Abstract
The Rev3 gene of Saccharomyces cerevisiae encodes the catalytic subunit of DNA polymerase ζ that is implicated in mutagenic translesion synthesis of damaged DNA. To investigate the function of its mouse homologue, we have generated mouse embryonic stem cells and mice carrying a targeted disruption of Rev3. Although some strain-dependent variation was observed, Rev3−/− embryos died around midgestation, displaying retarded growth in the absence of consistent developmental abnormalities. Rev3−/− cell lines could not be established, indicating a cell-autonomous requirement of Rev3 for long-term viability. Histochemical analysis of Rev3−/− embryos did not reveal aberrant replication or cellular proliferation but demonstrated massive apoptosis in all embryonic lineages. Although increased levels of p53 are detected in Rev3−/− embryos, the embryonic phenotype was not rescued by the absence of p53. A significant increase in double-stranded DNA breaks as well as chromatid and chromosome aberrations was observed in cells from Rev3−/− embryos. The inner cell mass of cultured Rev3−/− blastocysts dies of a delayed apoptotic response after exposure to a low dose of N-acetoxy-2-acetylaminofluorene. These combined data are compatible with a model in which, in the absence of polymerase ζ, double-stranded DNA breaks accumulate at sites of unreplicated DNA damage, eliciting a p53-independent apoptotic response. Together, these data are consistent with involvement of polymerase ζ in translesion synthesis of endogenously and exogenously induced DNA lesions.
In cancer genetics, the paradigm holds the sequential mutation of a series of oncogenes and tumor suppressor genes responsible for the evolutionary development of a normal cell into a fully malignant, metastasizing tumor (67). Most mutations are induced by nucleotide damage, originating from endogenous sources or inflicted by exogenous agents (49). Nucleotide damage that is not removed by DNA repair proteins generally leads to an arrest of the replication fork, due to the rigidity of the replicative polymerases, preventing incorporation of a nucleotide opposite a damaged template (51). To escape this arrest, cells possess multiple pathways that enable the completion of DNA replication despite the presence of replication-blocking DNA damage (3, 9).
Considerable progress has recently been made in identifying the actors in translesion synthesis, a pathway implicated in replicating damaged DNA. In both prokaryotes and eukaryotes, multiple polymerases have been identified that are capable of replicating DNA templates containing a variety of lesions. This enables the completion of replication and therefore safeguards cellular survival, albeit frequently at the expense of the introduction of mutations. Based on sequence homology and activity in vitro, most of the polymerases associated with translesion synthesis belong to the newly recognized Y superfamily of DNA polymerases (16, 21, 30, 32, 57, 70). The heterodimeric Saccharomyces cerevisiae polymerase ζ, comprised of the REV3 catalytic subunit and the REV7 processivity subunit (41-43), is an exception, since REV3 bears strong sequence similarity to classical B-type DNA polymerases (41, 53, 55). S. cerevisiae rev3 strains display a moderate hypersensitivity to UV light as well as a reversionless (rev) phenotype (45, 46; reviewed in reference 39). Reversion of almost all tested UV light-induced substitution mutations and of most frameshift mutations, as well as mutagenesis by other genotoxic agents, depends on REV3 (20, 24, 34, 39, 41, 43-46). In addition, most spontaneously occurring mutations depend on REV3 (26, 39, 59, 60, 71), suggesting a role for polymerase ζ in the mutagenic translesion replication of DNA damaged by endogenous sources, by spontaneous base decay, or as a consequence of fortuitous mutagenic replication of an undamaged template.
Scarce data exist for the molecular mechanism of translesion synthesis and mutagenesis in vivo and for the involvement of polymerase ζ in these processes, although it was shown previously that an S. cerevisiae rev3 mutant had lost translesion replication in vivo of a site-specific N-(deoxyguanosine-C8-yl)-N-acetyl-2-aminofluorene (dG-C8-AAF) adduct in a plasmid substrate (4). In vitro, purified polymerase ζ possesses a moderately processive polymerase activity on an undamaged template and weak, substrate-dependent, translesion synthesis activity (23, 25, 55). The lack of proofreading activity of the enzyme, resulting in the capability of extending an unpaired or a mispaired nucleotide, has led to a model in which S. cerevisiae polymerase ζ functions to extend translesion products generated by translesion synthesis polymerases from the Y family (25, 33, 35, 41, 56, 74).
To enable the study of the molecular basis of mutagenesis in cells from higher eukaryotes, we and others have identified homologues of REV3 from Drosophila melanogaster (11) and mammals (19, 36, 48, 52, 54, 65, 72). The putative mammalian REV3 proteins are considerably larger than their D. melanogaster and S. cerevisiae homologues (350 versus 240 versus 173 kDa, respectively), the difference being due mainly to a long stretch of intervening sequence in the middle of the gene. However, the high homology in the carboxy-terminal consensus polymerase domains suggests that these genes are true paralogues. In agreement, Rev3 antisense RNA expressed in a human cell line has been shown previously to reduce the induction of mutations by UV light, consistent with a role of REV3 in mutagenic translesion synthesis in mammalian cells (19). However, some residual REV3 activity may have persisted in these experiments, complicating the assessment of the precise role of polymerase ζ in translesion synthesis and DNA damage survival, as well as in spontaneous and induced mutagenesis.
To investigate the function of mammalian Rev3, we have generated cells and mice carrying a deletion of consensus polymerase domains in one allele of the gene. No Rev3−/− mice or cell lines could be obtained, suggesting that the gene is indispensable for long-term cellular survival. Rev3−/− embryos die around midgestation, showing normal DNA replication and cellular proliferation but generalized, p53-independent apoptosis. Rev3−/− blastocysts display a delayed hypersensitivity to N-acetoxy-2-acetylaminofluorene (NA-AAF), supporting the involvement of Rev3 in translesion synthesis of dG-C8-AAF adducts. Cytogenetic analysis of cells from Rev3-deficient embryos shows a significantly enhanced number of double-stranded DNA breaks and translocations. Together, these results support a role of polymerase ζ in translesion synthesis; in the absence of Rev3, unrepaired endogenous DNA damage triggers apoptosis via the accumulation of double-stranded DNA breaks.
MATERIALS AND METHODS
Generation of the targeting vectors and gene targeting.
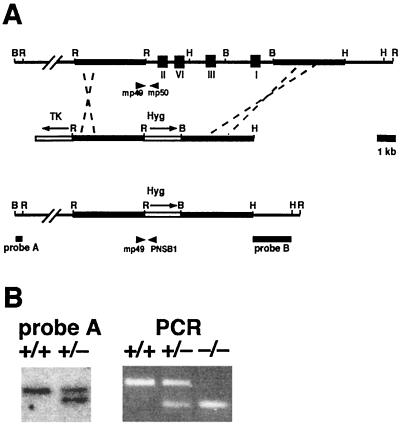
Genomic clones of the mouse Rev3 gene were isolated from a 129/SvEv BAC library (release II; Research Genetics) as previously described (65). A gene-targeting vector was constructed (Fig. 1A) that contained a phosphoglycerate kinase promoter (PGK)-hyg cassette replacing a 10-kbp fragment encoding the consensus polymerase domains I, II, III, and VI. In addition, a PGK-thymidine kinase gene cassette flanked genomic sequences to allow counterselection against random integration of the vector. The targeting vector was linearized with HindIII prior to electroporation.
FIG. 1.
Targeted disruption of the Rev3 gene. (A) (Top) Genomic Rev3 locus. I, II, III, and VI indicate the locations of consensus DNA polymerase domains. The thick line indicates the genomic region homologous to the targeting vector. (Middle) Targeting vector used to disrupt genomic Rev3. Hyg, PGK-hyg cassette; TK, PGK-thymidine kinase cassette. The arrows indicate the directions of transcription. (Bottom) Genomic Rev3hyg targeted locus. Probes A and B are probes used for analysis of gene targeting events. mp49 and mp50 are PCR primers used to amplify a 397-bp wild-type genomic fragment. mp49 and PNSB1 are PCR primers used to amplify a 275-bp targeted genomic fragment. Restriction sites: B, BamHI; H, HindIII; R, EcoRI. (B) (Left) Southern blot analysis of wild-type (+/+) and Rev3+/−(+/−) embryonic stem cell lines. Genomic DNA was digested with BamHI, and blots were hybridized with probe A. (Right) PCR analysis of wild-type (+/+), Rev3+/− (+/−), and Rev3−/− (−/−) embryos.
Culturing, electroporation, selection, and counterselection of subline IB10 of the 129/OLA-derived embryonic stem cell line E14 were performed as described previously (66).
Analysis of homologous recombinants.
Approximately 5 μg of DNA of expanded hygromycin-resistant, ganciclovir-resistant embryonic stem cell clones was digested with either EcoRI or BamHI. Following agarose gel electrophoresis, DNA was transferred to a Hybond-N+ membrane (Amersham) according to the alkaline blotting procedure as recommended by the manufacturer. Homologous disruption of the Rev3 gene was determined using a probe 5′ externally to the left arm of the targeting vector (probe A, Fig. 1A) or 3′ externally to the right arm (probe B, Fig. 1A). Hybridization of membranes containing BamHI-digested DNA with probe A resulted in a 23-kbp band representing the wild-type allele and a 20-kbp band for the targeted allele. Probe B, external to the right arm of the targeting DNA (Fig. 1A), was used on membranes containing EcoRI-digested DNA and produced a wild-type fragment of 16.5 kbp and an 8.5-kbp band representing the targeted allele.
An allele-specific multiplex PCR was developed to genotype mice and embryos. This PCR uses two gene-specific oligonucleotide primers (mp49, 5′-GTGCTGAGAAAGCTCATGTC-3′, and mp50, 5′-GATTGCCTTCCCTATCTGTC-3′) and a PGK promoter-specific oligonucleotide primer (PNSB1, 5′-CTAAAGCGCATGCTCCAGACT-3′) (Fig. 1A). The wild-type allele is represented by a PCR product of 397 bp, whereas the disrupted allele is represented by a PCR product of 275 bp. For PCR analysis, DNA from tails was isolated by incubating them overnight at 60°C in 50 mM Tris-HCl (pH 7.8)-12.5 mM MgCl2-100 μg of proteinase K/ml, followed by inactivation of the enzyme by boiling it for 5 min (65); DNA from yolk sac and cultured cells was isolated by incubating them for 1 h at 60°C in 10 mM Tris-HCl (pH 8.0)-2.5 mM MgCl2-0.45 μl of Nonidet P-40/ml-0.45 μl of Tween 20/ml-100 μg of proteinase K/ml, followed by boiling for 5 min. The multiplex PCR was performed in a total volume of 25 μl containing 1 to 3 μl of the DNA preparation, 200 μM deoxynucleoside triphosphates, 50 mM KCl, 0.1 mg of gelatin/ml, 0.2 mg of bovine serum albumin/ml, 50 μl of glycerol/ml, 10 pmol of each oligonucleotide primer, and 0.1 U of Amplitaq polymerase (Perkin-Elmer). After an initial denaturation step at 93°C for 5 min, 35 cycles of 30 s at 93°C, 30 s at 55°C, and 1 min at 72°C were performed in a Thermal Cycler (Perkin-Elmer).
Blastocyst culture, de novo embryonic stem cell derivation, and genotoxicity assays.
Isolation of blastocysts and de novo derivation of embryonic stem cell lines were performed essentially as described previously (28). Briefly, blastocysts of heterozygous mating pairs were isolated at 3.5 days postcoitum (dpc) (noon of the day of appearance of the vaginal plug is defined as 0.5 dpc) and cultured on irradiated mouse embryonic fibroblast (MEF) feeder layers in embryonic stem cell medium. To establish embryonic stem cell lines, the inner cell mass was disaggregated in trypsin-EDTA after 10 days of culture (13.5 dpc) and plated on irradiated MEF feeder layers. Approximately 7 days later, wells were scored for the growth of embryonic stem cells or of differentiated cell types.
Sensitivity of blastocysts to NA-AAF was determined as follows. After attachment to the gelatinized culture dish in the absence of feeder cells (at 5.5 to 6.5 dpc), blastocysts were pretreated with the deacetylase inhibitor paraoxon (3 nl/ml for 15 min) and subsequently with NA-AAF (25 μM for 30 min in the presence of paraoxon) to generate bulky dG-C8-AAF adducts (62). Survival of the inner cell mass was monitored up to 48 h after treatment, by visual inspection or by using a terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling (TUNEL) assay (see below). Genotyping was performed by PCR on DNA isolated from inner cell masses or from trophoblast cells, which were in all cases refractory to the NA-AAF treatment, as described above. All incubations were performed in a humidified incubator at 5% CO2 at 37°C.
Embryo histology and TUNEL assay.
Embryos from heterozygous matings were isolated, fixated in 4% paraformaldehyde, dehydrated in an isopropanol-NaCl series, and embedded in Paraplast (Sigma). Embryos were sectioned sagittally at 5 μm followed by staining with hematoxylin and eosin or by processing for immunostaining or TUNEL.
For labeling of S-phase cells in embryos with bromodeoxyuridine (BrdU), pregnant females were injected intraperitoneally with 100 μg of BrdU/g of body weight in phosphate-buffered saline and sacrificed 1 h later. BrdU incorporation in genomic DNA from embryo sections was detected using a monoclonal anti-BrdU antibody (Caltag) and a peroxidase-conjugated secondary anti-mouse antibody (Jackson Laboratories). Diaminobenzidine reagent (Sigma) was used for color development, and the slides were counterstained with hematoxylin (Sigma). Expression of the Ki67 cell proliferation marker (73) was detected with the primary antibody NCL-Ki67-MM1 (NovaCastra) in a protocol similar to that described for BrdU. p53 expression was determined using the CM5 antibody (Sanbio) and a biotin-conjugated anti-rabbit secondary antibody (Vector Laboratories) followed by addition of peroxidase-conjugated avidin (ABC-Elite kit; Vectastain). Diaminobenzidine reagent was used for color development; sections were counterstained with hematoxylin and eosin. The presence of apoptotic cells was determined using TUNEL staining as described previously (50); sections were counterstained with methyl green.
COBRA analysis of embryo-derived primary fibroblasts.
Embryos were isolated at 11.5 dpc from heterozygous matings between 129/OLA fathers and mothers of a mixed 129/OLA-C57BL/6 background. Primary embryonic cells were subsequently isolated by trypsinization, followed by seeding in Dulbecco modified Eagle medium plus 20% fetal calf serum. After 16 to 20 h of culture, embryonic cells were treated with demecolcine (Sigma; 25 ng/ml for 3 h) followed by trypsinization. Following a hypotonic treatment with 75 mM KCl, the cells were fixed with methanol and acetic acid (3:1 ratio). After two or three additional changes of fixative, the cell suspension was dropped on clean slides. The slides were air dried and aged for 3 days at room temperature prior to the mouse-specific COBRA staining procedure (14, 63). Genotyping for Rev3 was performed on DNA isolated from the yolk sac from each embryo.
RESULTS
Targeted disruption of the Rev3 gene.
Genomic clones encoding the consensus REV3 polymerase consensus boxes (65) (Fig. 1A) were obtained, and exon sequences were mapped (data not shown). A gene-targeting vector was constructed to enable the deletion of 10 kbp of the Rev3 gene, replacing the consensus boxes I, II, III, and VI with a hygromycin resistance cassette (Fig. 1A). To allow counterselection against random integration of the targeting construct, genomic sequences were flanked by a thymidine kinase cassette (Fig. 1A). Gene targeting in mouse embryonic stem cells by positive-negative selection resulted in a 10% targeting efficiency as analyzed with probes A (Fig. 1B) and B (data not shown).
Generation of Rev3 mutant mice and cells.
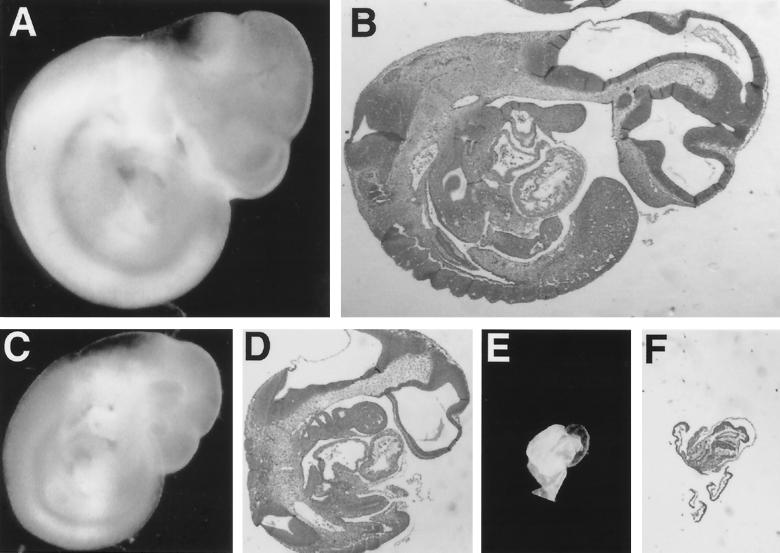
Three independently derived Rev3+/− embryonic stem cell clones were used for the generation of chimeric Rev3+/− mice. Germ line transmission of the targeted allele was obtained for all three clones, and Rev3+/− littermates were interbred after backcrossing to 129/Sv and to C57BL/6 mice. No homozygous (Rev3−/−) mutant offspring were obtained from the backcrosses (Table 1), whereas the heterozygous and wild-type littermates were born according to the expected (Mendelian) inheritance pattern, suggesting that Rev3 deficiency results in embryonic lethality. To investigate the fate of Rev3−/− embryos, pregnant females from backcrosses between Rev3 heterozygous mice of the C57BL/6 lineage were sacrificed at different stages of pregnancy. Analysis of embryos revealed a near-Mendelian ratio of live Rev3−/− embryos of up to 10.5 dpc. However, resorbing Rev3−/−, but not Rev3+/− or wild-type, embryos were found in significant numbers in pregnancies of 10.5 dpc and older; live Rev3−/− embryos older than 11.5 dpc were not found in these crosses between mice of predominantly the C57BL/6 background (Table 1). Remarkably, Rev3+/− embryos were somewhat underrepresented (Table 1); the cause of this remains unclear since Rev3+/− mice have no apparent phenotype. Surprisingly, in pure 129/OLA crosses and in crosses between pure 129/OLA and mixed 129/OLA-C57BL/6 backgrounds we observed viable Rev3−/− embryos of up to 15.5 dpc (data not shown), suggesting the presence of a strain-dependent genetic modifier of the phenotype. In all cases, nonresorbed Rev3−/− embryos displayed levels of growth retardation of 1 to 3 days (Fig. 2). Pathological examination of 10.5-dpc Rev3-deficient embryos, and histological sections from these embryos, revealed a spectrum of various developmental dysmorphias of internal organs and of external features (Fig. 2). The pleiotropy of this phenotype suggests the absence of a specific developmental defect. To investigate early embryonic development in vitro and to attempt to derive de novo Rev3−/− embryonic stem cell lines, blastocysts from heterozygous matings between mice of mixed backgrounds were isolated and cultured. Blastocysts were trypsinized after 10 days of growth and reseeded, after which the genotype was determined by PCR. Twenty-three cell lines were obtained from 41 such cultures. Among these 23 lines, none was Rev3 deficient whereas wild-type and heterozygous lines were present approximately according to the Mendelian ratio. In a separate experiment, blastocysts were sacrificed and genotyped after 7 days of culture. Rev3-deficient blastocysts derived from backcrosses between Rev3+/− C57BL/6 mice were not detected. However, Rev3−/− growing inner cell masses were obtained, nearly according to normal Mendelian distribution, from heterozygous crosses between mixed backgrounds (containing a minor C57BL/6 contribution) or when one parent was from the 129/OLA strain (Table 2). This result again supports the presence of strain-dependent modifiers of the Rev3 phenotype. The Rev3−/− inner cell mass outgrowths generally were smaller than heterozygous or wild-type blastocysts (see, e.g., Fig. 6; compare panels F and L). In addition, although cells adhere and grow for approximately 1 to 2 weeks, we have not succeeded in establishing fibroblast lines from 10.5-dpc Rev3−/− C57BL/6 or 13.5-dpc Rev3−/− mixed-background embryos whereas such lines were readily obtained from heterozygous and wild-type littermates (data not shown). Together, these results indicate that the absence of Rev3 results in a cell-autonomous phenotype rather than a specific embryonic defect.
TABLE 1.
Analysis of embryos from Rev3−/− crossesa
| No. of days of gestation | No. of fetuses with genotype:
|
No. of observed/ no. of expected Rev3−/− embryos | ||
|---|---|---|---|---|
| Rev3+/+ | Rev3+/− | Rev3−/− | ||
| 9.5 | 23 | 29 | 10 (6) | 0.64 |
| 10.5 | 36 | 67 | 22 (2) | 0.70 |
| 11.5 | 11 | 12 | 3 (3) | 0.46 |
| 12.5 | 5 | 3 | 0 (3) | 0 |
| 13.5 | 4 | 4 | 0 (0) | 0 |
| Weaned | 73 | 155 | 0 | 0 |
Pregnant females were sacrificed, and embryos were isolated. Crosses shown here were from backcrosses to the C57BL/6 background (generation F1 to F3). The given number of Rev3−/− embryos does not include the resorbing embryos, the number of which is shown in parentheses. All Rev3−/− embryos displayed growth retardation of between 1 and 3 days (Fig. 2). The ratio between the number of observed and the number of expected Rev3−/− embryos refers to the live embryos only.
FIG. 2.
Pleiotropic phenotype of Rev3−/− embryos. Shown are photographs of wild-type 10.5-dpc embryos (A and B) and Rev3−/− (C to F) littermates showing the growth retardation and morphology of Rev3−/− embryos. (A, C, and E) Whole embryos fixed in 4% paraformaldehyde. (B, D, and F) Sagittal sections stained with hematoxylin and eosin. All photographs were taken at the same magnification. (C and D) A Rev3−/− embryo with the size of an embryo of 9.5 dpc. However, external features such as the closure of the caudal neuropore and the development of the hind limb buds are consistent with an embryonic age of 10.5 dpc (38). (E and F) A dysmorphic Rev3−/− embryo displaying severe growth retardation. External features partially resemble those of a nonturned embryo of approximately 8.5 dpc (headfold stage [38]). Note that the development of the remaining part of the embryo is impaired more strongly.
TABLE 2.
Establishment of Rev3−/− blastocyst cultures from mice with a mixed 129/OLA and C57BL/6 background and sensitivity of the inner cell masses to paraoxon and NA-AAFa
| Treatment | No. surviving/total no. of blastocysts
|
||
|---|---|---|---|
| Rev3+/+ | Rev3+/− | Rev3−/− | |
| None | 12/13 | 21/22 | 6/8 |
| Paraoxon | 5/5 | 9/10 | 4/4 |
| Paraoxon + NA-AAF | 8/8 | 17/17 | 0/9 |
Blastocysts (3.5 dpc) were isolated and treated after attachment to the well (at 5.5 to 6.5 dpc). Survival was monitored at 48 h after treatment.
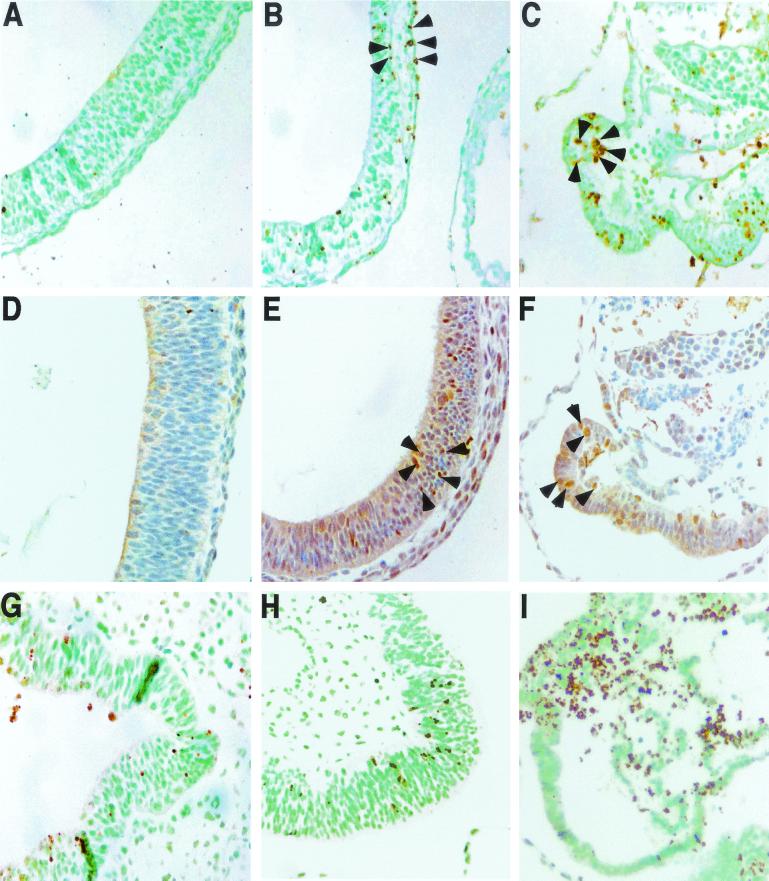
FIG. 6.
Apoptosis induced by NA-AAF in inner cell masses. TUNEL stainings of inner cell masses were visualized by transillumination, and the contrast was artificially enhanced to reveal apoptotic cells (dark spots). As a consequence of the illumination, trophoblast cells are not visible in most of the images. (A to D and G to J) Cells at 16 (A and G), 24 (B and H), 30 (C and I), and 41 (D and J) h after treatment. Note that at 30 h after treatment the inner cell mass of the Rev3−/− blastocyst has partially detached; the remaining part has largely stained black, indicating massive apoptosis. At 41 h most of the inner cell mass of the Rev3−/− blastocyst has detached. (E and K) TUNEL staining of wild-type and Rev3−/− blastocysts, 44 h after treatment with paraoxon alone, revealing similarly low levels of apoptosis for the two genotypes. (F and L) Phase-contrast images of TUNEL staining of a Rev3−/− and a wild-type blastocyst, 44 h after paraoxon treatment, illuminating the reduced size, but normal viability, of the Rev3−/− blastocyst. WT, wild type.
Normal replication and proliferation in the absence of Rev3.
To assess whether the Rev3-encoded DNA polymerase is important for genomic replication, we investigated sections from 10.5-dpc embryos by immunohistochemistry. To this aim, replicating DNA in embryos was pulse-labeled by injecting pregnant mothers with the nucleoside analog BrdU prior to sacrifice. Sections of Rev3−/− and wild-type embryos were subsequently stained for nuclear BrdU incorporation as a marker for genomic replication. BrdU staining revealed staining to a similar extent in most wild-type and Rev3−/− embryos (Fig. 3); only in Rev3−/− embryos that displayed a strong growth retardation was no BrdU incorporation detected (as exemplified in Fig. 3C), possibly because dwindling or stopped blood circulation interfered with BrdU incorporation.
FIG. 3.
Replication and cellular proliferation in Rev3−/− embryos. Wild-type embryos (A and D) and Rev3−/− littermates (B, C, E, and F) (all 10.5 dpc) were isolated after in vivo exposure to BrdU. Sagittal sections were immunostained with either anti-BrdU (A to C) or anti-Ki67 (D to F) antibody to investigate replication and cellular proliferation, respectively. Equivalent regions of the forebrain are shown, representative of staining in the rest of the embryo (except in panels C and F, where most of a severely growth-retarded embryo is shown). Note the lack of BrdU incorporation in the severely retarded embryo (C) (one out of four Rev3−/− embryos displayed no incorporation of BrdU), whereas staining for the proliferation marker Ki67 is apparent in all embryos (five embryos out of five mutants stained).
To investigate whether Rev3 is important for cellular proliferation, we stained adjacent sections of the same embryos for the presence of the proliferation marker Ki67, which is expressed in cells in most stages of the cell cycle (except in G0 and early G1 [73]). Similar levels of staining for Ki67 were detected in most cells within embryos of all genotypes (Fig. 3), demonstrating that cellular proliferation is not severely affected in the absence of Rev3. From these results it is apparent that Rev3 is essential neither for normal replication nor for normal cellular proliferation.
Apoptotic catastrophe and overexpression of p53 in Rev3-deficient embryos.
In the absence of Rev3, persistent DNA damage might induce apoptosis. This was investigated by staining sections of 10.5-dpc Rev3−/− and wild-type embryos for the presence of apoptotic cells, using a TUNEL assay. Although some variation was observed between individual embryos, Rev3−/− embryos invariably displayed prominent apoptosis in cells of all cell lineages, including the forebrain (Fig. 4B and C), as judged by positive TUNEL staining and nuclear compaction. In contrast, in wild-type embryos only occasional apoptotic cells were seen. Apoptosis was confirmed by staining for caspase 3 (data not shown).
FIG. 4.
Apoptosis in Rev3−/− embryos. (A to F) Sagittal sections of the developing forebrain from the same embryos as those shown in Fig. 3 were analyzed for apoptosis by TUNEL staining (A to C) or immunostained with an anti-p53 antibody (D to F). (A and D) Wild-type embryos; (B, C, E, and F) Rev3−/− embryos. Five mutant and three wild-type embryos were stained in this experiment. Equivalent regions of the forebrain are shown (except in panels C and F, where most of a severely growth-retarded embryo is shown). Rev3−/− embryos display extensive TUNEL staining (arrowheads in panels B and C), indicating massive apoptosis, accompanied by p53-positive cells (arrowheads in panels E and F). (G to I) TUNEL staining of sections of the neural folds (G and H) or the head (I) from three Rev3−/−; p53−/− embryos, displaying extensive apoptosis.
To address the involvement of p53 in the observed apoptosis, we stained sections from the same 10.5-dpc embryos for enhanced p53 expression. Indeed, strong nuclear p53 expression was seen in many cells from the Rev3−/− embryos whereas no significant numbers of p53-expressing cells were detected in wild-type littermates (Fig. 4, compare panel D with panels E and F). Confirming this observation, in Western blotting experiments enhanced overall p53 expression was seen in lysates from Rev3-deficient embryos (data not shown).
Inviability of Rev3−/− embryos does not depend on p53.
To determine the role of the observed p53 expression in causing a cell cycle arrest that may be involved in the observed developmental retardation and in the massive apoptosis in Rev3−/− embryos, we introduced a homozygous p53 truncation (C57BL/6 background [31]) in Rev3+/− mice of mixed backgrounds. Backcrossing of the latter mice yielded no live Rev3−/−; p53−/− progeny (based on Mendelian inheritance, the expected number should have been 17). Except for Rev3−/−; p53+/− and Rev3−/−; p53+/+, all other genotypes were found approximately according to Mendelian distribution (data not shown). Moreover, no obvious phenotypic differences were found between 10.5-dpc Rev3−/− embryos and Rev3−/−; p53−/− littermates (data not shown; four 10.5-dpc Rev3−/−; p53−/− embryos were investigated). To investigate whether the apoptosis observed in Rev3−/− embryos is dependent on p53, sections of 10.5-dpc Rev3−/−; p53−/− embryos were stained for apoptotic cells with caspase 3 staining (data not shown) and the TUNEL assay. These sections revealed persistence of apoptosis in Rev3−/−; p53−/− embryos (Fig. 4G to I). Together, these experiments show that p53 is not the major determinant of the growth retardation and is not responsible for the apoptotic catastrophe observed in Rev3-deficient embryos.
Enhanced frequency of chromosome aberrations in Rev3−/− embryo-derived cells.
Since Rev3 might be involved in translesion synthesis, persistent replication arrests opposite unrepaired endogenous DNA lesions, like abasic sites and oxidized nucleotides (48), could induce collapse of the replication fork leading to double-stranded DNA breaks during the subsequent S phase (40). These breaks are very efficient inducers of p53-dependent G1/S-phase cell cycle arrest and apoptosis (1) and, when misrepaired, precursors to chromatid exchanges and translocations (13). Therefore, defects in translesion synthesis could be accompanied by chromosomal instability. To obtain evidence for the involvement of Rev3 in translesion synthesis of endogenous DNA damage, we have therefore investigated chromosomal aberrancies in cells derived from 11.5-dpc Rev3−/− embryos of mixed background. This was done using a mouse-specific version of the COBRA chromosome painting method, enabling the identification of individual mouse chromosomes by painting each chromosome with a specific color. This analysis revealed a strong increase in chromosome and chromatid breaks and exchanges as precursors of translocations, as well as an enhanced frequency of translocations. Thus, 14% of Rev3−/− cells contained a chromosomal aberration versus 0.7% of cells from wild-type and heterozygous embryos (Table 3 and Fig. 5). These results are consistent with the predicted role of mouse polymerase ζ in translesion synthesis of endogenous DNA damage. In addition, the observed increase in chromosome and chromatid breaks may underlie the propensity of Rev3−/− embryonic cells for apoptosis.
TABLE 3.
Enhanced frequencies of all types of chromosomal aberrations derived from Rev3−/− embryos, as determined by COBRA analysisa
| Genotype | Embryo no. | No. of normal metaphases | No. of chromosome and chromatid breaks | No. of chromatid exchanges | No. of translocations | Total no. of aberrations |
|---|---|---|---|---|---|---|
| Wild type | 1 | 60 | 0 | 0 | 0 | 0 |
| Wild type | 2 | 39 | 1 | 0 | 0 | 1 |
| Rev3+/− | 3 | 50 | 0 | 0 | 0 | 0 |
| Rev3−/− | 4 | 43 | 2 | 3 | 2 | 7 |
| Rev3−/− | 5 | 32 | 1 | 1 | 0 | 2 |
| Rev3−/− | 6 | 41 | 3 | 3 | 1 | 7 |
Note that, as a consequence of the limited spatial resolution of the painted chromosomes, the actual number of chromosome and chromatid breaks probably is higher than that detected in this experiment.
FIG. 5.
Metaphase spreads from Rev3 mutant embryonic cells analyzed by mouse COBRA painting. The cell in the upper panels (A to C) contains a reciprocal translocation (indicated by arrowheads). The metaphase spread in the lower panels (D to F) contains an asymmetrical chromatid interchange (indicated by an arrowhead). Each set of panels contains a merged image of the ratio colors (A and D), a black and white image of the 4′,6′-diamidino-2-phenylindole-counterstained chromosomes (B and E), and the black and white image of the binary color (C and F).
Increased sensitivity of Rev3−/− blastocysts to NA-AAF.
In S. cerevisiae, translesion synthesis of replication-blocking dG-C8-AAF adducts depends on REV3 (4). To further investigate the possible involvement of the mouse Rev3 in translesion synthesis, we assayed whether NA-AAF (inducing dG-C8-AAF adducts in the presence of the deacetylase inhibitor paraoxon [62]) displays increased cytotoxicity toward Rev3−/− blastocysts. To this aim, blastocysts were isolated from heterozygous matings between 129/OLA mice and mixed 129/OLA-C57BL/6 background mice. After attachment and outgrowth of the inner cell mass, blastocysts were either mock treated, treated with paraoxon and NA-AAF, or treated with paraoxon alone. Survival of the inner cell mass was monitored after treatment. Subsequently, the blastocysts were genotyped using DNA isolated from the inner cell mass (when still present) or from nondividing trophoblast cells that in all cases appeared refractory to the toxic effects of NA-AAF. All Rev3−/− inner cell masses were detached at 48 h after treatment, whereas Rev3 heterozygous and wild-type inner cell masses all survived treatment (Table 2; also note that untreated Rev3−/− blastocysts survive to 10 days in culture [see above and Fig. 6L and M]). To determine whether detachment of Rev3-deficient inner cell masses is caused by NA-AAF-induced apoptosis, we performed an experiment in which blastocysts were fixed and stained for apoptosis by a TUNEL assay at various time points after treatment. Although some apoptosis was observed in blastocysts of all genotypes, mainly at early time points (Fig. 6A and B and 6G and H), in Rev3−/− blastocysts massive apoptosis initiated between 24 and 30 h after treatment (Fig. 6H and I), resulting in detachment of the inner cell masses between 30 and 41 h after treatment (Fig. 6I and J). The late occurrence of apoptosis suggests that NA-AAF-induced DNA damage needs to be processed to induce apoptosis. Overall, these data are supportive of a role of Rev3 in translesion synthesis of exogenous DNA damage.
DISCUSSION
Here we describe the generation and analysis of mouse strains carrying a targeted disruption of the Rev3 gene. In agreement with recent data from others (5, 12, 37, 69), Rev3−/− embryos die at midgestation. Embryonic death invariably was accompanied by enhanced levels of apoptosis in all cell lineages of the embryos. We have observed strain dependency of the phenotype of Rev3-deficient embryos: embryos with a high contribution of the C57BL/6 background all died before 12.5 dpc whereas embryos with a high contribution of the 129/OLA background survived up to 15.5 dpc. This strain-dependent difference in phenotype precludes correlation of the site of the Rev3 truncation (amino- or carboxy-terminal site) with the observed embryonic phenotype, as was suggested by others (37). Whereas Rev3−/− embryos of all genetic backgrounds invariably were retarded with respect to their heterozygous and wild-type littermates and frequently displayed morphological abnormalities, specific developmental defects were not found. This is in contrast with observations by others (12, 69) describing a defect in mesenchymal development. In agreement with others (69) we were unable to establish fibroblast lines from Rev3−/− embryos by using a protocol for spontaneous immortalization. In addition, we were unable to establish Rev3−/− embryonic stem cell lines from Rev3−/− blastocyst cultures, although the inner cell mass grew out and survived for 10 days in culture. These short-term blastocyst cultures could be established only when the strain background was mixed, again underscoring the strain dependency of the Rev3−/− phenotype. The latter may also explain the low incidence of viable Rev3−/− blastocyst cultures observed by others (5). From these data we conclude that Rev3 deficiency confers a cell-autonomous phenotype rather than a specific developmental defect. We believe that, since Rev3−/− embryos of a single genetic background die at different stages of embryonic development, displaying a wide range of abnormalities, it is likely that cell death is triggered by stochastic events in cells of all lineages within the embryo. The observation of normal viability of human cells expressing antisense Rev3 mRNA (19) may be explained by residual Rev3 activity.
BrdU incorporation, indicative of genomic replication, appeared normal in Rev3-deficient cells; also cellular proliferation, as judged by Ki67 expression, was not aberrant in Rev3−/− embryos. These results indicate that the embryonic phenotype of Rev3−/− embryos is not caused by a requirement for polymerase ζ in normal replication or cellular proliferation. To obtain evidence for the occurrence of double-stranded DNA breaks in Rev3-deficient cells, we performed an analysis of chromosomal aberrations in primary cells obtained from Rev3−/− embryos, using a mouse-specific chromosome painting method. Consistent with the notion that chromosome and chromatid breaks are precursors of translocations, we also observed an enhanced number of translocations in the mutant cells. To our knowledge, these results provide the first published evidence of chromosomal instability as a consequence of a putative defect in translesion synthesis.
Based on these results, we hypothesize that in Rev3−/− embryos arrested replication forks at the site of nonreplicated DNA damage are converted into double-stranded DNA breaks during the subsequent S phase (40). Thus, the Rev3−/− phenotype is consistent with a role for Rev3 in translesion replication of unrepaired endogenous DNA damage in mammalian cells. Furthermore, based on the reduced incidence of spontaneous mutations in S. cerevisiae rev3 strains, polymerase ζ is believed to play a role in translesion synthesis of endogenous DNA damage in yeast (26, 39, 59, 60, 71). Nevertheless, other mechanisms for the accumulation of double-stranded DNA breaks cannot be excluded at present. As an example, in S. cerevisiae, REV3 is implicated in mutagenesis associated with homology-dependent double-stranded break repair; however, repair itself is not measurably being affected (29).
In further agreement with a role for polymerase ζ in translesion synthesis, Rev3−/− blastocysts show massive apoptosis after exposure to a low dose of NA-AAF. Since mock-treated Rev3−/− blastocysts show a normal appearance with few apoptotic cells, we infer that apoptosis after NA-AAF treatment is a specific effect rather than a reflection of reduced viability of these blastocysts. NA-AAF-induced apoptosis is a late event, starting between 24 and 30 h after drug treatment. This late induction of apoptosis again is consistent with the requirement for processing of persistent DNA single-stranded regions to double-stranded DNA breaks, by replication fork collapse, during the S phase subsequent to the S phase with the initial replication arrest. Remarkably, earlier apoptosis was observed also in wild-type and Rev3−/− blastocysts (compare Fig. 6A and G with 6E and K, respectively), suggesting the presence of a second apoptotic pathway, common to the two genotypes.
In contrast to mouse Rev3 mutants, yeast Rev3 mutant cells display normal growth and viability (43, 44, 53), which may be a consequence of the small size of the yeast genome combined with the absence of apoptosis in yeast. Paralleling this difference in phenotype between yeast and mammals, yeast lif1 (the Xrcc4 homologue), ligase IV, and rad51 mutants, which accumulate double-stranded DNA breaks, have a near-normal viability (27, 61, 64, 68). In contrast, mouse embryos deficient for the Rad51, Xrcc4, DNA ligase IV, and ATR genes die by high levels of apoptosis, with the exception of the Rad51 mutant around midgestation (2, 6, 15, 17, 18, 47). The apoptotic phenotype is dependent on p53 in Rad51, Xrcc4, and DNA ligase IV-deficient embryos, the latter two being rescued to birth by p53 deficiency (15, 17, 18, 47). This is in marked contrast to Rev3−/−; p53 embryos that are not only not rescued to birth, as also found by others (68), but have the same morphological appearance as, and levels of apoptosis indistinguishable from, those of their Rev3−/− littermates. We infer that apoptosis in Rev3−/− embryos is caused by p53-independent damage signaling and effector pathways and that p53 expression in these embryos has no functional significance for the observed phenotype. Remarkably, it has been shown previously that hydroxyurea-induced replication arrest induces the accumulation of a form of p53 that is inactive in eliciting a cell cycle response (22). The presumed role of Rev3 in avoiding replication arrests is compatible with this result.
The recent finding that the putative mammalian homologue of Rev7 (54) (also called MAD2B or MAD2L2) displays homology with the mitotic spindle checkpoint protein Mad2 provides an alternative explanation for the inviability of Rev3-deficient mice. Like Mad2, Rev7 is involved in an anaphase arrest (8, 58). Interestingly, Mad2-deficient mouse embryos display an embryonic catastrophe similar to, albeit stronger than, that of the Rev3-deficient embryos (10). Also, it was found that apoptosis imparted by the mitotic spindle poison nocodazole is independent of p53 (7), compatible with the p53 independence of apoptosis in polymerase ζ-deficient embryos. Together, these data support the possibility that in the Rev3-deficient embryos an anaphase checkpoint is disrupted. Currently, we are addressing the molecular mechanisms inducing apoptosis in the Rev3-deficient embryos.
Acknowledgments
We acknowledge Cor Breukel, Peter Hohenstein, Ron Smit, Menno Kielman, and Anastasia Chtylik for the PGK-hyg and PGK-thymidine kinase cassettes and for help with immunohistochemical staining; Paul Lucassen for help with the TUNEL assay; and Joop Wiegant for help with the COBRA assay. Jacob Jansen is gratefully acknowledged for his intellectual contributions.
This work was supported financially by the Dutch Cancer Society (P.P.H.V.) and the Association for International Cancer Research (I.V.).
REFERENCES
- 1.Abraham, R. T. 2001. Cell cycle checkpoint signaling through the ATM and ATR kinases. Genes Dev. 15:2177-2196. [DOI] [PubMed] [Google Scholar]
- 2.Barnes, D. E., G. Stamp, I. Roswell, A. Denzel, and T. Lindahl. 1998. Targeted disruption of the gene encoding DNA ligase IV leads to lethality in embryonic mice. Curr. Biol. 8:1395-1398. [DOI] [PubMed] [Google Scholar]
- 3.Baynton, K., and R. P. Fuchs. 2000. Lesions in DNA: hurdles for polymerases. Trends Biochem. Sci. 25:74-79. [DOI] [PubMed] [Google Scholar]
- 4.Baynton, K., A. Bresson-Roy, and R. P. Fuchs. 1998. Analysis of damage tolerance pathways in Saccharomyces cerevisiae: a requirement for Rev3 DNA polymerase in translesion synthesis. Mol. Cell. Biol. 18:960-966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bemark, M., A. A. Khamlichi, S. L. Davies, and M. S. Neuberger. 2000. Disruption of mouse polymerase ζ (Rev3) leads to embryonic lethality and impairs blastocyst development in vitro. Curr. Biol. 10:1213-1216. [DOI] [PubMed] [Google Scholar]
- 6.Brown, E. J., and D. Baltimore. 2000. ATR disruption leads to chromosomal fragmentation and early embryonic lethality. Genes Dev. 14:397-402. [PMC free article] [PubMed] [Google Scholar]
- 7.Casenghi, M., R. Mangiacasale, M. Tuynder, P. Caillet-Fauquet, A. Elhajouji, P. Lavia, S. Mousset, M. Kirsch-Volders, and E. Cundari. 1999. p53-independent apoptosis and p53-dependent block of DNA rereplication following mitotic spindle inhibition in human cells. Exp. Cell Res. 250:339-350. [DOI] [PubMed] [Google Scholar]
- 8.Chen, J., and G. Fang. 2001. MAD2B is an inhibitor of anaphase-promoting complex. Genes Dev. 15:1765-1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cox, M. M., M. Goodman, K. N. Kreuzer, D. J. Sherratt, and K. L. Marians. 2000. The importance of repairing stalled replication forks. Nature 404:37-41. [DOI] [PubMed] [Google Scholar]
- 10.Dobles, M., V. Liberal, M. L. Scott, R. Benezra, and P. K. Sorger. 2000. Chromosome missegregation and apoptosis in mice lacking the mitotic checkpoint protein Mad2. Cell 101:635-645. [DOI] [PubMed] [Google Scholar]
- 11.Eeken, J. C. J., R. J. Romeijn, A. W. M. De Jong, A. Pastink, and P. H. M. Lohman. 2001. Isolation and genetic characterisation of the Drosophila homologue of (SCE)REV3, encoding the catalytic subunit of DNA polymerase ζ. Mutat. Res. 485:237-253. [DOI] [PubMed] [Google Scholar]
- 12.Esposito, G., I. Godindagger, U. Klein, M. L. Yaspo, A. Cumano, and K. Rajewsky. 2000. Disruption of the Rev3-encoded catalytic subunit of polymerase ζ in mice results in early embryonic lethality. Curr. Biol. 10:1221-1224. [DOI] [PubMed] [Google Scholar]
- 13.Flores-Rozas, H., and R. D. Kolodner. 2000. Links between replication, recombination and genome instability in eukaryotes. Trends Biochem. Sci. 25:196-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fodde, R., J. Kuipers, C. Rosenberg, R. Smits, M. Kielman, C. Gaspar, C. van Breukel, J. Wiegant, R. H. Giles, and H. Clevers. 2001. Mutations in the APC tumour suppressor gene cause chromosome instability. Nat. Cell Biol. 3:433-438. [DOI] [PubMed] [Google Scholar]
- 15.Frank, K. M., N. E. Sharpless, Y. Gao, J. M. Sekiguchi, D. O. Ferguson, C. Zhu, J. P. Manis, J. Horner, R. A. DePinho, and F. W. Alt. 2000. DNA ligase IV deficiency in mice leads to defective neurogenesis and embryonic lethality via the p53 pathway. Mol. Cell 5:993-1002. [DOI] [PubMed] [Google Scholar]
- 16.Friedberg, E. C., W. Feaver, and V. L. Gerlach. 2000. The many faces of DNA polymerases: strategies for mutagenesis and for mutational avoidance. Proc. Natl. Acad. Sci. USA 97:5681-5683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gao, Y., D. O. Ferguson, W. Xie, J. P. Manis, J. Sekiguchi, K. M. Frank, J. Chaudhuri, J. Horner, R. A. DePinho, and F. W. Alt. 2000. Interplay of p53 and DNA-repair protein XRCC4 in tumorigenesis, genomic stability and development. Nature 404:897-900. [DOI] [PubMed] [Google Scholar]
- 18.Gao, Y., Y. Sun, K. M. Frank, P. Dikkes, Y. Fujiwara, K. J. Seidl, J. M. Sekiguchi, G. A. Rathbun, W. Swat, J. Wang, R. T. Bronson, B. A. Malynn, M. Bryans, C. Zhu, J. Chaudhuri, L. Davidson, R. Ferrini, T. Stamato, S. H. Orkin, M. E. Greenberg, and F. W. Alt. 1998. A critical role for DNA end-joining proteins in both lymphogenesis and neurogenesis. Cell 95:891-902. [DOI] [PubMed] [Google Scholar]
- 19.Gibbs, P. E., W. G. McGregor, V. M. Maher, P. Nisson, and C. W. Lawrence. 1998. A human homolog of the Saccharomyces cerevisiae REV3 gene, which encodes the catalytic subunit of DNA polymerase zeta. Proc. Natl. Acad. Sci. USA 95:6876-6880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Glassner, B. J., L. J. Rasmussen, M. T. Najarian, L. M. Posnick, and L. D. Samson. 1998. Generation of a strong mutator phenotype in yeast by imbalanced base excision repair. Proc. Natl. Acad. Sci. USA 95:9997-10002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goodman, M. F., and B. Tippin. 2000. Sloppier copier DNA polymerases involved in genome repair. Curr. Opin. Genet. Dev. 10:162-168. [DOI] [PubMed] [Google Scholar]
- 22.Gottifredi, V., S.-Y. Shieh, Y. Taya, and C. Prives. 2001. p53 accumulates but is functionally impaired when DNA synthesis is blocked. Proc. Natl. Acad. Sci. USA 98:1036-1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guo, D., X. Wu, D. K. Rajpal, J.-S. Taylor, and Z. Wang. 2001. Translesion synthesis by yeast polymerase ζ from templates containing lesions of ultraviolet radiation and acetylaminofluorene. Nucleic Acids Res. 29:2875-2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Halas, A., H. Baranowska, Z. Policinska, and W. J. Jachymczyk. 1997. Involvement of the REV3 gene in the methylated base-excision repair system. Co-operation of two DNA polymerases, delta and Rev3p, in the repair of MMS-induced lesions in the DNA of Saccharomyces cerevisiae. Curr. Genet. 31:292-301. [DOI] [PubMed] [Google Scholar]
- 25.Haracska, L., I. Unk, R. E. Johnson, R. Johansson, P. M. J. Burgers, S. Prakash, and L. Prakash. 2001. Roles of yeast polymerases δ and ζ and of Rev1 in the bypass of abasic sites. Genes Dev. 15:945-954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harfe, B. D., and S. Jinks-Robertson. 2000. DNA polymerase ζ introduces multiple mutations when bypassing spontaneous DNA damage in Saccharomyces cerevisiae. Mol. Cell 6:1491-1499. [DOI] [PubMed] [Google Scholar]
- 27.Herrmann, G., T. Lindahl, and P. Schar. 1998. Saccharomyces cerevisiae LIF1: a function involved in DNA double-strand break repair related to mammalian XRCC4. EMBO J. 17:4188-4198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hogan, B., R. Beddington, F. Constantini, and E. Lacy. 1994. Manipulating the mouse embryo, p. 265-272. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 29.Holbeck, S. L., and J. N. Strathern. 1997. A role for REV3 in mutagenesis during double-strand break repair. Genetics 147:1017-1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hubscher, U., H. P. Nasheuer, and J. E. Syvaoja. 2000. Eukaryotic DNA polymerases, a growing family. Trends Biochem. Sci. 25:143-147. [DOI] [PubMed] [Google Scholar]
- 31.Jacks, T., L. Remington, B. O. Williams, E. M. Schmitt, S. Halachmi, R. T. Bronson, and R. A. Weinberg. 1994. Tumor spectrum analysis in p53-mutant mice. Curr. Biol. 4:1-7. [DOI] [PubMed] [Google Scholar]
- 32.Johnson, R. E., M. T. Washington, S. Prakash, and L. Prakash. 1999. Bridging the gap: a family of novel DNA polymerases that replicate faulty DNA. Proc. Natl. Acad. Sci. USA 96:12224-12226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Johnson, R. E., L. Haracska, S. Prakash, and L. Prakash. 2001. Role of DNA polymerase η in the bypass of a (6-4) TT photoproduct. Mol. Cell. Biol. 21:3558-3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Johnson, R. E., C. A. Torres-Ramos, T. Izumi, S. Mitra, S. Prakash, and L. Prakash. 1998. Identification of APN2, the Saccharomyces cerevisiae homolog of the major human AP endonuclease HAP1, and its role in the repair of abasic sites. Genes Dev. 12:3137-3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Johnson, R. E., M. T. Washington, L. Haracska, S. Prakash, and L. Prakash. 2000. Eukaryotic polymerases ι and ζ act sequentially to bypass DNA lesions. Nature 406:1015-1019. [DOI] [PubMed] [Google Scholar]
- 36.Kajiwara, K., H. Nagawawa, S. Shimizu-Nishikawa, T. Ookuri, M. Kimura, and E. Sugaya. 1996. Molecular characterization of seizure-related genes isolated by differential screening. Biochem. Biophys. Res. Commun. 219:795-799. [DOI] [PubMed] [Google Scholar]
- 37.Kajiwara, K., J. O-Wang, T. Sakurai, S. Yamashita, M. Tanaka, M. Sato, M. Tagawa, E. Sugaya, K. Nakamura, K. Nakao, M. Katsuki, and M. Kimura. 2001. Sez4 gene encoding an elongation subunit of DNA polymerase ζ is required for normal embryogenesis. Genes Cells 6:99-106. [DOI] [PubMed] [Google Scholar]
- 38.Kaufman, M. H. 1992. The atlas of mouse development. Academic Press Limited, London, United Kingdom.
- 39.Kunz, B. A., A. F. Straffon, and E. J. Vonarx. 2000. DNA damage-induced mutation: tolerance via translesion synthesis. Mutat. Res. 451:169-185 [DOI] [PubMed] [Google Scholar]
- 40.Kuzminov, A. 2001. Single-strand interruptions in replicating chromosomes cause double-strand breaks. Proc. Natl. Acad. Sci. USA 98:8241-8246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lawrence, C. W., and D. C. Hinkle. 1996. DNA polymerase ζ and the control of DNA damage induced mutagenesis in eukaryotes. Cancer Surv. 28:21-31. [PubMed] [Google Scholar]
- 42.Lawrence, C. W., G. Das, and R. B. Christensen. 1985. REV7, a new gene concerned with UV mutagenesis in yeast. Mol. Gen. Genet. 200:80-85. [DOI] [PubMed] [Google Scholar]
- 43.Lawrence, C. W., P. E. Nisson, and R. B. Christensen. 1985. UV and chemical mutagenesis in rev7 mutants of yeast. Mol. Gen. Genet. 200:86-91. [DOI] [PubMed] [Google Scholar]
- 44.Lawrence, C. W., T. O'Brien, and J. Bond. 1984. UV-induced reversion of his4 frameshift mutations in rad6, rev1, and rev3 mutants of yeast. Mol. Gen. Genet. 195:487-490. [DOI] [PubMed] [Google Scholar]
- 45.Lemontt, J. F. 1971. Mutants of yeast defective in mutation induced by ultraviolet light. Genetics 68:21-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lemontt, J. F. 1972. Induction of forward mutations in mutationally defective yeast. Mol. Gen. Genet. 119:27-42. [DOI] [PubMed] [Google Scholar]
- 47.Lim, D. S., and P. Hasty. 1996. A mutation in mouse rad51 results in an early embryonic lethal that is suppressed by a mutation in p53. Mol. Cell. Biol. 16:7133-7143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lin, W., X. Wu, and Z. Wang. 1999. A full-length cDNA of hREV3 is predicted to encode DNA polymerase ζ for damage-induced mutagenesis in humans. Mutat. Res. 433:89-98. [DOI] [PubMed] [Google Scholar]
- 49.Lindahl, T., and R. D. Wood. 1999. Quality control by DNA repair. Science 286:1897-1905. [DOI] [PubMed] [Google Scholar]
- 50.Lucassen, P. J., W. C. Chung, J. P. Vermeulen, M. Van Lookeren Campagne, J.-H. Van Dierendonck, and D. F. Swaab. 1995. Microwave-enhanced in situ end-labeling of fragmented DNA: parametric studies in relation to postmortem delay and fixation of rat and human brain. J. Histochem. Cytochem. 43:1163-1171. [DOI] [PubMed] [Google Scholar]
- 51.Minnick, D. T., and T. A. Kunkel. 1996. DNA replication errors, mutators and cancer. Cancer Surv. 28:3-20. [PubMed] [Google Scholar]
- 52.Morelli, C., A. J. Mungall, M. Negrini, G. Barbanti-Brodano, and C. M. Croce. 1998. Alternative splicing, genomic structure, and fine chromosome localization of REV3L. Cytogenet. Cell Genet. 83:18-20. [DOI] [PubMed] [Google Scholar]
- 53.Morrison, A., R. B. Christensen, J. Alley, A. K. Beck, E. G. Bernstine, J. F. Lemontt, and C. W. Lawrence. 1989. REV3, a Saccharomyces cerevisiae gene whose function is required for induced mutagenesis, is predicted to encode a nonessential DNA polymerase. J. Bacteriol. 171:5659-5667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Murakumo, Y., T. Roth, H. Ishii, D. Rasio, S. Numata, C. M. Croce, and R. Fishel. 2000. A human REV7 homolog that interacts with the polymerase ζ catalytic subunit hREV3 and the spindle assembly checkpoint protein hMAD2. J. Biol. Chem. 275:4391-4397. [DOI] [PubMed] [Google Scholar]
- 55.Nelson, J. R., C. W. Lawrence, and D. C. Hinkle. 1996. Thymine-thymine dimer bypass by yeast DNA polymerase ζ. Science 272:1646-1649. [DOI] [PubMed] [Google Scholar]
- 56.Nelson, J. R., C. W. Lawrence, and D. C. Hinkle. 1996. Deoxycytidyl transferase activity of yeast REV1 protein. Nature 382:729-731. [DOI] [PubMed] [Google Scholar]
- 57.Ohmori, H., E. C. Friedberg, R. P. Fuchs, M. F. Goodman, F. Hanaoka, D. Hinkle, T. A. Kunkel, C. W. Lawrence, Z. Livneh, T. Nohmi, L. Prakash, S. Prakash, T. Todo, G. C. Walker, Z. Wang, and R. Woodgate. 2001. The Y-family of DNA polymerases. Mol. Cell. 8:7-8. [DOI] [PubMed] [Google Scholar]
- 58.Pfleger, C. M., A. Salic, E. Lee, and M. W. Kirschner. 2001. Inhibition of Cdh1-APC by the MAD2-related protein MAD2L2: a novel mechanism for regulating Cdh1. Genes Dev. 15:1759-1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Quah, S. K., R. C. von Borstel, and P. J. Hastings. 1980. The origin of spontaneous mutation in Saccharomyces cerevisiae. Genetics 96:819-839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Roche, H., R. D. Gietz, and B. A. Kunz. 1994. Specificity of the yeast Rev3 delta antimutator and REV3 dependency of the mutator resulting from a defect (rad1 delta) in nucleotide excision repair. Genetics 137:637-646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schar, P., G. Herrmann, G. Daly, and T. A. Lindahl. 1997. Newly identified DNA ligase of Saccharomyces cerevisiae involved in RAD52-independent repair of DNA double-strand breaks. Genes Dev. 11:1912-1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shibutani, S., N. Suzuki, and A. P. Gollman. 1998. Mutagenic specificity of (acetylamino)fluorene-derived DNA adducts in mammalian cells. Biochemistry 37:12034-12041. [DOI] [PubMed] [Google Scholar]
- 63.Tanke, H. J., J. Wiegant, R. P. M van Gijlswijk, V. Bezrookove, H. Patteneir, R. J. Heetebrij, E. G. Talman, A. K. Raap, and J. Vrolijk. 1999. New strategy for multi-colour fluorescence in situ hybridization. COBRA: Combined Binary Ratio labeling. Eur. J. Hum. Genet. 7:2-11. [DOI] [PubMed] [Google Scholar]
- 64.Teo, S. H., and S. P. Jackson. 1997. Identification of Saccharomyces cerevisiae DNA ligase IV: involvement in DNA double-strand break repair. EMBO J. 16:4788-4795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Van Sloun, P. P., R. J. Romeijn, and J. C. Eeken. 1999. Molecular cloning, expression and chromosomal localisation of the mouse Rev3l gene, encoding the catalytic subunit of polymerase ζ. Mutat. Res. 433:109-116. [DOI] [PubMed] [Google Scholar]
- 66.Van Sloun, P. P., S. W. Wijnhoven, H. J. Kool, R. Slater, G. Weeda, A. A. Van Zeeland, P. H. Lohman, and H. Vrieling. 1998. Determination of spontaneous loss of heterozygosity mutations in Aprt heterozygous mice. Nucleic Acids Res. 26:4888-4894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Vogelstein, B., and K. Kinzler. 1993. The multistep nature of cancer. Trends Genet. 9:138-141. [DOI] [PubMed] [Google Scholar]
- 68.Wilson, T. E., U. Grawunder, and M. R. Lieber. 1997. Yeast DNA ligase IV mediates non-homologous DNA end joining. Nature 388:495-498. [DOI] [PubMed] [Google Scholar]
- 69.Wittschieben, J., M. K. Shivji, E. Lalani, M. A. Jacobs, F. Marini, P. J. Gearhart, I. Rosewell, G. Stamp, and R. D. Wood. 2000. Disruption of the developmentally regulated Rev3l gene causes embryonic lethality. Curr. Biol. 10:1217-1220. [DOI] [PubMed] [Google Scholar]
- 70.Woodgate, R. 1999. A plethora of lesion-replicating DNA polymerases. Genes Dev. 13:2191-2195. [DOI] [PubMed] [Google Scholar]
- 71.Xiao, W., T. Fontanie, S. Bawa, and L. Kohalmi. 1999. REV3 is required for spontaneous but not methylation damage-induced mutagenesis of Saccharomyces cerevisiae cells lacking O6-methylguanine DNA methyltransferase. Mutat. Res. 431:155-165. [DOI] [PubMed] [Google Scholar]
- 72.Xiao, W., T. Lechler, B. L. Chow, T. Fontanie, M. Agustus, K. C. Carter, and Y. F. Wei. 1998. Identification, chromosomal mapping and tissue-specific expression of hREV3 encoding a putative human DNA polymerase ζ. Carcinogenesis 19:945-949. [DOI] [PubMed] [Google Scholar]
- 73.Yu, C. C., A. L. Woods, and D. A. Levinson. 1992. The assessment of cellular proliferation by immunohistochemistry: a review of currently available methods and their applications. Histochem. J. 24:121-131. [DOI] [PubMed] [Google Scholar]
- 74.Yuan, F., Y. Zhang, D. K. Rajpal, X. Wu, D. Guo, M. Wang, J.-S. Taylor, and Z. Wang. 2000. Specificity of DNA lesion bypass by the yeast polymerase η. J. Biol. Chem. 275:8233-8239. [DOI] [PubMed] [Google Scholar]