Abstract
Large coiled-coil proteins are being found in increasing numbers on the membranes of the Golgi apparatus and have been proposed to function in tethering of transport vesicles and in the organization of the Golgi stack. Members of one class of Golgi coiled-coil protein, comprising giantin and golgin-84, are anchored to the bilayer by a single C-terminal transmembrane domain (TMD). In this article, we report the characterization of another mammalian coiled-coil protein, CASP, that was originally identified as an alternatively spliced product of the CUTL1 gene that encodes CCAAT-displacement protein (CDP), the human homologue of the Drosophila homeodomain protein Cut. We find that the Caenorhabditis elegans homologues of CDP and CASP are also generated from a single gene. CASP lacks the DNA binding motifs of CDP and was previously reported to be a nuclear protein. Herein, we show that it is in fact a Golgi protein with a C-terminal TMD and shares with giantin and golgin-84 a conserved histidine in its TMD. However, unlike these proteins, CASP has a homologue in Saccharomyces cerevisiae, which we call COY1. Deletion of COY1 does not affect viability, but strikingly restores normal growth to cells lacking the Golgi soluble N-ethylmaleimide-sensitive factor attachment protein receptor Gos1p. The conserved histidine is necessary for Coy1p's activity in cells lacking Gos1p, suggesting that the TMD of these transmembrane Golgi coiled-coil proteins is directly involved in their function.
INTRODUCTION
The Golgi apparatus occupies a central position in the eukaryotic secretory pathway and provides a sorting point for proteins delivered to, and from, a variety of organelles. The Golgi is itself comprised of several compartments or cisternae typically arranged in a stack (Warren and Malhotra, 1998; Glick, 2000). Although many components of the Golgi have been identified, it remains unclear how the stacked structure is maintained and how the specificity of vesicular traffic between individual cisternae is conferred. Vesicles are targeted to both the cis- and trans-cisternae from other organelles, and vesicles must also move between cisternae. Newly synthesized proteins arriving from the endoplasmic reticulum (ER) traverse the stack to the trans-side for sorting to the plasma membrane, endocytic compartments, and other post-Golgi structures. It is now widely believed that such proteins move through the stack by maturation of the cisternae, with this perhaps supplemented by some anterograde vesicular transport (Pelham and Rothman, 2000; Barr, 2002). In addition, retrograde transport between cisternae must exist to recycle resident modification enzymes and other membrane proteins to maintain their distribution within the Golgi stack. Because many of these residents are restricted to particular subsets of cisternae, there are likely to be mechanisms to allow vesicles to recycle specifically to many, if not all, of the cisternae within the stack.
The “golgins” are a class of molecules that have emerged as candidates to organize the structure and trafficking pathways of the Golgi (Fritzler et al., 1993, 1995; Seelig et al., 1994; Nakamura et al., 1995; Bascom et al., 1999). These large proteins are predicted to form coiled-coils over most of their length, and as such have a structure suitable either to tether vesicles before fusion, or to hold adjacent cisternae together in what has been termed a Golgi “matrix” or scaffold (Slusarewicz et al., 1994; Orci et al., 1998; Seemann et al., 2000a). Because there is a large number of such proteins, at least 15 in mammalian cells, of which 10 have clear homologues in Drosophila, it is possible that they perform more than one general function in the Golgi. Indeed, in vitro systems have provided evidence in support of both tethering and organizational functions for a number of these proteins. For instance, Uso1p, and its mammalian homologue p115, are required for ER-to-Golgi transport in vivo, and reconstitution of this process in vitro has shown that the requirement is at the stage of vesicle tethering to the cis-Golgi (Cao et al., 1998; Seemann et al., 2000b; Alvarez et al., 2001). Two other coiled-coil proteins, GM130 (or golgin-95) and giantin (or macrogolgin), have been shown to recruit p115 to Golgi membranes (Sonnichsen et al., 1998; Lesa et al., 2000; Puthenveedu and Linstedt, 2001). These proteins also seem to contribute to cisternal structure in vivo and are required in an in vitro assay system that reconstitutes the stacking of Golgi cisternae (Shorter and Warren, 1999; Puthenveedu and Linstedt, 2001).
As might be expected for proteins that confer specificity to vesicular transport, or Golgi organization, many golgins are restricted to a subset of cisternae, perhaps performing similar roles in different parts of the Golgi stack (Nakamura et al., 1995; Erlich et al., 1996). In some cases, amino acid sequences or domains have been identified that target these proteins to distinct loci. Most golgins are peripheral membrane proteins and often have short noncoiled-coil regions at either end of the protein that mediate targeting and other interactions. Thus, the C terminus of GM130 binds to GRASP65, a lipid-anchored protein on the cis-Golgi (Barr et al., 1998). Similarly, four of the golgins share a C-terminal GRIP domain that is sufficient to target them to the trans-Golgi (Barr, 1999; Kjer-Nielsen et al., 1999; Munro and Nichols, 1999). In contrast, two golgins, giantin and golgin-84, are integral membrane proteins anchored to the bilayer via a C-terminal transmembrane domain (TMD), although the functional significance of this is not yet understood (Linstedt et al., 1995; Bascom et al., 1999; Misumi et al., 2001).
Analysis of the targeting and function of Golgi coiled-coil proteins has mainly been performed in mammalian cells, where many were first identified. However, yeast has proven valuable in identifying and analyzing other proteins involved in membrane traffic. Saccharomyces cerevisiae has homologues of p115 and GRASP65 and also has a single GRIP-domain–containing protein, Imh1p (Sapperstein et al., 1995; Tsukada et al., 1999). However, neither of the two members of the transmembrane-anchored class of coiled-coil proteins, golgin-84 and giantin, has a clear homologue in the yeast genome.
In this article, we describe an investigation of a human coiled-coil protein, CDP/cut alternatively spliced product (CASP), that was originally identified as an alternatively spliced transcript from the CUTL1 gene that encodes CCAAT-displacement protein (CDP, also termed Cux in mice) (Lievens et al., 1997; Nepveu, 2001). CDP is a transcriptional repressor that was originally identified as a factor that bound to CCAAT elements in the promoter of a sea urchin histone gene, and thereby displaced CCAAT binding protein (Barberis et al., 1987). Cloning of human CDP revealed it to be the homologue of the Drosophila homeodomain protein Cut, and both proteins contain three “cut repeat” DNA-binding domains in addition to a homeobox (Neufeld et al., 1992; Ludlow et al., 1996). CDP has been found to repress expression of a wide range of genes during both the cell cycle and development, and analysis of Drosophila mutants and mouse knockouts shows that CDP/cut is involved in a diverse range of cell fate decisions (Blochlinger et al., 1991; Tufarelli et al., 1998; Ellis et al., 2001; Nepveu, 2001; Sinclair et al., 2001). In addition, loss of heterozygosity of the human CUTL1 locus has been observed in a subset of breast and uterine cancers, leading to the suggestion that it acts as a tumor suppressor (Zeng et al., 1999; Neville et al., 2001). During the characterization of human CDP, an alternatively spliced transcript was found that encodes a protein that lacks all of the DNA binding domains of CDP, but instead has a unique C-terminal region (Figure 1A). Called CASP, the function of this protein is unknown, but it was reported to be nuclear (Lievens et al., 1997; confusingly, the name CASP has also been applied to a recently identified scaffolding protein that binds to cytohesin; Mansour et al., 2002; however, this scaffolding protein is unrelated to the CASP described herein). In this article, we show that CASP is in fact a Golgi membrane protein with an overall structure similar to giantin and golgin-84. Moreover, it shares with these proteins hydrophilic residues in its TMD that are conserved between homologues in different species. However, unlike these proteins CASP has a homologue in S. cerevisiae. This has allowed us to examine the importance of the conserved TMD residues shared with giantin and golgin-84.
Figure 1.
A family of proteins related to CASP, the alternatively spliced product of the CUTL1 gene. (A) Diagrammatic representation of human CDP, CASP, and the product of the yeast ORF YKL179c. CDP and CASP share an N-terminal region, with the point of divergence indicated by an arrow. The cut repeat and homeobox (Hx) domains in CDP are shown, along with coiled-coil predictions for the proteins (Lupas et al., 1991). (B) Alignment of the C termini of homologues of CASP, golgin-84, and giantin from the indicated species. The putative TMD is boxed and the conserved histidine and tyrosine residues indicated by triangles. The CASP homologues are as in C, plus Candida albicans orf6.3753; S. pombe SPCC364.04c; Neurospora crassa 9G6.340. The giantin homologues are from ESTs, GenBank accession numbers BM486947 and BJ037357. (C) Species distribution of homologues of the indicated proteins, with the gene names given, and the number of residues in brackets. Cux-2 is a neuronal-specific isoform of CDP found in humans and mice that does not seem to have an alternative product analogous to CASP. (D) Structure of two C. elegans cDNAs from the region of the predicted genes Y54F10AM.4 and Y54F10AM.3 (cDNAs kindly provided by Yuji Kohara, National Institute of Genetics, Mishima, Japan). The positions in the genome of the exons present in the cDNAs are indicated. The cDNAs share exons but then diverge to encode the C. elegans homologues of CDP and CASP.
MATERIALS AND METHODS
Plasmids
Full-length and C-terminal regions of human CASP were polymerase chain reaction (PCR) amplified from cDNA, and cloned into COS cell vectors containing the cytomegalovirus promoter, and enhanced green fluorescent protein (CLONTECH, Palo Alto, CA) attached via a myc tag. A full-length cDNA for human CDP was assembled from IMAGE clone 4903642 (residues 749-1446; Human Genome Mapping Project Resource Center, Hinxton, United Kingdom), and PCR products were amplified from cDNA. All PCR products were checked by sequencing. CASP and CDP have two alternative first exons, resulting in the alternative N termini MAANVGSMFQYWKRFDLQQLQ or MLCVRGARLK (Rong Zeng et al., 2000). When CDP and CASP constructs described in this article have an intact N terminus, it is always the former version. Full-length CDP was cloned into a COS cell vector containing the cytomegalovirus promoter, with enhanced green fluorescent protein attached to the C terminus via a GAGAGA linker.
Antibody Production
A plasmid expressing a chimera of glutathione S-transferase (GST) and residues 472–618 of human CASP was constructed using an appropriate PCR product and pGEX-4T-2 (Amersham Biosciences, Piscataway, NJ). Fusion proteins were expressed in DH5α, and the resulting inclusion bodies were washed twice in 0.5% (vol/vol) Triton X-100, 1 mM EDTA in phosphate-buffered saline, resuspended in sample buffer, and separated by SDS-PAGE. The gel was stained briefly in Coomassie Blue, the protein band excised, eluted from the gel by electrophoresis, and recovered by ethanol precipitation. Sera from immunized rabbits were absorbed with GST-Sepharose and then antibodies affinity purified on the GST-CASP antigen coupled to cyanogen bromide-activated Sepharose (Amersham Biosciences).
Immunofluorescence of Mammalian Cells
COS cells were transfected using FuGene (Roche Applied Science, Indianapolis, IN), split onto glass slides, and fixed 30–48 h posttransfection with 4% (wt/vol) paraformaldehyde. Cells were permeabilized with 0.5% (vol/vol) Triton X-100 in PBS, blocked with 20% (vol/vol) fetal calf serum/0.25% (vol/vol) Tween 20 in PBS, and probed with antibodies in the same solution. Polyclonal antibodies against CASP, giantin (Seelig et al., 1994), and TGN46 (Prescott et al., 1997), and monoclonals against TGN38 (2F7.1; Affinity Bioreagents, Golden, CO) and β′-COP [23C (TCP-1); Stressgen, Victoria, British Columbia, Canada] were detected with species-specific Alexa-labeled secondary antibodies (Molecular Probes, Eugene, OR), the cells mounted in Fluoromount-G (Southern Biotechnology Associates, Birmingham, AL), and images obtained on a Radiance confocal microscope (Bio-Rad, Hercules, CA).
Preparation of Golgi Membranes and Analysis of CASP
Golgi membranes were prepared essentially as described previously (Slusarewicz et al., 1994; Hui et al., 1998). Briefly, rat livers were homogenized, equilibrated in phosphate-buffered 0.5 M sucrose, and layered on a phosphate-buffered 0.86 M sucrose cushion, overlaid with phosphate-buffered 0.25 M sucrose and centrifuged at 105,000 × g for 60 min at 4°C. The Golgi stacks were collected from the 0.5 M/0.86 M sucrose interface, diluted to 0.25 M-buffered sucrose, and pelleted by centrifugation at 6,000 × g for 20 min at 4°C. The pellet was washed once and aliquoted for storage at −20°C.
For large-scale immunoprecipitation of CASP, Golgi membranes (1 mg) were solubilized by 2-h rotation in lysis buffer (1% [wt/vol] digitonin, 20 mM HEPES-KOH, pH 7.4, 100 mM KCl, 1 mM EDTA, 1 mM phenylmethylsulfonyl fluoride). After centrifugation at 16,000 × g for 5 min, the supernatant was incubated overnight with either anti-CASP, or rabbit IgG covalently coupled to protein A beads, washed three times with 1 ml of lysis buffer, and once with 1 ml of 5 mM NH4Ac, pH 6.5. All steps were at 4°C. Bound proteins were eluted with 100 μl of 0.5 M acetic acid, pH 3.4, lyophilized, and resuspended in SDS sample buffer. After gel electrophoresis and brief staining with Coomassie Blue, the protein bands were excised, digested with trypsin, and analyzed by matrix-assisted laser desorption ionization mass spectrometry (Shevchenko et al., 1996).
Yeast Strains and Plasmids
The genotypes of yeast strains used are listed in Table 1. YKL179c was tagged at the C terminus in strain SEY6210 by using the PCR method and template plasmid p3xHA-His5 that contains three copies of the hemagglutinin (HA) epitope tag and Schizosaccharomyces pombe HIS5 gene (Baudin et al., 1993; Jungmann et al., 1999). YKL179c was deleted in the EUROSCARF parental haploid strain BY4742, and in the diploid strains vti1Δ and ykt6Δ by transformation with PCR products from p3xHA-His5, to give strains AGY01, AGY05, AGY06, and AGY07, respectively (Table 1). For other genes, AGY01 was mated with haploid EUROSCARF strains arl1Δ, cod3Δ, dor1Δ, gos1Δ, ric1Δ, rud3Δ, and sec22Δ (Brachmann et al., 1998). The resulting diploids were sporulated, tetrads dissected (Singer Instruments, Wachet, United Kingdom), and spores were incubated on nonselective plates at 30°C for 2 d before testing of marker segregation.
Table 1.
Yeast strains used in this study
| BY4741 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 |
| BY4742 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 |
| BY4743 | MATa/α his3Δ1/his3Δ1 leu2Δ0/leu2Δ0 lys2Δ0/LYS2 MET15/met15Δ0 ura3Δ0/ura3Δ0 |
| SEY6210 | MATα ura3-52 his3-Δ200 leu2-3,112 trp1-Δ901 lys2-801 suc2-Δ9 |
| AGY01 | BY4742 ykl179cΔ::HIS5Sp |
| AGY04 | BY4743 ykl006c-aΔ::kanMX4/YKL006c-a ykl179cΔ::HIS5Sp/YKL179c |
| AGY05 | BY4743 ymr197cΔ::kanMX4/YMR197c ykl179cΔ::HIS5Sp/YKL179c |
| AGY06 | BY4743 ykl196cΔ::kanMX4/YKL196c ykl179cΔ::HIS5Sp/YKL179c |
| APY02 | SEY6210 ykl179cΔ::YKL179c-HAx3-HIS5Sp |
| RDY198 | MATα ura3 leu2 sec35-1 (R. Duden, Univ. Cambridge, United Kingdom) |
| RSY279 | MATα ura3-52 his4-619 sec22-3 (R. Duden, Univ. Cambridge, United Kingdom) |
| RSY961 | MATα ura3 leu2 sec34-2 (R. Duden, Univ. Cambridge, United Kingdom) |
| arl1Δ | BY4741 ybr164cΔ::kanMX4 |
| cod3Δ | BY4741 ygl223cΔ::kanMX4 |
| dor1Δ | BY4741 yml071cΔ::kanMX4 |
| gos1-Δ1 | BY4741 yhl031cΔ::kanMX4 |
| gos1-Δ2 | SEY6210 gos1Δ::HIS3 (Lewis et al., 2000) |
| ric1Δ | BY4741 ylr039cΔ::kanMX4 |
| rud3Δ | BY4741 yor216cΔ::kanMX4 |
| sec22Δ | BY4741 ylr268wΔ::kanMX4 |
| sft1-11 | SEY6210 sft1Δ::LEU2 (CEN HIS3 sft1-11) (M. Lewis, MRC Laboratory of Molecular Biology, United Kingdom) |
| uso1-ΔC | SEY6210 ydl058wΔ::YDL058w(1-950-GFP)-kanMX4 |
| ypt6Δ | SEY6210 ypt6Δ::HIS3 ((M. Lewis, MRC Laboratory of Molecular Biology, United Kingdom) |
Full-length YKL179c was cloned into the galactose-inducible plasmid pAK, which is pRS416 (CENURA3; Sikorski and Hieter, 1989) containing a GAL1 promoter and an ADH1 terminator (gift of Robert Arkowitz, Université de Nice, France), to create pAK-YKL179c. The C terminus of YKL179c was modified by PCR to insert a GAGA linker and a 3xHA tag. For mutation of Y619 to L and H624 to L, PCR products generated using appropriate primers were cloned into the pAK-YKL179c plasmid and the amplified region checked by sequencing. The YKL179c ORF was also cloned into pRS426 (2 μ URA3) with a constitutive PHO5 promoter to create plasmid pRS426-YKL179c.
Yeast Immunoblotting and Immunofluorescence
Strains transformed with galactose-inducible plasmids were induced in log phase, and total protein samples were prepared by resuspending 1 A600unit/20 μl of SDS buffer, bead beating for 1 min at 4°C (425–600-μm glass beads; Sigma-Aldrich, St. Louis, MO), and denaturing at 80°C for 5 min. After gel electrophoresis, proteins transferred onto nitrocellulose were probed with mouse monoclonal 12CA5 to the HA epitope and horseradish peroxidase-conjugated secondary antibodies followed by enhanced chemiluminescence (Amersham Biosciences).
Immunofluorescence of formaldehyde fixed cells was carried out as described previously, except for the omission of extraction in methanol/acetone (Holthuis et al., 1998). Affinity-purified rabbit antisera against Anp1p (Jungmann and Munro, 1998) and Tlg1p (Lewis et al., 2000), and 12CA5 were detected with appropriate Alexa488 secondary antibodies (Molecular Probes) and images obtained with confocal microscopy.
RESULTS
CASP Defines a Conserved Family of Coiled-Coil Proteins, Related to Giantin and Golgin-84
The mammalian Golgi membrane proteins giantin and golgin-84 do not have clear homologues in yeast. However, when the C-terminal region of an Arabidopsis homologue of golgin-84 was used to search the GenBank database with the iterative program PSI-BLAST, homologues of the uncharacterized S. cerevisiae gene YKL179c were found to have a similarity only just below the default cut-off for significance (p = 0.005). This yeast gene encodes a protein that is predicted to contain a C-terminal TMD and extensive regions of coiled-coil, the same overall structure as giantin and golgin-84 (Figure 1A). However, the product of the YKL179c gene cannot be a distant yeast homologue of either of these proteins because it is clearly related to another family of proteins represented by a single gene in the genomes of higher eukaryotes (Figure 1, B and C). The human member of this family is the protein CASP that is produced from an alternatively spliced transcript from the CUTL1 gene that also encodes the transcriptional repressor CDP (Lievens et al., 1997) (Figure 1, A and C). CASP was reported to have homologues in mouse, chicken, and yeast, but the presence of a putative TMD near the C terminus was not noted (Lievens et al., 1997). In C. elegans, a predicted gene Y54F10AM.3 encodes a small protein related to the C terminus of CASP. However, the adjacent gene Y54F10AM.4 encodes a homologue of CDP. We thus sequenced two C. elegans expressed sequence tags containing the 3′ end of these two genes, and found that they share 5′ exons, indicating that the structure of the human CDP/CASP gene is conserved in C. elegans (Figure 1D).
Although most of the sequence of CASP, and of its relatives, is predicted to form regions of coiled-coil, this is not so for the N- and C-terminal regions. Database searches with either of these regions shows significant similarity to all known homologues in other species, but to no other proteins. Figure 1B shows an alignment of the C-terminal region of these homologues, and the program TMHMM predicts a TMD in this region for all of these proteins (Krogh et al., 2001). In addition, some of the TMD residues are conserved, and a tyrosine and histidine in the middle of the TMD are invariant across species. Histidine is of course charged and is, not surprisingly, very rare in the TMDs of single-span proteins (Landolt-Marticorena et al., 1993). Strikingly, comparing this putative TMD region to those of golgin-84 and giantin reveals that these two residues are also well conserved in both of these proteins (Figure 1B). These similarities suggest that CASP could be a Golgi membrane protein that has properties in common with golgin-84 and giantin but, unlike these proteins, is conserved in yeast. However, the original characterization of CASP reported that the protein was localized in the nucleus, although the data were not shown (Lievens et al., 1997). In the light of the above-mentioned homologies, and conservation of a putative TMD, we decided to reinvestigate the localization of CASP.
CASP Is a Golgi-localized Membrane Protein
To examine the subcellular distribution of CASP, polyclonal antibodies were raised against residues 472–618 of the human protein, a section not present in CDP. The affinity-purified antiserum recognized a single prominent band on blots of total cellular proteins, and of purified Golgi membranes, with an apparent size of 80 kDa (Figure 2B; our unpublished data). When this antiserum was used for immunofluorescence of various tissue culture cells, including COS, normal rat kidney, human embryonic kidney 293, and HeLa, in every case a juxtanuclear ribbon-like staining was observed that colocalized with Golgi markers (Figures 2A and 3; our unpublished data).
Figure 2.
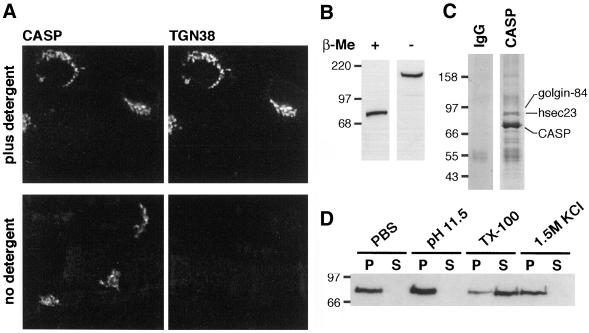
CASP behaves as a type II integral membrane protein of the Golgi apparatus. (A) Immunofluorescence localization of CASP in normal rat kidney fibroblasts. Cells fixed in 4% paraformaldehyde were permeabilized by freeze-thaw and either treated with 0.5% Triton X-100 in PBS (top) or left untreated (bottom). Cells were probed with antibodies against CASP and the Golgi protein TGN38 as described in MATERIALS AND METHODS, with the omission of Tween 20. (B) Anti-CASP protein blot of rat liver Golgi membranes (5 μg/lane) solubilized in SDS sample buffer with or without 5% β-mercaptoethanol (β-ME). (C) Coomassie Blue-stained protein gel of immunoprecipitates from rat liver Golgi membranes solubilized in digitonin, by using anti-CASP or a control IgG. Proteins identified by mass spectrometry are indicated. (D) Anti-CASP protein blot of rat liver Golgi membranes extracted with PBS containing either nothing (PBS), 1.5 M KCl, 100 mM sodium carbonate, pH 11.5 or 1% Triton X-100. Membranes (1 mg/ml) were extracted for 30 min at 4°C, centrifuged (105,000 × g for 40 min), and equivalent amounts of pellet (P) and supernatant (S) were analyzed by SDS-PAGE and protein blotting.
Figure 3.
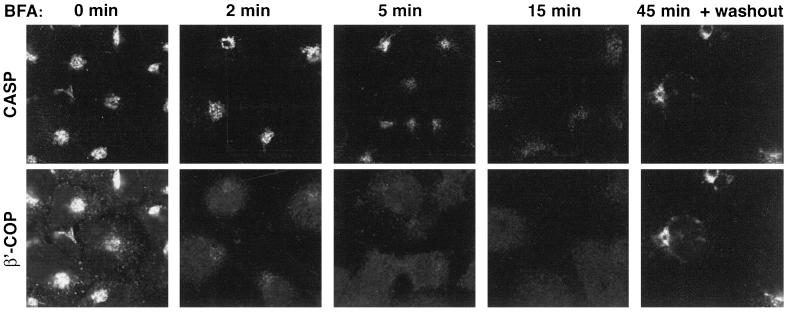
Redistribution of CASP after treatment with brefeldin A (BFA). Immunofluorescence micrographs of COS cells treated with 10 μg/ml BFA at 37°C for the times indicated, and then fixed, permeabilized, and stained with antibodies against CASP and β′-COP. In the right-hand panels cells were treated for 45 min with BFA, washed thoroughly, and allowed to recover for 60 min.
To examine the possibility that CASP is an integral membrane protein, Golgi membranes were extracted with various disruptive agents. Figure 2D shows that CASP remained Golgi-associated after treatment with high salt (1.5 M KCl), or high pH (carbonate pH 11.5), but upon addition of detergent (1% Triton X-100) much of the protein was solubilized. Thus, CASP behaves as an integral membrane protein. Moreover, when CASP was analyzed by protein blotting in nonreducing conditions, the gel mobility of the protein was reduced to an apparent molecular mass of 160–180 kDa (Figure 2B). This is consistent with a disulfide-linked dimer and implies that at least part of the molecule has been exposed to the oxidizing environment of the ER lumen. To confirm that CASP dimerizes with itself, rather than with another protein of a similar size, it was immunoprecipitated under nonreducing conditions from detergent-solubilized Golgi membranes. As shown in Figure 2C, the precipitate contained a single abundant protein, and mass spectrometric analysis of tryptic peptides confirmed that the band solely comprised CASP. Two other proteins present at substoichiometric levels were identified as golgin-84 and hSec23, a subunit of the COPII coat.
To investigate the orientation of CASP in the bilayer, the anti-CASP antiserum was used to probe cells in which the plasma membrane had been permeabilized by freeze-thaw, a procedure known to leave Golgi membranes intact (Seaman et al., 1993). The antiserum was raised against a region of CASP located on the N-terminal side of the predicted TMD and showed a clear signal both in the absence and presence of detergent (Figure 2A). In contrast, antibodies against a luminal epitope of the transmembrane protein TGN38 only gave a signal in the presence of detergent (Figure 2A). This indicates that CASP has a type II orientation with its C terminus in the Golgi lumen. There are two cysteine residues in the region C terminal to the putative TMD that could participate in the formation of intermolecular disulfide bridges in a CASP dimer.
Finally, the localization of CASP was examined after treatment with the Golgi-disrupting drug brefeldin A. Figure 3 shows that CASP began to redistribute to the ER after 2–5 min of drug treatment, kinetics slower than the Golgi peripheral coat protein COPI, but similar to that of known integral membrane proteins giantin and golgin-84 (Seelig et al., 1994; Bascom et al., 1999). After 5 min, intermediate tubules decorated with CASP were clearly visible, and by 15 min CASP was completely dispersed from the Golgi region. The dispersed signal was too weak to confirm that it corresponded to the ER, but it was at least clear that there was no concentration of staining in the vesicular tubular clusters labeled by GM130 and p115 (our unpublished data). On removal of the drug the Golgi reassembled and CASP returned. Taken together, these data indicate that CASP is a membrane protein of the Golgi apparatus with a large N-terminal region protruding into the cytoplasm. This is the same structure as found for both golgin-84 and giantin (Linstedt and Hauri, 1993; Bascom et al., 1999).
Identification of Regions of CASP Involved in Golgi Targeting
Although CASP is predicted to adopt a coiled-coil conformation over the majority of its length, there are regions of ∼150 residues at both the N and C termini that seem to be not coiled-coil. To determine whether either of these regions is sufficient to target CASP to the Golgi apparatus, green fluorescent protein (GFP)-tagged chimeras encoding CASP residues 1–137 [CASP(1–137)-GFP] or residues 519–678 [GFP-CASP(519–678)] were expressed in COS cells. The C-terminal region of CASP is sufficient to target GFP to the Golgi apparatus, whereas the N-terminal chimera was found diffusely distributed throughout the cytoplasm (Figure 4A; our unpublished data).
Figure 4.
The C terminus of CASP contains a TMD important for Golgi targeting. Confocal micrographs of COS cells expressing GFP fusions to the C-terminal 160 residues of CASP [GFP-CASP(519–678)] (A), or full-length CASP (GFP-CASP), or to CASP with a TMD mutation [GFP-CASP(Y624L)] (B). In A, the cells were fixed, permeabilized, and stained with antibodies against the Golgi marker protein giantin.
The TMD residues of CASP Y624 and H629 (numbering for human sequence) seem to be invariant across species, and to determine whether either is important for the intracellular localization of CASP, both were mutated independently to leucine in the context of the full-length protein. Wild-type GFP-CASP localized to the Golgi apparatus of transfected COS cells, providing further support for the proposed Golgi localization of the endogenous protein (Figure 4B). The mutant GFP-CASP(H629L) also localized to the Golgi apparatus, but in contrast, the Y624L mutant accumulated in the ER (Figure 4B; our unpublished data). Protein blot analysis of extracts prepared in the absence or presence of reducing agent showed that the mutant proteins accumulated to levels comparable with wild type and were still capable of forming disulfide-linked dimers (our unpublished data). This implies that the mutations do not affect protein folding, and indicate that Y624 may contribute to ER exit, but that H629 is not required for exit from the ER or localization to the Golgi.
CASP Does Not Seem to Interact with CDP In Vivo
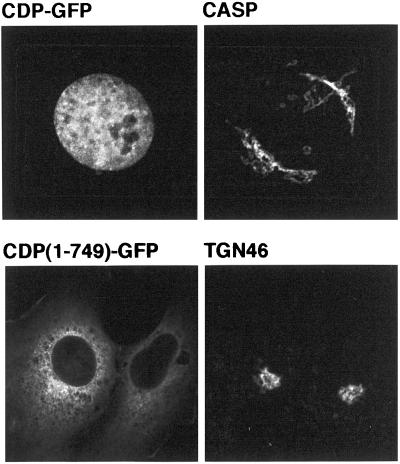
The results mentioned above indicate that CASP is localized to Golgi membranes, raising the question of what function it performs in this organelle. Because CASP shares >300 residues of coiled-coil with CDP, one possibility is that the proteins can heterodimerize, or associate by formation of four-helical bundles or lateral association of coiled-coils as occurs with filamentous myosins (McLachlan and Karn, 1982). CDP has previously been localized to the nucleus, as expected for a transcription factor (Ellis et al., 2001), but interaction with CASP could conceivably serve to sequester a proportion of CDP in the cytoplasm. However, when full-length CDP was expressed with GFP fused to its C terminus, only nuclear staining was observed with no detectable colocalization with endogenous CASP (Figure 5). It has been reported that a truncated version of mouse CDP that only extends to the second of the cut repeats, accumulates in the cytoplasm, presumably because the nuclear targeting signal is located near the C terminus (Ellis et al., 2001). When an N-terminal portion of human CDP comprising the coiled-coil region and the first cut repeat (residues 1–749) was expressed as a GFP fusion it also accumulated in the cytoplasm, but no Golgi localization was observed (Figure 5). These results suggest that CASP does not readily recruit CDP to Golgi membranes. Moreover, the nuclear targeting signal of CDP may be located in the C terminal region to ensure that the cell does not place both proteins in the nucleus.
Figure 5.
Subcellular localizations of CDP and CASP are distinct. Confocal micrographs of COS cells expressing either full-length CDP fused to GFP (CDP-GFP), or the N-terminal 749 residues fused to GFP [CDP(1–749)-GFP]. Cells were also stained with antibodies to endogenous CASP or TGN46.
Characterization of YKL179c, the Yeast Homologue of Mammalian CASP
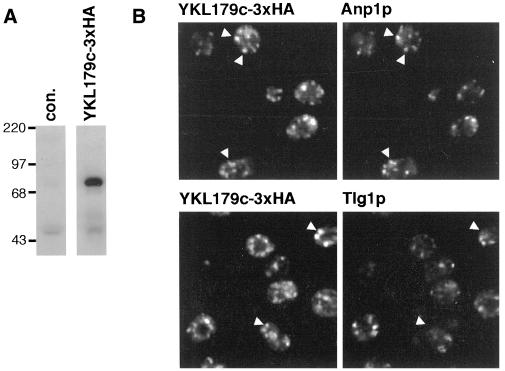
To investigate the function of CASP, we examined its homologue in the yeast S. cerevisiae. The protein is encoded by the open reading frame (ORF) YKL179c, that was shown to be nonessential in a global analysis of the yeast genome, but is otherwise uncharacterized (Winzeler et al., 1999). Initially, three copies of the HA epitope tag were inserted at the C terminus of the YKL179c ORF by homologous recombination. Probing blots of total cellular proteins with anti-HA antibodies revealed a 70-kDa protein, indicating that the gene is expressed under laboratory growth conditions (Figure 6A). Immunofluorescence showed that the tagged YKL179c was present in punctate structures characteristic of the yeast Golgi (Figure 6B). These showed substantial overlap with the early Golgi marker protein Anp1p, and little with the late Golgi t-soluble N-ethylmaleimide-sensitive factor attachment protein receptor (SNARE) Tlg1p (Holthuis et al., 1998; Jungmann and Munro, 1998). Because the protein encoded by YKL179c is related to CASP, and seems also to be in the Golgi apparatus, we suggest that the gene be named COY1, for CASP of yeast.
Figure 6.
Yeast ORF YKL179c is expressed under normal growth conditions and encodes a protein that localizes to the early Golgi. (A) Anti-HA protein blots of total protein from yeast strain APY02 in which the YKL179c ORF is followed by a triple HA epitope tag (YKL179c-3xHA) or from the parental strain SEY6210 (con.). (B) Confocal micrographs of APY02 double labeled with anti-HA monoclonal 12CA5 and antibodies against either the early Golgi protein Anp1p or the late Golgi protein Tlg1p.
Genetic Interactions of COY1/YKL179c and Genes Whose Products Are Involved in the Secretory Pathway
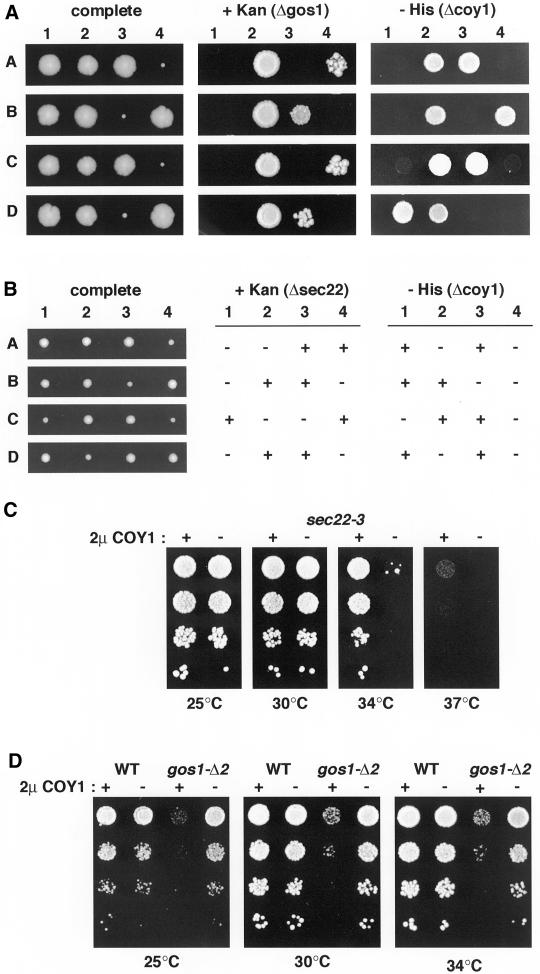
It has been reported that COY1/YKL179c is not required for viability, and we also deleted the gene in both the SEY6210 and BY4742 strain backgrounds and found no effect on growth, or temperature sensitivity (Winzeler et al., 1999; our unpublished data). However, a number of genes encoding well conserved Golgi proteins only show a growth phenotype when deleted in combination with mutations in other genes involved in membrane traffic (Tsukada et al., 1999; Bensen et al., 2000; Siniossoglou et al., 2000). This synthetic lethality may reflect redundancy of components acting in a given transport step, or the loss of a particular transport step being compensated for by the increased use of other steps. We therefore deleted COY1 from diploid strains lacking a number of genes involved in ER-Golgi and intra-Golgi traffic, and examined the viability of double mutants by sporulation and tetrad analysis. Loss of COY1 had no effect on the growth rate of spores that also lacked the nonessential genes RUD3 (a Golgi coiled-coil protein), COD3 and DOR1 (subunits of the COG [or Sec34/35] complex), ARL1 (an ARF-like GTPase), or RIC1 (the GTP exchange factor for Ypt6p) (VanRheenen et al., 1999; Siniossoglou et al., 2000; Whyte and Munro, 2001). In contrast, analysis of spores from diploids lacking a single copy of COY1 in combination with loss of either of the Golgi SNAREs Gos1p or Sec22p revealed a striking genetic interaction. As previously reported, spores that lack GOS1 alone are viable, but grow very slowly after germination (Figure 7A, e.g., A4; McNew et al., 1998). However, when the spores lacked COY1 in addition to GOS1, growth was restored to wild-type levels (Figure 7A, e.g., A2). This surprising result was reproduced with an independent Δgos1 allele, and the absence of Gos1p was confirmed by protein blotting (our unpublished data). A similar effect was seen with spores lacking SEC22, but the effect was more subtle because loss of Sec22p has a less severe effect on growth (Figure 7B). Deletion of COY1 did not restore growth to spores lacking the essential SNAREs Vti1p or Ykt6p. Sec22p and Gos1p are both implicated in ER-Golgi transport, and Gos1p is additionally involved in intra-Golgi traffic.
Figure 7.
Genetic interactions between COY1/YKL179c and the SNARE-encoding genes GOS1 and SEC22. (A) Tetrad analysis of spores from a gos1Δ::kanMx/GOS1, ykl179cΔ::HIS5/YKL179c diploid strain. Tetrads were dissected and incubated on YEPD plates at 30°C for 3 d (complete), and markers were then tested by replica plating on kanamycin (+ Kan) and minus-histidine plates (− His). Four representative tetrads are shown. The small segregants in each tetrad are Kanres, but segregants that are both Kanres and HIS+ do not display any growth or germination defect. (B) Tetrad analysis as in A of spores from a sec22Δ::kanMx/SEC22, ykl179cΔ::HIS5/ YKL179cdiploid strain (AGY03). Segregants that contained sec22Δ::kanMx alone showed a small but reproducible growth defect, which was not seen in those that also contained ykl179cΔ::HIS5. (C) Growth at the indicated temperatures of the temperature-sensitive sec22-3 strain containing the 2 μ plasmid pRS426 with (+) or without (−) full-length Coy1p expressed from a constitutive PHO5 promoter. Ten-fold serial dilutions of cells were spotted onto selective plates and incubated at the indicated temperatures for 3 d. (D) Growth of parental strain SEY6210 (WT) or gos1-Δ2 containing 2 μ plasmids as in C.
Overexpression of Coy1p Has Distinct Effects on Loss of ER-Golgi SNAREs Gos1p and Sec22p
To obtain further evidence for a role of Coy1p in Golgi function we examined the effect of overexpressing Coy1p in strains carrying temperature-sensitive or deletion mutations of various Golgi proteins involved in membrane traffic. Overexpression of Coy1p from a multicopy 2 μ plasmid had no effect on the temperature sensitivity or growth of ypt6Δ, sec35-1, or sec34-2, partially suppressed the temperature sensitivity of uso1ΔCsft1-11 and sec22-3 (Figure 7C; our unpublished data). In contrast, Coy1p overexpression severely compromised the viability of the gos1Δ-2 strain at all temperatures tested. Although strains lacking GOS1 grow slowly after sporulation, they eventually acquire near normal growth rates, suggesting some process of adaptation (McNew et al., 1998; Niedenthal et al., 1999). Nonetheless, overexpression of Coy1p slowed growth of cells lacking Gos1p but had no effect on the wild type, consistent with the removal of Coy1p improving growth in the absence of the SNARE (Figure 7D).
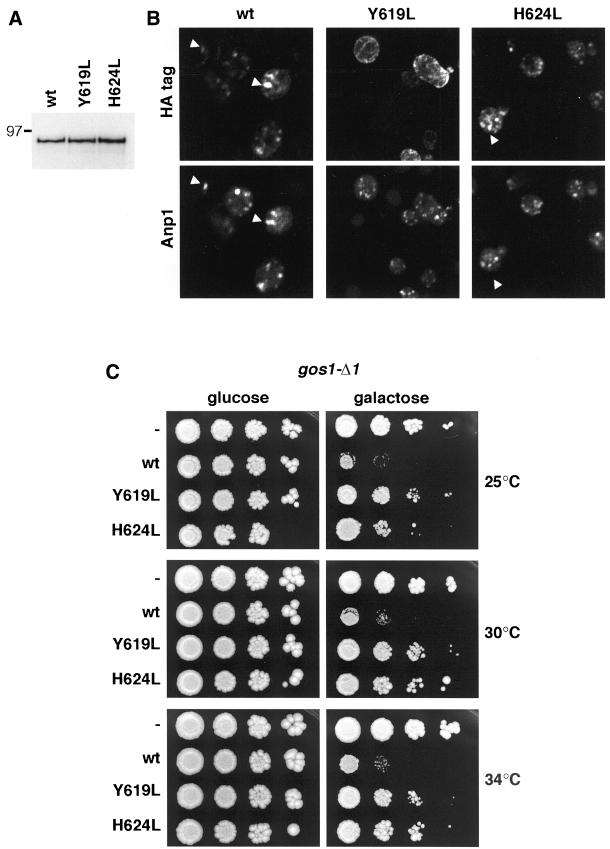
Conserved TMD Residues in Coy1p Are Important for Function
The slow growth phenotype induced by Coy1p when overexpressed in cells lacking the Golgi SNARE Gos1p provides an indirect assay for at least some aspect of Coy1p function. This assay was used to investigate the importance of the tyrosine and histidine residues conserved in the TMD. Versions of full-length Coy1p with either Y619 or H624 mutated to leucine were expressed from a low copy centromeric plasmid under the control of the galactose-inducible GAL1 promoter. We initially compared the levels and localization of wild-type and mutant proteins that were tagged at the C terminus with three copies of the HA epitope. Figure 8A shows that the mutations had no effect on the level of Coy1p that accumulated after 2 h of galactose induction. Immunofluorescence localization of the induced proteins showed that both wild type and Coy1p(H624L) were in punctate structures that colocalized with the Golgi marker Anp1p (Figure 8B). In contrast, Coy1p(Y619L) showed nuclear envelope and peripheral staining characteristic of the ER, consistent with the localization of human CASP carrying the equivalent mutation. The activity of the mutant proteins was then assayed in a gos1Δ strain, by comparing growth on glucose and galactose plates. When Coy1p expression was suppressed by glucose, the gos1-Δ1 strain grew well at all temperatures tested, whereas induction of wild-type Coy1p on galactose decreased viability as expected (Figure 8C). In contrast, induction of either Coy1p(Y619L) or Coy1p(H624L) only slightly inhibited growth (Figure 8C). These results indicate that the TMD of Coy1p is important for both efficient exit of the protein from the ER, and for the function of the protein because mutation of the conserved histidine affects activity without any apparent effect on localization. This suggests that the TMD of Coy1p, and hence its human homologue, serves a role in the function of the protein beyond that of providing an attachment to the lipid bilayer.
Figure 8.
Role of the conserved TMD residues in the Coy1p function. (A) Anti-HA protein blots of total protein from yeast expressing HA-tagged Coy1p (wt) or the same protein with the indicated TMD mutations. Proteins were expressed in strain gos1-Δ1 from CEN plasmids under the control of the galactose-regulated GAL1 promoter and induced with 2% galactose for 3 h at 30°C. (B) Confocal micrographs of yeast as in A. After induction as in B, cells were fixed, permeabilized, and stained with antibodies against the HA epitope and the Golgi marker Anp1p. (C) Effect of TMD mutants on the ability of Coy1p to perturb growth of the gos1-Δ1 strain. Cells were transformed with plasmids as in A but expressing Coy1p (wt) without an epitope tag, or the same protein with the indicated TMD mutants, or no gene (−). Yeast were grown overnight in media containing 2% raffinose, spotted onto selective plates containing either 2% glucose or 2% galactose, and incubated at the indicated temperatures for 2–3 d.
DISCUSSION
The results presented herein show that the two proteins produced by the human CUTL1 locus have strikingly different locations: a nuclear transcription factor CDP and a Golgi-localized membrane protein CASP. The choice between the two alternative transcripts encoding these proteins seems to be controlled by use of a site for transcript termination and poly(A) addition (Lievens et al., 1997; Rong Zeng et al., 2000; Figure 1D). If the termination site is used then the transcript is spliced to produce CDP, but if it is read through then the transcript includes further exons, which results in an alternative splice and the production of CASP. We have confirmed that this unusual gene structure is conserved in the nematode C. elegans, suggesting that it has persisted for a long period of evolution and raising the question of whether it has any functional significance. However, we have been unable to find any evidence that the region shared between CDP and CASP results in targeting of the former to the Golgi. When CASP was originally described it was reported to associate with CDP when the two proteins were translated together in vitro (Lievens et al., 1997). However, the authors stated that they did not observe any such association when the proteins were expressed in transfected cells. This would agree with our observations and suggests that mechanisms may exist in vivo to ensure that homodimeric coiled-coil domains only associate with translation products from the same polysome.
An origin for this unusual gene structure is suggested by the fact that CASP seems more evolutionarily ancient than CDP because only the former is present in plants and fungi. Moreover, both the C-terminal TMD and the N-terminal region of the protein are conserved between all CASP homologues. Thus, it is possible that during evolution the CDP DNA-binding domains were rearranged into a CASP intron, allowing the coiled-coil region to be shared by the two proteins. The resulting dimerization of CDP may have proved valuable and difficult to lose during evolution. Interestingly, the golgins GMAP-210 and golgin-84 have both been found to participate in oncogenic rearrangements that attach their N-terminal mostly coiled-coil regions to the tyrosine kinase domains of the platelet-derived growth factor receptor β and the Ret oncogene, respectively (Abe et al., 1997; Klugbauer et al., 1998). This presumably results in constitutive dimerization and altered activity of the tyrosine kinase domains, although in these cases the imposed dimerization is not of any evolutionary benefit.
Irrespective of the evolutionary history of CDP, it is clear that CASP has a function on the Golgi that is independent of CDP. This function seems likely to be related to that of giantin and golgin-84 because the proteins share an overall structure and conserved TMD residues. Indeed, all the eukaryotes shown in Figure 1C have at least one member of this trio, although none of the three is universal. Thus, in Drosophila, the cut locus does not seem to produce a homologue of CASP (although the N terminus of the cut protein has short regions predicted to form coiled-coils) but there is a homologue of golgin-84. In contrast, C. elegans has CASP, but no obvious golgin-84, whereas plants have both (Figure 1C). This suggests that the proteins may have been able to substitute for one another when lost during evolution. Only CASP shares an N-terminal domain with yeast Coy1p, but even so it cannot be assumed that the two proteins are precise functional homologues. Because mammals also have giantin and golgin-84, it is possible that yeast Coy1p additionally performs functions associated with these other proteins. Nonetheless, the results presented herein suggest that the TMD residues shared by the proteins serve in both efficient ER exit and in Golgi function, and indeed the tyrosine and histidine residues examined are predicted to be on opposite sides of an α-helix. Efficient ER exit may be important to prevent the protein forming interactions with other Golgi components before it is correctly localized.
What then can be said of the function of CASP? The lack of a growth phenotype when COY1 is deleted from yeast indicates that the protein is not essential for anterograde transport through the Golgi. However, a similar situation is found with the yeast homologue of GRASP65 and with the Golgi coiled-coil proteins Imh1p and Rud3p (Tsukada et al., 1999; VanRheenen et al., 1999; Winzeler et al., 1999). Thus, it may be that under normal growth conditions there is redundancy between vesicle tethers or between transport routes in the Golgi. Indeed, a number of nonessential Golgi proteins show genetic interactions with known trafficking proteins. For example, Imh1p shows synthetic lethality with the yeast rab GTPase Ypt6, or its exchange factors (Tsukada and Gallwitz, 1996; Siniossoglou et al., 2000). In this article, we find that COY1 shows genetic interactions that suggest a role in membrane traffic. In particular COY1 shows a striking interaction with the gene encoding the Golgi SNARE Gos1p, in that deletion of COY1 restores normal growth to gos1Δ cells, whereas overexpression has the opposite effect. This SNARE is well conserved in evolution with a homologue in mammals called GOS-28 or GS28 (Nagahama et al., 1996; Subramaniam et al., 1996; McNew et al., 1998). Antibodies to GOS-28 inhibit in vitro assays for both intra-Golgi transport and ER-to-Golgi transport, although the latter effect could be an indirect consequence of perturbed intra-Golgi traffic. In both yeast and mammals the protein has been found to form complexes with the Golgi t-SNARE Sed5p (syntaxin-5) (Hay et al., 1997; McNew et al., 1998). This syntaxin plays an essential role in ER-to-Golgi and intra-Golgi transport, and participates in the formation of at least two SNARE complexes, and possibly several (Banfield et al., 1995; Tsui et al., 2001). The individual components of these complexes do not all have the same distribution. Thus, in mammalian cells GOS-28 is found throughout the Golgi stack, whereas other syntaxin-5–interacting SNAREs such as rBet1 are concentrated toward the cis-side of the Golgi (Hay et al., 1998; Orci et al., 2000).
The results reported herein show that Coy1p has a deleterious effect on cells lacking Gos1p, which might suggest that the protein has a negative rather than positive role in transport. However, this effect does not preclude a positive role for Coy1p such as the tethering of vesicles, as proposed for other coiled-coil proteins. Vesicle tethering is likely to be beneficial only if it is productive. If a component required downstream of tethering is missing then the posttethering steps might be slowed and proteins such as SNAREs become sequestered, with the result that transport elsewhere in the Golgi would be compromised. Thus, if Coy1p and Gos1p act in the same transport step in intra-Golgi traffic then removal of Coy1p in a gos1Δ strain could reduce the tethering of vesicles that are dependent on Gos1p, and so release frustrated vesicles or sequestered trafficking components to function elsewhere in the Golgi in steps that are independent of Gos1p. It is at least unlikely that the toxicity of deleting GOS1 reflects Gos1p normally masking some toxic part of Coy1p, because increasing the levels of Coy1p is not detrimental to wild-type cells.
Our results also suggest that the function of CASP, and by implication those of giantin and golgin-84, involves a specific interaction with residues in the TMD. This is perhaps unexpected because all the functional evidence so far for giantin and other long coiled-coil proteins of the Golgi has shown that they are involved in attaching vesicles or soluble proteins to the Golgi (Nakamura et al., 1997; Cao et al., 1998; Linstedt et al., 2000). Functional residues in the TMD suggest an additional interaction within the lipid bilayer. So far, we have not found any other membrane protein associating stoichiometrically with either rat CASP or yeast Coy1p. A small fraction of the former protein coprecipitated with hSec23, and we have found a similar interaction in yeast (our unpublished data), which may reflect an interaction with the COPII coat required for ER exit, as was recently reported for another Golgi membrane protein Sys1p (Votsmeier and Gallwitz, 2001). Rat CASP also associated with a small amount of golgin-84, but because this protein is not present in yeast this interaction cannot be obligatory, and so its significance is unclear. It may be that a stoichiometric interaction with another membrane protein was destabilized by detergent solubilization, or alternatively any interactions may be transient. Such an interaction could serve to mediate communication between the tethering process and events in the cisternal bilayer. What is clear is that the TMD of CASP, and presumably those of giantin and golgin-84, is doing more than simply providing an anchor to the Golgi membrane. The identification of a member of this family in yeast now provides the opportunity to use genetic approaches to identify the components that interact with this class of proteins.
ACKNOWLEDGMENTS
We are grateful to Rob Arkowitz, Rainer Duden, Yuji Kohara, and Mike Lewis for generous provision of reagents, and to Hugh Pelham and James Whyte for comments on the manuscript
Footnotes
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E02–06–0349. Article and publication date are at www.molbiolcell.org/cgi/doi/10.1091/mbc.E02–06–0349.
REFERENCES
- Abe A, Emi N, Tanimoto M, Terasaki H, Marunouchi T, Saito H. Fusion of the platelet-derived growth factor receptor beta to a novel gene CEV14 in acute myelogenous leukemia after clonal evolution. Blood. 1997;90:4271–4277. [PubMed] [Google Scholar]
- Alvarez C, Garcia-Mata R, Hauri HP, Sztul E. The p115-interactive proteins GM130 and giantin participate in endoplasmic reticulum-Golgi traffic. J Biol Chem. 2001;276:2693–2700. doi: 10.1074/jbc.M007957200. [DOI] [PubMed] [Google Scholar]
- Banfield DK, Lewis MJ, Pelham HR. A SNARE-like protein required for traffic through the Golgi complex. Nature. 1995;375:806–809. doi: 10.1038/375806a0. [DOI] [PubMed] [Google Scholar]
- Barberis A, Superti-Furga G, Busslinger M. Mutually exclusive interaction of the CCAAT-binding factor and of a displacement protein with overlapping sequences of a histone gene promoter. Cell. 1987;50:347–359. doi: 10.1016/0092-8674(87)90489-2. [DOI] [PubMed] [Google Scholar]
- Barr FA. A novel Rab6-interacting domain defines a family of Golgi-targeted coiled-coil proteins. Curr Biol. 1999;9:381–384. doi: 10.1016/s0960-9822(99)80167-5. [DOI] [PubMed] [Google Scholar]
- Barr FA. The Golgi apparatus: going round in circles? Trends Cell Biol. 2002;12:101–104. doi: 10.1016/s0962-8924(01)02240-1. [DOI] [PubMed] [Google Scholar]
- Barr FA, Nakamura N, Warren G. Mapping the interaction between GRASP65 and GM130, components of a protein complex involved in the stacking of Golgi cisternae. EMBO J. 1998;17:3258–3268. doi: 10.1093/emboj/17.12.3258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bascom RA, Srinivasan S, Nussbaum RL. Identification and characterization of golgin-84, a novel Golgi integral membrane protein with a cytoplasmic coiled-coil domain. J Biol Chem. 1999;274:2953–2962. doi: 10.1074/jbc.274.5.2953. [DOI] [PubMed] [Google Scholar]
- Baudin A, Ozierkalogeropoulos O, Denouel A, Lacroute F, Cullin C. A simple and efficient method for direct gene deletion in Saccharomyces cerevisiae. Nucleic Acids Res. 1993;21:3329–3330. doi: 10.1093/nar/21.14.3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bensen ES, Costaguta G, Payne GS. Synthetic genetic interactions with temperature-sensitive clathrin in Saccharomyces cerevisiae. Roles for synaptojanin-like Inp53p and dynamin-related Vps1p in clathrin-dependent protein sorting at the trans-Golgi network. Genetics. 2000;154:83–97. doi: 10.1093/genetics/154.1.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blochlinger K, Jan LY, Jan YN. Transformation of sensory organ identity by ectopic expression of Cut in Drosophila. Genes Dev. 1991;5:1124–1135. doi: 10.1101/gad.5.7.1124. [DOI] [PubMed] [Google Scholar]
- Brachmann CB, Davies A, Cost GJ, Caputo E, Li J, Hieter P, Boeke JD. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 1998;14:115–132. doi: 10.1002/(SICI)1097-0061(19980130)14:2<115::AID-YEA204>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Cao XC, Ballew N, Barlowe C. Initial docking of ER-derived vesicles requires Uso1p and Ypt1p but is independent of SNARE proteins. EMBO J. 1998;17:2156–2165. doi: 10.1093/emboj/17.8.2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis T, Gambardella L, Horcher M, Tschanz S, Capol J, Bertram P, Jochum W, Barrandon Y, Busslinger M. The transcriptional repressor CDP (Cutl1) is essential for epithelial cell differentiation of the lung and the hair follicle. Genes Dev. 2001;15:2307–2319. doi: 10.1101/gad.200101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erlich R, Gleeson PA, Campbell P, Dietzsch E, Toh BH. Molecular characterization of trans-Golgi p230. A human peripheral membrane protein encoded by a gene on chromosome 6p12–22 contains extensive coiled-coil α-helical domains and a granin motif. J Biol Chem. 1996;271:8328–8337. doi: 10.1074/jbc.271.14.8328. [DOI] [PubMed] [Google Scholar]
- Fritzler MJ, Hamel JC, Ochs RL, Chan EK. Molecular characterization of two human autoantigens: unique cDNAs encoding 95- and 160-kD proteins of a putative family in the Golgi complex. J Exp Med. 1993;178:49–62. doi: 10.1084/jem.178.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritzler MJ, Lung CC, Hamel JC, Griffith KJ, Chan EK. Molecular characterization of Golgin-245, a novel Golgi complex protein containing a granin signature. J Biol Chem. 1995;270:31262–31268. doi: 10.1074/jbc.270.52.31262. [DOI] [PubMed] [Google Scholar]
- Glick BS. Organization of the Golgi apparatus. Curr Opin Cell Biol. 2000;12:450–456. doi: 10.1016/s0955-0674(00)00116-2. [DOI] [PubMed] [Google Scholar]
- Hay JC, Chao DS, Kuo CS, Scheller RH. Protein interactions regulating vesicle transport between the endoplasmic reticulum and Golgi apparatus in mammalian cells. Cell. 1997;89:149–158. doi: 10.1016/s0092-8674(00)80191-9. [DOI] [PubMed] [Google Scholar]
- Hay JC, Klumperman J, Oorschot V, Steegmaier M, Kuo CS, Scheller RH. Localization, dynamics, and protein interactions reveal distinct roles for ER and Golgi SNAREs. J Cell Biol. 1998;141:1489–1502. doi: 10.1083/jcb.141.7.1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holthuis JC, Nichols BJ, Dhruvakumar S, Pelham HR. Two syntaxin homologues in the TGN/endosomal system of yeast. EMBO J. 1998;17:113–126. doi: 10.1093/emboj/17.1.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui N, Nakamura N, Slusarewicz P, Warren G. Purification of rat liver Golgi stacks. In: Celis JE, editor. Cell Biology: A Laboratory Handbook. Vol. 2. Orlando, FL: Academic Press; 1998. pp. 46–55. [Google Scholar]
- Jungmann J, Munro S. Multi-protein complexes in the cis Golgi of Saccharomyces cerevisiae with α-1,6-mannosyltransferase activity. EMBO J. 1998;17:423–434. doi: 10.1093/emboj/17.2.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jungmann J, Rayner JC, Munro S. The Saccharomyces cerevisiae protein Mnn10p/Bed1p is a subunit of a Golgi mannosyltransferase complex. J Biol Chem. 1999;274:6579–6585. doi: 10.1074/jbc.274.10.6579. [DOI] [PubMed] [Google Scholar]
- Kjer-Nielsen L, Teasdale RD, van Vliet C, Gleeson PA. A novel Golgi-localization domain shared by a class of coiled-coil peripheral membrane proteins. Curr Biol. 1999;9:385–388. doi: 10.1016/s0960-9822(99)80168-7. [DOI] [PubMed] [Google Scholar]
- Klugbauer S, Demidchik EP, Lengfelder E, Rabes HM. Detection of a novel type of RET rearrangement (PTC5) in thyroid carcinomas after Chernobyl and analysis of the involved RET-fused gene RFG5. Cancer Res. 1998;58:198–203. [PubMed] [Google Scholar]
- Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- Landolt-Marticorena C, Williams KA, Deber CM, Reithmeier RA. Non-random distribution of amino acids in the transmembrane segments of human type I single span membrane proteins. J Mol Biol. 1993;229:602–608. doi: 10.1006/jmbi.1993.1066. [DOI] [PubMed] [Google Scholar]
- Lesa GM, Seemann J, Shorter J, Vandekerckhove J, Warren G. The amino-terminal domain of the Golgi protein giantin interacts directly with the vesicle-tethering protein p115. J Biol Chem. 2000;275:2831–2836. doi: 10.1074/jbc.275.4.2831. [DOI] [PubMed] [Google Scholar]
- Lewis MJ, Nichols BJ, Prescianotto-Baschong C, Riezman H, Pelham HR. Specific retrieval of the exocytic SNARE Snc1p from early yeast endosomes. Mol Biol Cell. 2000;11:23–38. doi: 10.1091/mbc.11.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lievens PM, Tufarelli C, Donady JJ, Stagg A, Neufeld EJ. CASP, a novel, highly conserved alternative-splicing product of the CDP/cut/cux gene, lacks cut-repeat and homeo DNA-binding domains, and interacts with full-length CDP in vitro. Gene. 1997;197:73–81. doi: 10.1016/s0378-1119(97)00243-6. [DOI] [PubMed] [Google Scholar]
- Linstedt AD, Foguet M, Renz M, Seelig HP, Glick BS, Hauri HP. A C-terminally-anchored Golgi protein is inserted into the endoplasmic reticulum and then transported to the Golgi apparatus. Proc Natl Acad Sci USA. 1995;92:5102–5105. doi: 10.1073/pnas.92.11.5102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linstedt AD, Hauri HP. Giantin, a novel conserved Golgi membrane protein containing a cytoplasmic domain of at least 350 kDa. Mol Biol Cell. 1993;4:679–693. doi: 10.1091/mbc.4.7.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linstedt AD, Jesch SA, Mehta A, Lee TH, Garcia-Mata R, Nelson DS, Sztul E. Binding relationships of membrane tethering components. The giantin N terminus and the GM130 N terminus compete for binding to the p115 C terminus. J Biol Chem. 2000;275:10196–10201. doi: 10.1074/jbc.275.14.10196. [DOI] [PubMed] [Google Scholar]
- Ludlow C, Choy R, Blochlinger K. Functional analysis of Drosophila and mammalian cut proteins in files. Dev Biol. 1996;178:149–159. doi: 10.1006/dbio.1996.0205. [DOI] [PubMed] [Google Scholar]
- Lupas A, Van Dyke M, Stock J. Predicting coiled coils from protein sequences. Science. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- McLachlan AD, Karn J. Periodic charge distributions in the myosin rod amino acid sequence match cross-bridge spacings in muscle. Nature. 1982;299:226–231. doi: 10.1038/299226a0. [DOI] [PubMed] [Google Scholar]
- McNew JA, Coe JG, Sogaard M, Zemelman BV, Wimmer C, Hong W, Sollner TH. Gos1p, a Saccharomyces cerevisiae SNARE protein involved in Golgi transport. FEBS Lett. 1998;435:89–95. doi: 10.1016/s0014-5793(98)01044-8. [DOI] [PubMed] [Google Scholar]
- Mansour M, Lee SY, Pohajdak B. The N-terminal coiled coil domain of the cytohesin/ARNO family of guanine nucleotide exchange factors interacts with the scaffolding protein CASP. J Biol Chem. 2002;227:32302–32309. doi: 10.1074/jbc.M202898200. [DOI] [PubMed] [Google Scholar]
- Misumi Y, Sohda M, Tashiro A, Sato H, Ikehara Y. An essential cytoplasmic domain for the Golgi localization of coiled-coil proteins with a COOH-terminal membrane anchor. J Biol Chem. 2001;276:6867–6873. doi: 10.1074/jbc.M010121200. [DOI] [PubMed] [Google Scholar]
- Munro S, Nichols BJ. The GRIP domain: a novel Golgi-targeting domain found in several coiled-coil proteins. Curr Biol. 1999;9:377–380. doi: 10.1016/s0960-9822(99)80166-3. [DOI] [PubMed] [Google Scholar]
- Nagahama M, Orci L, Ravazzola M, Amherdt M, Lacomis L, Tempst P, Rothman JE, Sollner TH. A v-SNARE implicated in intra-Golgi transport. J Cell Biol. 1996;133:507–516. doi: 10.1083/jcb.133.3.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura N, Lowe M, Levine TP, Rabouille C, Warren G. The vesicle docking protein p115 binds GM130, a cis-Golgi matrix protein, in a mitotically regulated manner. Cell. 1997;89:445–455. doi: 10.1016/s0092-8674(00)80225-1. [DOI] [PubMed] [Google Scholar]
- Nakamura N, Rabouille C, Watson R, Nilsson T, Hui N, Slusarewicz P, Kreis TE, Warren G. Characterization of a cis-Golgi matrix protein, GM130. J Cell Biol. 1995;131:1715–1726. doi: 10.1083/jcb.131.6.1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nepveu A. Role of the multifunctional CDP/Cut/Cux homeodomain transcription factor in regulating differentiation, cell growth and development. Gene. 2001;270:1–15. doi: 10.1016/s0378-1119(01)00485-1. [DOI] [PubMed] [Google Scholar]
- Neufeld EJ, Skalnik DG, Lievens PM, Orkin SH. Human CCAAT displacement protein is homologous to the Drosophila homeoprotein, cut. Nat Genet. 1992;1:50–55. doi: 10.1038/ng0492-50. [DOI] [PubMed] [Google Scholar]
- Neville PJ, Thomas N, Campbell IG. Loss of heterozygosity at 7q22 and mutation analysis of the CDP gene in human epithelial ovarian tumors. Int J Cancer. 2001;91:345–349. doi: 10.1002/1097-0215(200002)9999:9999<::aid-ijc1050>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- Niedenthal R, Riles L, Guldener U, Klein S, Johnston M, Hegemann JH. Systematic analysis of S. cerevisiae chromosome VIII genes. Yeast. 1999;15:1775–1796. doi: 10.1002/(SICI)1097-0061(199912)15:16<1775::AID-YEA496>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Orci L, Perrelet A, Rothman JE. Vesicles on strings: morphological evidence for processive transport within the Golgi stack. Proc Natl Acad Sci USA. 1998;95:2279–2283. doi: 10.1073/pnas.95.5.2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orci L, Ravazzola M, Volchuk A, Engel T, Gmachl M, Amherdt M, Perrelet A, Sollner TH, Rothman JE. Anterograde flow of cargo across the Golgi stack potentially mediated via bidirectional “percolating”COPI vesicles. Proc Natl Acad Sci USA. 2000;97:10400–10405. doi: 10.1073/pnas.190292497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelham HR, Rothman JE. The debate about transport in the Golgi–two sides of the same coin? Cell. 2000;102:713–719. doi: 10.1016/s0092-8674(00)00060-x. [DOI] [PubMed] [Google Scholar]
- Prescott AR, Lucocq JM, James J, Lister JM, Ponnambalam S. Distinct compartmentalization of TGN46 and β1,4-galactosyltransferase in HeLa cells. Eur J Cell Biol. 1997;72:238–246. [PubMed] [Google Scholar]
- Puthenveedu MA, Linstedt AD. Evidence that Golgi structure depends on a p115 activity that is independent of the vesicle tether components giantin and GM130. J Cell Biol. 2001;155:227–238. doi: 10.1083/jcb.200105005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rong Zeng W, Soucie E, Sung Moon N, Martin-Soudant N, Berube G, Leduy L, Nepveu A. Exon/intron structure and alternative transcripts of the CUTL1 gene. Gene. 2000;241:75–85. doi: 10.1016/s0378-1119(99)00465-5. [DOI] [PubMed] [Google Scholar]
- Sapperstein SK, Walter DM, Grosvenor AR, Heuser JE, Waters MG. p115 is a general vesicular transport factor related to the yeast endoplasmic reticulum to Golgi transport factor Uso1p. Proc Natl Acad Sci USA. 1995;92:522–526. doi: 10.1073/pnas.92.2.522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seaman MN, Ball CL, Robinson MS. Targeting and mistargeting of plasma membrane adaptors in vitro. J Cell Biol. 1993;123:1093–1105. doi: 10.1083/jcb.123.5.1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seelig HP, Schranz P, Schroter H, Wiemann C, Renz M. Macrogolgin: a new 376 kD Golgi complex outer membrane protein as target of antibodies in patients with rheumatic diseases and HIV infections. J Autoimmun. 1994;7:67–91. doi: 10.1006/jaut.1994.1006. [DOI] [PubMed] [Google Scholar]
- Seemann J, Jokitalo E, Pypaert M, Warren G. Matrix proteins can generate the higher order architecture of the Golgi apparatus. Nature. 2000a;407:1022–1026. doi: 10.1038/35039538. [DOI] [PubMed] [Google Scholar]
- Seemann J, Jokitalo EJ, Warren G. The role of the tethering proteins p115 and GM130 in transport through the Golgi apparatus in vivo. Mol Biol Cell. 2000b;11:635–645. doi: 10.1091/mbc.11.2.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevchenko A, Jensen ON, Podtelejnikov AV, Sagliocco F, Wilm M, Vorm O, Mortensen P, Shevchenko A, Boucherie H, Mann M. Linking genome and proteome by mass spectrometry: large-scale identification of yeast proteins from 2-dimensional gels. Proc Natl Acad Sci USA. 1996;93:14440–14445. doi: 10.1073/pnas.93.25.14440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shorter J, Warren G. A role for the vesicle tethering protein, p115, in the post-mitotic stacking of reassembling Golgi cisternae in a cell-free system. J Cell Biol. 1999;146:57–70. doi: 10.1083/jcb.146.1.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinclair AM, Lee JA, Goldstein A, Xing D, Liu S, Ju R, Tucker PW, Neufeld EJ, Scheuermann RH. Lymphoid apoptosis and myeloid hyperplasia in CCAAT displacement protein mutant mice. Blood. 2001;98:3658–3667. doi: 10.1182/blood.v98.13.3658. [DOI] [PubMed] [Google Scholar]
- Siniossoglou S, Peak-Chew SY, Pelham HR. Ric1p and Rgp1p form a complex that catalyzes nucleotide exchange on Ypt6p. EMBO J. 2000;19:4885–4894. doi: 10.1093/emboj/19.18.4885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slusarewicz P, Nilsson T, Hui N, Watson R, Warren G. Isolation of a matrix that binds medial Golgi enzymes. J Cell Biol. 1994;124:405–413. doi: 10.1083/jcb.124.4.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnichsen B, Lowe M, Levine T, Jamsa E, Dirac-Svejstrup B, Warren G. A role for giantin in docking COPI vesicles to Golgi membranes. J Cell Biol. 1998;140:1013–1021. doi: 10.1083/jcb.140.5.1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramaniam VN, Peter F, Philp R, Wong SH, Hong W. GS28, a 28-kilodalton Golgi SNARE that participates in ER-Golgi transport. Science. 1996;272:1161–1163. doi: 10.1126/science.272.5265.1161. [DOI] [PubMed] [Google Scholar]
- Tsui MM, Tai WC, Banfield DK. Selective formation of Sed5p-containing SNARE complexes is mediated by combinatorial binding interactions. Mol Biol Cell. 2001;12:521–538. doi: 10.1091/mbc.12.3.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukada M, Gallwitz D. Isolation and characterization of SYS genes from yeast, multicopy suppressors of the functional loss of the transport GTPase Ypt6p. J Cell Sci. 1996;109:2471–2481. doi: 10.1242/jcs.109.10.2471. [DOI] [PubMed] [Google Scholar]
- Tsukada M, Will E, Gallwitz D. Structural and functional analysis of a novel coiled-coil protein involved in Ypt6 GTPase-regulated protein transport in yeast. Mol Biol Cell. 1999;10:63–75. doi: 10.1091/mbc.10.1.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tufarelli C, Fujiwara Y, Zappulla DC, Neufeld EJ. Hair defects and pup loss in mice with targeted deletion of the first cut repeat domain of the Cux/CDP homeoprotein gene. Dev Biol. 1998;200:69–81. doi: 10.1006/dbio.1998.8950. [DOI] [PubMed] [Google Scholar]
- VanRheenen SM, Cao X, Sapperstein SK, Chiang EC, Lupashin VV, Barlowe C, Waters MG. Sec34p, a protein required for vesicle tethering to the yeast Golgi apparatus, is in a complex with Sec35p. J Cell Biol. 1999;147:729–742. doi: 10.1083/jcb.147.4.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Votsmeier C, Gallwitz D. An acidic sequence of a putative yeast Golgi membrane protein binds COPII and facilitates ER export. EMBO J. 2001;20:6742–6750. doi: 10.1093/emboj/20.23.6742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren G, Malhotra V. The organization of the Golgi apparatus. Curr Opin Cell Biol. 1998;10:493–498. doi: 10.1016/s0955-0674(98)80064-1. [DOI] [PubMed] [Google Scholar]
- Whyte JR, Munro S. The Sec34/35 Golgi transport complex is related to the exocyst, defining a family of complexes involved in multiple steps of membrane traffic. Dev Cell. 2001;1:527–537. doi: 10.1016/s1534-5807(01)00063-6. [DOI] [PubMed] [Google Scholar]
- Winzeler EA, Shoemaker DD, Astromoff A, Liang H, Anderson K, Andre B, Bangham R, Benito R, Boeke JD, Bussey H, et al. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science. 1999;285:901–906. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]
- Zeng WR, Watson P, Lin J, Jothy S, Lidereau R, Park M, Nepveu A. Refined mapping of the region of loss of heterozygosity on the long arm of chromosome 7 in human breast cancer defines the location of a second tumor suppressor gene at 7q22 in the region of the CUTL1 gene. Oncogene. 1999;18:2015–2021. doi: 10.1038/sj.onc.1202519. [DOI] [PubMed] [Google Scholar]