Abstract
Phosphate (Pi) deficiency is a major nutritional problem faced by plants in many agro-ecosystems. This deficiency results in altered gene expression leading to physiological and morphological changes in plants. Altered gene expression is presumed to be due to interaction of regulatory sequences (cis-elements) present in the promoters with DNA binding factors (trans-factors). In this study, we analyzed the expression and DNA-protein interaction of promoter regions of Pi starvation-induced genes AtPT2 and TPSI1. AtPT2 encodes the high-affinity Pi transporter in Arabidopsis, whereas TPSI1 codes for a novel gene induced in the Pi-starved tomato (Lycopersicon esculentum). Expression of AtPT2 was induced rapidly under Pi deficiency and increased with decreasing concentrations of Pi. Abiotic stresses except Pi starvation had no affect on the expression of TPSI1. DNA mobility-shift assays indicated that specific sequences of AtPT2 and TPSI1 promoter interact with nuclear protein factors. Two regions of AtPT2 and TPSI1 promoters specifically bound nuclear protein factors from Pi-sufficient plants. Interestingly, the DNA binding activity disappeared during Pi starvation, leading to the hypothesis that Pi starvation-induced genes may be under negative regulation.
Crop productivity is often limited by phosphate (Pi) availability (Barber, 1980). This limitation initiates a series of physiological and genetic changes leading to increased survival of plants under these conditions (Raghothama, 2000). Altered morphology, physiology, and biochemical pathways allow plants to cope with the nutrient deficiency (Plaxton and Carswell, 1999). It is becoming clear that a coordinated expression of genes during Pi starvation is the underlying factor in all these responses. Many genes including Pi transporters, phosphatases, RNases, β-glucosidase, and others of unknown function are induced during Pi starvation (Raghothama, 1999). Interestingly, the deficiency of Pi sets these molecular events in motion. One can envision that the Pi starvation-mediated signaling pathway results in specific interactions of trans-factors with conserved cis-elements in Pi deficiency-induced genes. This type of interaction is the basis for activation of suites of genes involved in Pi starvation rescue mechanism in yeast and bacteria.
In yeast, both positive and negative regulatory elements control the expression of Pi starvation-induced genes (Oshima, 1997). A key positive regulator, Pho4, controls the expression of multiple genes including phosphatases and Pi transporters. Pho4 has an amphipathic helix-loop-helix-type DNA binding domain that interacts with specific promoter sequences of PHO genes (Okamura et al., 2000). Under Pi sufficiency a complex of two negative regulators, Pho80 (cyclin) and Pho85 (cyclin-dependent protein kinase), render Pho4 inactive by hyper-phosphorylation, thus preventing its nuclear localization (O'Neill et al., 1996). During Pi deficiency the Pho81 another member of PHO regulon, inhibits the function of the Pho80/Pho85 complex, thus allowing Pho4 to interact with cis-elements (Ogawa et al., 1995; Lenburg and O'Shea, 1996). A similar type of gene regulation has also been described in Neurospora crassa (Metzenberg, 1998). In Escherichia coli, at least 30 genes are involved in scavenging Pi during the starvation response. PhoB, a regulator, and PhoR, a sensor, act in concert as a two-component system that responds to the cell's need for Pi (Wanner, 1997).
Plants have a much more complex system of regulating Pi uptake and homeostasis (Raghothama, 2000). It is presumed that intricate gene regulatory circuits similar to the PHO regulon may be involved in the Pi starvation response mechanism (Goldstein et al., 1989). A computer search of the plant DNA and protein databases revealed the presence of genes similar to PHO genes of yeast. In addition, the major molecular determinants of the Pi starvation response, Pi transporters and phosphatases, are activated in a manner very similar to that of yeast. The high-affinity Pi transporters is among the well-characterized Pi starvation-induced genes in plants (Muchhal et al., 1996; Mitsukawa et al., 1997; Smith et al., 1997). They are regulated at the level of transcription and expressed preferentially in roots (Muchhal and Raghothama, 1999; Karthikeyan et al., 2000). There are nine full-length sequences of high-affinity Pi transporter homologs in the Arabidopsis sequence databases. Thus far, the expression of only two genes (AtPT1 and AtPT2) has been described (Muchhal et al., 1996; Smith et al., 1997; Okumura et al., 1998). Another novel gene that is specifically induced under Pi starvation is TPS11 of tomato (Lycopersicon esculentum; Liu et al., 1997). Members of the TPSI1 family are also found in Medicago truncatulata (Burleigh and Harrison, 1997), Arabidopsis (Burleigh and Harrison, 1999; Martin et al., 2000), tobacco (Nicotiana tabacum), soybean (Glycine max), Lotus japonicas, and rice (Oryza sativa; expressed sequence tag databases). Expression of TPSI1 and its homologs has been extensively studied (Burleigh and Harrison, 1997, 1999; Liu et al., 1997; Biddinger et al., 1998). The promoter region of TPSI1 contains several conserved cis-elements found in yeast and N. crassa PHO genes (Liu et al., 1997). At present, very little is known about the transcriptional regulation of Pi starvation-induced genes in plants. The promoter regions of the Pi starvation-induced gene β-glucosidase showed specific interactions with protein factors (Malboobi et al., 1998). Some of the conserved cis-sequences in this promoter are similar to Pho4 and cAMP response element binding sites of PHO gene promoters of yeast.
Here, we report the detailed analysis of promoters of two Pi starvation-induced genes, AtPT2 and TPSI1 from Arabidopsis and tomato, respectively. The temporal and concentration-mediated expression of Arabidopsis Pi transporters is also analyzed in this study. The nature of the cis-elements present in both the promoters and their interactions with the nuclear protein factors are discussed. It is interesting that the data suggests a possible negative regulation of genes under Pi starvation.
RESULTS
Promoters of AtPT2 and TPSI1 Share Conserved Sequences
A 4.2-kb genomic DNA fragment containing AtPT2 gene was subcloned and sequenced (Mukatira et al., 1997). The transcription start site of the gene was mapped by primer extension analysis to the nucleotide 172 bp 5′ upstream of the translation initiation codon (Fig. 1). The 1,604-bp open reading frame of AtPT2 is flanked by a 172 and 163 bp of 5′- and 3′-untranslated region, respectively. Primer extension analysis of TPSI1 indicated that transcription start site is located 91 bp upstream of the translational start site ATG (Fig. 1). The complete sequences of TPSI1 and AtPT2 are accessible in GenBank database (accession nos. X99214 and AF022872, respectively). Promoter analysis of the TPSI1 gene using the transcription factor database revealed several protein binding domains and, more interestingly, a sequence (CACGTG) similar to the Pho4 binding domain. Pho4 is a positive regulator of the PHO regulon in yeast (Oshima, 1997). The cis-element CACGTG that binds with Pho4 is also a well-conserved stress regulatory element in plants. This element seems to be conserved in other Pi starvation-induced genes of Arabidopsis and M. truncatulata (Table I). Another interesting cis-element in the TPSI1 promoter region is the NIT-2 [TATCT(/A/G/T)] binding domain. The NIT-2 is a positive regulatory factor involved in nitrate acquisition in N. crassa. The AtPT2 promoter also contains two sequences similar to NIT-2 binding domain, in addition to other zinc finger protein binding sequences (Fig. 2; Table I). The other conserved element observed between the two promoters is ATGCCAT. The septamer is also found in the promoter region of another Pi transporter (AtPT5) of Arabidopsis. This palindromic sequence may be of some significance in the transcription factor interactions during Pi deficiency.
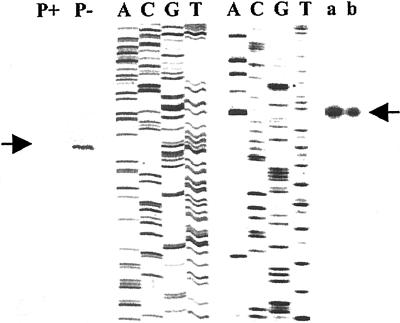
Figure 1.
Primer extension analysis transcription start sites of AtPT2 and TPSI1. The transcription start site was mapped to 172 and 91 bp upstream of the translational start site for AtPT2 and TPSI1, respectively. The total RNA isolated from Pi-sufficient (P+) and -deficient (P−) seedlings were used for mapping transcription start site of AtPT2. The poly(A+) RNA (a) and total RNA (b) isolated from Pi-starved roots of tomato were used to map TPSI1 transcriptional start site. DNA sequencing ladders were used to determine the size of the primer extended products.
Table I.
cis-Regulatory regions on the TPSI1 and Pi transporter promoters
| Sequence | Name of the Element |
|---|---|
| CACGTG/C | PHO element |
| -772 | TPSI1 |
| -373 | Mt4 |
| CAT(/G)A(/C)TG | Helix-loop-helix element |
| -889, -1143 | TPSI1 |
| -1951, -685 | MT4 |
| -341 | At4 |
| TATCA(/T)A(/T) | NIT 2 element |
| -230, -594, -1117, -1377 | TPSI1 |
| -500, -786 | Mt4 |
| -64, -1141, -2754 | At4 |
| -333, -737, -621, -1521, -2053 | AtPT1 |
| -1623, -2053, -2180 | AtPT2 |
Promoter analysis of TPSI1, AtPT1, and AtPT2 using the transfactor database (Heinemeyer et al., 1998) led to the identification of sequences found in various promoter elements of the yeast PHO regulon genes. Positions of the Pho4 regulatory domain, consensus Pho box-like domain (helix-loop-helix), and an NIT-2 regulatory domain were mapped 5′ of the putative start codon. The locations of these sequences are indicated along with a comparative analysis of the promoters of TPSI1 orthologs Mt4 and At4.
Figure 2.
Nucleotide sequence of promoter region of the AtPT2 gene. The putative transcription start site indicated by the bold letter is numbered +1. The promoter fragments interacting with nuclear factors are shown in bold. The underlined palindromic sequence ATGCCAT represents the conserved motif. The sequences showing similarity to the NIT-2 binding domain found in the regions of the promoter interacting with proteins are shaded.
Expression of TPSI1 and Pi Transporters Is Regulated by Pi
Our earlier studies provided evidence that TPSI1 and AtPT2 are Pi starvation-induced genes (Muchhal et al., 1996; Liu et al., 1997). In this study, the specificity of TPSI1 induction to Pi starvation and the response of AtPT2 to altered Pi concentration were further examined. Total RNA was extracted from leaves and roots of tomato plants subjected to different abiotic stresses including Pi starvation. RNA-blot analysis data clearly shows that TPSI1 is specifically induced in response to Pi starvation and that other abiotic stresses have little or no effect on expression of this gene (Fig. 3).
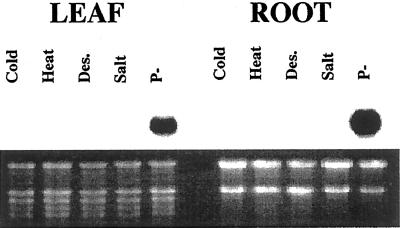
Figure 3.
TPSI1 is specifically induced under Pi starvation. Hydroponically grown tomato plants were exposed to 4°C (Cold), 37°C (Heat), desiccation (Des.), salt stress (Salt), and Pi starvation (P−) for 4 d. RNA extracted from leaves and roots was subjected to northern-blot analysis with 32P-labeled TPSI1 cDNA fragment as the probe. The ethidium bromide stained gel below shows uniform loading of RNA samples.
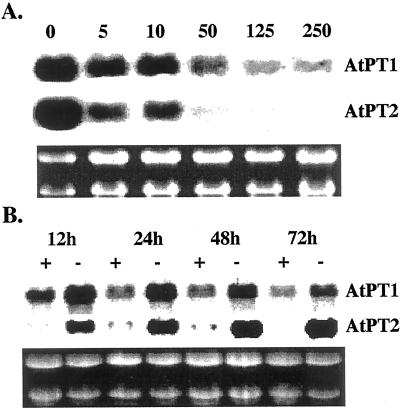
The effect of altered Pi levels in the medium on the expression of AtPT1 and AtPT2 was determined using liquid culture-grown Arabidopsis plants. The low levels of AtPT1 transcripts that were detectable at 250 μm Pi increased significantly with decreasing concentration of Pi. In contrast, the AtTP2 transcript levels were very low in plants supplemented with higher concentration of Pi. A distinct increase in AtPT2 transcripts was observed when the concentration of Pi in the medium was reduced below 50 μm (Fig. 4A). Both the genes were induced rapidly in response to Pi deficiency (Fig. 4B). Transcript abundance of both genes was noticeable following 12 h of Pi starvation. This confirmed our earlier observation that plants are able to respond rapidly to changes Pi levels by altering the expression of Pi starvation-induced genes.
Figure 4.
Concentration-dependent and temporal expression of Pi transporters. A, Arabidopsis plants grown in liquid culture for 7 d were transferred to medium containing indicated amounts (μm) of Pi. Five days after the treatment, plants were harvested and stored at −70°C. Total RNA extracted from the plants was subjected to northern analysis with AtPT1 and AtPT2 cDNAs as probes. The ethidium bromide-stained gel below shows uniform loading of RNA samples. B, Seven-day-old Arabidopsis plants grown in liquid culture were subject to Pi starvation for indicated time. Total RNA was extracted and subjected to northern analysis with labeled AtPT1 and AtPT2 cDNAs as probes
Nuclear Protein Factors Interact Specifically with the Promoter Regions of TPSI1 and AtPT2
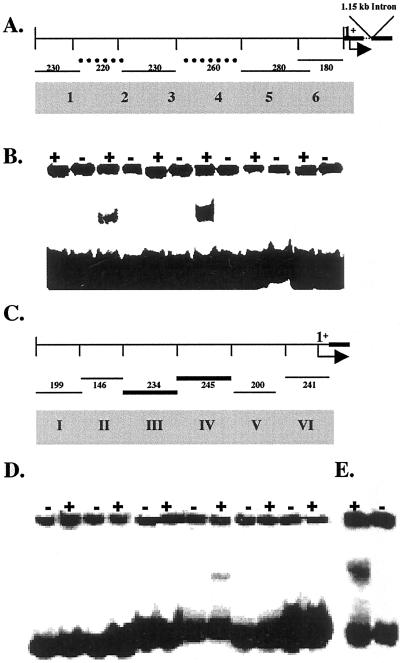
Eukaryotic gene expression is typified by the interaction of cis-acting regulatory sequences with the trans-acting factors. Binding of a factor may induce or inhibit gene expression. Subsequently, the release of transcription factor(s) from its target sequence could also result in altered gene expression. To understand the interaction of nuclear proteins with the promoter element(s) of the Pi starvation-induced genes, gel retardation experiments were performed. The DNA fragments used for binding assays encompassed 1.2 and 1.15 kb of the AtPT2 and TPSI1 promoters, respectively. Promoter fragments representing the 5′-regulatory regions of AtPT2 and TPSI1 were subject to electrophoretic gel mobility-shift assay. The nuclear extracts were incubated with double-stranded 32P-labeled DNA probes representing six fragments of AtPT2 promoter from Arabidopsis (Fig. 5A) and the TPSI1 promoter of tomato (Fig. 5C). To avoid non-specific binding, 2 μg of poly(dI-dC) was added to all reactions. The mobility-shift assays revealed the formation of specific DNA-protein complexes by fragments 2 and 4 of AtPT2, and fragment IV of TPSI1 promoters (Fig. 5, B and D). Ten micrograms of the nuclear extract was determined to be optimal for the formation of DNA-protein complexes of TPSI1 and AtPT2 promoter fragments. Because TPSI1 is also expressed in the leaves upon Pi starvation, nuclear proteins extracted from tomato leaf tissue were used in binding studies. These experiments revealed that nuclear proteins from leaves interacted with fragment III of TPSI1 promoter (Fig. 5E). Perhaps the most significant result from these studies is that only protein factors derived from Pi-sufficient plants interacted specifically with the promoters of these genes. This suggests that the DNA binding factors are conspicuously absent or incapable of binding to the promoter elements during Pi deficiency. These interactions were consistently observed over several replications with different protein extracts for both TPSI1 and AtPT2 promoter fragments.
Figure 5.
Nuclear protein factors are not associated with promoters during Pi starvation. A, Map of the different promoter fragments of AtPT2 used in gel-shift assays. Dotted lines indicate fragments that interacted with protein factors. The location of an intron and the transcriptional start site (1+) are also shown. B, Labeled upstream fragments (1–6) of the promoter of AtPT2 were incubated with nuclear protein extracts (10 μg) obtained from Pi-sufficient (+) and -deficient (−) Arabidopsis plants. The reaction mix was analyzed on a polyacrylamide gel. C, Map of the different promoter fragments of TPSI1 used in gel-shift assays. Bold lines indicate the fragments interacting with protein factors. The transcriptional start site is marked as +1. D, Labeled upstream fragments (I–VI) were mixed with nuclear extracts (10 μg) isolated from Pi-sufficient (+) and -deficient (−) tomato roots. The reaction mix was analyzed on a polyacrylamide gel. E, Fragment III of the TPSI1 promoter interacted with leaf proteins extracted from Pi-sufficient plants, whereas the other fragments showed no interaction (data not shown).
Nuclear Proteins Interact Specifically with Promoter Elements
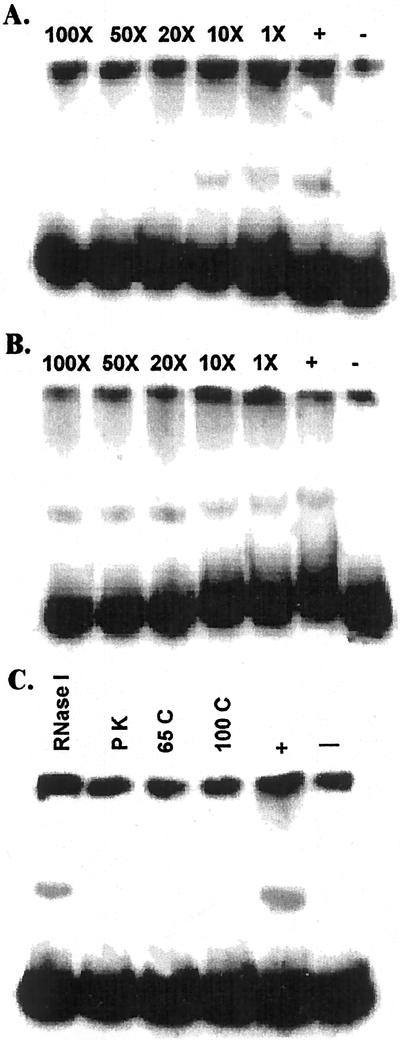
To confirm the specificity of interaction between the nuclear factors and DNA fragments, competition experiments were performed. Unlabeled fragments representing 1 to 100 times molar excess of the labeled probe were added to the DNA-protein interaction reaction mixture. A 20-fold molar excess of fragment IV of TPSI1 promoter was sufficient to reduce or inhibit the DNA-protein complex formation (Fig. 6A). A similar competition was also observed with labeled and unlabeled fragments of AtPT2 promoter (data not shown). A proportional decline in binding in response to increasing specific competitors indicates that the DNA-protein interaction is specific. The pBluescript plasmid digested with HhaI and Sau3A was added as a non-specific competitor to the reaction mix. A 100-fold molar excess of non-specific competitors had little or no effect on DNA-protein interaction of the promoter fragments of TPSI1 gene (Fig. 6B). A similar type of interaction was also observed with the promoter fragments of AtPT2 (data not shown). This further confirmed the specific binding of the nuclear factors with target sequences. After clarifying the specific nature of the DNA-protein binding, the proteinaceous nature of the binding factors was determined. The nuclear extract was subject to ProteinaseK and RNaseI treatment for 10 min before incubation with the promoter fragments. The nuclear proteins were also subjected to boiling and heated to 65°C for 10 min to denature proteins. The DNA-protein interaction was observed only with RNaseI treated and non-denatured proteins. Heat and ProteinaseK treatments that denature proteins completely abolished the binding with DNA fragments (Fig. 6C). These result shows that the binding factors are indeed proteinaceous in nature as expected of transcription factors.
Figure 6.
Nuclear proteins interact specifically with promoter fragments. A, Labeled DNA fragment IV of TPSI1 promoter was mixed with nuclear extracts obtained from Pi-starved (−) or -sufficient (+) Arabidopsis tissue. Indicated mass excess of unlabeled fragment IV was added as specific competitors. Increasing concentrations of the competitor progressively reduced the binding of the labeled fragment. B, Increasing quantities of pBluescript DNA digested with HhaI and Sau3A were added to the DNA (TPSI1 promoter fragment IV)-protein interaction mix. Lack of competition with increasing mass excess of non-specific competitor indicated that the interaction between the DNA and the nuclear proteins is specific. C, The DNA binding factors are proteinaceous in nature. Nuclear proteins from Pi-sufficient tomato roots were subject to RNaseI and ProteinaseK (PK) treatments and heat denaturation (65°C and 100°C) for 10 min. The treated nuclear proteins were used in gel retardation reactions with the TPSI1 promoter fragment IV. The treatments that denature proteins resulted in a loss of DNA-protein interaction, whereas RNaseI treatment did not affect binding.
DISCUSSION
Regulation of gene expression is in part due to the interaction of nuclear-localized transcription factors with the cis-elements of a gene. The genome sequencing efforts have shed light on the promoter region (cis-elements) of genes from various organisms. It has been hypothesized that a relatively small number of divergently oriented promoter elements regulate gene expression in plants (Somerville and Somerville, 1999). Because several genes share common cis-elements, coordinated expression of those genes may indeed be a valid assumption. This may be particularly true in case of genes involved in similar function(s) across the species and/or among genes that are regulated by signal transduction pathways triggered by a common stimulus (Oshima, 1997). The large scale sequencing efforts of various genomes shows that the protein coding regions are much more conserved across the genomes, whereas promoters are highly divergent. This is clearly evident in the open reading frames of plant Pi transporters. They share a high degree of nucleotide and amino acid sequence identity, whereas their promoters and 3′-untranslated regions are highly divergent. Because Pi transporters are among the most evolutionarily conserved proteins (Pao et al., 1998), divergences in the promoter are presumed to be important for differential gene expression, including tissue specificity.
In plants, Pi starvation results in coordinated expression of genes with diverse biological functions (Raghothama, 1999). It is likely that these genes may share some common regulatory sequences as observed in the Pi starvation-induced genes of microorganisms (Oshima, 1997; Metzenberg, 1998). Here we studied the expression of Pi starvation-induced genes and the interaction of their promoter with nuclear factors. The detailed analysis of TPSI1 expression under abiotic stresses further confirmed its specific responsiveness to Pi. Our earlier studies have suggested that among the different essential nutrients tested, only Pi deficiency leads to the expression of TPSI1 (Liu et al., 1997; Biddinger et al., 1998). The widespread distribution of TPSI1-like genes in many plant species and their induction under Pi starvation suggest a conserved role for this gene family in the stress response. The Pi transporter genes AtPT1 and AtPT2 exhibited different sensitivity to changing concentrations of Pi in the medium. AtPT1 was strongly induced when the Pi concentration in the medium was reduced to 125 μm, whereas distinct accumulation of AtPT2 transcripts was observed when Pi concentration was reduced below 50 μm (Fig. 4). The induction of both genes was rapid in response to Pi starvation. A similar type of temporal induction of AtPT1 (APT1) and enhanced uptake of Pi was also observed in hydroponically grown Arabidopsis plants (Dong et al., 1999). The responses of the two high-affinity transporters to changes in nutrient concentration is indicative of the plant's ability to differentially regulate one or more genes involved in nutrient acquisition based on its needs and availability of nutrients.
The data on gene regulation during Pi starvation indicate that the plants are always monitoring cellular Pi levels and that the response to any changes in Pi levels is rapid and specific. This specific response may result from interactions of Pi starvation-specific promoter elements (cis-elements) with nuclear protein factors (trans-factors) produced and/or activated under Pi deficiency. Such an interaction was examined in detail by DNA-protein interaction studies using gel-shift assays. Nuclear protein factors from seedlings of Arabidopsis grown under Pi-sufficient conditions interacted with two labeled fragments of the AtPT2 promoter. In the case of TPSI1 promoter, the cis-elements interacted with nuclear factors isolated from roots and leaves of Pi-sufficient plants. The DNA-protein interaction is specific, and the binding factors are proteinaceous in nature. Even 100-fold molar excess of non-specific DNA fragments did not affect the interactions. It is interesting that the binding is conspicuously absent in nuclear extracts obtained from Pi-deficient tissues. A lack of binding of nuclear proteins from Pi-deficient tissues was observed with the promoter elements of two different genes belonging to two different plant species. In another study, a similar type of protein-DNA interaction was observed with the promoter of a β-glucosidase isolated from Brassica nigra (Malboobi et al., 1998). This gene is specifically induced under Pi starvation, and it is presumed to play a role in Pi starvation rescue process. In addition, differential expression analysis of genes induced under Pi deficiency revealed that genes coding for transcription factors like MYB and b-ZIP proteins are down regulated during Pi starvation (personal communication, Drs. Carroll Vance [U.S. Department of Agriculture-Agricultural Research Service, University of Minnesota, St. Paul] and Deborah Allan [University of Minnesota]). Taken together, all of these observations point to a negative regulation of gene expression during Pi starvation. One can envision that protein factors that are abundant and able to bind promoters of Pi starvation-induced genes may suppress their transcription, whereas during Pi deficiency DNA binding proteins may be less abundant and/or incapable of associating with promoters due to post-translational modifications. In yeast the phosphorylation and dephosphorylation of the positive regulator Pho4 is the key step in activation of the pho regulon genes. Furthermore repressors are known to play important roles in plant gene expression as observed in the GCC box binding of AtERF3 and -4 promoters (Fujimoto et al., 2000) and negative regulation of AGAMOUS by leunig and apetala2 in Arabidopsis (Conner and Liu, 2000).
The DNA binding factors generally recognize the conserved or specific sequences in the promoter region of genes. The databases TRANSFAC (Wingender et al., 2000), PlantCARE (Plant cis-acting regulatory elements; Thijs et al., 2000), and TFSEARCH (Heinemeyer et al., 1998) are quite robust in identifying and analyzing putative trans-factor binding sites. Because the two genes under study are induced specifically in response to Pi starvation, they may have similar cis-regulatory regions. Sequence analyses of the promoter from Arabidopsis (AtPT2) and tomato (TPSI1) have revealed that although certain common elements may be shared, other putative regulatory elements are specific to each gene. The conserved sequence ATGCCAT is found in the promoter regions of both genes interacting with protein factors. The ATGCCAT sequence is palindromic, and hence it has a potential to interact with protein factors as commonly seen in the interactions between restriction enzymes and palindromic DNA sequences. Although the database revealed no known trans-factor binding to this sequence, it is certainly worthy of further examination because the AtPT2 and the TPSI1 promoters contain this conserved sequence (Fig. 2; Table I). The presence of the same conserved sequence in the promoter of another Pi transporter in Arabidopsis (AtPT5) further suggests a potential role for this element. Such interactions may not be uncommon among plant promoters because more than 1,500 transcription factor genes are presumed to exist in Arabidopsis (Riechmann and Ratcliffe, 2000).
Another common trans-factor binding region shared by both promoters is the NIT-2 binding domain, [TATCT(/A/G/T)], which is a positive regulatory domain of the nitrate uptake genes of N. crassa (Fu and Marzluf, 1990). In N. crassa, NIT-2 activates the expression of genes like nitrate and nitrite reductase, various transporters, and enzymes that are related to nitrogen metabolism (Tao and Marzluf, 1999). This master control gene of nitrogen regulatory circuitry encodes a GATA-type zinc finger DNA binding domain and redundant transcription activation domains. The TATCTA sequences are found exclusively in the protein interacting regions of the AtPT2 promoter (Fig. 1), whereas TATCAA binding NIT-2 domain is found in the TPSI1 promoter region interacting with protein factors from tomato. This element may be of significance in Pi starvation response because all of the nine Pi transporter promoters from Arabidopsis contain this element (data not shown). A major difference between the N and P response is that during the N catabolism cascade, NIT-2 activates the regulatory genes in N. crassa, whereas a lack of binding of protein factors to the promoter regions containing putative NIT-2-like binding sites appears to be associated with activation of Pi starvation-induced genes in plants.
Some of the other conserved elements in the TPSI1 promoter include the CACGTG(/T) sequences. This sequence has been determined as the Pho4 binding region in yeast (Ogawa et al., 1995) and the Pho box described in E. coli (Wanner, 1997). All of the known Pi starvation-induced genes belonging to the TPSI1 family of Arabidopsis, M. truncatulata, and tomato have Pho4 binding-conserved element in addition to other helix-loop-helix sequences (Table I) that are attributed to various DNA binding activities (Blackwell and Weintraub, 1990). Furthermore, the presence of Pho4 binding G-box-like elements in the members of TPSI1 family of genes and on β-glucosidase (Malboobi et al., 1998) may be indicative of a conserved role for this element in Pi starvation responses of bacteria, fungi and plants. These conserved domains could be used as targets for future characterization of gene expression under Pi starvation. Based on the results presented here and evidence obtained from others, it appears that protein factors interacting with promoter elements may be associated with down-regulation of genes under Pi sufficiency conditions. During Pi deficiency the genes coding for these DNA binding factors may themselves be down-regulated or the factors are incapable of binding to the promoter elements. The specific roles of the transcription factors and the DNA domains remain to be elucidated. The completion of Arabidopsis genome sequences and development of powerful bioinformatics tools should lead to the identification of these factors.
MATERIALS AND METHODS
Plant Materials
Arabidopsis (Columbia) plants grown in liquid culture were used for northern analysis and nuclear protein extraction. Seeds were sterilized by rinsing with 75% (v/v) eth-anol followed by treating them in 50% (v/v) commercial bleach solution for 10 min. Seeds were thoroughly washed with sterile water to remove residual bleach. Surface-sterilized seeds were vernalized for 48 h at 4°C. The liquid culture setup of Arabidopsis consisted of sets of sterile 250-mL flasks with 30 mL of Murashige and Skoog medium (pH 5.7) placed on a gyratory shaker set at 100 rpm under long-day conditions (16-h light/8-h dark). Approximately 30 vernalized seeds were dispersed into each flask. After 7 d, seedlings were rinsed with sterile water and treated in flasks containing Pi-sufficient (1.25 mm) or -deficient Murashige and Skoog medium for another 5 d. In some treatments the concentration of Pi in the medium was adjusted by substituting K2SO4 for KH2PO4. Treated plants were harvested, frozen in liquid nitrogen, and stored at −80°C. The frozen tissues were used for RNA isolation and nuclear protein extractions.
Tomato (Lycopersicon esculentum) plants were grown in hydroponic solution essentially as described earlier (Liu et al., 1997). Abiotic stress treatments were initiated by transferring hydroponically grown plants to 4°C and 37°C (root zone temperature) chambers for 4 d. Some of the plants removed from hydroponics were allowed to desiccate at room temperature for at least 3 h. To test the effect of salt on gene expression, plants were transferred to hydroponic solutions containing 150 mm NaCl for 4 d. Tissues harvested after the treatments were immediately frozen in liquid nitrogen and stored at −80°C for further analysis.
Northern Analysis
Total RNA was extracted from plants as described by Pawlowski et al. (1994). Ten micrograms of total RNA was electrophoretically separated on a 1% (w/v) denaturing formaldehyde agarose gel and blotted onto a BA-S nitrocellulose membrane following the manufacturer's instructions (Schleicher & Schull, Keene, NH). The prehybridization was done for 4 h in a solution containing 50% (w/v) formamide, 5× Denhardt's solution, 0.1% (w/v) SDS, 6× SSPE, and 150 μg/μL denatured salmon sperm DNA at 42°C. DNA labeled with [32P]dCTP using DECA prime II DNA labeling kit (Ambion, Austin, TX) was used to probe the filters. Hybridization was carried out with 106 cpm of probe per milliliter in a buffer identical to the prehybridization buffer at 42°C for 16 h. Filters were then washed twice in 2× SSC and 0.2% SDS at room temperature for 10 min, twice in 1× SSC and 0.1% (w/v) SDS at 50°C for 15 min, and finally for stringent washes once in 0.1× SSC and 0.1% (w/v) SDS at 60°C for 20 min before autoradiography.
Primer Extension Analysis of AtPT2 and TPSI1
The oligonucleotide complementary to the sequence 4 bp downstream of translational start site of AtPT2 was end-labeled with [γ-32P]ATP using T4 polynucleotide kinase. Similarly, an oligonucleotide complementary to the sequence 20 bp downstream of the translation start site of TPSI1 gene was used to map the transcription start site. Approximately 5 × 104 cpm of the labeled primer was used to extend 10 μg of total RNA extracted from either Arabidopsis or tomato. The RNA template was reverse transcribed at 37°C for 30 min, and loaded to an 8% (w/v) sequencing gel with a sequencing ladder as a size marker (Raghothama et al., 1993).
Analysis of Pi Starvation-Induced Gene Promoters
Promoters of Pi transporters and TPSI1 were analyzed using several web-based programs. The TFSEARCH database was used to predict transcription factor binding sites on TPSI1 and AtPT2 promoter sequences (http://www.cbrc.jp/research/db/TFSEARCH.html). Another database TRANSFAC (http://transfac.gbf.de/TRANSFAC) provided information about transcription factors, their binding sites, and DNA binding profiles (Wingender et al., 2000). This database contains information about the eukaryotic cis-acting regulatory DNA elements and trans-acting factors. There are 2,285 non-independent transcription factors listed in this database (Heinemeyer et al., 1998). The PlantCARE web site (http://sphinx.rug.ac.be:8080/PlantCARE) provided information on conserved motifs between the promoters (Thijs et al., 2000). The basic assumption in all these searches is that a cluster of regulated genes is controlled by the same transcription factors and that the genes of a given cluster share common regulatory motifs.
Nuclear Protein Extraction
Nuclei from Arabidopsis and tomato tissues were obtained essentially as described earlier (Lawton et al.1990). All steps were performed at 4°C. The nuclear protein extracts were prepared by the protocol described by Miskimins et al. (1985). The nuclear proteins were dialyzed against the buffer containing 50% (v/v) glycerol (10 mm HEPES, pH 8.0, 1 mm MgCl2, 1 mm dithiothreitol, 50 mm NaCl, and 0.8 mm phenylmethylsulfonylfluoride) with one change after 8 h. The nuclear protein was dispensed in to aliquots and frozen at −80°C. Protein concentrations were measured by the dye binding assay with bovine serum albumin as the standard (Sigma, St Louis).
Preparation of the DNA Fragments Used in DNA-Protein Interaction
TPSI1 (1.2 kb) and AtPT2 (1.15 kb) promoters were used for preparing DNA probes. They were prepared by a combination of PCR amplifications and digestions with restriction enzymes. The size of the promoter fragments used in binding studies varied from 160 to 240 bp.
Electrophoretic Mobility-Shift Assay
The promoter fragments of AtPT2 and TPSI1 were end-labeled with [γ-32P]ATP using T4 polynucleotide kinase. Aliquots of the nuclear extracts (5–10 μg) were incubated with 3 to 5 ng (approximately 20,000 cpm) of labeled promoter fragments at 30°C for 30 min. The reactions were carried out in a buffer containing 2 μg of poly(dI-dC) as a non-specific competitor, 500 mm NaCl, 100 mm Tris-Cl, pH 8.0, 10 mm dithiothereitol, 10 mm EDTA, and 5% (v/v) glycerol. Each labeled fragment was mixed with nuclear proteins from plants grown under Pi-sufficient (+) or -deficient (−) conditions. The interacting products were analyzed on 4% (w/v) polyacrylamide gels. The dried gels were exposed to x-ray films with intensifying screens for 12 to 24 h. For specific competition assays, indicated molar excess of unlabeled (cold) fragment was added to the binding reaction. Non-specific competitor fragments were generated by restriction enzyme digestion of pBluescript plasmid with HhaI and Sau3A and added to the corresponding assay mixes.
ACKNOWLEDGMENTS
We thank Drs. Peter B. Goldsbrough and David Rhodes for critically reviewing the manuscript.
Footnotes
This work was supported in part by the U.S. Department of Agriculture-National Research Initiative Competitive Grants Program (grant no. 00–35100–9370). This is journal paper no. 16,617 of the Purdue University Agricultural Research Program.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.010876.
LITERATURE CITED
- Barber SA. Soil-plant interactions in the phosphorus nutrition of plants. In: Khasawneh FE, Sample EC, Kamprath EJ, editors. Role of Phosphorus in Agriculture. Madison, WI: American Society of Agronomy; 1980. pp. 591–615. [Google Scholar]
- Biddinger EC, Liu C, Joly RJ, Raghothama KG. Physiological and molecular responses of aeroponically grown tomato plants to phosphorus deficiency. J Am Soc Hortic Sci. 1998;123:330–333. [Google Scholar]
- Blackwell TK, Weintraub H. Differences and similarities in DNA-binding preferences of MyoD and E2A protein complexes revealed by binding site selection. Science. 1990;250:1104–1110. doi: 10.1126/science.2174572. [DOI] [PubMed] [Google Scholar]
- Burleigh SH, Harrison MJ. A novel gene whose expression in Medicago truncatularoots is suppressed in response to colonization by vesicular-arbuscular mycorrhizal (VAM) fungi and to phosphate nutrition. Plant Mol Biol. 1997;34:199–208. doi: 10.1023/a:1005841119665. [DOI] [PubMed] [Google Scholar]
- Burleigh SH, Harrison MJ. The down-regulation of Mt4-like genes by phosphate fertilization occurs systemically and involves phosphate translocation to the shoots. Plant Physiol. 1999;119:241–248. doi: 10.1104/pp.119.1.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conner J, Liu Z. LEUNIG, a putative transcriptional corepressor that regulates AGAMOUSexpression during flower development. Proc Natl Acad Sci USA. 2000;97:12902–12907. doi: 10.1073/pnas.230352397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong B, Ryan PR, Rengel Z, Delhaize E. Phosphateuptake in Arabidopsis thaliana: dependence of uptake onthe expression of transporter genes and internal phosphate concentrations. Plant Cell Environ. 1999;22:1455–1461. [Google Scholar]
- Fu YH, Marzluf GA. NIT-2, the major positive-acting nitrogen regulatory gene of Neurospora crassa, encodes a sequence-specific DNA-binding protein. Proc Natl Acad Sci USA. 1990;87:5331–5335. doi: 10.1073/pnas.87.14.5331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimoto SY, Ohta M, Usui A, Shinshi H, Ohme-Takagi M. Arabidopsis ethylene-responsive element binding factors act as transcriptional activators or repressors of GCC box-mediated gene expression. Plant Cell. 2000;12:393–404. doi: 10.1105/tpc.12.3.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinemeyer T, Wingender E, Reuter I, Hermjakob H, Kel AE, Kel OV, Ignatieva EV, Ananko EA, Podkolodnaya OA. Databases on transcriptional regulation: TRANSFAC, TRRD, and COMPEL. Nucleic Acids Res. 1998;26:364–367. doi: 10.1093/nar/26.1.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein AH, Baertlein DA, Danon A. Phosphate starvation stress an experimental system for molecular analysis. Plant Mol Biol Rep. 1989;7:7–16. [Google Scholar]
- Karthikeyan AS, Uthappa M, Varadarajan DK, D'Urzo PM, Raghothama KG (2000) Phosphate transporter promoters drive reporter gene activity in response to phosphate starvation (abstract no. 974). Plant Biol Suppl 191
- Lawton KA, Raghothama KG, Goldsbrough PB, Woodson WR. Regulation of senescence-related gene expression in carnation flower petals by ethylene. Plant Physiol. 1990;93:1370–1375. doi: 10.1104/pp.93.4.1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenburg ME, O'Shea EK. Signaling phosphate starvation. Trends Biochem Sci. 1996;10:383–387. [PubMed] [Google Scholar]
- Liu C, Muchhal US, Raghothama KG. Differential expression of TPS1I,a phosphate starvation-induced gene in tomato. Plant Mol Biol. 1997;33:867–874. doi: 10.1023/a:1005729309569. [DOI] [PubMed] [Google Scholar]
- Malboobi MA, Hannoufa A, Tremblay L, Lefebvre DD. Towards an understanding of gene regulation during the phosphate starvation response. In: Lynch JP, Deikman J, editors. Phosphorous in Plant Biology: Regulatory Roles in Molecular, Cellular, Organismic, and Ecosystem Processes. Rockville, MD: The American Society of Plant Physiologists; 1998. pp. 215–226. [Google Scholar]
- Martin AC, del Pozo JC, Iglesias J, Rubio V, Solano R, de la Pena A, Leyva A, Paz-Ares J. Influence of cytokinins on the expression of phosphate starvation responsive genes in Arabidopsis. Plant J. 2000;24:559–567. doi: 10.1046/j.1365-313x.2000.00893.x. [DOI] [PubMed] [Google Scholar]
- Metzenberg RL. How Neurosopara crassagets its phosphorous. In: Lynch JP, Deikman J, editors. Phosphorous in Plant Biology: Regulatory Roles in Molecular, Cellular, Organismic, and Ecosystem Processes. Rockville, MD: The American Society of Plant Physiologists; 1998. pp. 181–191. [Google Scholar]
- Miskimins K, Roberts MD, McClelland A, Ruddle FH. Use of protein-blotting procedure and a specific DNA probe to identify nuclear proteins that recognize the promoter region of the transferrin receptor gene. Proc Natl Acad Sci USA. 1985;82:6741–6744. doi: 10.1073/pnas.82.20.6741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsukawa N, Okumura S, Shirano Y, Sato S, Kato T, Harashima S, Shibata D. Overexpression of an Arabidopsis thalianahigh-affinity phosphate transporter gene in tobacco cultured cells enhances cell growth under phosphate-limited conditions. Proc Natl Acad Sci USA. 1997;94:7098–7102. doi: 10.1073/pnas.94.13.7098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muchhal US, Pardo JM, Raghothama KG. Phosphate transporters from the higher plant Arabidopsis thaliana. Proc Natl Acad Sci USA. 1996;93:10519–10523. doi: 10.1073/pnas.93.19.10519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muchhal US, Raghothama KG. Transcriptional regulation of plant phosphate transporters. Proc Natl Acad Sci USA. 1999;96:5868–5872. doi: 10.1073/pnas.96.10.5868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukatira UT, Muchhal US, Raghothama KG. Cloning of Arabidopsis thaliana phosphate transporter gene, AtPT2 (accession no. AF022872) (PGR 97-163) Plant Physiol. 1997;115:1288. [Google Scholar]
- Ogawa N, Saitoh H, Miura K, Magbanua JP, Bun-ya M, Harashima S, Oshima Y. Structure and distribution of specific cis-elements for transcriptional regulation of PHO84 in Saccharomyces cerevisiae. Mol Gen Genet. 1995;249:406–416. doi: 10.1007/BF00287102. [DOI] [PubMed] [Google Scholar]
- Okamura H, Hanaoka S, Nagadoi A, Makino K, Nishimura Y. Structural comparison of the PhoB and OmpR DNA-binding/transactivation domains and the arrangement of PhoB molecules on the phosphate box. J Mol Biol. 2000;295:1225–1236. doi: 10.1006/jmbi.1999.3379. [DOI] [PubMed] [Google Scholar]
- Okumura S, Mitsukawa N, Shirano Y, Shibata D. Phosphate transporter gene family of Arabidopsis thaliana. DNA Res. 1998;5:261–269. doi: 10.1093/dnares/5.5.261. [DOI] [PubMed] [Google Scholar]
- O'Neill EM, Kaffman A, Jolly ER, O'Shea EK. Regulation of PHO4 nuclear localization by the PHO80-PHO85 cyclin-CDK complex. Science. 1996;271:209–212. doi: 10.1126/science.271.5246.209. [DOI] [PubMed] [Google Scholar]
- Oshima Y. The phosphatase system in Saccharomyces cerevisiae. Genes Genet Syst. 1997;72:323–334. doi: 10.1266/ggs.72.323. [DOI] [PubMed] [Google Scholar]
- Pao SS, Paulsen IT, Saier MH. Major facilitator superfamily. Microbiol Mol Biol Rev. 1998;62:1–34. doi: 10.1128/mmbr.62.1.1-34.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawlowski K, Kunze R, De Vries S, Bisseling T. Isolation of total, poly(A) and polysomal RNA from plant tissues. In: Gelvin SB, Shilperoort RA, editors. Plant Molecular Biology Manual. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1994. pp. 1–13. [Google Scholar]
- Plaxton WC, Carswell MC. Metabolic aspects of the phosphate starvation response in plants. In: Lerner HR, editor. Plant Responses to Environmental Stresses: From Phytohormones to Genome Reorganization. New York: Marcel Dekker; 1999. pp. 349–372. [Google Scholar]
- Raghothama KG. Phosphate acquisition. Annu Rev Plant Physiol Mol Biol. 1999;50:665–693. doi: 10.1146/annurev.arplant.50.1.665. [DOI] [PubMed] [Google Scholar]
- Raghothama KG. Phosphate transport and signaling. Curr Opin Plant Biol. 2000;3:182–187. [PubMed] [Google Scholar]
- Raghothama KG, Liu D, Nelson DE, Hasegawa PM, Bressan RA. Analysis of an osmotically regulated pathogenesis-related osmotin gene promoter. Plant Mol Biol. 1993;23:1117–1128. doi: 10.1007/BF00042346. [DOI] [PubMed] [Google Scholar]
- Riechmann JL, Ratcliffe OJ. A genomics perspective on plant transcription factors. Curr Opin Plant Biol. 2000;3:423–434. doi: 10.1016/s1369-5266(00)00107-2. [DOI] [PubMed] [Google Scholar]
- Smith FW, Ealing PM, Dong B, Delhaize E. The cloning of two Arabidopsisgenes belonging to a phosphate transporter family. Plant J. 1997;11:83–92. doi: 10.1046/j.1365-313x.1997.11010083.x. [DOI] [PubMed] [Google Scholar]
- Somerville C, Somerville S. Plant functional genomics. Science. 1999;285:380–383. doi: 10.1126/science.285.5426.380. [DOI] [PubMed] [Google Scholar]
- Tao Y, Marzluf GA. The NIT-2 nitrogen regulatory protein of Neurospora: expression and stability of NIT-2 mRNA and protein. Curr Genet. 1999;36:153–158. doi: 10.1007/s002940050485. [DOI] [PubMed] [Google Scholar]
- Thijs G, Rombauts S, Lescot M, Marchal K, De Moor B, Moreau Y, Rouzé P. Detection of cis-acting regulatory elements in plants: a Gibbs sampling approach. In Proceedings of the Second International Conference on Bioinformatics of Genome Regulation and Structure, ICG Akademgorodok Novosibirsk, Russia. Vol. 1. 2000. pp. 118–126. [Google Scholar]
- Wanner BL. Phosphate signaling and control of gene expression in Escherichia coli. In: Silver S, Walden W, editors. Metal Ions in Gene Regulation. Sterling, VA: Chapman & Hall; 1997. pp. 104–128. [Google Scholar]
- Wingender E, Chen X, Hehl R, Karas H, Liebich I, Matys V, Meinhardt T, Pruss M, Reuter I, Schacherer F. TRANSFAC: an integrated system for gene expression regulation. Nucleic Acids Res. 2000;28:316–319. doi: 10.1093/nar/28.1.316. [DOI] [PMC free article] [PubMed] [Google Scholar]