any of these proteins may act as a GABA aminotransferase to complete this stunt.
RNA Metabolism and Translation Apparatus
Proteins involved in mitochondrial RNA metabolism were also found in the Arabidopsis mitochondrial proteome. Among these, a DEAD box RNA helicase (163) was identified. A different protein of this family (AtSUV3) has recently been characterized in Arabidopsis mitochondria (Gagliardi et al., 1999). The function of such proteins in plant mitochondria remain unclear, but they are predicted to be involved in transcript unfolding during, for example, transcript maturation or degradation processes. In yeast, a homologous helicase, SUV3, is a component of the mitochondrial 3′- to 5′-RNA degradosome (Margossian et al., 1996). The presence of at least two proteins of this family in plant mitochondria might suggest that different helicases could be transcript specific, or alternatively, that they could be mechanism specific. For example, one could be involved in degradation and another in transcript maturation. Another protein showing a typical Gly-rich domain, which is characteristic of RNA-binding proteins, was also identified (2). This protein might be a component of the mitochondrial ribosome; it shows high similarity (data not shown) to the Arabidopsis mitochondrial ribosomal protein rps19 (Sanchez et al., 1996). Several members of the plant mitochondrial translational apparatus, the elongation factors Tu, G, and TS, were also found in this analysis (47, 94, 168, 102, 103, and 145). The mitochondrial EF-Tu protein has been characterized for Arabidopsis (Kuhlman and Palmer, 1995) and recently for maize (Choi et al., 2000). However, the putative functions of Q9SHD6 and O82505 as elongation factors G and TS are only based on homology to yeast mitochondrial proteins (Vambutas et al., 1991).
Comparisons of Targeting Prediction Programs Using Proteome Data
All the identified protein sequences were queried through three intracellular targeting prediction program: TargetP, Psort, and MitoProt. TargetP and Psort assess a number of potential targeting destinations. They identified 54 and 49 protein sequences, respectively, as mitochondrial targeted based on their N-terminal region (Table II). In this analysis, TargetP was used in its winner-takes-all mode without setting a specificity cut-off for targeting. MitoProt only predicts the likelihood of targeting to the mitochondria based on the probability that a presented sequence belongs to the group of known presequences based on 47 weighted characteristics. MitoProt identified 53 proteins as putatively mitochondrial based on a probability cut-off of >0.85. Notably, all three prediction programs only agreed on a subset of 32 of these protein sequences as being likely targeted to mitochondria. TargetP and Psort identified 16 and 13 sequences, respectively, as chloroplast targeted, agreeing on only nine of this set. The disputed sequences in most cases were assessed as mitochondrial by the other prediction program. This set included a range of well-known mitochondrial proteins such as subunits of succinly CoA ligase (3), pyruvate dehydrogenase complex (132), cytochrome c oxidase (151), and the mitochondrial elongation factor Tu (47, 94, and 168). A remaining 20 to 30 proteins sequence, depending on the program, were not considered to contain mitochondrial targeting sequences by the three prediction programs. This set of protein sequences included two aconitase protein sequences, the four prohibitin protein sequences, the two porin sequences, the TOM40 import pore protein sequence, and the adenylate kinase protein sequence. All these proteins are known to be mitochondrial, but their location (predominantly but not exclusively in the outer membrane and intermembrane space) does not require classical import by cleavable N-terminal extensions. Of the 10 proteins of unknown function identified in this study, a total of six contained putative N-terminal targeting sequences. The remaining four (125, 143, 78, and 124) do not contain clearly identifiable targeting sequences; however, as we have seen above, this may not preclude their mitochondrial localization.
GRAVY Scores, Protein Solubility, and Membrane Localization
A GRAVY score is a single value indication of the overall hydropathicity of a protein sequence based on the Kyte and Doolittle algorithms—the more positive the score the more hydrophobic the overall sequence (Kyte and Doolittle, 1982). The addition of thiourea as a chaotropic agent, the advent of new nonionic sulfobetaine detergents, and the use of the noncharged reductant tributylphosphine have all be credited with allowing more hydrophobic proteins to enter two-dimensional gels for separated and analysis (Chevallet et al., 1998; Herbert et al., 1998; Rabilloud, 1998). To date, proteins with GRAVY scores of approximately +0.25 to +0.30 are considered to represent the limit of these two-dimensional gel solubility improvements (Herbert, 1999). Our analysis has revealed several proteins with positive GRAVY scores of 0.000 to +0.165; however, these are unlikely proteins to be considered “highly hydrophobic.” They include malate dehydrogenase (56 and 202), the α-subunit of succinyl CoA ligase (3), and the ATP-dependent protease (65), which were designated in the soluble protein sample in Figure 2, as well as a peripheral membrane protein (26) and two unassigned proteins (130 and 144). The integral membrane proteins identified such as porin (57, 96, 14, and 128), prohibitins (13, 156, 166, and 169), and processing peptidases (25, 24, and 17) did not have positive GRAVY scores. Our DMA localization study (Fig. 2; Table II) showed that many of the major proteins shown to be soluble were subunits of the TCA cycle and the major HSPs of the 60 and 70 classes. The subunits of the F1Fo ATP synthase were all in the peripheral membrane class (Table II). So overall, although the localization of the identified proteins as soluble, peripheral membrane, or integral membrane are largely in agreement with expectation, the GRAVY scores do not correlate well with these DMA localizations.
DISCUSSION
A number of reports have presented two-dimensional gel arrays of mitochondria protein profiles from pea (Humphery-Smith et al., 1992), potato (Solanum tuberosum; Colas des Francs-Small et al., 1992), Arabidopsis (Davy de Virville et al., 1998), and maize (Dunbar et al., 1997). Most of these reports have highlighted changes that occur in this profile in different tissues from the same plant or during developmental changes within the same cell type. However, identifications of the different protein spots in these profiles have been limited by the need for cross-reacting antibodies or sufficient material and finance for N-terminal sequencing. The result has been a rather limited understanding of many of the major proteins that are found in the mitochondrial profile. Recently, Prime et al. (2000) identified six mitochondrial membrane-associated proteins in their Arabidopsis organelle membrane preparations using a combination of Edman degradation and MS/MS-based sequencing. Here, we show that the availability of full Arabidopsis genome sequence now enables the use of less expensive, high-throughput identifications of even very low abundance plant proteins from two-dimensional gel profiles by peptide mass fingerprinting using MALDI-ToF MS data. This opens the way for full subproteome analyses in plants. We have used a cell suspension culture in our experiments, which has the advantages of lower cell heterogeneity, higher cytoplasmic contents, and greater mass yields than a whole plant Arabidopsis system. However, extension of this research into whole plants and into specific differentiated plant organs has the potential to identify other proteins associated with metabolic pathways not evident in cell culture.
Identifying Functions in Plant Mitochondria
Based on the identifications we have obtained, our attempts to minimize contamination of our mitochondrial samples from plastid and peroxisomal material has been largely successful (Fig. 1). We have not identified any photosystem proteins, components of the Calvin cycle, or major peroxisomal proteins (Table II). By analysis of the major proteins of Arabidopsis mitochondria, we have begun to gain a more global appreciation of which proteins are present and thus which metabolic operations are possible in plant mitochondria. This analysis also provides insight into the relative abundance of different proteins that may provide the framework for determining turnover rates of different enzymes and the putative maximal fluxes of metabolic pathways. It is clear that from the results presented, the TCA cycle, the electron transport chain, and HSP60/HSP70s dominate the list of identifications (Table II) and the highly abundant proteins visualized on the two-dimensional gel profile (Fig. 3). The studies of these components in the plant mitochondrial research literature are significant, no doubt aided by this abundance and the extensive knowledge of these metabolic processes from mammals. Relatively little work has focused on the amino acid catabolism by plant mitochondria that is highlighted by the identifications in Table II. In addition, we know little about the potential presence of a GABA shunt, the possibility of P450 reactions, or the role of the different classes of chaperonins in plant mitochondria (Table II). Furthermore, more extensive investigations will be required to identify the status and importance of the unknown function proteins identified in this study.
A variety of mitochondrial proteins that are known to be present in significant abundance were not observed in this study. These include nearly all of the proteins encoded in the mitochondrial genome and a range of inner membrane carriers. In both of these cases, the hydrophobicity and basic nature of the protein sequences place them outside the resolving ability of current two-dimensional electrophoresis. The complex I, ATP synthase, cytochrome b/c1 complex, and cytochrome oxidase subunits encoded in the mitochondrial genome have GRAVY scores greater than +0.3 in almost every case, and the ribosomal proteins in the mitochondrial genome are more soluble in nature but have pI values of 10.4 to 11.3. The inner membrane carriers have pI values in excess of 9.5 and are also hydrophobic containing six transmembrane domains.
Peptide Mass Fingerprinting in Arabidopsis
Starting with a total of 170 spectra, we had 155 high-quality spectra for DB searching. A total of 91 spectra were matched to 77 different predicted protein sequences, providing a percentage hit rate of 59%. In the set of 77 protein sequences, only 11% were unknown proteins based on sequence comparison searches. In yeast and bacterial systems, peptide mass fingerprinting has yielded an approximately 90% hit rate (Shevchenko et al., 1996), and 30% to 40% of open reading frames (ORFs) identified by genomic sequencing are assigned as unknown function by sequence comparisons (The Arabidopsis Genome Initiative, 2000). Although our data are only preliminary, an explanation of what lies behind these statistics seen in this Arabidopsis study is required to further our analysis of the mitochondrial proteome of plants.
The inability to match 40% of spectra to predicted ORFs is likely to be due to at least two factors. First, post-translational modifications to proteins will alter the apparent masses of peptides and preclude matching to DB entries. The presence of such modifications is evident from the differences in apparent molecular mass and pI that were recorded for protein spots that were matched to the same gene product (Table II). Such modifications can be recognized by MS/MS-based analysis of peptide sequences, and sequence tag information afforded by these MS technologies will help to identify heavily modified proteins in the future. Second, matching to translated ORFs requires that intron-exon boundaries have been accurately determined, are in frame, and that all ORFs in the genomic sequence have been recognized. Searches of redundant DBs of translated ORFs such as GENpept and TE readily illustrate that intron-exon boundaries are not always identified accurately in Arabidopsis sequences. This phenomenon will be exacerbated in the case of unknown function proteins where sequence alignments to characterized orthologs do not provide a ready check of these splicing sites and frame shifts. This might explain the low overall hit rate and the even lower hit rate on unknown function translated ORFs in our data (Table II). A solution to this problem would be searches of peptide mass data not only against predicted translated ORFs, but also against six frame translations of the full genomic sequence of Arabidopsis. This would not only improve peptide mass fingerprinting searches, but will also act as a tool for the improvement of Arabidopsis ORF identification and correct construction from raw genomic sequence to supplement the use of expressed sequence tag sequence.
Future Mitochondrial Proteome Prediction
If the plant mitochondrial proteome is defined as the native proteins encoded on the mitochondrial genome and all the proteins encoded in the nucleus that could possibly be targeted to this organelle, then its elucidation is not a trivial matter. Comparison of MitoProt, Psort, and TargetP suggests that no one program currently provides a complete, high confidence analysis of the nuclear encoded components of the plant mitochondrial proteome. The small use of characterized plant mitochondrial proteins in the establishment of these tools and the abundant evidence of mitochondrial targeting by means other than N-terminal extensions do exacerbate these problems. The experimental approach we have outlined here provides direct evidence of proteins that are localized in mitochondria. However, this experimental proteome is limited to those proteins that accumulate to significant levels, that are expressed in the tissue sample used, and that are able to remain soluble during sample handling and separation. Further analysis of low-abundance proteins will also no doubt reveal low levels of contamination of mitochondrial samples with other cellular compartments. We are currently working toward reducing these experimental limitations in a number of ways. The separation of mitochondria into outer membrane, inner membrane, inter-membrane space, and matrix compartments reduces the complexity and increases the amount of low-abundance proteins. Comparison of the proteome across developmental stages and following environmental stimuli will identify proteins that are not found under the current cell growth regime. Improvements in isoelectric focusing (IEF)/SDS-PAGE, the use of blue-native-PAGE/SDS-PAGE, and the advent of non-gel based chromatography for protein separation will also help to improve the separation of hydrophobic proteins and further the experimentally identifiable mitochondrial proteome, especially those components encoded in the mitochondrial genome. As these experimental approaches are explored, they will then provide the badly needed basic data for the establishment of high confidence bioinformatic based detection of the full mitochondrial proteome.
MATERIALS AND METHODS
Maintenance of Cell Culture
A heterotrophic Arabidopsis cell culture, established from callus of cv Erecta stem explants, has been maintained for over 9 years by weekly subculture. Media used for this cell culture was Murashige and Skoog basal media supplemented with 3% (w/v) Suc, 0.5 mg/L naphthaleneacetic acid, and 0.05 mg/L kinetin (May and Leaver, 1993). The cell cultures were maintained in the dark at 22°C in an orbital shaker (150 rpm). At 6 to 7 d, each flask (120 mL) contained 8 to 10 g fresh weight of cells, and growth was approximately in the middle of the log phase. Subculture of 20 mL of culture to 100 mL of fresh media began the cycle again.
Mitochondrial Isolation
A total of 1.0 to 1.2 L of 7-d cell culture was filtered through gauze to remove media and was then ground by mortar and pestle, 30 g at a time. Grinding of each 30-g aliquot was performed in 100 mL of grinding medium (0.3 m mannitol, 50 mm sodium pyrophosphate, 0.5% [w/v] bovine serum albumin [BSA], 0.5% [w/v] polyvinylpyrrolidone-40, 2 mm EGTA, and 20 mm Cys, pH 8.0). Filtered cell extract was separated by centrifugation at 1,000g for 5 min at 4°C and the supernatant was centrifuged again at 18,000g for 15 min. The resulted organelle pellet was washed by repeating the 1,000 and 18,000g centrifugation steps. The final organelle pellet was resuspended in mannitol wash buffer (0.3 m mannitol, 0.1% [w/v]) BSA, and 10 mm TES (N-tris[hydroxymethyl]methyl-2-aminoethanesulfonic acid]-NaOH, pH 7.5) and loaded onto a Percoll step gradient consisting of 1:4:2 ratio, bottom to top, of 40% Percoll:23% Percoll:18% Percoll in mannitol wash buffer. The gradients were centrifuged for 45 min at 40,000g, and mitochondria were present as an opaque band at the 23%:40% interface. This band was aspirated, concentrated, and washed by centrifugation at 15,000g for 15 min and was then loaded onto a self-forming Percoll gradient containing 28% Percoll in Suc wash buffer (0.3 m Suc, 0.1% [w/v] BSA, and 10 mm TES-NaOH, pH 7.5). After centrifugation at 40,000g for 30 min, mitochondria banded near the top of the gradient and peroxisomal material banded near the bottom of the gradient. The mitochondrial band was aspirated and again washed and concentrated by two centrifugation steps at 15,000g for 15 min in mannitol wash buffer.
Purity Measurements
Fractions of 1 mL each were collected from top to bottom of the Percoll step gradient and the Percoll self-forming gradient. The activities of the following marker enzymes were measured in each fraction: cytochrome c oxidase (mitochondrion), catalase (peroxisome), alkaline pyrophosphatase (plastid), and alcohol dehydrogenase (cytosol). Enzyme assays were as detailed in the following references: cytochrome c oxidase and catalase (Neuberger, 1985), alkaline pyrophosphatase (Gross and ap Rees, 1986), and alcohol dehydrogenase (Smith and ap Rees, 1979).
Respiratory Measurements
Oxygen consumption was measured in a Clark-type oxygen electrode in 1 mL of reaction medium containing 0.3 m mannitol, 10 mm TES-KOH, pH 7.5, 5 mm KH2PO4, 10 mm NaCl, 2 mm MgSO4, and 0.1% (w/v) BSA. Pyruvate (5 mm), malate (0.5 mm), succinate (10 mm), NADH (1 mm), ADP (0.5 mm), KCN (0.5 mm), and nPG (0.05 mm) were added as indicated to modulate oxygen consumption rates. Cytochrome c oxidase activity was measured as ascorbate (5 mm), cytochrome c (25 μm)-dependent oxygen consumption in the presence of 0.05% (w/v) Triton X-100. Outer membrane integrity was assayed as the latency of cytochrome c oxidase activity (Neuberger, 1985),
Fractionation of Mitochondria
Mitochondria proteins were fractionated into soluble, membrane, integral membrane, and peripheral membrane samples. Approximately 10 mg of mitochondrial protein was incubated in 1 mL of 20 mm TES (pH 7.5), freeze-thawed in liquid nitrogen three times, and centrifuged for 25 min at 20,000g. The supernatant represented soluble proteins. One-half of the pellet was retained as the total membrane fraction. The other one-half of the pellet was resuspended in 1 mL of 100 mm Na2CO3 (pH of approximately 12), incubated for 20 min on ice, and again centrifuged for 25 min at 20,000g (Fujiki et al., 1982). The pellet was retained as integral membrane proteins, and the supernatant contained the peripheral membrane proteins. Protein samples were prepared for two-dimensional electrophoresis and loaded relative to the amount of total mitochondrial protein from which each fraction was derived to allow direct comparisons between whole mitochondrial samples and the different fractions.
Two-Dimensional Gel Electrophoresis
Mitochondria protein samples (500 μg) were acetone-extracted by the addition of acetone to a final concentration of 80% (v/v) at −80°C, samples were stored at −20°C for 4 h, and then were then centrifuged at 20,000g for 15 min. The pellets were resuspended in an IEF sample buffer consisting of 6 m urea, 2 m thiourea, 2% (w/v) CHAPS {3-[(3-cholamidopropyl)dimethylammonio]propanesulfonate}, 2% (v/v) ampholytes (3–10), 2 mm tributylphosphine, and 0.001% (w/v) bromphenol blue. Aliquots of 330 μL were used to re-swell dried 180-mm, pH 3 to 10 nonlinear immobilized pH gradient strips (Immobiline DryStrips, Amersham Pharmacia Biotech) overnight and then IEF was performed for 19.5 h, reaching a total of 49 KVh at 20°C on a flat-bed electrophoresis unit (Multiphor II, Amersham Pharmacia Biotech). Immobilized pH gradient strips were then transferred to an equilibration buffer consisting of 50 mm Tris-HCl (pH 6.8), 4 m urea, 2% (w/v) SDS, 0.001% (w/v) bromphenol blue, and 100 mm mercaptoethanol and were incubated for 20 min with rocking. The equilibrated strips were then slotted into central single wells of 4% (w/v) acrylamide stacking gels above 0.1 × 18.5 × 20 cm, 12% (w/v) acrylamide, 0.1% (w/v) SDS-polyacrylamide gels. Strips were overlaid with 0.5% (w/v) agarose in SDS-PAGE running buffer. Gel electrophoresis was performed at 25 mA per gel with circulating cooling and was completed in 5 h. Proteins were visualized by colloidal Coomassie (G250) staining. Molecular weight and pI standards from Amersham Pharmacia Biotech were used to confirm fixed pH gradient positioning on first dimension separation and to identify apparent molecular masses on second dimension separation.
MALDI-ToF MS for Peptide Fingerprint Analysis
The majority of the in-gel digestion and MS was performed as a service by The Australian Proteome Analysis Facility (Sydney, Australia) using a Micromass ToFSpec 2E machine. Additional analyses were performed by the authors with the aid of Dr. Richard Lipscombe (The University of Western Australia, Perth, Australia). Protein spots of interest were excised from gels and placed in wells of a 96-titer plate. Destain solution (50% [v/v] acetonitrile, 25 mm NH4HCO3) was added (50 μL) to samples for 45 min, removed, and replenished once. Destained gel slices were dried at 50°C for 20 min and digested at 37°C in 10 μL of 25 mm NH4HCO3 containing 12.5 μg/mL trypsin overnight. Acetonitrile (10 μL) containing 1% (v/v) trifluoroacetic acid was added to each gel slice and incubated for 15 min. Supernatant aliquots of 1 μL were added directly to matrix (α-cyano-4-hydroxycinnamic acid [8 mg/mL], 50% [v/v] acetonitrile, and 1% [v/v] trifluoroacetic acid) and air dried onto a MALDI plate for MS analysis using an Voyager-DE Pro (Applied Biosystems, Foster City, CA).
Identification of DB Entry Matches and Sequence Analysis
MALDI-ToF MS analyses provided a set of 15 to 30 peptide masses from each trypsinated protein sample. These masses were used, via MS-Fit-based software, to identify hits in translated GB, TE, and SWISS-PROT libraries, as well as in the The Arabidopsis Information Resource-translated ORFs of Arabidopsis. Matching was performed at ±50 ppm from the input masses and hits were assessed by peptide number matching (5–16), coverage (typically greater than 25%), and the Molecular Weight Search score. All data were then cross-matched by comparison of the predicted molecular weight and pI of the predicted protein with the observed molecular mass and pI of the excision site on two-dimensional gels. TargetP (http://www.cbs. dtu.dk/services/TargetP/), MitoProt (http://www.mips. biochem.mpg.de/cgi-bin/proj/medgen/mitofilter), and Psort (http://psort.nibb.ac.jp/) predictions, and GRAVY scores were determined using full-length predicted protein sequences. Molecular mass, pI, and GRAVY scores were determined using the ProtParam program on an ExPASy website (http://au.expasy.org).
ACKNOWLEDGMENTS
This research has been facilitated by access to the Australian Proteome Analysis Facility established under the Australian Government's Major National Research Facilities Program. We acknowledge the kind help of Dr. Richard Lipscombe with MALDI-ToF analysis.
Footnotes
A.H.M. was supported by an Australian Research Council Australian Postdoctoral Fellowship. This work was also supported by the Biotechnology and Biological Sciences Research Council (to C.J.L.) and by the University of Western Australia Small Grants Scheme (to A.H.M.).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.010387.
LITERATURE CITED
- Bartoli C, Pastori GM, Foyer C. Ascorbate biosynthesis in mitochondria is linked to the electron transport chain between complexes III and IV. Plant Physiol. 2000;123:335–343. doi: 10.1104/pp.123.1.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busch KB, Fromm H. Plant succinic semialdehyde dehydrogenase: cloning, purification, localization in mitochondria, and regulation by adenine nucleotides. Plant Physiol. 1999;121:589–597. doi: 10.1104/pp.121.2.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevallet M, Santoni V, Poinas A, Rouquie D, Fuchs A, Kieffe S, Rossignol M, Lunardi J, Garin J, Rabilloud T. New zwitterionic detergents improve the analysis of membrane proteins by two-dimensional electrophoresis. Electrophoresis. 1998;19:1901–1909. doi: 10.1002/elps.1150191108. [DOI] [PubMed] [Google Scholar]
- Choi K, Roh K, Kim J, Sim W. Genomic cloning and characterization of mitochondrial elongation factor Tu (EF-Tu) gene (tufM) from maize (Zea mays L.) Gene. 2000;257:233–242. doi: 10.1016/s0378-1119(00)00404-2. [DOI] [PubMed] [Google Scholar]
- Claros MG, Vincens P. Computational method to predict mitochondrially imported proteins and their targeting sequences. Eur J Biochem. 1996;241:779–786. doi: 10.1111/j.1432-1033.1996.00779.x. [DOI] [PubMed] [Google Scholar]
- Colas des Francs-Small C, Ambard-Bretteville F, Darpas A, Sallantin M, Huet J-C, Pernollet J-C, Remy R. Variation of the polypeptide composition of mitochondria isolated from different potato tissues. Plant Physiol. 1992;98:273–278. doi: 10.1104/pp.98.1.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui X, Wise R, Schnable P. The rf2 nuclear restorer of male-sterile T-cytoplasm maize encodes a putative aldehyde dehydrogenase. Science. 1996;272:1334–1336. doi: 10.1126/science.272.5266.1334. [DOI] [PubMed] [Google Scholar]
- Davy de Virville J, Alin M-F, Aaron Y, Remy R, Guillot-Salomon T, Cantrel C. Changes in functional properties of mitochondria during growth cycle of Arabidopsis thaliana cell suspension cultures. Plant Physiol Biochem. 1998;36:347–356. [Google Scholar]
- Day D, Neuberger M, Douce R. Biochemical characterization of chlorophyll-free mitochondria from pea leaves. Aust J Plant Physiol. 1985;12:219–228. [Google Scholar]
- Dunbar B, Elthon T, Osterman J, Whitaker B, Wilson S. Identification of plant mitochondrial proteins: a procedure linking two-dimensional gel electrophoresis to protein sequencing from PVDF membranes using a fastblot cycle. Plant Mol Biol Rep. 1997;15:46–61. [Google Scholar]
- Emanuelsson O, Nielsen H, Brunak S, von Heijne G. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J Mol Biol. 2000;300:1005–1016. doi: 10.1006/jmbi.2000.3903. [DOI] [PubMed] [Google Scholar]
- Fujiki Y, Fowler S, Shio H, Hubbard AL, Lazarow PB. Polypeptide and phospholipid composition of the membrane of rat liver peroxisomes: comparison with endoplasmic reticulum and mitochondrial membranes. J Cell Biol. 1982;93:103–110. doi: 10.1083/jcb.93.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagliardi D, Kuhn J, Spadinger U, Brennicke A, Leaver C, Binder S. An RNA helicase (AtSUV3) is present in Arabidopsis thaliana mitochondria. FEBS Lett. 1999;458:337–342. doi: 10.1016/s0014-5793(99)01168-0. [DOI] [PubMed] [Google Scholar]
- Gangloff S, Marguet D, Lauquin GM. Molecular cloning of the yeast mitochondrial aconitase gene (ACO1) and evidence of the synergistic regulation of expression by glucose plus glutamate. Mol Cell Biol. 1990;10:3551–3561. doi: 10.1128/mcb.10.7.3551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray M, Burger G, Lang Mitochondrial evolution. Science. 1999;283:1476–1481. doi: 10.1126/science.283.5407.1476. [DOI] [PubMed] [Google Scholar]
- Gross T, ap Rees T. Alkaline inorganic pyrophosphatase and starch synthesis in amyloplasts. Planta. 1986;167:140–145. doi: 10.1007/BF00446381. [DOI] [PubMed] [Google Scholar]
- Gueguen V, Macherel D, Jaquinod M, Douce R, Bourguignon J. Fatty acid and lipoic acid biosynthesis in higher plant mitochondria. J Biol Chem. 2000;275:5016–5025. doi: 10.1074/jbc.275.7.5016. [DOI] [PubMed] [Google Scholar]
- Halperin T, Zheng B, Itzhaki H, Clarke AK, Adam Z. Plant mitochondria contain proteolytic and regulatory subunits of the ATP-dependent Clp protease. Plant Mol Biol. 2001;45:461–468. doi: 10.1023/a:1010677220323. [DOI] [PubMed] [Google Scholar]
- Hatzfeld Y, Maruyama A, Schmidt A, Noji M, Ishizawa K, Saito K. β-Cyanoalanine synthase is a mitochondrial cysteine synthase-like protein in spinach and Arabidopsis. Plant Physiol. 2000;123:1163–1171. doi: 10.1104/pp.123.3.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbert B. Advances in protein solubilisation for two-dimensional electrophoresis. Electrophoresis. 1999;20:660–663. doi: 10.1002/(SICI)1522-2683(19990101)20:4/5<660::AID-ELPS660>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- Herbert BR, Molloy MP, Gooley AA, Walsh BJ, Bryson WG, Williams KL. Improved protein solubility in two-dimensional electrophoresis using tributyl phosphine as reducing agent. Electrophoresis. 1998;19:845–851. doi: 10.1002/elps.1150190540. [DOI] [PubMed] [Google Scholar]
- Humphery-Smith I, Colas des Francs-Small C, Ambart-Bretteville F, Remy R. Tissue-specific variation of pea mitochondrial polypeptides detected by computerized image analysis of two-dimensional electrophoresis gels. Electrophoresis. 1992;13:168–172. doi: 10.1002/elps.1150130134. [DOI] [PubMed] [Google Scholar]
- Jenner H, Winning B, Millar AH, Tomlinson K, Leaver CJ, Hill SA. NAD malic enzyme and the control of carbohydrate metabolism in potato tubers. Plant Physiol. 2001;126:1139–1149. doi: 10.1104/pp.126.3.1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhlman P, Palmer J. Isolation, expression, and evolution of the gene encoding mitochondrial elongation factor Tu in Arabidopsis mitochondria. Plant Mol Biol. 1995;29:1057–1070. doi: 10.1007/BF00014977. [DOI] [PubMed] [Google Scholar]
- Kyte J, Doolittle R. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Laloi M. Plant mitochondrial carriers: an overview. Cell Mol Life Sci. 1999;56:918–944. doi: 10.1007/s000180050484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindemann P, Luickner M. Biosynthesis of pregnane derivatives in somatic embryos of Digitalis lanata. Phytochemistry. 1997;46:507–513. [Google Scholar]
- Lutziger I, Oliver DJ. Molecular evidence of a unique lipoamide dehydrogenase in plastids: analysis of plastidic lipoamide dehydrogenase from Arabidopsis thaliana. FEBS Lett. 2000;484:12–16. doi: 10.1016/s0014-5793(00)02116-5. [DOI] [PubMed] [Google Scholar]
- Margossian S, Li H, Zassenhaus H, Butow R. The DExH box protein Suv3p is a component of a yeast mitochondrial 3′-to-5′ exoribonuclease that suppresses group I intron toxicity. Cell. 1996;84:199–209. doi: 10.1016/s0092-8674(00)80975-7. [DOI] [PubMed] [Google Scholar]
- May MJ, Leaver CJ. Oxidative stimulation of glutathione synthesis in Arabidopsis thaliana suspension cultures. Plant Physiol. 1993;103:621–627. doi: 10.1104/pp.103.2.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai K, Kanehisa M. A knowledge base for predicting protein localization sites in eukaryotic cells. Genomics. 1992;14:897–911. doi: 10.1016/S0888-7543(05)80111-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuberger M. Preparation of plant mitochondria, criteria for assessment of mitochondrial integrity and purity, survival in vitro. In: Douce R, Day D, editors. Higher Plant Cell Respiration. Berlin: Springer-Verlag; 1985. pp. 7–24. [Google Scholar]
- Nijtmans LGJ, de Jong L, Sanz MA, Coates PJ, Berden JA, Back JW, Muijsers AO, van der Spek H, Grivell LA. Prohibitins act as a membrane-bound chaperone for the stabilization of mitochondrial proteins. EMBO J. 2000;19:2444–2451. doi: 10.1093/emboj/19.11.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peltier J-B, Friso G, Kalume D, Roepstorff P, Nilsson F, Adamska I, van Wijk K. Proteomics of the chloroplast: systematic identification and targeting analysis of lumenal and peripheral thylakoid proteins. Plant Cell. 2000;12:319–341. doi: 10.1105/tpc.12.3.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peyret P, Perez P, Alric M. Structure, genomic organization, and expression of the Arabidopsis thaliana aconitase gene: plant aconitase show significant homology with mammalian iron-responsive element-binding protein. J Biol Chem. 1995;270:8131–8137. doi: 10.1074/jbc.270.14.8131. [DOI] [PubMed] [Google Scholar]
- Prime T, Sherrier D, Mahon P, Packman L, Dupree P. A proteomic analysis of organelles from Arabidopsis thaliana. Electrophoresis. 2000;21:3488–3499. doi: 10.1002/1522-2683(20001001)21:16<3488::AID-ELPS3488>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- Rabilloud T. Use of thiourea to increase the solubility of membrane proteins in two-dimensional electrophoresis. Electrophoresis. 1998;19:758–760. doi: 10.1002/elps.1150190526. [DOI] [PubMed] [Google Scholar]
- Rasmusson AG, Svensson AS, Knoop V, Grohmann L, Brennicke A. Homologues of yeast and bacterial rotenone-insensitive NADH dehydrogenases in higher eukaryotes: two enzymes are present in potato mitochondria. Plant J. 1999;20:79–87. doi: 10.1046/j.1365-313x.1999.00576.x. [DOI] [PubMed] [Google Scholar]
- Rebeille F, Macherel D, Mouillon JM, Garin J, Douce R. Folate biosynthesis in higher plants: purification and molecular cloning of a bifunctional 6-hydroxymethyl-7,8-dihydropterin pyrophosphokinase/7,8-dihydropteroate synthase localized in mitochondria. EMBO J. 1997;16:947–957. doi: 10.1093/emboj/16.5.947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saisho D, Nambara E, Naito S, Tsutsumi N, Hirai A, Nakazono M. Characterization of the gene family for alternative oxidase from Arabidopsis thaliana. Plant Mol Biol. 1997;35:585–596. doi: 10.1023/a:1005818507743. [DOI] [PubMed] [Google Scholar]
- Sanchez H, Fester T, Kloska S, Schroder W, Schuster W. Transfer of rps19 to the nucleus involves the gain of an RNP-binding motif which may functionally replace RPS13 in Arabidopsis mitochondria. EMBO J. 1996;15:2138–2149. [PMC free article] [PubMed] [Google Scholar]
- Santoni V, Rouquie D, Doumas P, Mansion M, Boutry M, Degand H, Dupree P, Packman L, Sherrier J, Prime T. Use of a proteome strategy for tagging proteins present at the plasma membrane. Plant J. 1998;16:633–641. doi: 10.1046/j.1365-313x.1998.00335.x. [DOI] [PubMed] [Google Scholar]
- Shevchenko A, Jensen ON, Podtelejnikov AV, Sagliocco F, Wilm M, Vorm O, Mortensen P, Boucherie H, Mann M. Linking genome and proteome by mass spectrometry: large-scale identification of yeast proteins from two-dimensional gels. Proc Natl Acad Sci USA. 1996;93:14440–14445. doi: 10.1073/pnas.93.25.14440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjoling S, Glaser E. Mitochondrial targeting peptides in plants. Trend Plant Sci. 1998;3:136–140. [Google Scholar]
- Smith A, ap Rees T. Pathways of carbohydrate fermentation in the roots of marsh plants. Planta. 1979;146:327–334. doi: 10.1007/BF00387805. [DOI] [PubMed] [Google Scholar]
- Snedden WA, Koutsia N, Baum G, Fromm H. Activation of a recombinant petunia glutamate decarboxylase by calcium/calmodulin or by a monoclonal antibody which recognizes the calmodulin binding domain. J Biol Chem. 1996;271:4148–4153. doi: 10.1074/jbc.271.8.4148. [DOI] [PubMed] [Google Scholar]
- Solish S, Picado-Leonard J, Morel Y, Kuhn R, Mohandas T, Hanukoglu I, Millar W. Human adrenodoxin reductase: two mRNAs encoded by a single gene on chromosome 17cen to q25 are expressed on steroidogenic tissues. Proc Natl Acad Sci USA. 1988;85:7104–7108. doi: 10.1073/pnas.85.19.7104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steglich G, Neupert W, Langer T. Prohibitins regulate membrane protein degradation by the m-AAA protease in mitochondria. Mol Cell Biol. 1999;19:3435–3442. doi: 10.1128/mcb.19.5.3435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Arabidopsis Genome Initiative. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- Vambutas A, Ackerman S, Tzagoloff A. Mitochondrial translational-initiation and elongation factors in Saccharomyces cerevisiae. Eur J Biochem. 1991;201:643–652. doi: 10.1111/j.1432-1033.1991.tb16325.x. [DOI] [PubMed] [Google Scholar]