Abstract
Soybean (Glycine max L. Merr.) contains two related and abundant proteins, VSPα and VSPβ, that have been called vegetative storage proteins (VSP) based on their pattern of accumulation, degradation, tissue localization, and other characteristics. To determine whether these proteins play a critical role in sequestering N and other nutrients during early plant development, a VspA antisense gene construct was used to create transgenic plants in which VSP expression was suppressed in leaves, flowers, and seed pods. Total VSP was reduced at least 50-fold due to a 100-fold reduction in VSPα and a 10-fold reduction in VSPβ. Transgenic lines were grown in replicated yield trials in the field in Nebraska during the summer of 1999 and seed harvested from the lines was analyzed for yield, protein, oil, and amino acid composition. No significant difference (α = 0.05) was found between down-regulated lines and controls for any of the traits tested. Young leaves of antisense plants grown in the greenhouse contained around 3% less soluble leaf protein than controls at the time of flowering. However, total leaf N did not vary. Withdrawing N from plants during seed fill did not alter final seed protein content of antisense lines compared with controls. These results indicate that the VSPs play little if any direct role in overall plant productivity under typical growth conditions. The lack of VSPs in antisense plants might be partially compensated for by increases in other proteins and/or non-protein N. The results also suggest that the VSPs could be genetically engineered or replaced without deleterious effects.
Large quantities of C, N, and other nutrients are mobilized from mature organs to younger organs during plant development. This recycling of material from older source tissues conserves assimilates and can contribute substantially to the nutrient requirements of sink tissues, such as developing leaves and seeds (Feller and Keist, 1986). Amino acids that are derived from the turnover of proteins are important components of the translocation pool. Although most proteins are eventually broken down in senescent tissue, the majority of these have important metabolic functions, and their amino acids are salvaged only after the proteins are no longer needed. On the other hand, some proteins have characteristics that suggest they have a primary, if not exclusive, role in nutrient reserve storage. The best studied examples are the storage proteins that accumulate specifically in developing seeds (Derbyshire et al., 1976).
Storage proteins have also been identified in vegetative tissues. Soybeans (Glycine max L. Merr.) contain two abundant and well-characterized proteins that have been called vegetative storage proteins (VSP) based on a number of factors (for review, see Staswick, 1994). VSPα and VSPβ are glycoproteins of around 27 kD (Wittenbach, 1983) that are derived from the genes VspA and VspB, respectively. The proteins are about 80% identical in amino acid sequence. They are not related to the seed-specific reserves (Staswick, 1988), but like other storage proteins they accumulate in vacuoles to relatively high level (Franceschi et al., 1983).
VSPα/β are synthesized abundantly in sink organs and are preferentially degraded when these organs transition to sources. For example, VSPα and VSPβ comprise 10% to 15% of the total soluble protein in young leaves and seed pod walls, but are only about 1% of the protein in these organs when they are mature. VSP constitutes about 45% of the protein that is degraded in seed pods during seed development (Staswick, 1989), thus contributing substantially to the amino acid pool available for translocation to maturing seeds. The preferential breakdown of VSPs in a source-dependent manner before the general turnover of other proteins has suggested they have an important role in the temporary storage of amino N. The cellular localization of VSPs in leaf paraveinal mesophyll, tissue thought to play an important role in temporary nutrient storage (Franceschi et al., 1983; Franceschi and Giaquinta, 1983), also supports the idea that VSPs are temporary storage reserves. A striking feature of the soybean VSPα and VSPβ is their dramatic over-accumulation in mature leaves when developing sink tissues, such as seed pods, are removed. In this case VSPs can accumulate to as much as 50% of leaf protein, whereas total protein level remains constant (Wittenbach, 1983). This extraordinary abundance of VSP in sink-deprived plants suggests that the VSPs sequester unused amino acids that would ordinarily be translocated to developing seeds. Vsp gene expression also correlates with the availability of N provided to the plant (Staswick et al., 1991). Taken together, these findings suggest that VSPs serve as transient reserves that can buffer the availability of N and perhaps other nutrient elements. The presence of similar proteins in several distantly related wild perennial soybeans strengthens the case for an important metabolic function (Staswick, 1997).
Despite the many indications that soybean VSPα and VSPβ are important storage reserves, the evidence for this is entirely correlative. It has not been possible to test VSP function directly in soybean because null lines are not known to exist. This investigation was designed to test whether VSPα and VSPβ play a critical role in soybean development and plant productivity by examining the effect of nearly eliminating the expression of the soybean VspA and VspB genes in transgenic plants. If VSPs serve as important storage reserves, then decreasing VSPα and VSPβ accumulation in vegetative tissues might be expected to alter seed development and/or seed protein accumulation. It is also possible that these VSPs have another biological function. The protein sequences are about 45% identical to a tomato acid phosphatase (Williamson and Colwell, 1991) and acid phosphatase activity has been reported for the purified soybean proteins (DeWald et al., 1992). If VSPα and VSPβ play an important catalytic role, then an aberrant phenotype might also be observed in vegetative tissues. Contrary to these expectations, the near elimination of VSPα and VSPβ in vegetative tissues had no detectable effect on plants grown in the greenhouse or in the field.
RESULTS
Vsp Genes Are Efficiently Down-Regulated by an Antisense Construct
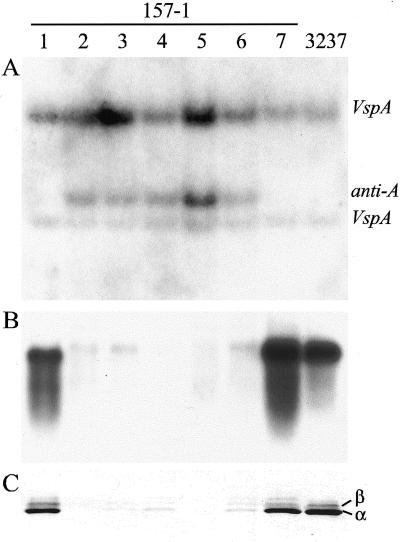
The first 578 nucleotides of cDNA pVSPA were used in antisense orientation under the direction of the cauliflower mosaic virus 35S promoter to obtain plants that were down-regulated for Vsp gene expression. Five of the seven independently transformed lines that were characterized exhibited decreased level of VSP in the T0 generation when soluble proteins or RNA from seed pods was analyzed (not shown). Down-regulation was followed in the next generation, and an example of the results from T1 progeny of one down-regulated line (157-1) is shown in Figure 1. As expected, the VspA cDNA probe used for Southern-blot hybridization detected the endogenous VspA and VspB genes in all plant samples. T1 individuals 1 and 7 lacked the transgene, whereas individuals 2 through 6 contained a single band corresponding to the VspA antisense insert (Fig. 1A). Subsequent analysis of T2 progeny showed that T1 plant 5 was homozygous for the transgene, whereas progeny 2, 3, 4, and 6 were hemizygous (not shown). These results indicate a single locus insert in the original transformant. Consistent with the genomic composition, T1 individuals 2 through 6 had greatly reduced levels of Vsp mRNA (Fig. 1B) and protein (Fig. 1C), whereas 1 and 7 contained levels of each that were similar to the non-transformed parent.
Figure 1.
Segregation of VSP down-regulation in progeny of transgenic line 157-1. DNA, total RNA, and soluble protein were extracted from expanding leaves taken from nodes 5 or 6. Individuals from the 157-1 T1 generation are shown in lanes 1 through 7 and non-transformed Asgrow 3237 is in the last lane. A, XbaI-digested genomic DNA hybridized with VspA cDNA insert. The bands labeled VspA and VspB correspond to the endogenous genes of size 9 and 5 kb, respectively. Anti-A, transgene insert. B, RNA-blot hybridization with VspA probe. C, Immunodetection of VSP after SDS-PAGE. α and β, VSP bands.
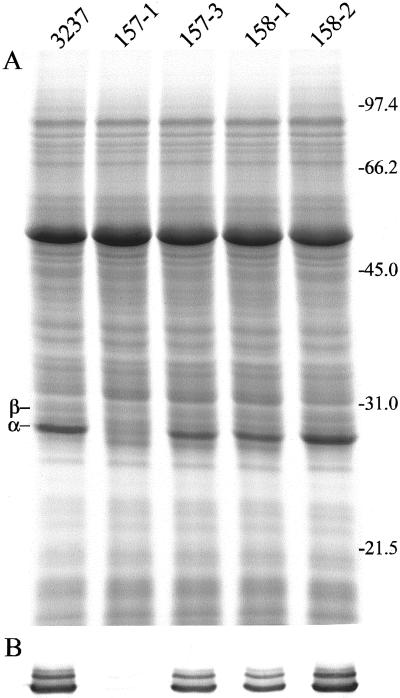
Homozygous lines for four of the seven original transformants were identified and subjected to more detailed study. The amount of VSP in young leaves was analyzed by SDS-PAGE as shown in Figure 2. Compared with the non-transformed parental line, 157-1 contained essentially no detectable VSP protein (Fig. 2A). On the other hand, transformant 158-2 showed essentially no down-regulation for the two VSP polypeptides, thus serving as a control line that had undergone the transformation process. Lines 157-3 and 158-1 were intermediate in VSP level. These results were confirmed with VSP antisera by western-blot analysis. Only faint bands corresponding to the two VSP polypeptides were detected in line 157-1, and lines 157-3 and 158-1 were intermediate in staining intensity (Fig. 2B). Quantitative estimates of VSP levels relative to wild type were determined by comparing a dilution series of the protein extract from the Asgrow 3237 control with those of 157-1, 157-3, and 158-1 on western blots. VSPα in 157-1 was reduced about 100-fold and VSPβ about 10-fold relative to the non-transformed control (not shown). Although the efficiency of VSPβ down-regulation was less than that for VSPα, the normal level of VSPβ in young leaves is at least 10-fold below that of VSPα. Therefore, total VSP reduction in 157-1 was >50-fold. In 157-3 and 158-1, VSPα and VSPβ were each reduced by about 50%.
Figure 2.
Analysis of protein composition in young expanding leaves of VSP antisense plants. Soluble protein was isolated from non-transformed Asgrow 3237 and homozygous transformed lines 157-1, 157-3, 158-1, and 158-2. The position of molecular mass markers in kilodaltons and the VSPα and VSPβ bands is indicated to the right and left, respectively. A, SDS-PAGE. B, Western blot with VSP antisera.
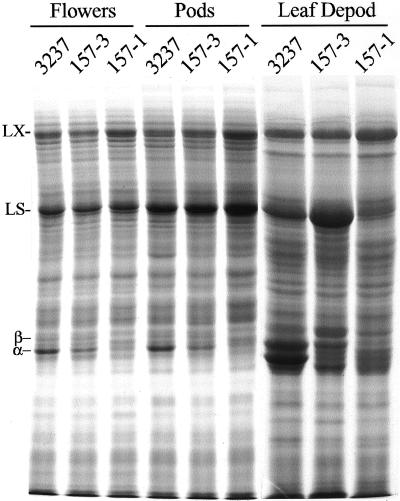
To determine whether the antisense construct was effective in other plant tissues we examined flowers and seed pods, organs in which VSPs normally accumulate to high level. Figure 3 shows that down-regulation in seed pods and flowers of lines 157-1 and 158-1 was consistent with that found in young leaves. This indicated that the VspA antisense construct was effective in VSP suppression throughout the development of vegetative portions of the plant. To further examine the efficacy of the construct in down-regulation, field-grown plants were depodded for 5 to 6 weeks. This treatment stimulates VSP accumulation of up to 50% of soluble protein in leaves. Leaves of 157-1 and 158-1 retained high chlorophyll levels, and leaves remained on plants for an extended period of time, similar to the depodded control line. Protein analysis of mature leaves from these plants by SDS-PAGE revealed the same pattern of VSP accumulation as seen in other plant parts. In depodded non-transformed plants both VSPα and VSPβ accumulated to very high levels, whereas both polypeptides were markedly reduced in line 157-1 and intermediate in 158-1 (Fig. 3). These results demonstrate that the single locus antisense transgene insertion in line 157-1 was an efficient suppressor of Vsp gene expression, even under conditions where Vsp mRNA normally accumulates to as much as 16% of total mRNA (Staswick, 1989).
Figure 3.
Protein composition of flowers, seed pods, and mature leaves of depodded plants. VSP bands are marked as α and β. LX, Lipoxygenase; LS, Rubisco large subunit.
VSP Down-Regulation Lowers Leaf Soluble Protein Content but Not Total N
Total soluble protein in expanding leaves harvested from greenhouse-grown plants at the time of flowering was quantified by the modified Lowery assay. For this experiment, a second down-regulated line (168-5) showing a strong reduction in leaf VSP levels similar to that of 157-1 was included. Results summarized in Table I show that down-regulated lines 157-1 and 168-5 had mean extractable protein levels of 73.8 and 72.8 μg/mg fresh weight, respectively. This is 2.3% and 3.6% lower than the non-transformed control (As3237). The ANOVA test indicated significance at the 0.05 level between the down-regulated lines and the controls, whereas there was no significant difference between the two down-regulated lines or between As3237 and the transformant not showing reduced VSP level (158-2). In contrast to soluble protein however, there was no significant difference in total leaf N levels in either field-grown or greenhouse-grown plants (not shown).
Table I.
Soluble protein in young leaves of transgenic and control plants
| Line | Protein |
|---|---|
| μg mg fresh wt−1a | |
| Controls | |
| 3237 | 75.5* ± 0.4 |
| 158-2 | 75.9* ± 0.4 |
| Down-regulated | |
| 157-1 | 73.8** ± 0.4 |
| 168-5 | 72.8** ± 0.5 |
* and **, Means with one asterisk or two asterisks indicate significantly different (P = 0.05).
Mean ± se (n = 16).
Although total leaf protein was decreased around 3% in antisense plants, this is somewhat less than would be expected from the nearly complete removal of VSPs, which comprise 6% to 15% of soluble protein in expanding leaves (Wittenbach, 1983). One possible reason for the discrepancy might be a compensatory increase in other proteins. Some members of the abundant lipoxygenase gene family (LOX, about 90 kD; Fig. 2) are regulated similarly to VSPα and VSPβ and thus might serve as temporary storage reserves (Tranbarger et al., 1991; Grimes et al., 1992). Analysis of the results from SDS-PAGE did not reveal a marked change in the relative abundance of specific proteins that could be visualized from young leaves (Fig. 2). An apparent minor elevation in the intensity of a doublet band just above the 31-kD marker was not consistently observed in repeated analyses. Compensation for VSP loss in expanding leaves might also occur by a minor increase in a number of other proteins, a result not detectable in this assay. On the other hand, LOX proteins did appear to be reproducibly increased in flowers, pods, and mature leaves from depodded plants of line 157-1 compared with the control (Fig. 3, compare lanes 1 and 3). In the depodded sample of line 158-1, substantially more Rubisco large subunit (approximately 50 kD) was present than in the control or line 157-1. Rubisco is normally lost from the soluble extract of depodded plants but the decline can vary somewhat in its timing. Therefore, the difference seen for this intermediate down-regulated line is probably not attributable to the antisense expression. Consistent with this, levels of Rubisco did not differ substantially between the control and the strongly down-regulated line 157-1. These results suggest that loss of VSPs is partially compensated for by an increase in LOX in some tissues and possibly by minor elevation of a number of other proteins.
VSP Down-Regulation Does Not Influence Seed Yield or Protein Composition
Down-regulated lines were grown in the field along with controls during the summer of 1999. Specific phenotypic measurements were not taken during the growing season, but no obvious visual differences in plant growth or development were observed. Transgenic plants grown in the greenhouse also did not exhibit any unusual characteristics. The seed yield of field plots for antisense plants was determined and compared with controls. The results shown in Table II indicate that yield calculated on a kilograms per hectare basis ranged from an average of 3167 to 3974. The analysis of variance F-test indicated no significant yield difference (α = 0.05, P = 0.37) between the down-regulated lines (157-1, 157-3, and 158-1) and either the non-transformed control (3237) or the transformed control (158-2) that did not exhibit noticeable down-regulation. Seed harvested from these field-grown plants was also analyzed for total protein and oil. Table II shows that average protein ranged from 390 to 405 g/kg, but there was no significant difference among these lines (α = 0.05, P = 0.31). Seed oil content ranged from 202 to 210 g/kg. Again, down-regulated lines and the controls did not differ significantly (α = 0.05, P = 0.15).
Table II.
Seed yield, protein, and oil content of VSP transgenic lines
| Line | Yield | Protein | Oil |
|---|---|---|---|
| kg ha−1a | g kg−1b | ||
| 157-1 | 3,424 ± 225 | 399 ± 46 | 202 ± 26 |
| 157-3 | 3,947 ± 290 | 402 ± 29 | 202 ± 23 |
| 158-1 | 3,572 ± 57 | 390 ± 24 | 210 ± 22 |
| 158-2 | 3,167 ± 145 | 405 ± 58 | 202 ± 36 |
| 3237 | 3,371 ± 315 | 402 ± 25 | 206 ± 13 |
Mean ± se (n = 6 except n = 3 for 158-1).
Mean ± se (n = 4).
Although no difference in total seed protein content was observed, eliminating major vegetative proteins such as VSPα and VSPβ might influence overall seed amino acid composition, because these proteins contribute to the pool of material translocated to developing seeds. Therefore, the amino acid composition was determined for two lines; the strongly down-regulated 157-1 and the non-transformed control Asgrow 3237. Table III summarizes the values determined for each amino acid expressed as a percentage of total amino acids recovered. The lack of VSP accumulation in 157-1 had no significant effect (α = 0.05) on the relative proportion of any amino acid in the harvested seeds, although the statistical P values for Tyr, Cys, and Glu (0.051, 0.079, and 0.086, respectively) approached significance at this level. Together these results indicate that dramatically decreasing the expression of Vsp genes throughout the soybean plant had no detectable effect on the traits of primary interest for soybean production.
Table III.
Seed amino acid composition in transgenic and control plants
| Amino Acid | Transgenic 157-1 | Control 3237 |
|---|---|---|
| %a | ||
| Asp | 11.52 ± 2 | 11.52 ± 0 |
| Thr | 3.97 ± 2 | 3.93 ± 4 |
| Ser | 4.40 ± 6 | 4.36 ± 14 |
| Glu | 18.15 ± 1 | 18.35 ± 6 |
| Pro | 4.76 ± 1 | 4.82 ± 3 |
| Gly | 4.39 ± 1 | 4.35 ± 1 |
| Ala | 4.40 ± 0 | 4.38 ± 2 |
| Cys | 1.85 ± 1 | 1.81 ± 0 |
| Val | 5.05 ± 5 | 5.01 ± 6 |
| Met | 1.54 ± 1 | 1.50 ± 1 |
| Ile | 4.70 ± 4 | 4.71 ± 6 |
| Leu | 7.97 ± 3 | 7.95 ± 1 |
| Tyr | 3.75 ± 0 | 3.72 ± 1 |
| Phe | 5.25 ± 2 | 5.25 ± 1 |
| His | 2.82 ± 0 | 2.83 ± 2 |
| Lys | 6.86 ± 1 | 6.82 ± 4 |
| Arg | 7.49 ± 5 | 7.58 ± 7 |
| Trp | 1.12 ± 7 | 1.11 ± 7 |
Percent total amino acids. Mean ± se (n = 2).
One possible explanation for the lack of effect on seed yield or protein content could be that adequate levels of nutrients were available from the soil in the field throughout plant development, decreasing the dependence on nutrients accumulated and stored away earlier. To test whether VSPs are an important storage reservoir under less optimal nutrient conditions, plants were grown in sand in the greenhouse and supplied with N in a complete nutrient solution until flowering. N was then withdrawn from one-half of the plants by watering with the same solution lacking N, whereas the other one-half continued receiving N throughout seed maturation. The hypothesis was that plants not receiving N during seed development would be more dependent on previously assimilated N. Within 2 to 3 weeks of N withdrawal, plants showed strong symptoms of N chlorosis, whereas plants provided N maintained a normal dark green color until seed maturation. Seed protein was determined from a sample of the total seed of five individual plants for each treatment. The results summarized in Table IV show that, as expected, withdrawal of N at the time of flowering significantly decreased final seed protein level in both the non-transformed control and the down-regulated line 157-1. However, there was no significant difference in seed protein level between the control and 157-1 for the +N or −N treatment. This indicates that accumulating N in VSPs as a reserve during vegetative growth does not influence seed protein level, even under N limitation during seed development.
Table IV.
Total protein seed protein in transgenic plants grown under different N treatments
| Treatment | Protein |
|---|---|
| % dry wta | |
| Minus N | |
| 157-1 | 33.7* |
| 3237 | 33.4* |
| Plus N | |
| 157-1 | 37.3** |
| 3237 | 39.1** |
* and **, Means with one asterisk or two asterisks indicate significantly different (P = 0.05).
Mean ± se (n = 5).
DISCUSSION
The results presented here demonstrate the efficient down-regulation of soybean Vsp genes. Notably, the antisense strategy was very effective even though these are abundantly expressed genes. VspA was chosen for the antisense construct because the protein from this gene is typically more highly accumulated than that of VspB. However, the similarity in gene sequence resulted in down-regulation of both Vsp genes by the single antisense construct, albeit less efficiently for VspB. Vsp transcripts can amount to as much as 16% of the mRNA in depodded plants (Staswick, 1989), yet even after this treatment accumulation of VSP was strongly suppressed in line 157-1. One other strongly down-regulated line (168-5) was also grown in the field and analyzed in the same manner as the lines reported here. Its level of down-regulation was similar to 157-1 but it was not included in the reported data because analysis of individual field-grown plants revealed that it was still segregating for VSP down-regulation at that time. However, yield and seed composition data from the pooled, harvested seed of 168-5 was consistent with that found for the other lines evaluated here. The hypothesis that accumulation of VSPs during vegetative growth would increase seed protein accumulation if N was subsequently limiting during seed development was also not shown to be true.
Given the hypothesized role of VSPs as an important source of mobilized nutrients for developing plant organs, our results are surprising. Evidence indicates these VSPs are evolutionarily conserved at least in some plants. In addition to soybean, they occur in several perennial Glycine species (Staswick, 1997) and in certain other legumes that have leaf paraveinal mesophyll tissue (Klauer et al., 1996). Related proteins have also been characterized in Arabidopsis (Berger et al., 1995) and in the seed pods of common bean (Zhong et al., 1997). Thus there is reason to think the VSPs have an important role in plant biology.
Our findings cannot completely rule out the possibility that VSPs are indeed storage reserves that contribute to nutrient flux under some unusual environmental condition. However, we have demonstrated that under typical growth conditions VSPs are not essential for maximal seed production or seed protein accumulation. It appears that VSP loss is partially compensated for by increase in other proteins; LOX in particular, but perhaps other proteins as well. Additionally, non-protein forms of N apparently help maintain total leaf N to similar levels in the absence of VSPs.
Soybean VSPs might be involved in a presently unknown role, such as ameliorating a biotic or abiotic stress not encountered in this study. Our results suggest that any enzymatic activity attributable to the soybean VSPs is not essential, since we observed no deleterious effect on plant growth and development. Although related to acid phosphatases, there is presently no direct evidence for a functional role being associated with this activity. The reported specific activity of VSPα and VSPβ is low (DeWald et al., 1992; Staswick et al., 1994), and it was recently noted that VSPα and VSPβ lack a nucleophilic Asp residue that is proposed to have catalytic function (Penheiter, 1998) and that is present in other members of this acid phosphatase gene family. Interestingly, soybeans contain another VSP-like gene that is expressed exclusively in soybean root nodules (Penheiter et al., 1997). In contrast to VSPα and VSPβ the nodule protein is relatively low in abundance, exhibits a high acid phosphatase specific activity, and has the nucleophilic Asp. This suggests that the function of the nodule acid phosphatase is chiefly enzymatic and not as a reserve protein. Taken together these findings suggest that the enzymatic activity associated with VSPα and VSPβ may be a remnant of an earlier gene function, as has been shown for a number of storage proteins (Van Damme et al., 2000).
It was previously suggested that the VSPs might be engineered for altered amino acid composition as a means to modify the amino acid pool that is translocated to developing seeds during seed development (Staswick, 1988). This could be useful in tandem with seed protein genes that are engineered for altered amino acid composition to improve seed nutritional content. The fact that VSPs were essentially eliminated in this study without negative consequences to plant productivity suggests that genetically engineered VSPs would not compromise an essential metabolic function. Furthermore, although their absence was not deleterious, VSPs with an altered amino acid composition may still influence the availability of specific amino acids when these VSP genes are expressed at high level.
MATERIALS AND METHODS
Vector Construction, Plant Transformation, and Analysis of Gene Expression
A 0.6-kb cDNA fragment extending from the 5′ end to the BamHI site of pVSPA (Staswick, 1988) was fused in antisense orientation with the enhanced cauliflower mosaic virus 35S promoter at the BamHI site in the vector pRTL2 (Carrington and Freed, 1990). The resulting 1.5-kb PvuII fragment containing the antisense cassette was excised and used to replace the GUS cassette of binary vector pPTN101 by blunt end ligation into the EcoRI-HindIII sites. The resulting vector, pVARTN-6, was mobilized into the Agrobacterium strain EHA105 (Hood et al., 1993) by triparental mating (Ditta et al., 1980). Soybean (Glycine max L. Merr.) genotype Asgrow 3237 was transformed by an Agrobacterium-mediated cotyledonary node transformation system utilizing the bar gene as a selectable marker coupled with glufosinate as a selective agent (Zhang et al., 1999).
Transgene integration in putative transformants was verified by Southern-blot hybridization as previously described (Zhang et al., 1999) with a probe generated by excising the 0.6-kb XbaI-BamHI VspA fragment from the vector pVARTN-6 or from the full-length cDNA insert from pVSPA. T1 progeny from primary (T0) transformants were grown in a greenhouse and allowed to self. T1 plants that were homozygous for the transgene were identified by screening T1-derived T2 progeny for the selectable marker in the greenhouse with a leaf paint assay (Zhang et al., 1999). The herbicide Liberty (100 mg/L) was painted onto the upper surface of a primary or first trifoliate leaf of young seedlings using a cotton swab. At least seven T2 progeny were assayed for each T1 plant. The herbicide tolerance segregation patterns were evaluated 5 d after leaf-painting.
The down-regulation of VSP genes in transgenic soybean was analyzed by SDS-PAGE and western blots. Total soluble protein was isolated from the various tissues in extraction buffer (40 mm Tris-HCl, pH 7.5, 10 mm EDTA, 5 mm dithiothreitol, 0.01 mm phenylmethylsulfonylfluoride, and 1% polyvinlypolypyrolidone). Protein concentration was determined with a Bio-Rad DC Protein Assay (Hercules, CA), and proteins were electorophoresed on 12% polyacrylamide gels as described previously (Staswick, 1989) and stained with Coomassie Brilliant Blue R. Alternately, proteins were electrophoretically transferred onto nitrocellulose membranes and then developed with polyclonal antisera raised in rabbits against VSPα/β (Staswick, 1989). Northern-blot hybridization was performed as described earlier using 20 μg of total RNA obtained from young leaf tissue for each sample (Zhang et al., 1998).
Soluble leaf protein levels were determined from expanding leaves of 16 greenhouse-grown plants for each line at the time of flowering. One centimeter discs were punched from leaves 2 to 3 cm in length, avoiding the midrib. Discs were weighed, ground in extraction buffer in a mortar and protein was quantified as described above.
Plant Growth and Trait Analysis
T2 or T3 seed derived from homozygous T1 lines was grown for trait analysis in a field nursery located on the East Campus of the University of Nebraska-Lincoln during the summer of 1999. The herbicide leaf paint assay was applied to field-grown plants to confirm homozygosity of the transgene in the lines tested. Homozygosity for VSP down-regulation was also verified by SDS-PAGE analysis of individual plants grown from seed harvested from the field-grown material. Field-grown leaves, immature seed pods, and flowers were harvested and frozen at −80°C for subsequent analysis by SDS-PAGE. The experimental design for analysis of yield performance was a randomized complete block with six replications for all lines except 158-1, for which seed was only available for three replications. The experimental units (field plots) were single rows 1.52 m long, spaced 0.76 m apart. The test was bordered by soybean cultivars of the same maturity and growth type.
Mature plants were gathered from each plot and bulk-threshed. The weight of harvested seed was adjusted to the standard 13% seed moisture content and converted to kilograms per hectare units of yield. The yield, protein, oil, and amino acid data were subjected to an analysis of variance F-test to determine if yield differences among lines were significant (α = 0.05). Total protein and oil content of dry seed was determined by the Soil Plant Analytical Laboratory (University of Nebraska, Lincoln) from 150 g of dry powdered seed for each of four replicate field plots for each line (three replicates for 158-1). Amino acid composition was determined from 70 g of powdered seed from each of two field replicate plots for each line by the Experiment Station Chemical Laboratories (University of Missouri, Columbia). The amide group of Asn and Gln was hydrolyzed during the assay and values for these amino acids are included in the total for Asp and Glu, respectively.
Plants were also grown in a greenhouse at 25°C during the winter of 2001 with supplemental light giving a 14-h day/10-h night cycle. Plants were grown individually in 25-cm plastic pots in washed sand to limit nodulation. Nutrients were supplied daily with or without N from a solution described previously (Staswick et al., 1991). After germination plants were watered with 250 mL of one-half-strength nutrient solution with 2.5 mm NH4NO3 for 2 weeks. The N source was then increased to 5 mm for 4 weeks, and then the daily volume was increased to 400 mL/pot until flowering. Additional water was provided as needed to maintain moisture level. After flowering, one-half of the pots were watered with the same nutrient solution and the remainder with nutrient solution lacking N.
ACKNOWLEDGMENTS
The authors would like to acknowledge the technical assistance of Drs. Martha Rowe and Aiqiu Xing.
Footnotes
This research was supported in part by a grant from The North Central Soybean Research Program. This is journal series paper no. 13,115 from the Nebraska Agriculture Research Division.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.010638.
LITERATURE CITED
- Berger S, Bell E, Sadka A, Mullet JE. Arabidopsis thaliana Atvsp is homologous to soybean VspA and VspB, genes encoding vegetative storage protein acid phosphatases, and is regulated similarly by methyl jasmonate, wounding, sugars, light and phosphate. Plant Mol Biol. 1995;27:933–942. doi: 10.1007/BF00037021. [DOI] [PubMed] [Google Scholar]
- Carrington J, Freed DD. Cap-independent enhancement of translation by a plant potyvirus 5′ nontranslated region. J Virol. 1990;64:1590–1597. doi: 10.1128/jvi.64.4.1590-1597.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derbyshire E, Wright DJ, Boulter D. Legumin and vicilin, storage proteins of legume seeds. Phytochemistry. 1976;15:3–24. [Google Scholar]
- DeWald DB, Mason HS, Mullet JE. The soybean vegetative storage proteins VSPα and VSPβ are acid phosphatases active on polyphosphates. J Biol Chem. 1992;267:15958–15964. [PubMed] [Google Scholar]
- Ditta G, Stanfield S, Corbin D, Helinski D. Broad host range DNA cloning system for gram-negative bacteria: construction of a gene bank of Rhizobium meliloti. Proc Natl Acad Sci USA. 1980;77:7347–7351. doi: 10.1073/pnas.77.12.7347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feller U, Keist M. Senescence and nitrogen metabolism in annual plants. In: Lambers H, Neetson JJ, Stulen I, editors. Fundamental, Ecological and Agricultural Aspects of Nitrogen Metabolism. Dordrecht, The Netherlands: Martinus Nijhoff; 1986. pp. 219–234. [Google Scholar]
- Franceschi VR, Giaquinta RT. Specialized cellular arrangements in legume leaves in relation to assimilate transport and compartmentation: comparison of the paraveinal mesophyll. Planta. 1983;159:415–422. doi: 10.1007/BF00392077. [DOI] [PubMed] [Google Scholar]
- Franceschi VR, Wittenbach VA, Giaquinta RT. Paraveinal mesophyll of soybean leaves in relation to assimilate transfer and compartmentation. III. Immunohistochemical localization of specific glycopeptides in the vacuole after depodding. Plant Physiol. 1983;72:586–589. doi: 10.1104/pp.72.2.586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimes HD, Koetje DS, Franceschi VR. Expression, activity and cellular accumulation of methyl jasmonate-responsive lipoxygenase in soybean seedlings. Plant Physiol. 1992;100:433–443. doi: 10.1104/pp.100.1.433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hood EE, Gelvin SB, Melchers LS, Hoekema A. New Agrobacterium helper plasmids for gene transfer to plants. Transgenic Res. 1993;2:208–218. [Google Scholar]
- Klauer SF, Franceschi VR, Ku MSB, Zhang D. Identification and localization of vegetative storage proteins in legume leaves. Am J Bot. 1996;83:1–10. [Google Scholar]
- Penheiter AR. Biochemical and genetic characterization of an acid phosphatase/5′-nucleotidase from soybean root nodules. PhD thesis. Lincoln: University of Nebraska; 1998. [Google Scholar]
- Penheiter AR, Duff SMG, Sarath G. Soybean root nodule acid phosphatase. Plant Physiol. 1997;114:597–604. doi: 10.1104/pp.114.2.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staswick PE. Soybean vegetative storage protein structure and gene expression. Plant Physiol. 1988;87:250–254. doi: 10.1104/pp.87.1.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staswick PE. Developmental regulation and the influence of plant sinks on vegetative storage protein gene expression in soybean leaves. Plant Physiol. 1989;89:309–315. doi: 10.1104/pp.89.1.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staswick PE. Storage proteins of vegetative plant tissues. Annu Rev Plant Physiol Plant Mol Biol. 1994;45:303–322. [Google Scholar]
- Staswick PE. The occurrence and gene expression of vegetative storage proteins and a Rubisco complex protein in several perennial soybean species. J Exp Bot. 1997;48:2031–2036. [Google Scholar]
- Staswick PE, Huang J, Rhee Y. Nitrogen and methyl jasmonate induction of soybean vegetative storage protein genes. Plant Physiol. 1991;96:130–136. doi: 10.1104/pp.96.1.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staswick PE, Papa C, Huang J-F, Rhee Y. Purification of the major soybean leaf acid phosphatase that is increased by fruit removal. Plant Physiol. 1994;104:49–57. doi: 10.1104/pp.104.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tranbarger TJ, Franceschi VR, Hildebrand DF, Grimes HD. The soybean 94-kilodalton vegetative storage protein is a lipoxygenase that is localized in paraveinal mesophyll cell vacuoles. Plant Cell. 1991;3:973–987. doi: 10.1105/tpc.3.9.973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Damme EJM, Hao Q, Barre A, Rouge P, Van Leuven F, Peumans WJ. Major protein of resting rhizomes of Calystegia sepium (Hedge Bindweed) closely resembles plant RNases but has no enzymatic activity. Plant Physiol. 2000;122:433–445. doi: 10.1104/pp.122.2.433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson VM, Colwell G. Acid phosphatase-1 from nematode resistant tomato. Plant Physiol. 1991;97:139–146. doi: 10.1104/pp.97.1.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittenbach VA. Purification and characterization of a soybean leaf storage glycoprotein. Plant Physiol. 1983;73:125–129. doi: 10.1104/pp.73.1.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Aiqiu X, Staswick P, Clemente TE. The use of glufosinate as a selective agent in Agrobacterium-mediated transformation of soybean. Plant Cell Tiss Org Cult. 1999;56:37–46. [Google Scholar]
- Zhang Z, Coyne DP, Vidaver AK, Mitra A. Expression of human lactoferrin cDNA confers resistance to Ralstonia solanacearum in transgenic tobacco plants. Phytopathology. 1998;88:730–734. doi: 10.1094/PHYTO.1998.88.7.730. [DOI] [PubMed] [Google Scholar]
- Zhong PY, Tanaka T, Yamauchi D, Minamikawa T. A 28-kilodalton pod storage protein of French bean plants: purification, characterization, and primary structure. Plant Physiol. 1997;113:479–485. doi: 10.1104/pp.113.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]