Abstract
Hypoxia is an important factor that elicits numerous physiological and pathological responses. One of the major gene expression programs triggered by hypoxia is mediated through hypoxia-responsive transcription factor hypoxia-inducible factor 1 (HIF-1). Here, we report the identification and cloning of a novel HIF-1-responsive gene, designated RTP801. Its strong up-regulation by hypoxia was detected both in vitro and in vivo in an animal model of ischemic stroke. When induced from a tetracycline-repressible promoter, RTP801 protected MCF7 and PC12 cells from hypoxia in glucose-free medium and from H2O2-triggered apoptosis via a dramatic reduction in the generation of reactive oxygen species. However, expression of RTP801 appeared toxic for nondividing neuron-like PC12 cells and increased their sensitivity to ischemic injury and oxidative stress. Liposomal delivery of RTP801 cDNA to mouse lungs also resulted in massive cell death. Thus, the biological effect of RTP801 overexpression depends on the cell context and may be either protecting or detrimental for cells under conditions of oxidative or ischemic stresses. Altogether, the data suggest a complex type of involvement of RTP801 in the pathogenesis of ischemic diseases.
Oxygen is an essential factor for viability and normal function of living organisms, and its level is closely monitored by intracellular mechanisms. Under hypoxic conditions, oxygen-sensing machinery activates a transcription factor known as hypoxia-inducible factor 1 (HIF-1). This factor switches on a series of genes participating in compensatory mechanisms that support cell survival in a potentially lethal microenvironment. One group of HIF-1 target genes involved in the adaptive response facilitates O2 delivery to oxygen-deprived tissues. It includes, e.g., genes coding for erythropoietin (stimulates production of erythrocytes), heme-oxygenase 1 (mediates O2 binding to heme), vascular endothelial growth factor (VEGF; triggers new vasculature formation), and inducible nitric oxide synthase (participates in local blood vessel dilation) (25, 27, 29, 32, 41). Another group of HIF-1-dependent genes acts to compensate for the inhibition of oxidative phosphorylation that occurs when oxygen is lacking. It includes genes coding for glycolytic enzymes (e.g., lactate dehydrogenase [LDH], phosphoglyceromutase, and others) and for glucose transporters (e.g., Glut1) (11a, 13, 40).
Prolonged oxygen deprivation is detrimental for cells and may result in their death through either apoptotic or necrotic mechanisms (reviewed in reference 28). Paradoxically, like the adaptive response to hypoxia, hypoxia-dependent apoptosis was shown to be HIF-1 dependent. Cells with genetically deleted HIF-1α appeared to be resistant to hypoxia-triggered apoptosis (6). Moreover, HIF-1α was demonstrated to mediate hypoxia-induced delayed neuronal death in a stroke model (15). While HIF-1-dependent genes participating in the adaptive response to hypoxia are widely characterized, genes mediating its proapoptotic function remain largely unknown. One of the proapoptotic genes, Nip3, was only recently characterized as HIF-1 dependent (5).
Since alterations in gene expression caused by hypoxia underlie the pathogenesis of several major diseases (i.e., stroke, cancer, myocardial infarction, and retinopathy), we were interested in the identification of novel hypoxia-responsive genes, regardless of whether they participate in the adaptive or in the apoptotic response. This task was tackled by employing a microarray hybridization technique to investigate the hypoxia-dependent gene expression in rat glioma C6 cells. Several previous studies demonstrating a potent HIF-1 response in C6 cells supported the choice of this cell system (10, 18).
Here, we report the identification and cloning of a novel HIF-1-dependent gene, RTP801. This gene is ubiquitously expressed in multiple human tissues at low levels. However, in response to hypoxia its transcription is rapidly and sharply increased both in vitro and in vivo. Inducible overexpression of RTP801 promoted the apoptotic death of differentiated neuronal PC12 cells and led to their sensitization to hypoxic and oxidative stress. Similarly, liposomal delivery of RTP801 to mouse lungs resulted in apoptosis of parenchymal cells. However, inducible overexpression of RTP801 in dividing PC12 and MCF7 breast carcinoma cells made them resistant to both hypoxia and H2O2-induced apoptosis via dramatic suppression of the treatment-induced increase in the intracellular concentration of reactive oxygen species (ROS). Thus, like HIF-1 itself, RTP801 possesses both pro- and antiapoptotic activities depending on the cell context.
MATERIALS AND METHODS
Cell culture.
C6 rat glioma cells and MCF7-Tet-Off (Clontech) human epithelial breast carcinoma cells were maintained in Dulbecco modified Eagle medium (DMEM) supplemented with 10% fetal calf serum (Gibco-BRL), 20 U of penicillin/ml, and 20 μg of streptomycin/ml. Neomycin (100 μg/ml) was also added to the latter cell type.
PC12 Tet-Off (Clontech) rat pheochromocytoma cells were maintained in DMEM supplemented with 8% fetal calf serum, 8% horse serum (HyClone), 20 U of penicillin/ml, 20 μg of streptomycin/ml, and 100 μg of neomycin/ml.
Plasmids.
The pcDNA3-RTP801 construct was prepared by subcloning the open reading frame (ORF) of human RTP801 cDNA (nucleotides 216 to 907; accession no. AF335324) into the KpnI/NotI sites of expression plasmid pcDNA3 (Invitrogen). The cDNA fragment was obtained by PCR amplification using human RTP801-specific primers 5′-GGGGTACCATGCCTAGCCTTTGG-3′ and 5′-TAAAGCGGCCGCTCACAACATGTCAATGAGCAGCTG-3′ with the added KpnI and NotI restriction sites, respectively.
The tetracycline-repressible pSHTet-RTP801 construct was prepared as follows. A rat RTP801 cDNA fragment (nucleotides 190 to 878; accession no. AF334162) was obtained by PCR amplification of template cDNA with rat RTP801-specific primers 5′-CCATCGATGGATTACAAGGACGACGACGATAAGCCTAGCCTTTGG-3′ and 5′-CGAAGCTTCAACACTCTTCAATG-3′ flanked by ClaI and HindIII restriction sites, respectively. The forward primer included 30 bp coding for the Flag tag. The PCR fragment was subcloned into ClaI/HindIII sites of the pTet-Splice vector (Gibco-BRL). Then, the fragment containing Flag-RTP801 and the tetracycline-responsive element of pTet-Splice was excised by double digestion with XhoI and HindIII. The XhoI site was filled in, and the resulting blunt/HindIII insert was ligated into SmaI/HindIII sites of pSHTetSVPLA, a vector based on the pGEM3 backbone. It contains the multiple cloning site derived from pBluescript (Stratagene) and the 870-bp XhoI-BamHI (filled in) fragment from pMSG (Pharmacia) subcloned into the HincII site of the polylinker. This fragment includes the intron and late polyadenylation signal of simian virus 40 t antigen.
pBluescript-rRTP801, used for the preparation of riboprobes for in situ hybridization, contained the rat RTP801 cDNA fragments encompassing nucleotides 585 to 1095 inserted into the EcoRI site.
Preparation of cDNA microarray.
A cDNA microarray containing 1,847 cDNA fragments was constructed with clones from subtracted cDNA libraries derived from C6 glioma cells enriched for hypoxia-responsive mRNAs. Specifically, mRNA prepared from C6 cells cultured under hypoxic conditions (0.5% O2, 5% CO2) for 4 and 16 h was subjected to bidirectional subtraction (PCR-Select cDNA subtraction kit; Clontech) followed by cloning into pBluescript. The array also contained a set of control genes whose response to hypoxia in C6 glioma (and other cells) was documented previously. They included, e.g., VEGF, glucose transporter 1 (Glut1), and LDH.
Preparation of probes for hybridization to the cDNA microarray.
Nuclei from C6 cells cultured under either normoxia (control) or hypoxia conditions (4 or 16 h) were obtained in a single fractionation procedure as previously described (33). Total or nuclear RNA was used for the preparation of probes for microarray hybridization. Probe labeling was performed by reverse transcription using 50 μg of template RNA and an oligo(dT) 18-mer as a primer. Control RNA and RNA from treated cells were labeled with Cy3-dCTP and Cy5-dCTP (Amersham), respectively. The following sets of probes were utilized for microarray hybridizations: (i) total RNA at normoxia (Cy3) and total RNA after 4 h of hypoxia (Cy5); (ii) total RNA at normoxia (Cy3) and total RNA after 16 h of hypoxia (Cy5); (iii) nuclear RNA at normoxia (Cy3) and nuclear RNA after 4 h of hypoxia (Cy5); (iv) nuclear RNA at normoxia (Cy3) and nuclear RNA after 16 h of hypoxia (Cy5). Hybridizing, washing, and scanning of the slides were performed as previously described (37). cDNA clones displaying differential expression were sequenced, and their identities were defined.
Stable transfections.
MCF7-Tet-Off and PC12-Tet-Off cells (2.5 × 105; both from Clontech) were cotransfected with 2.5 μg of pSHTet-RTP801 and 0.5 μg of pTK-Hyg (Clontech) plasmid DNA using Lipofectamine reagent (Gibco-BRL). The empty vector was used as a control. Cells were then grown in selection medium containing 100 μg of hygromycin (Roche)/ml and 1 μg of tetracycline or doxocycline (Sigma)/ml. Seventy-two hours after tetracycline removal, the selected cell clones were screened for inducible expression of RTP801 by Northern blot analysis and by immunoblotting with anti-Flag antibodies (Sigma).
In vitro cell treatments and viability assays.
MCF7-Tet-Off and PC12-Tet-Off parental cells and their RTP801 and control transfectants were seeded into 24-well dishes at densities of 104 and 3 × 104 cells per well, respectively, in either the presence or absence of 1 μg of tetracycline/ml. After 72 h of cultivation, the cells were exposed either to hypoxia (glucose-free medium; 5% CO2, 0.5% O2, 37°C), to oxidative stress (0.5 to 1 mM H2O2), or to serum starvation (0.1% serum) for 24 h. For all assays, both differentiated and nondifferentiated PC12 cells were used. For induction of neuronal differentiation, the cells were seeded on polylysine-coated plates in the presence of nerve growth factor (50 ng/ml) and 1 μg of tetracycline/ml. Each type of treatment was followed by an assessment of cell viability (neutral red uptake), cell apoptosis (APOPercentage apoptosis assay), and cell death (LDH activity in conditioned medium of treated cells).
The neutral red assay was conducted as described before (43) with neutral red dye from Sigma. All measurements were performed in triplicate at a λ of 540 nm.
An LDH leakage assay was performed using a cytotoxicity detection kit (Molecular Biochemicals) according to the manufacturer's protocol. LDH activity was measured at a λ of 492 nm. LDH release was defined as the ratio of LDH activity in the medium to LDH activity corresponding to total cell death and was expressed in percent. All measurements were performed in triplicate.
Quantitative detection of apoptotic cells was performed with the APOPercentage apoptosis assay kit (Biocolor Ltd., Belfast, Northern Ireland) according to the manufacturer's instructions. For the assay, the cells were seeded at 104 cells per well in a 96-well plate.
For inhibition of apoptotic death, caspase inhibitor Boc-D (OMe)-FMK (100 μM; Alexis Biochemicals) was added to differentiated RTP801-expressing PC12 cells either in the presence or in the absence of 1 μg of tetracycline/ml. After 72 h of cultivation, LDH activity was measured.
Assessment of apoptosis in vivo.
In histological sections, apoptotic cells were detected by terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling (TUNEL). The assay was performed using the ApopTag peroxidase in situ apoptosis detection kit (Intergen Company) according to manufacturer's protocol.
Measurement of generation of intracellular ROS.
RTP801-transfected and control MCF7-Tet-Off and PC12-Tet-Off cells were seeded into 96-well dishes at densities of 103 and 3 × 103 cells per well, respectively, either in the presence or in the absence of 1 μg of tetracycline/ml. After 72 h of cultivation, the cells were incubated in DMEM containing 2′,7′-dichlorofluorescein (DCFH)-diacetate (10 μM; Molecular Probes) for 30 min at 37°C and then were exposed either to hypoxia (glucose-free medium, 5% CO2, 0.5% O2, 37°C) or to oxidative stress (0.5 to 1 mM H2O2) for 24 h. Cell fluorescence was determined by microplate fluorometer (excitation, 485 nm; emission, 538 nm) before and following hypoxia or H2O2 treatment and normalized according to cell number.
EMSA and supershift analysis.
Electrophoretic mobility shift assays (EMSA) were performed on nuclear extracts from embryonic stem (ES) cells. The extracts were prepared as described by Schreiber et al. (38), with slight modifications. Specifically, buffers A and C contained a Complete protease inhibitor cocktail (Roche Diagnostics) and 1 mM orthovanadate. The double-stranded oligonucleotides used included (i) the HIF-1-binding site of the transferrin receptor gene (TR-HRE) promoter (30) (5′-CGCGAGCGTACGTGCCTCAGG-3′; TR-HBS), (ii) a fragment of the RTP801 promoter region (nucleotides −454 to −434 of the mouse sequence shown in Fig. 3B [5′-ACGTTGCGAACGTGCGCCCGG-3′]; RTP801-HRE), and (iii) a mutated version of oligonucleotide ii (5′-ACGTTGCGAACTAGTGCCCGG-3′, RTP801-MHRE). The binding reactions were performed as described before (45). For supershift experiments, 1 μg of monoclonal antibodies against HIF-1α (NB 100-105; Novus Biologicals) or control anti-Flag monoclonal antibody M5 (Sigma) was added to the reaction mixture before the addition of labeled oligonucleotides. For the binding competition experiment, unlabeled oligonucleotides were added into the reaction mixture in a 100-fold excess. The reaction mixture was incubated for 15 min at 4°C before and after addition of labeled oligonucleotides. DNA-protein complexes were analyzed in a gradient (4 to 10%) polyacrylamide gel in a Bio-Rad minigel device with 0.5× Tris-borate-EDTA at 100 V and 4°C. The gel was vacuum dried and exposed to Kodak film. Visual inspection of the free probe band at the bottom of the gel confirmed that equivalent amounts of radiolabeled probe were used for all samples (data not shown).
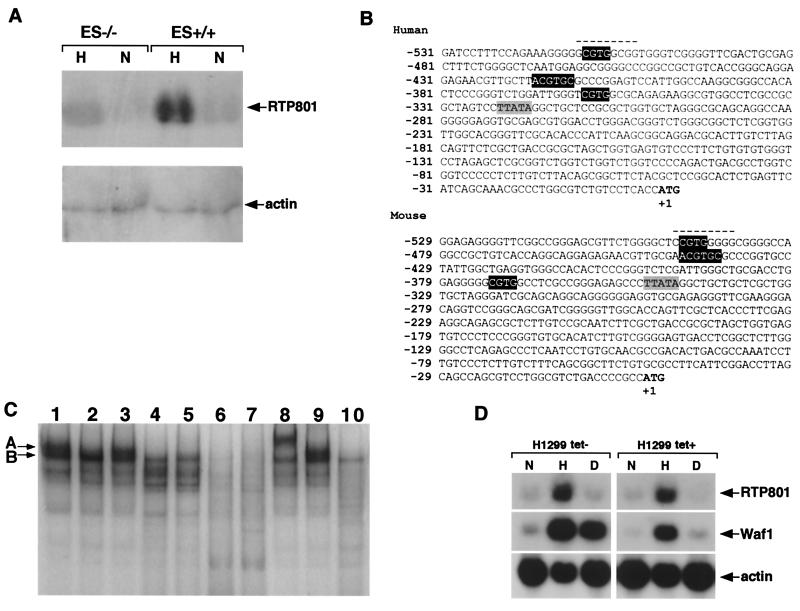
FIG. 3.
Transcriptional regulation of RTP801. (A) Northern blot analysis of RTP801 transcription in wild-type mouse ES cells (ES+/+) and in HIF-1α null mouse ES cells (ES−/−) cultured under normoxic (N) or hypoxic conditions (H) for 16 h. Total RNA (15 μg) was loaded in each slot. (B) Nucleotide sequences of immediate upstream genomic regions of mouse and human RTP801 orthologues. The initiation ATG codon is in boldface, and the position of T is counted as +1. The TATA box is shaded gray. White letters in black background, putative HRE; dashed line, putative Egr-1 binding site. (C) EMSA and supershift analysis of mouse RTP801 promoter region. All the binding reactions except for those whose mixtures are loaded in lanes 2, 4, and 5 were performed with nuclear extracts prepared from wild-type ES cells cultured under hypoxic conditions for 16 h. The reaction mixture loaded in lane 2 contains nuclear extract prepared from wild-type ES cells cultured in normoxia, whereas reaction mixtures loaded in lanes 4 and 5 contain nuclear extracts from HIF-1α−/− ES cells maintained in normoxic and hypoxic conditions, respectively. Lane 1, 32P-TR-HRE oligonucleotide; lanes 2 to 5, 32P-RTP801-HRE oligonucleotide; lane 6, 32P-RTP801-HRE oligonucleotide and the excess of nonlabeled RTP801-HRE oligonucleotide; lane 7, 32P-RTP801-HRE oligonucleotide and the excess of nonlabeled TR-HRE oligonucleotide; lane 8, 32P-RTP801-HRE oligonucleotide and anti-HIF-1α antibodies; lane 9, 32P-RTP801-HRE oligonucleotide and anti-Flag antibodies; lane 10, 32P-RTP801-MHRE oligonucleotide. For details see Results and Materials and Methods. (D) Northern blot analysis demonstrating the p53 independence of hypoxic transactivation of RTP801. H1299 is a human lung carcinoma p53-negative cell line that was engineered to express the wild-type p53 under the control of a tetracycline-repressible promoter. The cells were cultured either in the absence (left) or presence (right) of tetracycline to induce (left) or to suppress (right) p53 expression, respectively. Both p53-positive and p53-negative H1299 cells were maintained either under normal (N) or hypoxic (H) conditions or in the presence of doxorubicin (D). Total RNA (15 μg) derived from each experiment was analyzed by Northern blotting using the probes for human RTP801 and for Waf1 (as a positive control for p53-dependent transactivation).
Permanent middle cerebral artery occlusion (MCAO) stroke model.
The stroke model using a spontaneously hypertensive rat strain (SHR) was prepared as previously described (26). Experimental animals were sacrificed 0.5, 1, 2, 12, 24, 48, and 72 h after the operation (two animals per time point). The brains were removed, fixed in formalin, and embedded in paraffin, and coronal sections were prepared for further use in in situ hybridization with 35S-UTP-labeled rat RTP801-, c-fos-, and VEGF-specific sense and antisense riboprobes.
In situ hybridization.
Rat RTP801 radioactive riboprobes were produced from the pBluescript-rRTP801 vector (see above) with either T7 (antisense probe) or T3 (sense probe) as previously described (21). In situ hybridization was performed according to a previously published protocol (12). The exposed slides were developed in Kodak D-19 developer, fixed in Kodak fixer, and counterstained with hematoxylin-eosin. The microphotographs were taken with a Zeiss Axioscop-2 microscope equipped with a Spot RT charge-coupled device camera (Diagnostic Instruments).
Liposome preparation.
Large unilamellar vesicles (LUV) were prepared by mixing N(1-(2,3-dioleoyloxy)propyl)-N,N,N-trimethylammonium chloride (DOTAP) and cholesterol at a molar ratio of 1:1 in tert-butanol and freeze drying overnight. The lyophilized cake was hydrated with 5% dextrose and vortexed for several minutes to form large cationic multilamellar vesicles (MLV). LUV were downsized from the MLV with the LipoSofast (Avestin) extrusion system by passaging 15 times through a 0.4-μm-pore-size filter and 15 times through a 0.05-μm pore-size filter (Poretics), successively. The concentration of each type of lipid was 20 mM. The size of the DOTAP/cholesterol LUV cationic liposome was around 100 nm. Plasmid DNA was mixed with spermidine-free base (Sigma) at a spermidine/plasmid DNA molar ratio of 1: 6.16 in 5% dextrose. The mixture was allowed to stand at room temperature for 10 min. Then the spermidine-DNA complex was added to the DOTAP/cholesterol LUV cationic liposome at a DOTAP/DNA molar ratio of 5.2:1, and the mixture was incubated at room temperature for 10 to 15 min before use.
Nucleotide sequence accession numbers.
The cDNA sequences reported in this study are in the GenBank under accession no. AF334162, AF335324, and AF335325.
RESULTS
Cloning and structural characterization of a novel hypoxia-responsive gene.
To identify hypoxia-regulated genes in rat C6 glioma cells, we employed a cDNA microarray hybridization technique. To optimize the screening process, the cDNA microarray was prepared from clones from a subtracted cDNA library from C6 glioma mRNA enriched for hypoxia-regulated genes. Moreover, to efficiently monitor the acute hypoxia-dependent changes in gene expression, the probes for the microarray analysis were derived not only from total cellular RNA but also from nuclear RNA. Early detection of transcriptional activation or suppression of stable mRNA species by total cellular RNA probes is hampered by the presence of preexisting RNA molecules. Being independent of RNA stability, nuclear RNA probes help to overcome this obstacle by reflecting only the changes in de novo RNA synthesis. The up-regulation of novel gene RTP801, detected by a nuclear RNA probe, was extremely sharp: 14-fold after 4 h of hypoxia and 37-fold after 16 h (Fig. 1A). This prompted us to further analyze this gene.
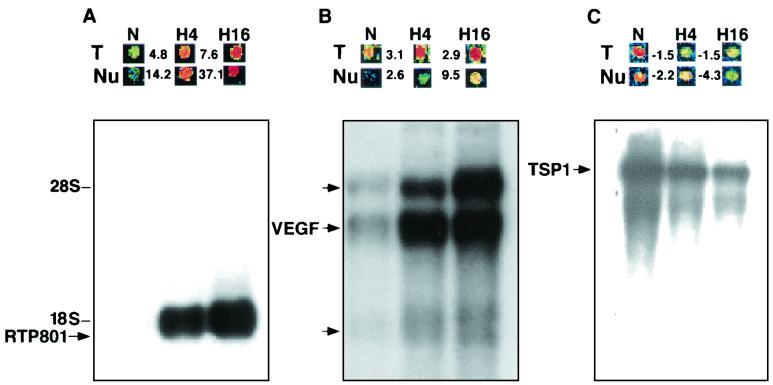
FIG. 1.
Regulation of RTP801 expression by hypoxia. (A) RTP801; (B) VEGF; (C) TSP1 (thrombospondin 1). N, normoxia; H4, 4 h of hypoxia; H16, 16 h of hypoxia. (Top) Microarray images of hybridization signals. T, total-RNA-derived probe; Nu, nuclear-RNA-derived probe. The numbers indicate the fold increase (+) and decrease (−) in gene expression under hypoxic (H4, H16) conditions versus that under normoxic (N) conditions. (Bottom) Northern analysis of gene expression using 2 μg of poly(A) RNA extracted from C6 cells cultured under either normoxic (N) or hypoxic (H4, H16) conditions. The three probes for RTP801 and the VEGF, and TSP1 genes (used to demonstrate that the normoxic slot does not contain less RNA than the hypoxic one) were consequently hybridized to the same membrane.
The results of microarray hybridization were confirmed by Northern blot analysis of C6 RNA. The gene coding for RTP801 was expressed as a single mRNA species of 1.8 kb. Its response to hypoxia was much more prominent than that of the VEGF gene, a well-known hypoxia-induced gene (Fig. 1A and B). Sequencing the arrayed RTP801 cDNA fragment revealed that it belongs to a previously unidentified gene.
Rat RTP801 full-length cDNA was cloned from the C6 hypoxia-specific cDNA library prepared in λZAP. The human orthologue of RTP801 was identified in GenBank as a single expressed sequence tag (EST), clone 364073 (accession no. AA021122). The human and the rat cDNAs are highly homologous (85% identity), and their single ORFs code for putative proteins of 232 and 229 amino acids, respectively (expected molecular mass, 25 kDa) (Fig. 2A). The amino acid conservation reaches 90%. In an in vitro translation assay, both rat and human RTP801 cDNAs gave rise to protein products with an estimated size of 35 kDa according to their mobility in gels (not shown). A search of the Ensembl database indicated that the RTP801 gene is localized at human chromosome 10q24.33.
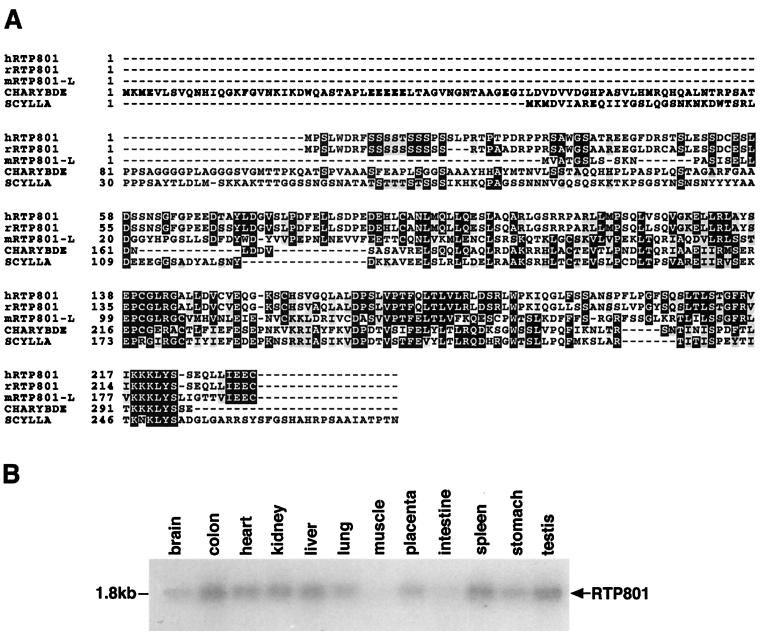
FIG. 2.
Sequence analysis of RTP801 cDNA and protein. (A) Multiple alignment of rat and human RTP801 putative proteins, two Drosophila proteins encoded by scylla and charybde, and mouse RTP801L. The alignment was performed by using the Prettybox program of the Genetics Computer Group sequence analysis package. (B) Northern blot analysis of RTP801 expression in various human tissues. A human 12-tissue Northern blot membrane was purchased from Origene (HB2010).
Analysis of the putative amino acid sequence of RTP801 did not reveal any known structural domains or functional motifs. Both the rat and the human proteins are leucine rich (17%) and contain conserved 9- or 10-serine stretches at their N termini. A search of the public databases revealed that RTP801 is homologous to two Drosophila melanogaster genes with unknown function, scylla (accession no. AF221110) and charybde (accession no. AF221109), which are designated in the GenBank as Hox targets. By extensive search of public mammalian EST database, we were also able to discover two overlapping mouse EST clones (accession no. AA647389 and AA472607) containing an ORF substantially similar to that of RTP801. In silico contigization of these ESTs enabled the identification of a new mouse gene product (designated RTP801L for RTP801-like) that displayed an overall 65% similarity (35% identity) to both rat and human RTP801 on the protein level. The results of contigization were confirmed by reverse transcription-PCR and the sequencing of the obtained cDNA fragment. The multiple alignment of protein sequences of human (hRTP801) and rat (rRTP801) RTP801 as well as of mouse RTP801L and Drosophila scylla and charybde protein products is shown in Fig. 2A. The four proteins are most similar at their C termini, potentially indicating the presence of a conserved domain not previously described. Within the N-terminal unique portion of RTP801, there is a cluster of negatively charged amino acids (positions 61 to 101 of rRTP801) absent from RTP801L and from both Drosophila proteins.
Low-level ubiquitous expression of RTP801 was evident by Northern blot analysis using poly(A) RNA from multiple human tissues (Fig. 2B). The lowest expression was detected in brain, skeletal muscle, and intestine.
RTP801 is a HIF-1-responsive gene.
The kinetics of RTP801 response to hypoxia as detected by microarray hybridization, resembled those of known HIF-1 targets: genes for VEGF and glycolytic enzymes. This raised the possibility that RTP801 is also a HIF-1-dependent gene. To test this hypothesis, we compared RTP801 mRNA induction by hypoxia in wild-type and HIF-1α−/− mouse ES cells (6). As is evident from Fig. 3A, unlike normal ES cells, which displayed strong hypoxic stimulation of RTP801, the HIF-1α−/− ES cells failed to induce RTP801 expression under similar conditions. This suggests that, at least in ES cells, hypoxia-dependent stimulation of RTP801 is under the control of HIF-1α.
To further confirm the HIF-1 dependence of RTP801, we tested whether its expression can be triggered by alternative stimuli known to activate the HIF-1 response. It was recently shown that treatment of cells with H2O2 is sufficient to promote HIF-1 stabilization (7). Iron chelators, e.g., diferoxamine mesylate, have also been shown to activate a hypoxia stress response pathway via HIF-1 (45). As expected, the addition of either hydrogen peroxide or DFO to cells of various types elicited a rapid and strong up-regulation of RTP801 in parallel with the observed increase in the amount of nuclear HIF-1α protein (not shown).
HIF-1 stimulates transcription of its target genes by binding to a distinct nucleic acid motif, named hypoxia-responsive element (HRE). The putative regulatory regions immediately upstream to the first exons of mouse and human RTP801 orthologues were searched for the presence of an HRE(s) with the Genomatix software. A mouse genomic clone was obtained from a mouse genomic λ phage library, while a human genomic clone containing the 5′ flanking region of RTP801 was identified by a database search (accession no. AC006186). The HRE consensus sequence was previously described as either 5′-(G/C/T)ACGTGC(G/C)-3′ (29) or 5′-RCGTG-3′ (43a). In both mouse and human DNA, three positionally conserved short HRE consensus motifs were detected within the 1,000 bp preceding the first ATG codon (Fig. 3B). However, only one of them, starting at positions −422 and −440 of human and mouse DNA, respectively, fit the extended consensus. Several additional potential HRE sites (the short consensus) were found within the more distant upstream region, but their positions were not conserved between mouse and human DNA (not shown).
To prove the direct involvement of HIF-1 in the regulation of RTP801 transcription under hypoxic conditions, we performed EMSA with an oligonucleotide containing the extended HRE consensus sequence derived from the mouse RTP801 promoter region (nucleotides −454 to −434; Fig. 3B) RTP801-HRE. A known HIF-1 binding oligonucleotide derived from the transferrin receptor gene promoter (TR-HRE) (30) and an RTP801-specific oligonucleotide with the mutated core HRE sequence (RTP801-MHRE) were used as positive and negative controls, respectively. As evident from Fig. 3C, addition of a 32P-labeled TR-HRE oligonucleotide to nuclear extract of ES cells cultured under hypoxic conditions resulted in the formation of two major complexes, A and B (lane 1). Two similarly migrating complexes were formed when an RTP801-specific oligonucleotide was added to the same nuclear extract (lane 3); however formation of complex A was abolished when RTP801-MHRE was used (lane 10), indicating the dependence of complex A on the presence of HRE core sequences. Formation of complex A was also inhibited when a radiolabeled RTP801-HRE probe was added to the nuclear extract from ES cells cultured in normoxic conditions (lane 2) or to nuclear extracts from HIF-1α−/− ES cells regardless of whether they were maintained in normoxia or hypoxia (lanes 4 and 5, respectively). These results point out that complex A is hypoxia dependent and potentially contains HIF-1α. The specificity of the formation of complex A on RTP801-HRE is proven by its competitive inhibition with an excess of the same nonlabeled oligonucleotide (lane 6), while competitive inhibition with an excess of cold TR-HRE (lane 7), a known HIF-1 binding sequence, further supports the suggestion that complex A formed with RTP801-HRE is HIF-1 dependent. We next performed a supershift analysis of the observed complexes using the anti-HIF-1α antibodies. Their addition to the binding reaction mixture with the radiolabeled RTP801-HRE resulted in a complete supershift of complex A (lane 8), whereas nonspecific anti-Flag antibodies did not influence the mobilities of any of the observed complexes (lane 9). Thus, the EMSA and supershift analysis have proven that hypoxia regulation of RTP801 is mediated via direct binding of HIF-1α-containing transcription complexes to its promoter.
p53 is known to be stabilized by forming a physical complex with HIF-1 (2) and to mediate HIF-1-dependent hypoxia-induced delayed neuronal death (15). Therefore, we assessed whether hypoxic regulation of RTP801 is also p53 dependent. For this, we analyzed the response of RTP801 to hypoxia in several p53-negative (SCOV3, H1299, PC3) and p53-positive (MCF7, HT1080) cell lines. The results clearly indicated that hypoxic regulation of RTP801 is preserved regardless of the p53 status of the cells. An example of p53-independent activation of RTP801 transcription by hypoxia in H1299 cells is shown in Fig. 3D. Moreover, known p53-activating stimuli, such as doxorubicin (Fig. 3D) and UV and gamma irradiation (not shown), failed to enhance the expression of RTP801. Thus, while hypoxic regulation of RTP801 is HIF-1 dependent, it appears to be p53 independent.
The expression of RTP801 is induced in the MCAO model of stroke.
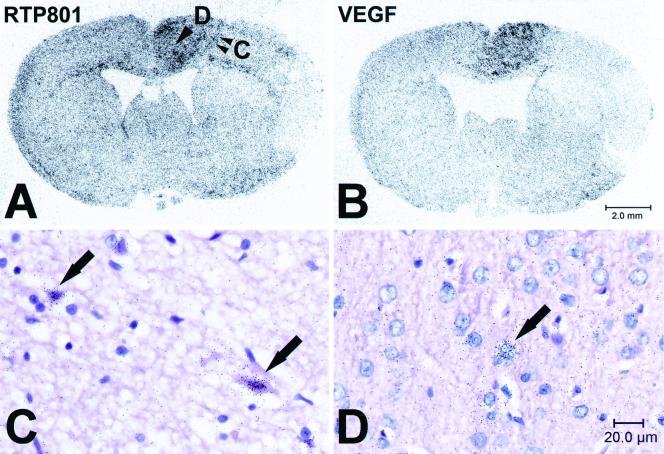
We next studied whether expression of RTP801 in vivo is induced under pathological conditions that result from ischemia or are accompanied by it. For a thorough analysis, we chose a widely used rat model of stroke produced by permanent MCAO. The injury of brain tissue in stroke results from a combination of pathophysiological processes that develop both within the ischemic core and within the surrounding peri-infarction area (penumbra) (11). Coronal sections of rat brains fixed at different time points (from 30 min to 72 h) after the MCAO were hybridized with the 35S-labeled riboprobe complement to RTP801 mRNA. A c-fos-specific probe served as a positive control for delineation of the peri-infarction area at early time points following MCAO (8, 9, 16), while a VEGF-specific probe was used as a positive control for delineation of peri-infarction ischemic areas at later time points (31). In control brain sections, the RTP801 riboprobe produced a low-intensity signal in cells of neuronal and glial origin. The enhanced expression of RTP801 in neuronal and glial cells within the injured hemisphere was sustained at all analyzed postinsult time points although a certain spatial redistribution of the hybridization signal was observed over the time course. Soon after occlusion (0.5 to 2 h), the RTP801-specific signal was localized within distant peri-infarction areas that also displayed a prominent expression of c-fos (not shown). Expression of VEGF at this time was still not evident. Twenty-four hours after the MCAO, accumulation of RTP801 mRNA occurred in VEGF-positive areas of the injured brain (Fig. 4A and B). It was significant within the eosinophilic neurons at the very boundary of the ischemic core (Fig. 4A and C), although, in more distant cortex areas, RTP801-positive neurons looked morphologically normal (Fig. 4A and D). At 48 and 72 h after the MCAO, the RTP801-expressing neurons did not display any evident signs of ischemic injury. In addition, expression of RTP801 could also be detected in endothelial cells within the necrotic zone (not shown). A complex expression pattern of RTP801 in the MCAO stroke model suggests that, besides hypoxia, other factors regulate its expression.
FIG. 4.
In situ hybridization analysis of RTP801 expression in the rat MCAO model 24 h after artery occlusion. (A and B) Microphotographs of distribution of radioactive hybridization signal specific to RTP801 (A) and VEGF (B) mRNA in coronal sections of rat brain 24 h after the MCAO (right hemisphere). Brain regions one (infarct boundary) and two (more-distant area) (arrowheads) are enlarged in panels C and D, respectively. (C and D) Microphotographs of coronal rat brain sections, bright field, with hematoxylin-eosin staining. Arrows, red neurons (C) and morphologically normal neurons (D) expressing RTP801. The RTP801-specific hybridization signal appears as black dots concentrated over expressing cells.
Inducible expression of RTP801 protects target cells from hypoxia- and H2O2-induced cell death via suppression of the generation of ROS.
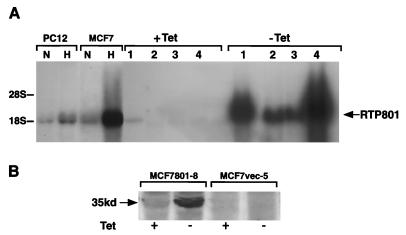
To assess the effect of acute up-regulation of RTP801 in target cells, we employed an in vitro-inducible expression system. Human epithelial breast carcinoma MCF7-Tet-Off and rat pheochromocytoma PC12-Tet-Off cells, both expressing the tetracycline-repressible transactivator (tTA), were transfected with Flag-tagged RTP801 driven by a tetracycline-responsive promoter element (TRE). Control cells were transfected with an empty vector. We isolated three clones of MCF7 cells (MCF801-8, MCF801-11, and MCF801-12) and one clone of PC12 cells (PC801-10) in which tetracycline-inducible exogenous RTP801 expression exceeded the hypoxic-endogenous-expression levels (Fig. 5).
FIG. 5.
Analysis of MCF7-Tet-Off and PC12-Tet-Off transfectants expressing RTP801 cDNA under the control of a tetracycline-repressible promoter. (A) Northern blot analysis of tetracycline-dependent expression of RTP801 in stably transfected cell clones (15 μg of total RNA per lane). Lane 1, PC801-10; lane 2, MCF801-11; lane 3, MCF801-12; lane 4, MCF801-8. Lanes N and H contain 15 μg of total cellular RNA extracted from parental MCF7-Tet-Off cells or PC12-Tet-Off cells cultured under normoxic and hypoxic conditions, respectively. (B) Western blot analysis with anti-Flag antibodies (Sigma) of expression of RTP801-Flag in total protein extracts from MCF801-8 and control cells. The position of the 35-kDa RTP801-specific band is indicated. For induction of RTP801 expression, cells usually cultured in the presence of 1 μg of tetracycline/ml and 100 μg of hygromycin/ml were washed with phosphate-buffered saline and seeded directly in culture medium without tetracycline for 72 h.
RTP801-transfected and control cells cultured either in the presence or in the absence of tetracycline for 72 h were subsequently subjected to either hypoxia (no glucose, 5% CO2, 0.5% O2) or H2O2 (0.5 to 1 mM) for an additional 24 h. After incubation, the majority of control cells displayed the shrunken morphology, became detached from the dish, and were positively stained with the APOPercentage kit dye, indicating the apoptotic nature of the ongoing cell death (not shown). In contrast, all MCF7 and PC12 cell clones expressing exogenous RTP801 were protected from apoptosis induced by hypoxia or H2O2 treatment. They preserved normal morphology and remained attached to the culture dish. Quantitatively, the observed protection from apoptosis was coordinately demonstrated by measuring LDH release (Fig. 6A and C) and neutral red or APOPercentage dye uptake (not shown). After 48 h of H2O2 treatment the antiapoptotic effect elicited by RTP801 expression was reduced and was completely abolished when H2O2 was applied for 72 h. For hypoxia in combination with glucose deprivation, RTP801-mediated protection was stable and lasted for at least 72 h of treatment (not shown).
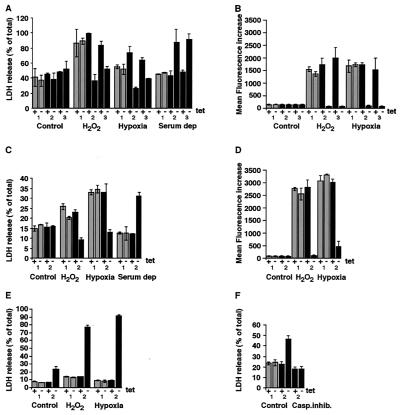
FIG. 6.
Analysis of viability of RTP801-transfected cells. (A) Assessment of cytotoxic effect of H2O2 treatment, hypoxia and glucose deprivation, and serum deprivation conditions in control and RTP801-expressing MCF7-Tet-Off cells. (B) Assessment of the influence of H2O2, hypoxia and glucose deprivation, and RTP801 expression on generation of ROS in MCF7-Tet-Off cells. For panels A and B, 1 indicates data for MCF7-vector (a clone of MCF7-Tet-Off cells transfected with the empty pSHTet vector), 2 indicates data for MCF801-8, and 3 indicates data for MCF801-12. (C) Assessment of cytotoxic effect of H2O2 treatment, hypoxia and glucose deprivation, and serum deprivation conditions in control and RTP801-expressing PC12-Tet-Off cells. (D) Assessment of the influence of H2O2, hypoxia and glucose deprivation, and RTP801 expression on generation of ROS in PC12-Tet-Off cells. (E) Assessment of cytotoxic effect of H2O2 treatment and hypoxia and glucose deprivation in control and RTP801-expressing differentiated PC12-Tet-Off cells. (F) Inhibition of RTP801-induced cytotoxicity in differentiated PC12-Tet-Off cells by Boc-D (OMe)-FMK. Casp.inhib., pan-caspase inhibitor. For panels C to F, 1 indicates data for PC12-vector (a clone of PC12-Tet-Off cells transfected with the empty pSHTet vector) and 2 indicates data for PC801-10. For experimental details, see Results and Materials and Methods. Data are means of three independent experiments performed in triplicate.
The proapoptotic nature of both types of applied treatments may be connected to the associated changes in generation of ROS within the target cells. Therefore, RTP801 antiapoptotic activity may stem from the potential activation of intracellular mechanisms that abolish generation of ROS. To test this hypothesis, we used the fluorescence-based detection of DCFH oxidation by ROS. For the assay, DCFH was added to the control cells and to the cells where RTP801 expression was induced for 72 h. The measurements of fluorescence were performed 30 min later or following 24 h of cell incubation either with H2O2 or in hypoxic conditions with no glucose as detailed in Materials and Methods. It is evident from Fig. 6B and D that, indeed, both H2O2 and hypoxia with no glucose led to a sharp increase in intracellular ROS in all control MCF7 and PC12 cells. This rise of ROS was almost completely abolished in cells in which expression of RTP801 was induced by tetracycline removal. Induction of RTP801 expression in nontreated cells did not influence the concentration of ROS.
We concluded that preceding expression of RTP801 suppresses generation of ROS in cells exposed to H2O2 and hypoxia, thus protecting them from apoptosis.
Inducible expression of RTP801 in differentiated PC12 cells promotes their apoptosis and sensitizes them to hypoxia- and H2O2-triggered cell death.
The responses of cycling and nondividing differentiated cells to hypoxic stress may be different. Since up-regulation of endogenous RTP801 was observed in hypoxic neurons in vivo, we tested the influence of inducible expression of RTP801 on nondividing neuron-like differentiated PC12 cells. To promote neuronal differentiation, the cells were treated with nerve growth factor for 5 days in the presence of tetracycline. As a result, almost all the cells displayed the typical flattened morphology and outgrowth of processes. Seventy-two hours after tetracycline removal, the cells were subjected to hypoxia and glucose deprivation or H2O2 treatments, and the apoptotic response was evaluated after an additional 24 h by measuring the LDH release (Fig. 6E) and neutral red uptake. Unlike dividing cells, differentiated PC12 cells expressing exogenous RTP801 were significantly more sensitive to hypoxia and H2O2 than their control counterparts (Fig. 6E). Moreover, induction of RTP801 expression by tetracycline removal was by itself sufficient to elicit the death response in these cells. Since RTP801-mediated cytotoxicity was completely abolished by addition of pan-caspase inhibitor Boc-D (OMe)-FMK (see Materials and Methods) (Fig. 6F), we concluded that induction of RTP801 expression in differentiated PC12 cells leads to apoptosis via activation of caspases.
Differentiated PC12 cells are maintained under low-serum conditions. To test whether reduced concentration of serum may render RTP801 proapoptotic, we transferred MCF7 and nondifferentiated PC12 cells in which expression of RTP801 was induced for 72 h into the medium containing 0.1% serum. Remarkably, both MCF7 and PC12 cells that expressed exogenous RTP801 appeared much more sensitive to serum starvation than control cells (Fig. 6A and C).
Thus, under certain conditions expression of RTP801 may be detrimental to cells. There was no influence of RTP801 on cell sensitivity to other apoptotic triggers, e.g., doxorubicin, UV irradiation, and taxol (not shown). In differentiated PC12 cells too, the sensitivity to UV irradiation did not appear to be affected by RTP801 overexpression (not shown).
Liposomal delivery of RTP801 to mouse lungs elicits apoptosis of parenchyma cells.
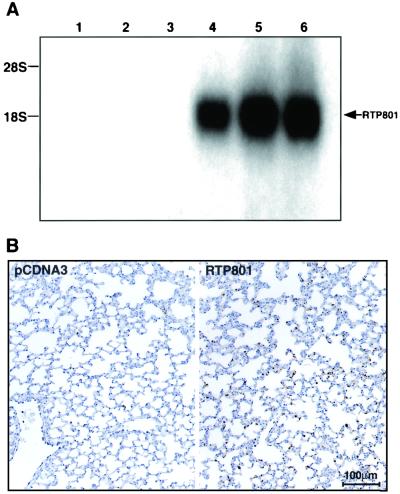
To assess the consequences of acute overexpression of RTP801 in vivo, cationic liposomes were used for the delivery of pcDNA3-RTP801 plasmid DNA into mouse lungs. The empty pcDNA3 vector and pcDNA3-p53 expression construct served as negative and positive controls, respectively, for potential apoptotic response. Each of the three liposome complexes containing 50 μg of plasmid DNA was administered to six mice intravenously. Twenty-four hours postinjection, the mice were sacrificed and their lungs were removed for further evaluation. Northern blot analysis revealed high exogenous expression of RTP801 in RNA extracted from the lungs of pCDNA3-RTP801-injected mice but not in RNA from the lungs of control mice (Fig. 7A).
FIG. 7.
Liposomal delivery of RTP801 cDNA into mouse lungs. (A) Northern blot analysis of RNA (15 μg per lane) extracted from lungs of mice injected with liposomes containing either pcDNA3 DNA (lanes 1 to 3) or pcDNA3-RTP801 (lanes 4 to 6). The position of the RTP801-specific band is indicated. (B) TUNEL staining of representative histological sections of control and RTP801-expressing mouse lungs. The experiment was repeated twice with independent liposomal preparations.
Paraffin sections of lung samples were processed for TUNEL staining. Lungs of mice injected with liposomes expressing RTP801 contained a large number of TUNEL-positive cells (Fig. 7B, lower right). The severity of the apoptotic response was in direct correlation with the intensity of the RTP801 hybridization signal. Mice injected with the empty vector were generally TUNEL negative (Fig. 7B, lower left). Only a few TUNEL-positive cells were evident in the lungs of mice injected with p53 liposomes (not shown).
DISCUSSION
Cellular response to hypoxia plays an important role in numerous biological processes, such as hematopoiesis (1), wound healing (36), ischemic stroke, myocardial infarction, retinopathy, and carcinogenesis (reviewed in reference 41a). Hypoxia-responsive genes may thus be important targets for the development of drugs modulating these major normal and pathological conditions. Therefore, we were interested in the identification of novel genes whose expression is altered in response to changing oxygen concentrations.
One such gene, designated RTP801, was identified by us as sharply up-regulated in C6 rat glioma cells in response to hypoxia. Its enhanced transcription in hypoxic tissues in vivo, for example, in permanent MCAO model of stroke, was also documented. Human and rat full-length cDNAs for RTP801 were cloned and found to represent a novel gene encoding a protein without any defined structural domains. A search of public databases revealed that RTP801 is homologous to two Drosophila homeobox target genes, scylla and charybde (Fig. 2A). The homology between RTP801 and the two identified Drosophila genes is maximal within regions encoding the C-terminal regions of the associated proteins, implying that the RTP801 sequence encodes a previously uncharacterized protein domain. This suggestion is further supported by the identification of a putative RTP801-like protein, designated RTP801L, in the mammalian genome. The maximal similarity between putative proteins RTP801 and RTP801L also resides within their C termini. The pattern of expression of RTP801L in embryogenesis is different from that of RTP801 (A. Faerman and E. Feinstein, unpublished observation). Moreover, its transcription is not responsive to a reduced oxygen concentration both in vitro and in vivo (H. Kalinski, A. Faerman, and E. Feinstein, unpublished observation). Overall, the data suggest that the product of the newly cloned RTP801 cDNA belongs to a protein family not previously described that participates in various cellular processes.
The predicted ORF of RTP801 codes for a protein of 25 kDa. However, all utilized expression systems gave rise to a recombinant protein that migrated on gels with a mobility corresponding to ∼35 kDa. This size discrepancy is unlikely to be due to posttranslational phosphorylation or glycosylation since the 35-kDa protein was detected not only in mammalian cells but also when the RTP801 ORF (from both rats and humans) was either translated in vitro or expressed in bacteria. The reason for the observed gel mobility shift is thus unclear.
Our initial experiments strongly suggested that RTP801 is a HIF-1-responsive gene, since mouse ES HIF-1α−/− cells failed to induce its expression in response to hypoxia. This suggestion was also supported by the presence of three evolutionarily conserved HREs in close vicinity (as judged by the position of the TATA box) to the initiation sites of both human and mouse mRNAs. Moreover, when the oligonucleotide containing one of these potential HRE elements was used in an EMSA, it promoted the formation of a specific complex (complex A in Fig. 3C). The formation of this complex was dependent on the presence of (i) a core HRE sequence within the labeled oligonucleotide, (ii) hypoxic conditions of cell cultivation, and (iii) the presence of the HIF-1α gene in the genomes of the cells used for the preparation of nuclear extracts. Overall, the experimental data confirm that RTP801 is a new member of the growing family of direct HIF-1 target genes. HIF-1α knockout in mice leads to embryonic lethality at the late stages of fetal development (19). Therefore, it will be interesting to evaluate the potential modifications in the RTP801 expression pattern in early, still-viable, HIF-1α null embryos. In addition, such an analysis will shed light on whether embryonic expression of RTP801 is regulated solely by HIF-1α or whether other regulatory routes (e.g., homeobox genes) are also involved.
As is evident from Fig. 3A, HIF-1α−/− ES cells still express the residual hypoxia-regulated levels of RTP801, potentially indicating involvement of additional transcription factors in the regulation of RTP801 expression. These may be other members of the HIF family capable of binding to the same HREs as HIF-1α. In this regard, it is worth noting that, in the EMSA experiments, the utilized RTP801-HRE formed additional specific complexes (migrating faster than complex B) that were abolished by mutating the core HRE sequence (Fig. 3C, lane 10). It is also possible that transcription factors belonging to other families contribute to regulation of the RTP801 gene as well. For example, we have found a conserved binding site for Egr-1 in the upstream regions of both mouse and human RTP801 orthologues (Fig. 2B). This factor was recently demonstrated to mediate the ischemic stress induction of numerous cytokines and chemokines (44). Egr-1 binding sites were found in the regulatory sequences of a range of genes relevant to vascular homeostasis and dysfunction (20); this finding may be in agreement with the observed increased expression of RTP801 in endothelial cells within the stroke core.
The potential complexity of transcriptional regulation of RTP801 is further underscored by its complex temporal and spatial expression pattern in the MCAO model. It seems conceivable that the early (30 min) peri-infarction activation of RTP801 stems not from the reduced oxygen supply but rather from the developing excitotoxicity and spreading depression. The latter is known to induce transcription of immediate-early genes (9, 16, 17), in agreement with the observed colocalization of RTP801- and c-fos-specific hybridization signals and the absence of up-regulation of hypoxia-specific marker VEGF (31, 34) at this time point. Only later on did the RTP801 hybridization signal become spatially colocalized with that of VEGF, suggesting a hypoxia-dependent regulation.
HIF-1 has a dual role in the cellular response to hypoxia. It is an important mediator of hypoxia-induced cell death (6, 15), and it seems to play an equally important role in mediating hypoxia-induced ischemic tolerance (3, 45). Like HIF-1 itself, RTP801 had a dual effect on the target cells. We have demonstrated that the prior expression of RTP801 in MCF7 and PC12 cells prevented apoptosis triggered by hypoxia and no glucose or H2O2 treatments via complete abolishment of the generation of ROS that would otherwise be caused by both types of treatment. It is not clear at this stage whether RTP801 has an antioxidant activity by itself or whether it somehow stimulates this adaptive response. The latter possibility seems more plausible, since (i) RTP801 does not have conserved cysteine residues, whose presence is typical for proteins with direct antioxidant activity, and (ii) its protective effect is transient. Other products of HIF-1 target genes, e.g., erythropoietin (22) and heme-oxygenase 1 (23, 24, 42), were also shown to elicit a protective effect in target cells against similar cytotoxic stimuli. However, under other conditions tested, overexpression of RTP801 turned out to be detrimental. First, nondividing neuron-like differentiated PC12 cells were killed by the very fact of enhanced expression of RTP801 through activation of caspases (moreover, these cells became much more sensitive to hypoxia and H2O2 cytotoxicity). Second, both MCF7 and PC12 cells overexpressing RTP801 died from serum starvation unlike their control counterparts. And third, the liposomal delivery of RTP801 to mouse lungs elicited a prominent apoptotic response on the target cells. The observed proapoptotic activity of RTP801 seems to be in line with the indirect correlative evidence obtained from the in situ hybridization studies of RTP801 using the MCAO model. At the brain infarct boundary in the MCAO model of stroke, the RTP801-specific mRNA was concentrated within the so-called eosinophilic, or “red,” neurons, located close to the necrotic core (Fig. 4A and C). These cells are not shrunken, and their nuclei are not pyknotic although they contain a clumped chromatin. The pathogenesis and the fate of red neurons remain uncertain, but they are regarded as possibly representing early apoptotic neurons or neurons suffering from hypoxia (14, 35).
Currently, the intracellular pathways affected by RTP801 overexpression are unknown. The only common feature that could be traced is that RTP801 overexpression caused the apoptosis-resistant phenotype in cycling cells and apoptosis sensitivity in growth arrested cells. Indeed, serum starvation is known to synchronize MCF7 cells in G0/G1 (46), and differentiated PC12 and lung parenchyma cells are also nondividing. The observed differences in the consequences of RTP801 overexpression in cycling and resting cells may be potentially explained by the underlying differences in their energy metabolism and demand. Specifically, the proliferative state is associated with a significant increase in aerobic glycolysis, and these cells are able to suppress phorbol myristate acetate-induced generation of ROS, probably due to increased concentrations of pyruvate, an effective ROS scavenger (4). The fact that overproduction of RTP801 in serum-starved neuron-like PC12 cells confers their increased sensitivity to hypoxic or oxidative injury may have certain implications for stroke, where due to the lack of blood supply the brain neurons suffer not only from ischemia but also from deprivation of growth factors.
Overall, the data lead us to conclude that newly identified direct HIF-1α target gene RTP801 participates not only in protective HIF-1-dependent molecular pathways but also in its proapoptotic effects. Accordingly, it will be important to monitor the changes in gene expression in general and changes in the expression of pro- and antiapoptotic genes specifically following the tetracycline induction of RTP801 in cycling and arrested cells. It will also be interesting to study the response of RTP801 null and transgenic mice to ischemic injury. Experiments with transgenic mice overexpressing exogenous RTP801 in brain, retina, and heart are ongoing. We believe that further investigation of RTP801 function will lead to the opening of new avenues in the treatment of hypoxia- and ischemia-associated diseases.
Acknowledgments
We are grateful to H. Ovadia and R. Lecker for preparation of MCAO rats, to Y. Barenholz for preparation of liposomes, to M. Oren for the gift of pcDNA3-p53 expression construct, to M. Levinson for the help with growth and treatment of ES cells and to A. Gudkov, T. Byk, and S. Kachalsky for critical reading of the manuscript and helpful discussions. We also thank E. Berent, L. Novak, and A. Bar-On for excellent technical assistance, D. Ben-Avraam and R. Rosenfeld for their help with the protein alignment, and Mitsubishi-Tokyo Pharmaceuticals Inc. for support.
REFERENCES
- 1.Adelman, D. M., E. Maltepe, and M. C. Simon. 1999. Multilineage embryonic hematopoiesis requires hypoxic ARNT activity. Genes Dev. 13:2478-2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.An, W. G., M. Kanekal, M. C. Simon, E. Maltepe, M. V. Blagosklonny, and L. M. Neckers. 1998. Stabilization of wild-type p53 by hypoxia-inducible factor 1alpha. Nature 392:405-408. [DOI] [PubMed] [Google Scholar]
- 3.Bergeron, M., J. M. Gidday, A. Y. Yu, G. L. Semenza, D. M. Ferriero, and F. R. Sharp. 2000. Role of hypoxia-inducible factor-1 in hypoxia-induced ischemic tolerance in neonatal rat brain. Ann. Neurol. 48:285-296. [PubMed] [Google Scholar]
- 4.Brand, K. A., and U. Hermfisse. 1997. Aerobic glycolysis by proliferating cells: a protective strategy against reactive oxygen species. FASEB J. 11:388-395. [DOI] [PubMed] [Google Scholar]
- 5.Bruick, R. 2000. Expression of the gene encoding the proapoptotic Nip3 protein is induced by hypoxia. Proc. Natl. Acad. Sci. USA 97:9082-9087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carmeliet, P., Y. Dor, J. M. Herbert, D. Fukumura, K. Brusselmans, M. Dewerchin, M. Neeman, F. Bono, R. Abramovitch, P. Maxwell, C. J. Koch, P. Ratcliffe, L. Moons, R. K. Jain, D. Collen, and E. Keshet. 1998. Role of HIF-1alpha in hypoxia-mediated apoptosis, cell proliferation and tumour angiogenesis. Nature 394:485-490. [DOI] [PubMed] [Google Scholar]
- 7.Chandel, N. S., D. S. McClintock, C. E. Feliciano, T. M. Wood, J. A. Melendez, A. M. Rodriguez, and P. T. Schumacker. 2000. Reactive oxygen species generated at mitochondrial complex III stabilize hypoxia-inducible factor-1-alpha during hypoxia: a mechanism of O2 sensing. J. Biol. Chem. 275:25130-25138. [DOI] [PubMed] [Google Scholar]
- 8.Christensen, T., M. B. Jorgensen, and N. H. Diemer. 1993. Impairment of Fos protein formation in the rat infarct borderzone by MK-801, but not by NBQX. Acta Neurol. Scand. 87:510-515. [DOI] [PubMed] [Google Scholar]
- 9.Collaco-Moraes, Y., B. S. Aspey, J. de Belleroche, and M. J. Harrison. 1994. Focal ischemia causes an extensive induction of immediate early genes that are sensitive to MK-801. Stroke 25:1855-1861. [DOI] [PubMed] [Google Scholar]
- 10.Damert, A., E. Ikeda, and W. Risau. 1997. Activator-protein-1 binding potentiates the hypoxia-induciblefactor-1-mediated hypoxia-induced transcriptional activation of vascular-endothelial growth factor expression in C6 glioma cells. Biochem. J. 327:419-423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dirnagl, U., C. Iadecola, and M. A. Moskowitz. 1999. Pathobiology of ischaemic stroke: an integrated view. Trends Neurosci. 22:391-397. [DOI] [PubMed] [Google Scholar]
- 11a.Ebert, B. L., J. D. Firth, and P. J. Ratcliffe. 1995. Hypoxia and mitochondrial inhibitors regulate expression of glucose transporter-1 via distinct cis-acting sequences. J. Biol. Chem. 270:29083-29090. [DOI] [PubMed]
- 12.Faerman, A., and M. Shani. 1997. Transgenic mice: production and analysis of expression. Methods Cell Biol. 52:373-403. [DOI] [PubMed] [Google Scholar]
- 13.Firth, J. D., B. I. Ebert, C. W. Pigh, and P. J. Ratcliffe. 1994. Oxygen-regulated control elements in phosphoglycerate kinase 1 and lactate dehydrogenase A genes: similarities with the erythropoietin 3′ enhancer. Proc. Natl. Acad. Sci. USA 91:6496-6500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garcia, J. H., Y. Yoshida, H. Chen, Y. Li, Z. G. Zhang, J. Lian, S. Chen, and M. Chopp. 1993. Progression from ischemic injury to infarct following middle cerebral artery occlusion in the rat. Am. J. Pathol. 142:623-635. [PMC free article] [PubMed] [Google Scholar]
- 15.Halterman, M. W., C. C. Miller, and H. J. Federoff. 1999. Hypoxia-inducible factor-1alpha mediates hypoxia-induced delayed neuronal death that involves p53. J. Neurosci. 19:6818-6824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Honkaniemi, J., B. A. States, P. R. Weinstein, J. Espinoza, and F. R. Sharp. 1997. Expression of zinc finger immediate early genes in rat brain after permanent middle cerebral artery occlusion. J. Cereb. Blood Flow Metab. 17:636-646. [DOI] [PubMed] [Google Scholar]
- 17.Hossmann, K. A. 1996. Periinfarct depolarizations. Cerebrovasc. Brain Metab. Rev. 8:195-208. [PubMed] [Google Scholar]
- 18.Ikeda, E., M. G. Achen, G. Breier, and W. Risau. 1995. Hypoxia-induced transcriptional activation and increased mRNA stability of vascular endothelial growth factor in C6 glioma cells. J. Biol. Chem. 270:19761-19766. [DOI] [PubMed] [Google Scholar]
- 19.Iyer, N. V., L. E. Kotch, F. Agani, S. W. Leung, E. Laughner, R. H. Wenger, M. Gassmann, J. D. Gearhart, A. M. Lawler, A. Y. Yu, and G. L. Semenza. 1998. Cellular and developmental control of O2 homeostasis by hypoxia-inducible factor1α. Genes Dev. 12:149-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Khachigian, L., V. Linder, A. Williams, and T. Collins. 1996. Erg-1-induced endothelial gene expression: common theme in vascular injury. Science 271:1427-1431. [DOI] [PubMed] [Google Scholar]
- 21.Komarova, E. A., K. Christov, A. I. Faerman, and A. V. Gudkov. 2000. Different impact of p53 and p21 on the radiation response of mouse tissues. Oncogene 19:3791-3798. [DOI] [PubMed] [Google Scholar]
- 22.Konishi, Y., D. H. Chui, H. Hirose, T. Kunishita, and T. Tabira. 1993. Trophic effect of erythropoiethin and other hematopoietic factors on the cholinergic neurons in vitro and in vivo. Brain Res. 606:29-35. [DOI] [PubMed] [Google Scholar]
- 23.Le, W. D., W. J. Xie, and S. H. Appel. 1999. Protective role of heme oxygenase-1 in oxidative stress-induced neuronal injury. J. Neurosci. Res. 56:652-658. [DOI] [PubMed] [Google Scholar]
- 24.Lee, P. J., J. Alam, G. W. Wiegand, and A. M. Choi. 1996. Overexpression of hemeoxygenase in human pulmonary epithelial cells results in cell growth arrest and increased resistance to hyperoxia. Proc. Natl. Acad. Sci. USA 93:10393-10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee, P. J., B. H. Jiang, B. Y. Chin, N. V. Iyer, J. Alam, G. L. Semenza, and A. M. Choi. 1997. Hypoxia-inducible factor-1 mediates transcriptional activation of the heme oxygenase-1 gene in response to hypoxia. J. Biol. Chem. 272:5375-5381. [PubMed] [Google Scholar]
- 26.Leker, R. R., A. Teichner, H. Ovadia, H., E. Keshet, E. Reinherz, and T. Ben-Hur. 2001. Expression of endothelial nitric oxide synthase in the ischemic penumbra: relationship to expression of neuronal nitric oxide synthase and vascular endothelial growth factor. Brain Res. 909:1-7. [DOI] [PubMed] [Google Scholar]
- 27.Levy, A. P., N. S. Levy, S. Wegner, and M. A. Goldberg. 1995. Transcriptional regulation of the rat vascular endothelial growth factor gene by hypoxia. J. Biol. Chem. 270:13333-13340. [DOI] [PubMed] [Google Scholar]
- 28.Lipton, P. 1999. Ischemic cell death in brain neurons. Physiol. Rev. 79:1431-1568. [DOI] [PubMed] [Google Scholar]
- 29.Liu, Y., S. R. Cox, T. Morita, and S. Kourembas. 1995. Hypoxia regulates vascular endothelial growth factor gene expression in endothelial cells. Identification of a 5′ enhancer. Circ. Res. 77:638-643. [DOI] [PubMed] [Google Scholar]
- 30.Lok, C. N., and P. Ponka. 1999. Identification of a hypoxia response element in the transferrin receptor gene. J. Biol. Chem. 274:24147-24152. [DOI] [PubMed] [Google Scholar]
- 31.Marti, H. J., M. Bernaudin, A. Bellail, H. Schoch, M. Euler, E. Petit, and W. Risau. 2000. Hypoxia-induced vascular endothelial growth factor expression precedes neovascularization after cerebral ischemia. Am. J. Pathol. 156:965-976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Melillo, G., T. Musso, A. Sica, L. S. Taylor, G. W. Cox, and L. Varesio. 1995. A hypoxia-responsive element mediates a novel pathway of activation of the inducible nitric oxide synthase promoter. J. Exp. Med. 182:1683-1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meyuhas, O., Y. Biberman, P. Pierandrei-Amaldi, and F. Amaldi. 1996. Isolation and analysis of polysomal RNA, p. 65-81. In. P. A. Krieg (ed.), A laboratory guide to RNA: isolation, analysis and synthesis. Wiley-Liss, Inc., New York, N.Y.
- 34.Plate, K. H., H. Beck, S. Danner, P. R. Allegrini, and C. Wiessner. 1999. Cell type specific upregulation of vascular endothelial growth factor in an MCA-occlusion model of cerebral infarct. J. Neuropathol. Exp. Neurol. 58:654-666. [DOI] [PubMed] [Google Scholar]
- 35.Rosenblum, W. 1997. Histopathologic clues to the pathways of neuronal death following ischemia/hypoxia. Neurotrauma 14:313-326. [DOI] [PubMed] [Google Scholar]
- 36.Scheid, A., R. H. Wenger, H. Christina, I. Camenisch, A. Ferenc, U. G. Stauffer, M. Gasmann, and M. Meuli. 2000. Hypoxia-regulated gene expression in fetal wound regeneration and adult wound repair. Pediatr. Surg. Int. 16:232-236. [DOI] [PubMed] [Google Scholar]
- 37.Schena, M., D. Shalon, R. Heller, A. Chai, P. O. Brown, and R. W. Davis. 1996. Parallel human genome analysis: microarray-based expression monitoring of 1000 genes. Proc. Natl. Acad. Sci. USA 93:10614-10619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schreiber, E., P. Matthias, M. M. Muller, and W. Schaffner. 1989. Rapid detection of octamer binding proteins with “mini-extracts,” prepared from a small number of cells. Nucleic Acids Res. 17:6419-6423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Semenza, G. L., B. H. Jiang, S. W. Leung, R. Passantino, J. P. Concordet, P. Maire, and A. Giallongo. 1996. Hypoxia response elements in the aldolase A, enolase 1, and lactate dehydrogenase A gene promoters contain essential binding sites for hypoxia-inducible factor 1. J. Biol. Chem. 271:32529-32537. [DOI] [PubMed] [Google Scholar]
- 40.Semenza, G. L., P. H. Roth, H. M. Fang, and G. L. Wang. 1994. Transcriptional regulation of genes encoding glycolytic enzymes by hypoxia-inducible factor 1. J. Biol. Chem. 269:23757-23763. [PubMed] [Google Scholar]
- 41.Semenza, G. L., and G. L. Wang. 1992. A nuclear factor induced by hypoxia via de novo protein synthesis binds to human erythropoietin gene enhancer at a site required for transcriptional activation. Mol. Cell. Biol. 12:5447-5454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41a.Semenza, G. L., F. Agani, D. Feldser, N. Iyer, L. Kotch, E. Laughner, and A. Yu. 2000. Hypoxias, HIF-1, and the pathophysiology of common human diseases. Adv. Exp. Med. Biol. 475:123-130. [DOI] [PubMed]
- 42.Takizawa, S., H. Hirabayashi, K. Matsushima, K. Tokuoka, and Y. Shinohara. 1998. Induction of heme-oxygenase protein protects neurons in cortex and striatum, but not in hippocampus, against transient forebrain ischemia. J. Cereb. Blood Flow Metab. 18:559-569. [DOI] [PubMed] [Google Scholar]
- 43.Wallach, D. 1984. Preparations of lymphotoxin induce resistance to their own cytotoxic effect. J. Immunol. 132:2464-2469. [PubMed] [Google Scholar]
- 43a.Wang, G. L., B. H. Jiang, E. A. Rue, and G. L. Semenza. 1995. Hypoxia-inducible factor 1 is a basic-helix-loop-helix-PAS heterodimer regulated by cellular O2 tension. Proc. Natl. Acad. Sci. USA 92:. 5510-5514. [DOI] [PMC free article] [PubMed]
- 44.Yan, S.-F., T. Fujita, J. Lu, K. Okada, Y. S. Zou, N. Mackman, D. Pinsky, and D. M. Stern. 2000. Egr-1, a master switch coordinating upregulation of divergent gene families underlying ischemic stress. Nat. Med. 6:1355-1361. [DOI] [PubMed] [Google Scholar]
- 45.Zaman, K., H. Ryu, D. Hall, K. O'Donovan, K. I. Lin, M. P. Miller, J. C. Marquis, J. M. Baraban, G. L. Semenza, and R. R. Ratan. 1999. Protection against oxidative stress-induced apoptosis in cortical neuronal cultures by iron chelators is associated with enhanced DNA binding of hypoxia-inducible factor-1 and ATF-1/CREB and increased expression of glycolytic enzymes, p21 (waf1/cip1) and erythropoietin. J. Neurosci. 19:9821-9830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zwijsen, R. M., R. Klompmaker, E. B. Wientjens, P. M. Kristel, B. van der Burg, and R. J. Michalides. 1996. Cyclin D1 triggers autonomous growth of breast cancer cells by governing cell cycle exit. Mol. Cell. Biol. 16:2554-2560. [DOI] [PMC free article] [PubMed] [Google Scholar]